Abstract
The chromatin factors Hmgb1 and Hmgb2 have critical roles in cellular processes, including transcription and DNA modification. To identify the function of Hmgb genes in embryonic development, we generated double mutants of Hmgb1;Hmgb2 in mice. While double null embryos arrest at E9.5, Hmgb1-/-;Hmgb2+/- embryos exhibit a loss of digit5, the most posterior digit, in the forelimb. We show that Hmgb1-/-;Hmgb2+/- forelimbs have a reduced level of Shh signaling, as well as a clear downregulation of Wnt and BMP target genes in the posterior region. Moreover, we demonstrate that hmgb1 and hmgb2 in zebrafish embryos enhance Wnt signaling in a variety of tissues, and that double knockdown embryos have reduced Wnt signaling and shh expression in pectoral fin buds. Our data show that Hmgb1 and Hmgb2 function redundantly to enhance Wnt signaling in embryos, and further suggest that integrating Wnt, Shh and BMP signaling regulates the development of digit5 in forelimbs.
INTRODUCTION
The vertebrate limb is formed from a limb bud, a protrusion from the main body, which elongates and undergoes morphogenesis to construct a stereotypical limb structure. The development of the limb requires coordinated cell proliferation and differentiation, which depend on highly regulated actions of signaling pathways, such as those initiated by Wnt, Shh, Bone morphogenetic protein (BMP) and Fibroblast growth factor (FGF) (Johnson and Tabin, 1997; Capdevila and Izpisua Belmonte, 2001; Niswander, 2003; Tamura et al., 2008; Duboc and Logan, 2009; Towers and Tickle, 2009; Zeller et al., 2009).
Wnt signaling in particular is known to have multiple roles in the early process of vertebrate limb development, including driving downstream genetic programs and coordinating the functions of signaling centers (Yang, 2003; Grigoryan et al., 2008). Wnt/ß-catenin signaling triggers Fgf10 expression in the lateral plate mesoderm during limb initiation in chick and zebrafish (Kawakami et al., 2001; Ng et al., 2002). Then, Fgf10-dependent Wnt signaling in the overlying surface ectoderm regulates formation of the apical ectodermal ridge (AER), a specialized epithelial structure required for distal outgrowth and proximal-distal patterning (Mariani et al., 2008) and for Fgf8 expression in the AER (Kengaku et al., 1998; Kawakami et al., 2001; Ng et al., 2002; Barrow et al., 2003; Soshnikova et al., 2003). Wnt7a from the dorsal ectoderm induces Lmx1b in the underlying mesenchyme and specifies dorsal fate of the limb mesenchyme (Riddle et al., 1995; Vogel et al., 1995; Chen et al., 1998).
Wnt7a from the dorsal ectoderm also plays a role coordinating the function of signaling centers by maintaining Shh expression in the posterior mesenchyme, a region called the zone of polarizing activity (Parr and McMahon, 1995; Yang and Niswander, 1995). Wnt7a-/- mouse limb bud and dorsal ectoderm-removed chick limb bud fail to maintain Shh expression in the posterior mesenchyme. Implanting Wnt7a-expressing cells into dorsal ectoderm-removed limb bud rescued Shh expression. A previous study in the chick limb bud suggested that the Wnt7a-maintenance of Shh is mediated by a specific frizzled receptor (Kawakami et al., 2000). Shh has multiple roles during limb development (Zeller et al., 2009); Shh instructs anterior-posterior digit pattern formation (Riddle et al., 1993) and promotes proliferation of digit progenitors (Towers et al., 2008; Zhu et al., 2008), and is essential for normal development of the autopod (Chiang et al., 1996). Since Wnt signaling is involved in such a variety of processes during early processes of limb development, its activity needs to be under tight control, and, thus, alterations in Wnt signaling disrupts normal development of the limb (Grigoryan et al., 2008). However, it is not well understood how each tissue elicits a proper level of Wnt signaling in order to regulate the development of the limb.
