Abstract
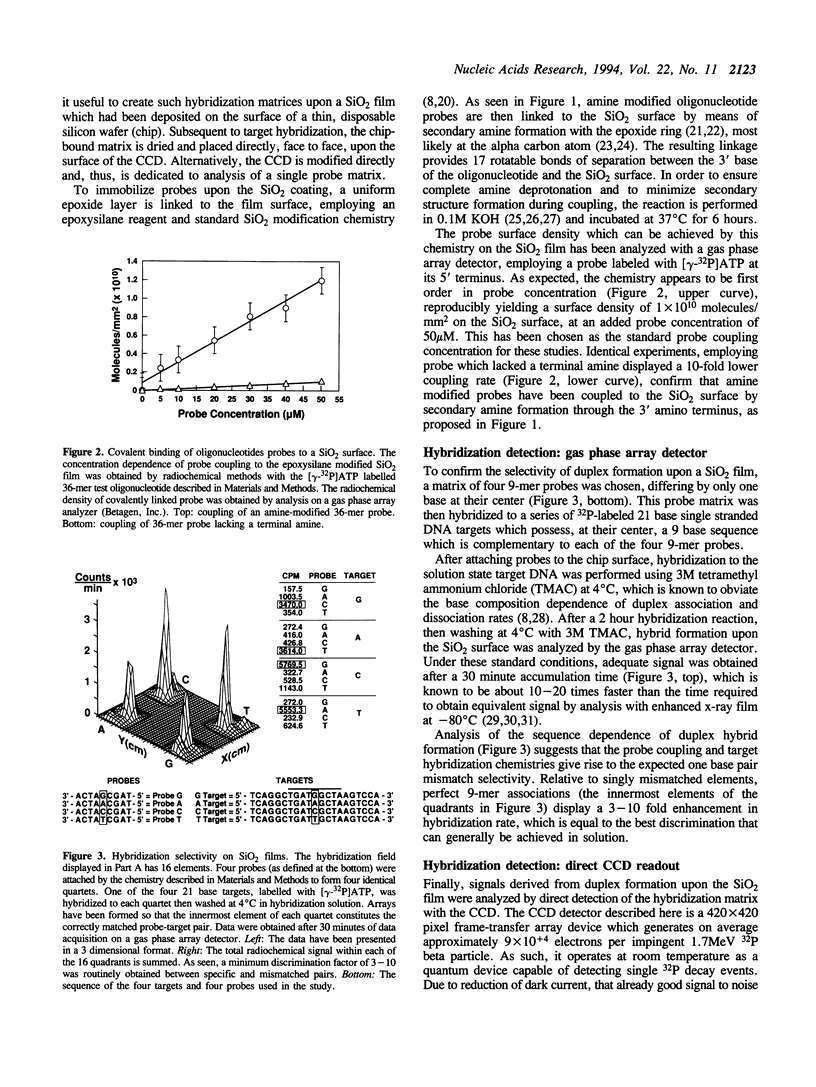
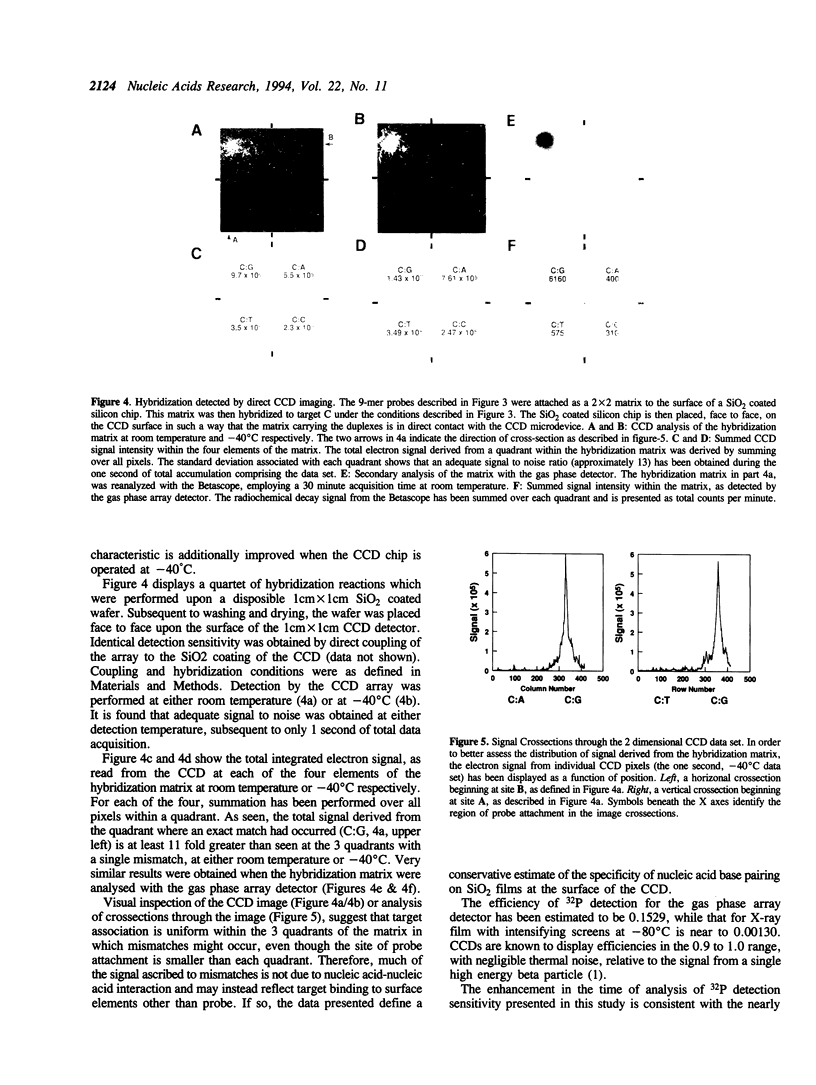
A method is described for the detection of DNA hybrids formed on a solid support, based upon the pairing of oligonucleotide chemistry and the technologies of electronic microdevice design. Surface matrices have been created in which oligonucleotide probes are covalently linked to a thin SiO2 film. 32P labeled target nucleic acid is then hybridized to this probe matrix under conditions of high stringency. The salient feature of the method is that to achieve the highest possible collection efficiency, the hybridization matrix is placed directly on the surface of a charge coupled device (CCD), which is used to detect 32P decay from hybridized target molecules (1, Eggers, M.D., Hogan, M.E., Reich, R.K., Lamture, J.B., Beattie, K.L., Hollis, M.A., Ehrilich, D.J., Kosicki, B.B., Shumaker, J.M., Varma, R.S., Burke, B.E., Murphy, A., and Rathman, D.D., (1993), Advances in DNA Sequencing Technology, Proc. SPIE, 1891, 13-26). Two implementations of the technology have been employed. The first involves direct attachment of the matrix to the surface of a CCD. The second involves attachment of the matrix to a disposible SiO2 coated chip, which is then placed face to face upon the CCD surface. As can be predicted from this favorable collection geometry and the known characteristics of a CCD, it is found that as measured by the time required to obtain equivalent signal to noise ratios, 32P detection speed by the direct CCD approach is at least 10 fold greater than can be obtained with a commercial gas phase array detector, and at least 100 fold greater than when X-ray film is used for 32P detection. Thus, it is shown that excellent quality hybridization signals can be obtained from a standard hybridization reaction, after only 1 second of CCD data acquisition.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bains W., Smith G. C. A novel method for nucleic acid sequence determination. J Theor Biol. 1988 Dec 7;135(3):303–307. doi: 10.1016/s0022-5193(88)80246-7. [DOI] [PubMed] [Google Scholar]
