Abstract
Background
Tuning of the olfactory system of male moths to conspecific female sex pheromones is crucial for correct species recognition; however, little is known about the genetic changes that drive speciation in this system. Moths of the genus Ostrinia are good models to elucidate this question, since significant differences in pheromone blends are observed within and among species. Odorant receptors (ORs) play a critical role in recognition of female sex pheromones; eight types of OR genes expressed in male antennae were previously reported in Ostrinia moths.
Methodology/Principal Findings
We screened an O. nubilalis bacterial artificial chromosome (BAC) library by PCR, and constructed three contigs from isolated clones containing the reported OR genes. Fluorescence in situ hybridization (FISH) analysis using these clones as probes demonstrated that the largest contig, which contained eight OR genes, was located on the Z chromosome; two others harboring two and one OR genes were found on two autosomes. Sequence determination of BAC clones revealed the Z-linked OR genes were closely related and tandemly arrayed; moreover, four of them shared 181-bp direct repeats spanning exon 7 and intron 7.
Conclusions/Significance
This is the first report of tandemly arrayed sex pheromone receptor genes in Lepidoptera. The localization of an OR gene cluster on the Z chromosome agrees with previous findings for a Z-linked locus responsible for O. nubilalis male behavioral response to sex pheromone. The 181-bp direct repeats might enhance gene duplications by unequal crossovers. An autosomal locus responsible for male response to sex pheromone in Heliothis virescens and H. subflexa was recently reported to contain at least four OR genes. Taken together, these findings support the hypothesis that generation of additional copies of OR genes can increase the potential for male moths to acquire altered specificity for pheromone components, and accordingly, facilitate differentiation of sex pheromones.
Introduction
Evolution of genes responsible for sex pheromone communication in moths is an attractive model for investigating the relationship between the divergence of genes and mechanisms of speciation. Release of sex pheromones from female moths is believed to play a critical role in species recognition [1]–[2]. However, little is known about the changes that have occurred in the genomes of newly derived species that use pheromones different from the ancestral one.
Sex pheromones of moths are usually a blend of a few compounds synthesized from common fatty acids through desaturation, chain shortening and other modifications [3], and some mutations of genes involved in the pheromone biosynthesis pathway cause changes in pheromone composition [4]. An important question is how the pheromone recognition system in males can adapt rapidly to the changes that have occurred in the female pheromone biosynthetic pathway.
The genus Ostrinia, which includes the European corn borer, Ostrinia nubilalis, and the Asian corn borer, Ostrinia furnacalis, is an excellent model for studying the evolution of the pheromone biosynthesis and recognition systems, since various species show distinct differentiation in sex pheromones despite a relatively short period after speciation [5]. Sex pheromones of nine Ostrinia species have been characterized to date and six compounds have been found as the components of female sex pheromones (Fig. S1) [6]. O. nubilalis and its closely related congener, Ostrinia scapulalis, are unique in showing intraspecific variations in their pheromone blends; Z-type, I-type (hybrid), and E-type (Fig. S1) [7]–[9]. A major advantage of using the Ostrinia moths is that genetical analysis can be conducted by interspecific crosses and intraspecific crosses between different pheromone races. For example, crosses between E- and Z-strains of O. nubilalis revealed that the genetic factor responsible for the difference in female pheromone blend production is autosomal [10]–[11]. Recently, this autosomal locus was shown to be an allelic variation in a fatty-acyl reductase gene which is specifically expressed in the pheromone gland [4].
Similar autosomal loci controlling the pheromone blend were identified by crossing other combinations of Ostrinia species [12]–[13]. Although both O. scapulalis and O. furnacalis have genes encoding Δ11-desaturase and Δ14-desaturase, key enzymes in pheromone biosynthesis, only Δ11-desaturase is functionally expressed in O. scapulalis, [14] and so is Δ14-desaturase in O. furnacalis [15]. We have shown that transcription of mRNA from these desaturase genes occurs species specifically [16], which might be under the control of an autosomal locus responsible for the difference in pheromone blend.
By contrast, the locus responsible for male behavioral response to sex pheromone was reported to be Z-linked in O. nubilalis [10]–[11], [17]. The most likely candidates for this locus are pheromone receptors. A lepidopteran pheromone receptor was first identified as an odorant receptor (OR) specific to bombykol, a pheromone component of the silkworm, Bombyx mori [18]–[20]; ORs of four other moths showed responses to major pheromone components of their own [21]–[22]. It was reported recently that a locus responsible for the differential male response to pheromone compounds between Heliothis subflexa and H. virescens was linked to four OR genes [23].
We have subsequently reported isolation of genes encoding male-specific (OR1, 3–6, 8) and non-male-specific (OR7) ORs from eight Ostrinia moth species [24]–[25]. These OR genes show high similarity to known lepidopteran sex pheromone receptor subfamily genes [24]–[25]. When co-expressed with an Or83b homologue (OscaOR2), some ORs (OscaOR1, 3–5) of E-type O. scapulalis were observed to respond to several pheromone components used by Ostrinia moths; however, the specific response of males to their own pheromone blend could not be explained by the specificity of the observed ORs alone [24]–[25]. Wanner and colleagues independently reported isolation of five OR genes from Z-race O. nubilalis, four of which were consistent with those in our reports [26]; however, the gene names (hereafter, abbreviated as OnOr1–5) were not identical to ours (hereafter, abbreviated as OnubOR1–8) except for the OR2 gene. The one not found in O. scapulalis, OnOr6, was reported to be highly specific for (Z)-11-tetradecenyl acetate, the main component of the Z-race O. nubilalis pheromone blend [26].
Here, we report the chromosomal mapping and genomic organization of the OR genes described above. We screened an O. nubilalis BAC library for clones containing OR genes using cDNA sequences from O. scapulalis, which we subsequently used for FISH analysis in O. nubilalis and sequence determination. At least seven OR genes were in tandem arrays on the Z chromosome of O. nubilalis; a 181-bp direct repeat sequence was conserved among four of them. This is the first report of clustering of lepidopteran sex pheromone receptor subfamily genes. The chromosomal region where the cluster was located, determined by FISH analysis, was orthologous to BmOr1, a sex pheromone receptor gene of B. mori.
Results
Isolation of O. nubilalis BAC clones containing OR genes
To characterize the genomic organization of the O. nubilalis OR genes, we isolated BAC clones containing OR genes from an O. nubilalis BAC library by PCR-based screening in the same manner as described previously [27]. Since we had started the screening before determining cDNA sequences of the O. nubilalis OR genes, we used the O. scapulalis OR genes for designing primers. Consequently, we could isolate one or more positive clones from the library for each gene (Table S1).