We have recently demonstrated that HMGB2, a chromatin factor, facilitates Wnt/ß-catenin signaling both in vitro and in adult articular cartilage (Taniguchi et al., 2009a). HMGB2 is a member of the high mobility group box (HMG) family, which are nuclear proteins, characterized by two basic HMG-box domains. Among four HMGB proteins, HMGB1, 2, and 3 have an additional long acidic C-terminal tail (Bustin, 1999). Biochemical studies have been performed mainly with HMGB1 and HMGB2, and demonstrated that HMGB1 and HMGB2 bind to DNA without sequence specificity (Bianchi and Agresti, 2005; Grasser et al., 2007; Stros et al., 2007). HMGB1 and HMGB2 have a role in formation of nucleoprotein complexes through altering chromatin structure that allow for binding of other factors (Pil and Lippard, 1992; Paull et al., 1993) and facilitating diverse DNA modifications (Agresti and Bianchi, 2003; Stros, 2010). They are also known to regulate various activities, such as transcription, replication and DNA repair (Bianchi and Agresti, 2005).
Given that Hmgb genes are involved in these fundamental cellular processes, it was expected that they play critical roles in animal development and physiology. The in vivo function of Hmgb genes in a mammalian model has been studied by expression analysis and mutant mouse phenotype analysis. During embryonic development, Hmgb1 is reported to be ubiquitously and highly expressed (Pauken et al., 1994), and Hmgb2 is also known to be highly and widely expressed by Northern blot analysis (Ronfani et al., 2001). Contrary to this, expression of Hmgb3 is only demonstrated by reverse-transcription-polymerase chain reaction of whole embryos (Vaccari et al., 1998), and expression of Hmgb4 has not been studied. In adult tissues, Hmgb1 is reported to have ubiquitous and high level expression by whole tissue extract assays (Mosevitsky et al., 1989; Muller et al., 2004); however, other Hmgb genes show restricted expression. Hmgb2 is highly expressed in thymus and testis (Ronfani et al., 2001) as well as articular cartilage surface, which contains a progenitor cell population (Taniguchi et al., 2009b). Hmgb3 is highly expressed in hematopoietic stem cells in the bone marrow (Nemeth et al., 2003), and Hmgb4 expression is limited to testis (Catena et al., 2009). These expression analyses suggest that Hmgb1 may have roles broadly in embryonic development and in adult tissues, and other Hmgb genes may have redundant functions depending on the stages of embryos and adults, as well as specific tissues.
Functional studies of Hmgb genes have been carried out with targeted mutations in mice. Hmgb1-/- mice are born without significant morphological defects, but die within a day by hypoglycemia (Calogero et al., 1999), which is caused by insufficient activation of a glucocorticoid receptor. Hmgb2-/- mice show defects in spermatogenesis (Ronfani et al., 2001) as well as failure to maintain articular cartilage homeostasis in adults (Taniguchi et al., 2009a; Taniguchi et al., 2009b). Hmgb3-/- mice exhibit erythrocythemia (Nemeth et al., 2005). These studies clarify that Hmgb1, 2, 3 are not required for embryonic development, except that Hmgb1-/- limb long bones show delay in endochondral ossification (Taniguchi et al., 2007). Normal embryonic development in these mutant mice is likely due to functional compensation based on their structural similarity. This is particularly likely the case for Hmgb1 and Hmgb2, given their high expression in embryos and shared biochemical characteristics.
In order to address the functional redundancy of Hmgb1 and Hmgb2 during embryonic development and to test whether Hmgb genes modulate Wnt/ß-catenin signaling and/or other signaling pathways in the developing limb, we generated double mutants of Hmgb1; Hmgb2. Consistent with the hypothesis of functional redundancy, Hmgb1; Hmgb2 double null mutants arrested at E9.5. Further analysis suggests that the functions of Hmgb1 and Hmgb2 are not completely equivalent in the development of the autopod. Analysis of the phenotype in the developing limb revealed a lack of the most posterior digit, digit 5, in the Hmgb1-/-; Hmgb2+/- forelimb, and this is associated with downregulation of Shh expression and Wnt and BMP target genes. Further analysis with zebrafish embryos also supports enhancement of Wnt/ß-catenin signaling by hmgb genes. Thus, our data suggest that Hmgb1 and Hmgb2 are required for maintaining a proper level of Shh expression in the limb through enhancing Wnt/ß-catenin signaling, and further suggest that Shh, Wnt and BMP signaling at the posterior region regulate the development of digit 5 in the forelimb.