- Cate R. L., Ehrenfels C. W., Wysk M., Tizard R., Voyta J. C., Murphy O. J., 3rd, Bronstein I. Genomic Southern analysis with alkaline-phosphatase-conjugated oligonucleotide probes and the chemiluminescent substrate AMPPD. Genet Anal Tech Appl. 1991 May;8(3):102–106. doi: 10.1016/1050-3862(91)90044-r. [DOI] [PubMed] [Google Scholar]
- Conner B. J., Reyes A. A., Morin C., Itakura K., Teplitz R. L., Wallace R. B. Detection of sickle cell beta S-globin allele by hybridization with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1983 Jan;80(1):278–282. doi: 10.1073/pnas.80.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. M., Georgalis A. M., Atchison B. A. Direct sequencing of double-stranded PCR products incorporating a chemiluminescent detection procedure. Biotechniques. 1993 May;14(5):824–828. [PubMed] [Google Scholar]
- Drmanac R., Drmanac S., Labat I., Crkvenjakov R., Vicentic A., Gemmell A. Sequencing by hybridization: towards an automated sequencing of one million M13 clones arrayed on membranes. Electrophoresis. 1992 Aug;13(8):566–573. doi: 10.1002/elps.11501301115. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Drmanac S., Strezoska Z., Paunesku T., Labat I., Zeremski M., Snoddy J., Funkhouser W. K., Koop B., Hood L. DNA sequence determination by hybridization: a strategy for efficient large-scale sequencing. Science. 1993 Jun 11;260(5114):1649–1652. doi: 10.1126/science.8503011. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Strezoska Z., Labat I., Drmanac S., Crkvenjakov R. Reliable hybridization of oligonucleotides as short as six nucleotides. DNA Cell Biol. 1990 Sep;9(7):527–534. doi: 10.1089/dna.1990.9.527. [DOI] [PubMed] [Google Scholar]
- Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991 Feb 15;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Karger A. E., Weiss R., Gesteland R. F. Digital chemiluminescence imaging of DNA sequencing blots using a charge-coupled device camera. Nucleic Acids Res. 1992 Dec 25;20(24):6657–6665. doi: 10.1093/nar/20.24.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K. R., Lysov YuP, Khorlin A. A., Ivanov I. B., Yershov G. M., Vasilenko S. K., Florentiev V. L., Mirzabekov A. D. A method for DNA sequencing by hybridization with oligonucleotide matrix. DNA Seq. 1991;1(6):375–388. doi: 10.3109/10425179109020793. [DOI] [PubMed] [Google Scholar]