The OnubOR2 gene, a single O. nubilalis ortholog of OscaOR2, was localized on one BAC clone, 07H10. Four BACs containing exons 7–8 of the previously reported OnOr6 gene [26] were identified, two of which also contained exons 1–3 (Table S1). Additional OR genes were co-localized on two other groups of BACs using primers derived from O. scapulalis cDNAs, indicating that they were clustered. One group of three clones was found to contain both of the OscaOR1/3 genes and six clones were isolated by primers for the OscaOR4–8 genes (Table S1). With the exception of OnOr6 which had already been published [26], we named each gene found on the O. nubilalis BAC clones according to the species from which the primer sequences were derived (Table S2).
Chromosomal locations of OR genes revealed by BAC-FISH analysis
Although the O. nubilalis OR genes were localized to four BAC contigs, this did not necessarily mean that these clones were separately located. To identify chromosomal locations of the OnubOR1–8 and OnOr6 genes, we performed BAC-FISH analysis of the clones, 07H10, 14B20, 44E03 and 50D02, representing the OnubOR2, OnubOR5/7, OnOr6 and OnubOR1/3 genes, respectively (Table S2). We also selected 32P24 as a probe since the size of the introns of the OnubOR7 gene on this clone was different from other clones.
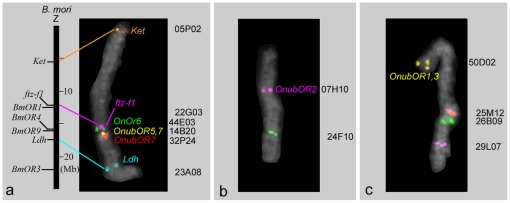
We previously reported the existence of significant synteny between B. mori, and the tobacco hornworm, Manduca sexta, by BAC-FISH [27]. In a parallel study, we isolated BACs containing O. nubilalis orthologs of B. mori genes [28] which could be used as specific probes for each chromosome. We first examined whether any OR genes were Z-linked since both the Resp locus responsible for male behavioral response to sex pheromone [11], [17] and the BmOr1 gene encoding a major sex pheromone receptor in B. mori are located on the Z chromosome [18]. BACs containing Z-linked O. nubilalis genes encoding kettin (Ket), FTZ-F1 (ftz-f1) and lactate dehydrogenase (Ldh) were used as markers specific for the Z chromosome (Table S2). Signals of the 14B20, 32P24 and 44E03 probes were detected from neighboring positions on approximately one third of the Z chromosome, distal to the ftz-f1 gene, indicating that the OnubOR4–8 and OnOr6 genes comprised a large gene cluster (Fig. 1A). The locations of 14B20 and 32P24 were not identical (Fig. 1A), suggesting that these clones contained different copies of the OnubOR7 genes.
Figure 1. Identification of the chromosomal position of O. nubilalis OR genes by BAC-FISH analysis.
BAC probe codes are shown on the right of each chromosome image (for details, see Table S2). Lines indicate correspondence of orthologous genes between B. mori and O. nubilalis. A) Comparison of the Z chromosomes. A vertical bar on the left side represents the B. mori Z chromosome drawn to relative scale taken from Kaikobase (http://sgp.dna.affrc.go.jp/KAIKObase/). B,C) Images of O. nubilalis chromosomes orthologous to B. mori chromosomes 16(B) and 23(C).
Two other clones containing the OnubOR2 and OnubOR1/3 genes were found on autosomes. Since sequences of Or83b co-receptor genes are well conserved among lepidopterans [22], we speculated that the chromosomal locations of OnubOR2 and its B. mori ortholog, BmOr2, were also conserved. Since the BmOr2 gene was mapped onto chromosome 16 of B. mori, we used 24F10 harboring two genes (EL929838 and EL929540) whose B. mori orthologs are located on this linkage group as markers (Table S2) together with 07H10 containing the OnubOR2 gene in a FISH analysis. As expected, 07H10 and 24F10 were co-localized on the same chromosome (Fig. 1B).
For mapping of the OnubOR1 and 3 genes, genetic analysis was necessary since their B. mori orthologs had not been identified by sequence comparison [25]–[26]. Recombination by crossing-over does not occur in lepidopteran oogenesis so that genes on the same chromosome always co-segregate. This makes it possible to test whether markers belong to the same linkage groups by using backcross progeny from a mating of an F1 female and a homozygous parental male (termed “BF1”). Using twenty-four BF1 progeny between O. nubilalis and O. scapulalis, we found that the OnubOR1 and 3 genes co-segregated with a gene (Accession no. FS438672) whose B. mori orthologs are located on B. mori chromosome 23 (Table S2). Therefore, three BACs physically mapped onto the O. nubilalis counterpart of B. mori chromosome 23 were used as probes for FISH analysis with 50D02 representing the OnubOR1 and 3 genes (Table S2). The signal from 50D02 was detected near the end of the chromosome where the signals from three positive controls were located (Fig. 1C).
Duplication of the OR5 group genes
To characterize the genomic organization of the OR genes, we sequenced the BAC clones (08K04 and 11K16) that contained O. nubilalis OR genes other than OnubOR2 and OnOR6.
In the previous report, we found multiple OR5 and OR6 genes for each Ostrinia species; however, OR5 and OR6 genes form a single clade without separating into OR5 and OR6 groups [25]. Thus, we classified these genes as variants of a newly defined OR5 group, which was also useful for avoiding confusion with the OnOr6 gene independently reported by Wanner et al. [26]. We also identified the O. scapulalis orthologue of the OnOr6 gene and confirmed that it was transcribed in male antennae.