RESULTS
Combined activities of Hmgb1 and Hmgb2 are required for the development of digit 5 in the mouse forelimb
Previous studies have shown that Hmgb1-/- embryos show delayed endochondral ossification in long bones during limb development without other morphological defects (Taniguchi et al., 2007). Hmgb2-/- mice did not exhibit any defects in limb development (Ronfani et al., 2001). Based on their structural conservation as well as ubiquitous and high level expression in the developing limb bud (Supplemental Fig. 1), we hypothesized that functional redundancy between Hmgb1 and Hmgb2 might allow for development of normal morphology in Hmgb1-/- and Hmgb2-/- limbs. In order to address this possibility, we generated Hmgb1; Hmgb2 double mutant embryos. At E10.5, we obtained 7 Hmgb1-/-; Hmgb2-/- embryos out of 257 embryos (2.7%); however, they were arrested at E9.5 (Table 1). At E11.5 and E12.5, we did not obtain any Hmgb1-/-; Hmgb2-/- embryos out of 66 embryos collected. This suggests that Hmgb1 and Hmgb2 have redundant functions and are required for development of the mouse embryo beyond E9.5.
Table 1.
Summary of number of Hmgb1; Hmgb2 embryos collected at E10.5
| Hmgb1; Hmgb2 | number | (%) |
|---|---|---|
| KO; KO | 7* | 2.7 |
| KO; Het | 27 | 10.5 |
| Het; KO | 19 | 7.4 |
| KO; WT | 20 | 7.8 |
| WT; KO | 16 | 6.2 |
| Het; Het | 80 | 31.1 |
| Het; WT | 57 | 22.1 |
| WT; Het | 20 | 7.8 |
| WT; WT | 11 | 4.3 |
| Total | 257 | 100 |
Embryos are developmentally arrested at E9.5
KO, Het and WT indicates -/-, +/- and +/+, respectively.
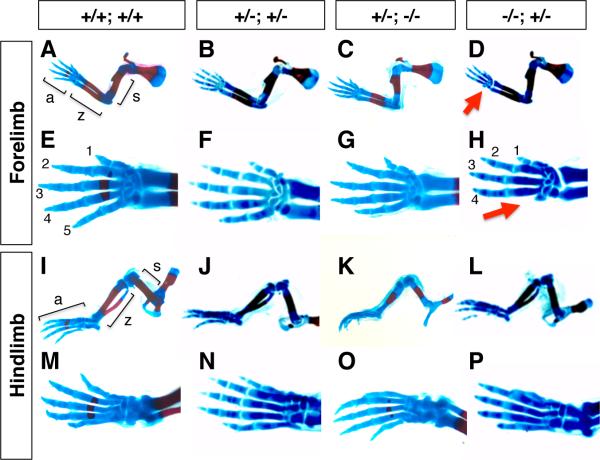
In order to address their function in limb development, we examined limb skeletons of an allelic series of Hmgb1; Hmgb2 at E13.5-16.5 (Fig. 1). We found that Hmgb1-/-; Hmgb2+/- embryos showed a defect in the development of the most posterior digit, digit 5, in the forelimb (Fig.1D), while we did not observe significant defects in other areas of the body. Based on the morphology of remaining elements, the lost skeletal elements are the phalanges and metacarpal for digit 5, while other elements appeared to be normal (Fig.1H). This phenotype was observed only in the forelimb but not in the hindlimb (Fig.1 I-P). Other allelic combinations either showed no skeletal phenotype or showed the same defect at very low frequencies (Table 2). This result suggests that Hmgb1 and Hmgb2 are functionally redundant for limb development, and that the function of Hmgb1 is more important than that of Hmgb2 for the development of digit 5 in the forelimb.
Fig. 1. Skeletal phenotype of the Hmgb1-/-; Hmgb2+/- limb.