- Kiseleva V. I., Kolesnik T. B., Turchinsky M. F., Wagner L. L., Kovalenko V. A., Plaksin DJu, Koukalová B., Kuhrová V., Brábec V., Poverenny A. M. Trans-diamminedichlorplatinum (II)-modified probes for detection of picogram quantities of DNA. Anal Biochem. 1992 Oct;206(1):43–49. doi: 10.1016/s0003-2697(05)80008-9. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Maskos U., Southern E. M. Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesised in situ. Nucleic Acids Res. 1992 Apr 11;20(7):1679–1684. doi: 10.1093/nar/20.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Orosz J. M., Wetmur J. G. DNA melting temperatures and renaturation rates in concentrated alkylammonium salt solutions. Biopolymers. 1977 Jun;16(6):1183–1199. doi: 10.1002/bip.1977.360160603. [DOI] [PubMed] [Google Scholar]
- Pevzner P. A., Lysov YuP, Khrapko K. R., Belyavsky A. V., Florentiev V. L., Mirzabekov A. D. Improved chips for sequencing by hybridization. J Biomol Struct Dyn. 1991 Oct;9(2):399–410. doi: 10.1080/07391102.1991.10507920. [DOI] [PubMed] [Google Scholar]
- Southern E. M., Maskos U., Elder J. K. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992 Aug;13(4):1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- Strezoska Z., Paunesku T., Radosavljević D., Labat I., Drmanac R., Crkvenjakov R. DNA sequencing by hybridization: 100 bases read by a non-gel-based method. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10089–10093. doi: 10.1073/pnas.88.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Shank P. R. X-Ray Intensifying Screens Greatly Enhance the Detection by Autoradiography of the Radioactive Isotopes 32P and 125I. Anal Biochem. 1978 May;86(1):184–192. doi: 10.1016/0003-2697(78)90333-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Coyne M. Y., Will S. G., Levenson C. H., Kawasaki E. S. Single-base mutational analysis of cancer and genetic diseases using membrane bound modified oligonucleotides. Nucleic Acids Res. 1991 Jul 25;19(14):3929–3933. doi: 10.1093/nar/19.14.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]