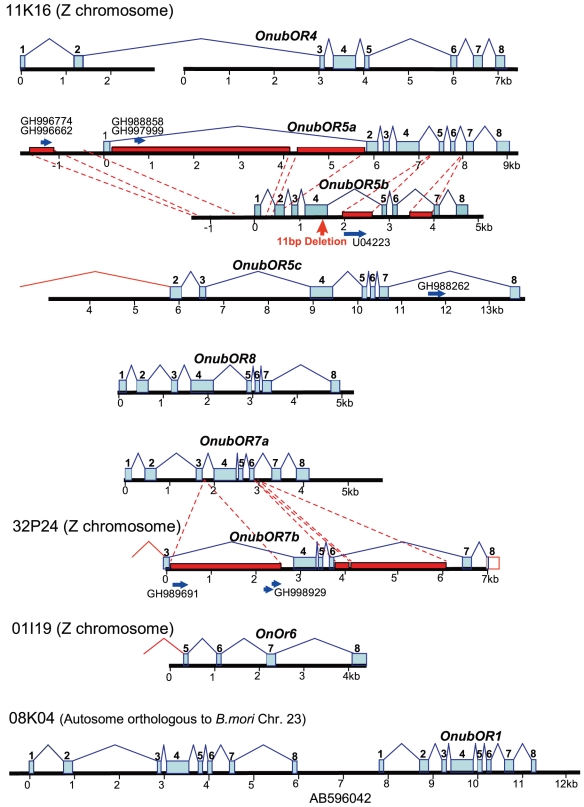
Sequencing of the BAC clone 11K16 revealed three apparently independent copies of the OnubOR5 gene. Exons for two of them were nearly identical to the OscaOR5, OnubOR6 [25] and OnOr1 [26] genes, so we designated them as OnubOR5a and 5b. The OnubOR5b gene contained an 11-bp deletion in exon 4 which caused a frameshift mutation leading to a truncated product (Fig. 2).
Figure 2. Structure of O. nubilalis OR genes determined in this study.
Numbers under bars indicate distance from putative transcriptional start sites (kbp) when all the exons and introns are identified. Blue squares represent exons and dotted squares indicate unidentified exons. Dotted lines show the correspondence of conserved sequences between the OnubOR5a/b, 7a/b genes and red squares indicate sequences solely appearing in one of the genes. Blue arrows represent putative repetitive elements found in O. nubilalis ESTs and genomic sequences.
Significant similarity was observed between the OnubOR5a and 5b genes including introns and flanking regions (Fig. 2). Non-conserved sequences were inserted into the upstream region and intron 1 of the OnubOR5a genes and introns 4 and 6 of the OnubOR5b genes (Fig. 2). Some of the additional sequences showed high partial similarity to O. nubilalis ESTs and genomic sequences, suggesting insertion of repetitive elements (Fig. 2). For example, the insertion into intron 4 of the OnubOR5b gene showed high similarity to intron 2 of the alpha-amylase gene (Accession no. U04223) and to an EST of the spruce budworm, Choristoneura fumiferana (Accession no. FC952039). Since the superfamily Tortricoidea to which C. fumiferana belongs is estimated to have diverged from advanced Lepidoptera soon after the lepidopteran radiation [29]–[30], the evolutionary origin of the insertion sequence is old. However, no similar sequences were found in the B. mori genome database nor in published sequences of other lepidopteran species.
The third copy, designated as OnubOR5c, showed a relatively low degree of similarity to the known Ostrinia OR5 genes; nevertheless, it was included in the OR5 clade (Fig. 3). Neither deletions nor nonsense mutations were found in it. In addition, we could not identify exon 1 of the gene, although more than 5 kb of genomic sequences was determined for the putative region corresponding to it (Fig. 2).
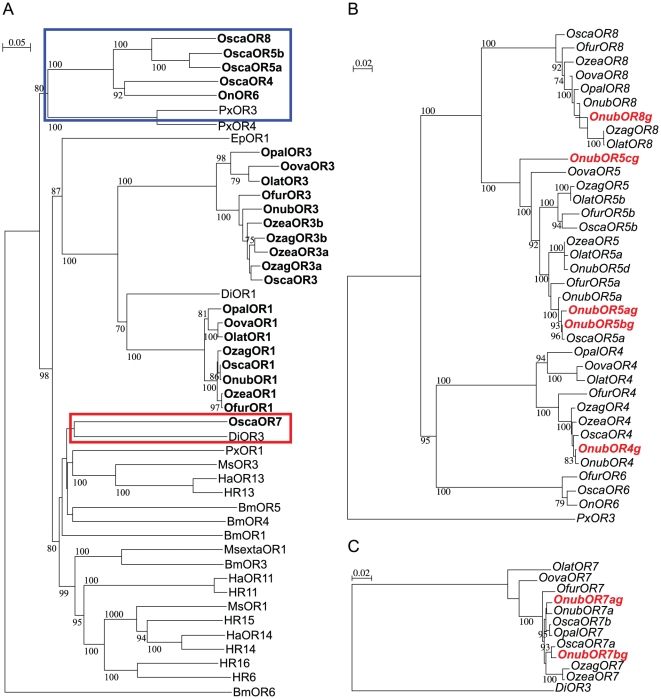
Figure 3. Phylogenetic relationship of lepidopteran OR genes belonging to the sex pheromone receptor subfamily.
Blue and red boxes in panel A indicate regions separately shown in panels B and C, detailed phylogenetic trees of OR4, 5, 8 and OnOr6 genes (B) and OR7 genes (C) of Ostrinia moths. Detailed information of genes used for comparison is listed in Table S4.
Tandem repeats spanning exon 7 and intron 7 of the OnubOR5/8 genes
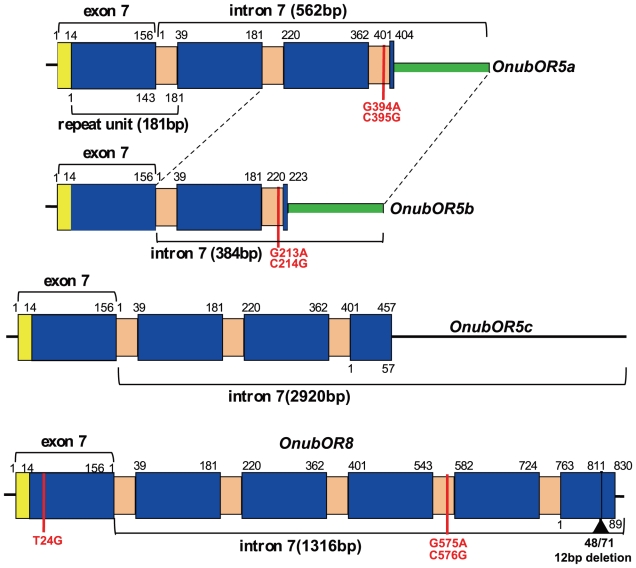
We found 181-bp tandem repeats located from exon 7 to the 5′- end of intron 7 of the OnubOR5a,b,c genes. The repeat unit was composed of a 143-bp portion identical to nucleotides 14–156 of exon 7 and a 38-bp portion unique to the repeat. The OnubOR5a,b,c genes contained three, two and three complete repeats followed by a truncated exon 7-like sequence (Fig. 4).
Figure 4. The structure of the 181-bp repeat sequences.