Alcian Blue and Alizarin red-stained E16.5 forelimbs (A-H) and hindlimbs (I-P) of Hmgb1; Hmgb2 mutants are shown. Genotypes of Hmgb1; Hmgb2 are indicated on the top: (A, E, I, M) +/+;+/+, (B, F, J, N) +/–;+/–, (C, G, K, O) +/–;–/–, (D, H, L, P) –/–;+/–. AD and I-L show lateral views of entire forelimb and hindlimb skeletons, respectively. E-H and M-P show dorsal views of the autopod. In (A) and (I) the stylopod, zeugopod and autopod are indicated as s, z and a, and in (E) and (H) digits are indicated with 1-5. The loss of digit 5 is indicated by red arrows in (D) and (H).
Table 2.
Summary of skeletal phenotype at E13.5-16.5
| Hmgb1; Hmgb2 | Number of embryos | Number with the phenotype | Frequency (%) |
|---|---|---|---|
| KO; KO | 0 | 0 | - |
| KO; Het | 16 | 13 | 81.3 |
| Het; KO | 11 | 3 | 27.3 |
| KO; WT | 19 | 2 | 10.5 |
| WT; KO | 9 | 0 | 0 |
| Het; Het | 25 | 0 | 0 |
| Het; WT | 16 | 0 | 0 |
| WT; Het | 8 | 0 | 0 |
| WT; WT | 7 | 0 | 0 |
| Total | 111 | 18 |
Phenotype is defined as lack of digit 5 in the forelimb.
KO, Het and WT indicates -/-, +/- and +/+, respectively.
Early onset of the digit 5 defect in the Hmgb1-/-; Hmgb2+/- mutant forelimb bud
To further characterize the loss of digit 5 in the Hmgb1-/-; Hmgb2+/- mutant embryo, we examined cartilage formation at earlier stages as well as region-specific molecular markers in the developing forelimb bud. At E13.5, the loss of digit 5 was evident by Alcian Blue staining (Fig 2A, B). This defect was detectable even at an earlier stage, E11.5. Chondrogenic condensation for digits 1-4 was visible by detecting Sox9, while the posterior region including the digit 5 primordia was lacking in the Hmgb1-/-; Hmgb2+/- forelimb bud (Fig. 2C, D). Correlating with normal development of the anterior skeletal elements shown in Fig.1, Pax9 expression, which marks the anterior region of the limb bud, was normal in the Hmgb1-/-; Hmgb2+/- forelimb bud (Fig. 2E, F). Hoxd13 that marks the autopod region is also detected, suggesting normal specification of the autopod region. A slight expansion in the anterior region where Hoxd13 is not expressed, as well as the disappearance of the most posterior Hoxd13-expressing structure suggests a slight effect on the anterior-posterior patterning by this stage (Fig. 2G, H). These results demonstrate that the posterior autopod region is specifically affected in the Hmgb1-/-; Hmgb2+/- forelimb bud by E11.5, and suggest that molecular alterations that cause the phenotype have taken place in earlier stages.
Fig. 2. Specific defects in the posterior development in the Hmgb1-/-; Hmgb2+/- limb.
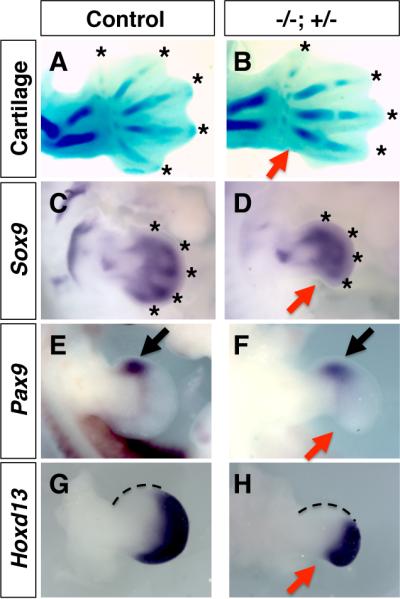
Control (A, C, E, G) and Hmgb1-/-; Hmgb2+/- (B, D, F, H) forelimb buds at E13.5 (A, B) and E11.5 (C-H). Alcian Blue staining (A, B) and Sox9 staining (C, D) visualized the loss of digit 5 in Hmgb1-/-; Hmgb2+/- forelimb bud (red arrows). Digit tips are marked by asterisks. Pax9 expression is normal in both control (E) and mutant (F) forelimb bud (black arrows). Hoxd13 expression in the autopod region is detected in both control (G) and mutant (H) forelimb bud. The posterior defect is visible in the mutant limb bud at E11.5 (red arrows). The Hoxd13-non-expressiong domain showed a slight expansion (marked by dotted line). All panels are dorsal views with the anterior to the top.
Late onset and reduced level of Shh signaling in the Hmgb1-/-; Hmgb2+/- limb bud
The lack of digit 5 is reminiscent of downregulation of Shh signaling (Zhu et al., 2008). Thus, we first examined the expression of Shh. Normal Shh expression in the posterior mesenchyme of the forelimb bud starts at E9.75 (Echelard et al., 1993; Charite et al., 2000). At E10.0 (28 somite stage), we did not detect Shh expression in the Hmgb1-/-; Hmgb2+/- forelimb bud, while Shh is expressed in the control limb bud (Fig. 3A, B). Consistent with this, the expression of Gli1, a target of Shh signaling in the Hmgb1-/-; Hmgb2+/- forelimb bud at 27 and 28 somite-stage embryos was significantly low compared to control embryos (Fig. 3C, D). These results indicate that the onset of Shh expression is delayed in the Hmgb1-/-; Hmgb2+/- forelimb bud.
Fig. 3. Downregulation of Shh signaling in the Hmgb1-/-; Hmgb2+/- forelimb bud.
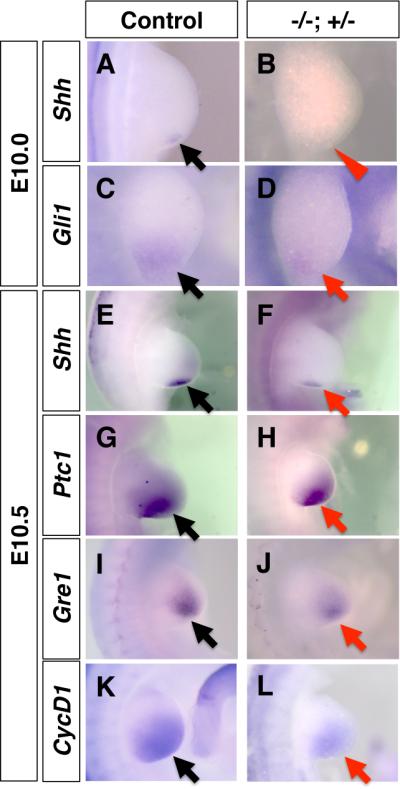
Control (A, C, E, G, I, K) and Hmgb1-/-; Hmgb2+/- (B, D, F, H, J, L) forelimb buds at E10.0 (A-D) and at E10.5 (E-L). Shh (A, B, E, F), Gli1 (C, D), Ptc1 (G, H), Gre1 (I, J) and CycD1 (K, L) expression are shown. Compared to the expression in the control forelimb bud (black arrows), expression in the mutant forelimb bud is not initiated (red arrowhead in B) or downregulated (red arrows). All panels are dorsal views with the anterior to the top.
At E10.5 we detected Shh expression in the posterior mesenchyme in the Hmgb1-/-; Hmgb2+/- limb bud, though it was clearly lower than the control limb bud (Fig. 3E, F). Consistent with this, Patched1 (Ptc1), another target of Shh signaling (Ingham and McMahon, 2001), and Gremlin1, whose maintenance requires Shh signaling (Nissim et al., 2006; Benazet et al., 2009) are downregulated (Fig. 3G-J). Furthermore, CyclinD1, which is known to be regulated by both Shh signaling and Wnt/ß-catenin signaling (Shtutman et al., 1999; Tetsu and McCormick, 1999; Towers et al., 2008) is also expressed at a lower level (Fig. 3K, L). These results demonstrate that Shh signaling is initiated with a delay and is not activated to the normal level in the Hmgb1-/-; Hmgb2+/- forelimb bud.