Yellow squares indicate exon 7 sequences not included in the repeat. Blue squares indicate sequences shared with exon7 and repeats in intron 7. Pink squares indicate intron7-specific sequences. Green squares indicate sequences conserved among the OnubOR5a,b genes. Red vertical lines show base substitutions. Black vertical line indicates a putative deletion in the last truncated repeat of the OnubOR8 gene.
The OR4,5,8 genes of several Ostrinia moths and the OnOr6 gene formed a definite clade which did not include any known OR genes of other Lepidoptera (Fig. 3A) [25]–[26]. This raised the possibility that the OnubOR4,8 and OnOr6 genes were also duplicated or contained the 181-bp repeat, so we determined their genomic sequences. Consequently, we found that the OnubOR8 gene contained five complete 181-bp repeats and one that was truncated (Fig. 4). No repeats were found in the OnubOR4 gene, nor did we find additional copies of the OnubOR4,8 genes within clone 11K16 (Fig. 2). We also determined the genomic sequence containing exons 5–8 of the OnOr6 gene by PCR amplification of BAC clone 01I19 (Fig. 2); however, the 181-bp repeat was not present. We also confirmed that the O. scapulalis orthologue of the OnOr6 gene was transcribed in male antennae, and designated it as OscaOR6 (Fig. 3B).
It is noteworthy that there was one base substitution in exon 7 of the OnubOR8 gene despite the identical repeat sequence and a 2-bp base substitution was conserved among all of the OnubOR5a,b and 8 genes (Fig. 4). No significant similarity was observed between exon 8 of the OnubOR8 and OnubOR5a,b,c genes, in contrast to extensive similarities through exons 1–7.
Genome sequencing encompassing exons 3 and 4 of the OR5b gene
To confirm whether deletions found in the BAC sequence of 11K16 occurred in other individuals, we determined genomic sequences encompassing exons 3–4 of the OnubOR5b genes using other BAC clones, as well as in one female and four male BF1 progeny from the crosses between O. nubilalis and O. scapulalis used for linkage analysis. The sequence of 53I05 was identical to 11K16. Altogether, we found one allele from clone 14B20 and six alleles from BF1 progeny, which were not identical to 11K16 (Fig. S2). The 11-bp deletion was not observed in other alleles; however, four of them harbored one of three indels in exon 4 causing frameshift mutations (Fig. S2).
Genome organization of the OnubOR7 genes
As described above, two copies of the OnubOR7 genes were found separately on clones 11K16 and 32P24. The sequence of the gene on 11K16 was determined in the same manner as described above, and eight exons corresponding to the cDNA sequence of the OscaOR7 gene were found, which we designated as OnubOR7a (Fig. 2).
Direct amplification was carried out on the gene on 32P24, designated as OnubOR7b, using primers designed from exon sequences of the OscaOR7 gene. Six exons (putative exons 3–8) and five introns (putative introns 3–7) were determined. Sequence identity of the OnubOR7b gene to the OscaOR7 gene was clearly higher than that of the OnubOR7a gene (Fig. 3C). We had previously identified a transcript nearly identical to the OnubOR7a gene from male antennae of O. scapulalis (Fig. 3C); however, we had interpreted it as an intraspecific variation of a single locus. We renamed the previously reported one as OscaOR7a and designated the other as OscaOR7b (Table S3), since it was likely that two types of OR7 genes were expressed in O. scapulalis.
The genomic sequences of the OnubOR7a and b genes were highly conserved including most of the introns; however, two observed differences in intron size were due to insertions detected in introns 3 and 6 of the OnubOR7b gene (Fig. 2). Interestingly, portions of both ends of intron 3 were very similar to O. nubilalis ESTs (Accession nos. GH989691 and GH998929) (Fig. 2). Direct repeats on the 3′- end also showed significant similarity to numerous genomic sequences in B. mori and to ESTs of a butterfly, Bicyclus anynana, suggesting that this insertion was a transcribed repeated sequence with conserved motifs (Fig. 2).
Genome sequence of the OnubOR1 and OnubOR3 genes
Since the OscaOR1 and OscaOR3 genes show significant sequence similarity [25] and their O. nubilalis orthologs were co-localized on the same BAC clones (Table S1), we speculated that they were also created through a gene duplication and remained closely located. Thus, we carried out extensive sequencing to construct a single contig. Ultimately, we determined approximately 13.8-kb genomic sequence and revealed that the OnubOR3 gene was located in a 1.85-kb interval upstream of the OnubOR1 genes (Fig. 2).
Tandem array of the OR genes on the Z chromosome
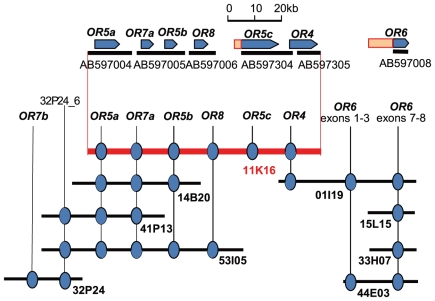
Finding a tandem array of the OnubOR1 and 3 genes strengthened our speculation that the OR genes on 11K16 might also be in a tandem array. To identify the complete organization of the OR gene cluster, we continued to sequence the intergenic regions between the OR genes described above. Consequently, we determined two scaffolds of more than 42.2-kb and 27.9-kb for clone 11K16, the former (Accession nos. AB597004–AB597006) containing the OnubOR5a-7a-5b-8 genes and the latter (Accession nos. AB597304–AB597305) containing the OnubOR5c-4 genes in tandem arrays (Fig. 5). Since the 5′-end of AB597004 and the 3′-end of AB597305 were consistent with the distal ends of clone 11K16, the order of the six OR genes was OnubOR5a-7a-5b-8-5c-4 (Fig. 5). We carried out PCR amplification against clones 01I19, 11K16 and 44E03 to confirm whether they overlapped, and found that clone 01I19 contained the OnubOR4 gene (Fig. 5). Judging from the overlaps with other BAC clones, the overall gene order was OnubOR5a-7a-5b-8-5c-4-OnOr6 (Fig. 5).
Figure 5. The order of O. nubilalis OR genes on the Z chromosome.
Pentagons represent the position, length and transcriptional orientation of OR genes. Bars under the pentagons indicate the sequenced contigs of clones 11K16 and 01I19.
A genomic marker, 32P24_6, designed from the genomic sequence of a randomly selected subclone of 32P24 was also positive for 41P13 and 53I05, indicating that the OnubOR7b gene was located upstream of the OR gene cluster (Fig. 5). This result was consistent with the FISH results indicating that the OnubOR7b and OnOr6 genes were on opposite ends of the OR gene cluster (Fig. 1).