Normal activity of AER-FGF and Wnt7a
Signaling pathways in the developing limb bud form feedback loops to maintain the activity of each other. Shh expression in the posterior mesenchyme is maintained by FGFs emanating from the AER (Laufer et al., 1994; Niswander et al., 1994; Lewandoski et al., 2000; Moon and Capecchi, 2000) and Wnt7a emanating from the dorsal ectoderm (Parr and McMahon, 1995; Yang and Niswander, 1995). In order to address if downregulation of Shh expression at E10.5 is caused by downregulation of AER-FGF signal and/or Wnt7a from the dorsal ectoderm, we examined their activity in the Hmgb1-/-; Hmgb2+/- forelimb bud.
We first examined Fgf8 expression in the AER because Fgf8 is the central factor for limb development among four Fgf genes expressed in the AER (Mariani et al., 2008). In the Hmgb1-/-; Hmgb2+/- forelimb bud, Fgf8 is expressed similar to control limb bud, which contrasts with the lower level of Shh expression (Fig. 4A-D). Since the sum of AER-FGF function is important for normal limb development, we next monitored if AER-FGF activity is altered by examining expression of Fgf10 and Mkp3. It has been demonstrated that expression of these genes depends on FGF activity from the AER (Ohuchi et al., 1997; Eblaghie et al., 2003; Kawakami et al., 2003; Mariani et al., 2008). We detected expression of both genes comparable to control limb bud (Fig. 4E-H). To further evaluate AER-FGF activity, we examined phosphorylated ERK1/2 (pERK1/2) on sectioned samples. A previous study has demonstrated that pERK1/2 in developing mouse limb buds depends on FGF signaling (Corson et al., 2003). We detected a normal level of pERK1/2 in the control and the Hmgb1-/-; Hmgb2+/- forelimb bud (Fig. 4I, J). The pERK1/2-positive cell layers were 10.4 ± 0.89 (n=5) in control and 10.2 ± 0.82 (n=5) in the Hmgb1-/-; Hmgb2+/- limb bud. These data suggests that AER-FGF activity is not significantly affected in the Hmgb1-/-; Hmgb2+/- forelimb bud.
Fig. 4. Normal signaling by AER-FGF and Wnt7a from the dorsal ectoderm.

Control (A, B, E, G, I, K, L) and Hmgb1-/-; Hmgb2+/- (C, D, F, H, J, L, M, N) forelimb buds at E10.5. Fgf8 in the AER and Shh in the posterior mesenchyme are co-stained (A-D). Fgf10 (E, F) and Mkp3 (G, H) expression in response to AER-FGF are shown. Despite the downregulation of Shh (A-D), Fgf8 expression in the AER (A-D) and expression of Fgf10 (E, F) and Mkp3 (G, H) are normal. (I, J) pERK1/2 staining on sections is also normal. Dotted lines show the boundary of the pERK1/2 signal. Patchy signals are derived from blood cells and were positive in all fluorescent channels. Lmx1b expression in the control (K, L) and mutant (M, N) forelimb bud shows normal Wnt7a signaling into dorsal mesenchyme. All panels except for B, D, I, J, L and N are dorsal views, and B, D, L and N are distal views with the anterior to the top, and I and J are section images with the AER to the bottom.
Dorsal ectoderm produces WNT7a, which has been shown to be required for maintenance of Shh expression (Parr and McMahon, 1995; Yang and Niswander, 1995). To examine if reduced Shh expression in the Hmgb1-/-; Hmgb2+/- forelimb bud at E10.5 is due to lowered Wnt7a activity, we monitored expression of Lmx1b, a gene regulated by WNT7a from the dorsal ectoderm (Riddle et al., 1995; Vogel et al., 1995). We detected normal expression of Lmx1b (Fig. 4K-N), suggesting that reduced Shh expression in the Hmgb1-/-; Hmgb2+/- limb is unlikely due to reduction of Wnt7a function from the dorsal ectoderm.
Taken together, these results suggest that the lower level of Shh expression in the Hmgb1-/-; Hmgb2+/- forelimb bud is not caused by alteration in AER-FGF or WNT7a from the dorsal ectoderm.