Discussion
The number and chromosomal distribution of insect OR genes vary among species [31]–[32]. The first sequenced insect, Drosophila melanogaster, has 62 OR genes which are scattered throughout the genome [33]. In contrast, other sequenced Diptera, the mosquitoes, Anopheles gambiae and Aedes aegypti, have more OR genes, 79 and 131, respectively, which are in many cases clustered [34]–[35]. There are also significantly many clustered OR genes, 163 and 299, in the honeybee, Apis mellifera, and the red flour beetle, Tribolium castaneum, representing Hymenoptera and Coleoptera [36]–[37].
B. mori was recently reported to have 66 OR genes [38], many of which are dispersed throughout the genome (Table S4). Most clusters contain pairs and triplets, but neither the BmOr1 nor the BmOr3 genes encoding pheromone receptors are clustered (Table S4). We found at least eight genes in the cluster on the Z chromosome described here (Fig. 5), which is the largest lepidopteran OR gene cluster reported to date. By linkage analysis using two congeneric moths, H. subflexa and H. virescens, Gould and colleagues revealed that the locus responsible for male response to female sex pheromone contains at least four OR genes (HR6,14–16 in Fig. 3A) [23]. These ORs are more closely related to each other than to any other Heliothis ORs [23], suggesting that the tightly linked genes were generated by gene duplication. These observations reinforce the attractive hypothesis that significant differences in sex pheromone blends of the Ostrinia and Heliothis moths are associated with the clustered OR genes.
It is likely that mutations in genes of the female pheromone biosynthesis pathway causing changes in pheromone blend must precede those of the male response system [39]. For example, Lassance and colleagues showed that intraspecific variations in the sex pheromone blend of O. nubilalis could be explained by the sequence divergence of a fatty-acyl reductase gene that affected its substrate specificity [4]. Pre- or immediate adaptation of male moths to such sudden changes is critical for emergence of a population using novel pheromone blends, and it is unlikely that a great change in the specificity of pheromone receptors is caused by mutations at the same time as those in the pheromone biosynthesis pathway.
On the other hand, there are “rare” Ostrinia males (3–5%) that possess the ability to respond to a pheromone blend which is not effective for most individuals in the population [15], [40]–[41]; moreover, significant differences in olfactory neuron responsiveness have been observed between rare and normal males [42]–[43]. Tandemly arrayed genes are thought to be subject to birth-and-death evolution, in which new genes are created by duplication. Some of these are retained in the genome as functional genes, but others are inactivated or eliminated from the genome [44]. Adaptation to a novel pheromone blend might be gained by altered specificity of newly created genes which are not initially under high selective pressure.
Genome sequencing revealed the presence of novel OR genes, the OnubOR5b and 5c genes, which had not been detected by RT-PCR analysis. As described above, the OnubOR5b gene seemed to be nonfunctional because insertions or deletions causing frameshift and truncation of the product occurred independently in several alleles (Fig. S2). Deletions similar to the OnubOR5b gene were observed in the strain used in the previous report [25]. Ninety-bp and 5-bp deletions were located in exon 4 of the OnubOR5 (renamed OnubOR5d) and OnubOR8 genes, respectively
The incongruence of the phylogenetic trees between the OR5/8 genes and the mitochondrial gene cytochrome oxidase II (COII) (Fig. S1) or other OR genes (Fig. 3A) also supports the hypothesis that the OR5/8 genes are undergoing a process of birth-and-death evolution. It is likely that the major OR5/8 genes expressed in each species are not necessarily orthologous due to gene duplication and independent gene inactivation. Since detailed sequencing was performed only for the clone 11K16 in this study, other OR genes might be located outside of it. In fact, we could not find an O. nubilalis ortholog of the OscaOR5b gene (Fig. 3B) in any of the BAC clones we investigated.
It is possible that gene duplications occurred via the 181-bp repeat which was conserved among the OR5/8 genes (Fig. 4), since the OR4 gene lacking the repeat seems to be relatively stable and all the Ostrinia species examined retained functional OR4 genes during speciation [25]. A number of inactivated copies of Δ11-desaturase genes were reported in O. nubilalis and O. furnacalis, and unequal crossover via a retrotransposon, ezi, was proposed to cause gene duplication [45]. Thus, we came up with a similar hypothesis that the 181-bp repeat enhances duplication of OR genes containing it. The presence of a 2-bp substitution which is conserved among the tandemly linked OnubOR5a,b and 8 genes is strong evidence for unequal crossovers via the 181-bp repeat, since it is very rare that such substitutions occurr independently. Recently, Heckel described the potential involvement of tandem gene duplication for the immediate creation of a novel phenotype in the sexual communication system [46]. The results reported here may be the first evidence supporting this idea. Further analysis of OR gene clusters including those of Ostrinia moths will reveal more details of the birth-and-death process of the OR5/8 genes including ones which have been inactivated.
The OR7 genes were also duplicated and both genes were transcribed in O. scapulalis (Fig. 3C). However, the mechanism of duplication seems to be different from that of the OR5/8 genes, since repeats like the OR5/8 genes were not present (Fig. 2). In the previous report, Osca7a showed no significant response to any of the pheromone compounds used by the Ostrinia moths, even though the OR7 genes were more similar to the sex pheromone receptor genes of other Lepidoptera than the male-specific Ostrinia OR genes [25]. The DiOR3 gene of the cotton caterpillar moth, Diaphania indica, similar to the OR7 genes (Fig. 3A), was also expressed in antennae of both males and females [22]. Since D. indica belongs to Pyraloidea, the same superfamily as the genus Ostrinia, the DiOR3 and Ostrinia OR7 genes might have lost a male-specific expression pattern in the lineage leading to Pyraloidea.
The OR gene cluster revealed by FISH analysis was located at approximately one third of the distance from the end of the Z chromosome near the ftz-f1 gene (Fig. 1A) which was consistent with the position of the BmOr1 gene (Table S1, S3). Thus, it is possible that the OR7 genes in the Ostrinia OR gene cluster and the BmOr1 gene evolved from a common ancestral gene, although evidence of other orthologous genes located in the neighboring chromosomal region will be needed to verify this hypothesis. On the other hand, no OR genes similar to OR4,5,8 or OnOr6 have been isolated from other advanced Lepidoptera including B. mori, which has been fully sequenced [47]. PxOR3 and PxOR4 isolated from the diamondback moth, Plutella xylostella, are the only reported OR genes which formed a clade with the OR4,5,8 and OnOr6 genes (Fig. 3A, B). Since Yponomeutoidea, which includes P. xylostella, is thought to have diverged earlier than the split of Pyraloidea from other advanced Lepidoptera [29]–[30], it is likely that this type of OR gene was lost or inactivated in the lineage leading to B. mori.
The OnubOR1 and 3 genes are autosomal (Fig. 1) and thus do not directly determine male pheromone behavioral response in O. nubilalis since the locus responsible for the response is reported to be Z-linked [10]–[11], [17]. This is consistent with our previous results indicating that OscaOR1 and OlatOR1 specifically responded to (E)-11-tetradecenol, a single pheromone component of O. latipennis [24], and OscaOR3 responded to a wide variety of pheromone components used in the Ostrinia moths [25]. Similarly, the locus responsible for electrophysiological response in O. nubilalis was previously thought to be autosomal [10]; however, a recent genetic study showed that the response of pheromone sensitive sensilla may be affected by both autosomal and Z-linked genes [48]. Therefore, the OnubOR1 and 3 genes might also be involved in pheromone recognition.
The Ostrinia OR1 genes showed higher similarity to the DiOR1 gene which encodes a putative sex pheromone receptor of D. indica [22], compared with the Ostrinia OR3 genes. This raises a question whether the locus responsible for male pheromone response is autosomal or sex-linked in D. indica or O. latipennis. As seen with OscaOR1 and OlatOR1, the existence of receptors specifically responding to a pheromone compound is necessary but not sufficient to explain species specificity in the pheromone communication system.
A remaining question is whether the OR gene cluster reported here is equivalent to the Resp locus responsible for the Ostrinia male behavioral response to sex pheromone. In molecular genetic mapping experiments, Dopman and colleagues reported that the gene order is Ket-Tpi (triosephosphate isomerase)-Ldh-Resp, with Ket at one end and Ldh in the middle of the map [17]. However, our FISH results indicated that Ldh was near the end of the Z chromosome far removed from Ket (Fig. 1). The fact that polymorphisms in Tpi are consistent with the behavior of E- and Z-type pheromones [17] also suggests that markers were incorrectly ordered in the genetic map and Resp is actually located near Tpi which resides between Ket and Ldh. These results are not in conflict with our assumption that the OR gene cluster plays a critical role in determining male behavioral response to sex pheromone. Recent findings that male electrophysiological response is affected by Z-linked genes [48] strengthens the possibility that the Resp locus is equivalent to the OR gene cluster.
Detailed genetic dissection is needed to reveal the relationship between the OR gene cluster and the Resp locus; however, the available data are insufficient to identify the gene responsible for the Ostrinia male behavioral response. The existence of tightly linked multiple male-specific pheromone receptor genes will make it difficult to clarify which factor is definitive in determining male behavioral response to sex pheromone, as reported in the Heliothis moths [23]. In addition, it is still unclear how signals mediated by multiple ORs with narrow or broad specificity to pheromone compounds [25]–[26] are finally recognized as the definitive stimulus. Integrated approaches combining genetics, genomics, evolutionary biology and neurobiology are needed to reveal the detailed mechanisms underlying this complex trait.
Materials and Methods
PCR-based screening of the BAC library
An O. nubilalis BAC library, ON_Ba (average insert size 125kb, 36,864 clones), was obtained from the Clemson University Genomics Institute (Clemson, SC, USA). PCR-based screening of the library is described elsewhere [28]. The first screening was performed against DNA pools derived from 96 plates, using a mixture of 384 BAC-DNAs for each plate, followed by a second screening against DNA pools for 24 columns and 16 rows, each composed of mixtures of BAC-DNAs located in the same column or row. Primers used for the study were designed from cDNA sequences of OscaOR1-8 of O. scapulalis and the OnOR6 of O. nubilalis genes [24]–[26] (Table S1). As an exception, a single set of primer pairs was designed for the OscaOR5 and 6 genes, since sequence similarity between them was too high.
Crossing experiments
F1 females from matings between Z-type O. scapulalis females (originally collected in Matsudo, Japan) and Z-race O. nubilalis males (originally collected in Darmstadt, Germany) were backcrossed with Z-type O. scapulalis males, and genomic DNA was extracted from individual larvae and adults of the resultant BF1 progeny using an AquaPure Genomic DNA kit (Bio-Rad, Hercules, CA). DNA samples of twelve larvae and twelve adults (6 males and 6 females) were used for genotyping with the same PCR primers used for BAC isolation [28] in the same manner as described previously [49].
BAC-FISH analysis
We followed the procedure described previously [27] for multi-color BAC-FISH. Chromosome spreads were prepared from pachytene spermatocytes of O. nubilalis larvae. BAC-DNA was extracted with a Plasmid Midi kit (QIAGEN, Hilden, Germany) and labeled with a fluorochrome using a Nick Translation System (Invitrogen Carlsbad, CA, USA). For each probe one of four fluorochromes [Green-dUTP, Orange-dUTP, Red-dUTP (Abbott Molecular Inc., Des Plaines, IL, USA), and Cy5-dUTP (GE Healthcare UK, Buckinghamshire, UK)], was used.
Sequence determination of BAC clones
A shotgun library was constructed from equal amounts of BAC-DNAs from clones 08K04 and 11K16. Sonicated BAC-DNA ranging around 2 kb was inserted into the Hinc II site of plasmid pUC119. Shotgun clones were aliquoted into six 384-well microplates and DNA pools were made for 24 columns and 16 rows representing clones located in the same column or row. PCR screening was carried out against column and row DNA pools directly as described above.
Shotgun clones were first screened with primers used for BAC isolation. DNA templates were prepared using a DNA isolation kit (Kurabo, Japan), and sequenced with an ABI-3730xl DNA analyzer. Genome sequences assembled from overlapped shotgun clones isolated with the same primers were then used to design new primers to isolate clones located in the neighboring region. Remaining gaps were filled by direct PCR amplification of BAC-DNAs. PCR amplification was also performed against BAC clones, 32P24 and 01I19, to amplify flanking regions of exons of the OnubOR7b and OnOr6 genes. The resultant PCR products were cloned using the pGEM-T easy Vector System (Promega, Madison, MA, USA) and used as sequencing templates.
In all, 229 end- or internal sequences of 143 shotgun clones and six PCR products were used for assembly of the 42.2-kb scaffold containing the OnubOR5a-7a-5b-8 genes. Similarly, 168 sequences of 102 shotgun clones and three PCR products were used to assemble the 27.9-kb scaffold containing the OnubOR5c-4 genes. The 13.8-kb genomic sequence containing the OnubOR3-1 genes was constructed from 64 end-sequences of 41 shotgun clones. All sequences were submitted to DDBJ/GenBank/EBI Data Bank with accession numbers AB597004–AB597008, and AB597304–AB597305.
Phylogenetic analysis
Deduced amino acid sequences (Fig. 3A) or nucleotide sequences (Fig. 3B, C) were used for comparison. The nucleotide and amino acid sequences were aligned using Clustal X [50]. The phylogenetic tree was constructed with the neighbor-joining method using the software PHYLIP 3.66 [51]. Branch support was assessed by the bootstrap test with 1000 re-samplings.
Sequence determination of exons 3 and 4 of the OR5b gene
Amplification of genomic fragments containing exons 3 and 4 of the OnubOR5b gene was performed using BACs, 14B20, 41P13, 53I05 or genomic DNAs of BF1 progeny described above with PCR primers 5′-CGTTCACAGGTCCGTAGTA and 5′-ACTTTATCTCAGGCGACAT. Amplified fragments were then cloned using the pGEM-T easy Vector System (Promega) and used as templates for sequencing. All of the PCR products amplified from 41P13 corresponded to a portion of the OnubOR5a gene.
Supporting Information
Phylogenetic relationships (left) and sex pheromone blends (right) of Ostrinia species. The phylogenetic tree was constructed based on mitochondrial COII gene sequences. The numbers near branches indicate bootstrap values. The size of circles represents a rough blend ratio. Z-type, I-type (hybrid), and E-type females of O. scapulalis and O. nubilalis produce mixtures of 3:97, 64:36, and 99:1 (E)- and (Z)-11-tetradecenyl acetates, respectively
(PDF)
Alignment of allelic genomic sequences encompassing exons 3 and 4 of the OnubOR5b gene (Accession Nos. AB596043–AB596049). Gray-shaded boxes indicate exons 3 and 4. Yellow-shaded letters indicate insertions or deletions causing frameshift mutations. Asterisks indicate nucleotides conserved among all the alleles.
(DOC)
O. nubilalis BACs isolated by PCR-based screening.
(DOC)
BAC probes used for FISH analysis.
(DOC)
Detailed information of genes appearing in Figure 3 and summary of renaming of OR genes previously reported in ref. 25. “g” was added to names of genes deduced from genomic sequences.
(XLS)
Chromosomal distribution of OR genes in B. mori.
(DOC)
Acknowledgments
We are grateful to Dr. M. R. Goldsmith for critical reading of the manuscript and 3 anonymous reviewers for helpful suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and grants 22380040/19208005 from Japan Society for the Promotion of Science (JSPS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Symonds MRE, Elgar MA. The evolution of pheromone diversity. Trends Ecol Evol. 2008;23:220–228. doi: 10.1016/j.tree.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Cardé RT, Haynes KF. Structure of the pheromone communication channel in moths. In: Cardé RT, Millar JG, editors. Advances in Insect Chemical Ecology. Cambridge, UK: Cambridge University Press; 2004. pp. 283–332. [Google Scholar]
- 3.Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones –an overview of biosynthesis and endocrine regulation. Insect Biochem Mol Biol. 1999;29:481–514. doi: 10.1016/s0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 4.Lassance J-M, Groot AT, Liénard MA, Antony B, Borgwardt C, et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466:486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 5.Kim CG, Hoshizaki S, Huang YP, Tatsuki S, Ishikawa Y. Usefulness of mitochondrial COII gene sequences in examining phylogenetic relationships in the Asian corn borer, Ostrinia furnacalis, and allied species (Lepidoptera: Pyralidae). Appl Entomol Zool. 1999;34:405–412. [Google Scholar]
- 6.Ishikawa Y, Takanashi T, Kim C-G, Hoshizaki S, Tatsuki S, et al. Ostrinia spp. in Japan: their host plants and sex pheromones. Entomol Exp Appl. 1999;91:237–244. [Google Scholar]
- 7.Roelofs WL, Du JW, Tang XH, Robbins PS, Eckenrode CJ. Three European corn borer populations in New York based on sex pheromones and voltinism J Chem Ecol. 1985;11:829–836. doi: 10.1007/BF01012071. [DOI] [PubMed] [Google Scholar]
- 8.Ohno S, Huang YP, Tatsuki S, Honda H, et al. A sex pheromone component novel to Ostrinia identified from Ostrinia latipennis (Lepidoptera: Crambidae). Chemoecology. 2000;10:143–147. [Google Scholar]
- 9.Huang YP, Takanashi T, Hoshizaki S, Tatsuki S, Ishikawa Y. Female sex pheromone polymorphism in adzuki bean borer, Ostrinia scapulalis, is similar to that in European corn borer, O. nubilalis. J Chem Ecol. 2002;28:533–539. doi: 10.1023/a:1014540011854. [DOI] [PubMed] [Google Scholar]
- 10.Roelofs WL, Glover T, Tang XH, Sreng I, Robbins P, et al. Sex pheromone production and perception in European corn borer moths is determined by both autosomal and sex-linked genes. Proc Natl Acad Sci USA. 1987;84:7585–7589. doi: 10.1073/pnas.84.21.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dopman EB, Bogdanowicz SM, Harrison RG. Genetic mapping of sexual isolation between E and Z pheromone strains of the European corn borer (Ostrinia nubilalis). Genetics. 2004;167:301–309. doi: 10.1534/genetics.167.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabata J, Ishikawa Y. Genetic basis to divergence of sex pheromones in two closely related moths, Ostrinia scapulalis and O. zealis. J Chem Ecol. 2005;31:1111–1124. doi: 10.1007/s10886-005-4251-3. [DOI] [PubMed] [Google Scholar]
- 13.Takanashi T, Huang YP, Takahasi R, Hoshizaki S, Tatsuki S, et al. Genetic analysis and population survey of sex pheromone variation in the adzuki bean borer moth, Ostrinia scapulalis. Biol J Linn Soc. 2005;84:143–160. [Google Scholar]
- 14.Fukuzawa M, Fu X, Tatsuki S, Ishikawa Y. cDNA cloning and in situ hybridization of Δ11-desaturase, a key enzyme of pheromone biosynthesis in Ostrinia scapulalis (Lepidoptera: Crambidae). J Insect Physiol. 2006;52:430–435. doi: 10.1016/j.jinsphys.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Roelofs WL, Liu WT, Hao GX, Jiao HM Rooney AP, et al. Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci USA. 2002;99:13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai R, Fukuzawa M, Nakano R, Tatsuki S, Ishikawa Y. Alternative suppression of transcription from two desaturase genes is the key for species specific sex pheromone biosynthesis in two Ostrinia moths. Insect Biochem Mol Biol. 2009;39:62–67. doi: 10.1016/j.ibmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Dopman EB, Pérez L, Bogdanowicz SM, Harrison RG. Consequences of reproductive barriers for genealogical discordance in the European corn borer. Proc Natl Acad Sci USA. 2005;102:14706–14711. doi: 10.1073/pnas.0502054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, et al. The sex pheromone receptor in the silkworm moth Bombyx mori. Proc Natl Acad Sci USA. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krieger J, Grosse-Wilde E, Gohl T, Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori. Eur J Neurosci. 2005;21:2167–2176. doi: 10.1111/j.1460-9568.2005.04058.x. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 21.Grosse-Wilde E, Gohl T, Bouché E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- 22.Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, et al. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur J Neurosci. 2008;28:893–902. doi: 10.1111/j.1460-9568.2008.06429.x. [DOI] [PubMed] [Google Scholar]
- 23.Gould F, Estocka M, Hillierb NK, Powella B, Groota AT, et al. Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc Natl Acad Sci USA. 2010;107:8660–8665. doi: 10.1073/pnas.0910945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura N, Nakagawa T, Tatsuki S, Touhara K, Ishikawa Y. A male specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int J Biol Sci. 2009;5:319–330. doi: 10.7150/ijbs.5.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura N, Nakagawa T, Touhara K, Ishikawa Y. Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem Mol Biol. 2010;40:64–73. doi: 10.1016/j.ibmb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE. 2010;5:e8685. doi: 10.1371/journal.pone.0008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasukochi Y, Tanaka-Okuyama M, Shibata F, Yoshido A, Marec F, et al. Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping. PLoS ONE. 2009;4:e7465. doi: 10.1371/journal.pone.0007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasukochi Y, Tanaka-Okuyama M, Kamimura M, Nakano R, Naito Y, et al. Isolation of BAC clones containing conserved genes from libraries of three distantly related moths: a useful resource for comparative genomics of lepidoptera. J Biomed Biotech. 2011;2011:165894. doi: 10.1155/2011/165894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regier JC, Zwick A, Cummings MP, Kawahara AY, Cho S, et al. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol Biol. 2009;9:280. doi: 10.1186/1471-2148-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mutanen M, Wahlberg N, Kaila L. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc R Soc B. 2010;277:2839–2848. doi: 10.1098/rspb.2010.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- 33.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 35.Bohbot J, Pitts RJ, Kwon H-W, Rützler M, Robertson HM, et al. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol Biol. 2007;16:525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engsontia P, Sanderson AP, Cobb M, Walden KK, Robertson HM, et al. The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881–890. doi: 10.1016/j.cub.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 39.Roelofs WL, Rooney AP. Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc Natl Acad Sci USA. 2003;100:9179–9184. doi: 10.1073/pnas.1233767100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linn CE, O'Connor M, Roelofs WL. Silent genes and rare males: a fresh look at pheromone blend response specificity in the European corn borer moth, Ostrinia nubilalis. J Insect Sci. 2003;3:15. [PMC free article] [PubMed] [Google Scholar]
- 41.Linn CE, Musto CJ, Roelofs WL. More rare males in Ostrinia: response of Asian corn borer moths to the sex pheromone of the European corn borer. J Chem Ecol. 2007;33:199–212. doi: 10.1007/s10886-006-9204-y. [DOI] [PubMed] [Google Scholar]
- 42.Domingue MJ, Musto CJ, Linn CE, Roelofs WL, Baker TC. Evidence of olfactory antagonistic imposition as a facilitator of evolutionary shifts in pheromone blend usage in Ostrinia spp. (Lepidoptera: Crambidae). J Insect Physiol. 2007;53:488–496. doi: 10.1016/j.jinsphys.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Domingue MJ, Musto CJ, Linn CE, Roelofs WL, Baker TC. Altered olfactory receptor neuron responsiveness in rare Ostrinia nubilalis males attracted to the O. furnacalis pheromone blend. J Insect Physiol. 2007;53:1063–1071. doi: 10.1016/j.jinsphys.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue B, Rooney AP, Kajikawa M, Okada N, Roelofs WL. Novel sex pheromone desaturases in the genomes of corn borers generated through gene duplication and retroposon fusion. Proc Natl Acad Sci USA. 2007;104:4467–4472. doi: 10.1073/pnas.0700422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckel DG. Smells like a new species: Gene duplication at the periphery. Proc Natl Acad Sci USA. 2010;107:9481–9482. doi: 10.1073/pnas.1004511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Olsson SB, Kesevan S, Groot AT, Dekker T, Heckel DG, et al. Ostrinia revisited: Evidence for sex linkage in European Corn Borer (Hubner) pheromone reception. BMC Evol Biol. 2010;10:285. doi: 10.1186/1471-2148-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasukochi Y. A simple and accurate method for generating co-dominant markers: an application of conformation-sensitive gel electrophoresis on the silkworm linkage analysis. Mol Gen Genet. 1999;261:796–802. doi: 10.1007/s004380050023. [DOI] [PubMed] [Google Scholar]
- 50.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 51.Felsenstein J. PHYLIP (Phylogeny Inference Package) version. 2006;3.66 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships (left) and sex pheromone blends (right) of Ostrinia species. The phylogenetic tree was constructed based on mitochondrial COII gene sequences. The numbers near branches indicate bootstrap values. The size of circles represents a rough blend ratio. Z-type, I-type (hybrid), and E-type females of O. scapulalis and O. nubilalis produce mixtures of 3:97, 64:36, and 99:1 (E)- and (Z)-11-tetradecenyl acetates, respectively
(PDF)
Alignment of allelic genomic sequences encompassing exons 3 and 4 of the OnubOR5b gene (Accession Nos. AB596043–AB596049). Gray-shaded boxes indicate exons 3 and 4. Yellow-shaded letters indicate insertions or deletions causing frameshift mutations. Asterisks indicate nucleotides conserved among all the alleles.
(DOC)
O. nubilalis BACs isolated by PCR-based screening.
(DOC)
BAC probes used for FISH analysis.
(DOC)
Detailed information of genes appearing in Figure 3 and summary of renaming of OR genes previously reported in ref. 25. “g” was added to names of genes deduced from genomic sequences.
(XLS)
Chromosomal distribution of OR genes in B. mori.
(DOC)