Reduced Wnt/ß-catenin signaling and BMP signaling in the posterior region of the Hmgb1-/-; Hmgb2+/- forelimb bud
Our recent analysis has demonstrated that Hmgb2 enhances Wnt/ß-catenin signaling (Taniguchi et al., 2009a). The apparently normal expression of Lmx1b in the Hmgb1-/-; Hmgb2+/- forelimb bud suggests normal Wnt7a signaling in the dorsal mesenchyme (Fig. 4K-N). There remains the possibility that uncharacterized Wnt7a signaling via ß-catenin in the posterior mesenchyme is affected in the Hmgb1-/-; Hmgb2+/- forelimb bud, while Wnt7a signaling through a ß-catenin-independent pathway maintains normal expression of Lmx1b (Kengaku et al., 1998). To address this, we examined expression of Msx2. While Msx2 is a known target of BMP signaling in the limb (Pizette and Niswander, 1999; Khokha et al., 2003; Michos et al., 2004), studies have further demonstrated that in the presence of BMP signaling, Wnt/ß-catenin signaling directly regulates the Msx2 promoter via the Lef1-ß-catenin pathway and facilitates Msx2 expression (Willert et al., 2002; Hussein et al., 2003). In the Hmgb1-/-; Hmgb2+/- forelimb bud, expression of Msx2 in the posterior mesenchyme is significantly downregulated, while anterior expression is not altered (Fig. 5A-D), suggesting that Wnt/ß-catenin signaling and/or BMP signaling is downregulated in the Hmgb1-/-; Hmgb2+/- forelimb bud. To confirm the effect on BMP signaling, we examined expression of Msx1, another well-recognized target of BMP signaling (Khokha et al., 2003; Michos et al., 2004). The expression of Msx1 is also downregulated in the posterior mesenchyme, while anterior expression remained unaffected (Fig. 5E, F). These results suggest that BMP signaling is downregulated in the posterior mesenchyme in the Hmgb1-/-; Hmgb2+/- forelimb bud.
Fig. 5. Reduced Wnt/ß-catenin signaling and BMP signaling in the posterior Hmgb1-/-; Hmgb2+/- forelimb bud.
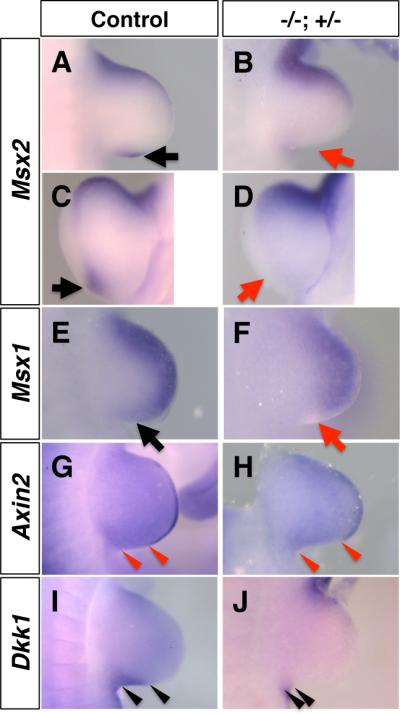
Control (A, C, E, G, I) and Hmgb1-/-; Hmgb2+/- (B, D, F, H, J) forelimb buds at E10.5, stained with Msx2 (A-D), Msx1 (E, F), Axin2 (G, H) and Dkk1 (I, J). Msx2 expression in the posterior region is lost in Hmgb1-/-; Hmgb2+/- limbs (B, D, red arrows), compared to control limbs (A, C, black arrows). (E, F) Msx1 expression in the posterior region is also lost in Hmgb1-/-; Hmgb2+/- limbs (F, red arrow), compared to control limbs (E, black arrow). (G, H) The posterior boundary of Axin2 expression in the AER, and the posterior boundary of the forelimbs are indicated by arrowheads. The domain without Axin2 expression is longer in the Hmgb1-/-; Hmgb2+/- limb bud (H), compared to the control limb bud (G). (I, J) The posterior Dkk1 expression domain in the forelimb bud is indicated by arrowheads. The Dkk1-expressing domain is smaller in the Hmgb1-/-; Hmgb2+/- limb bud (J), compared to control limb bud (I). All panels except for C and D are dorsal views, and C and D are distal-posterior views, with the anterior to the top.
We next examined expression of known targets of Wnt/ß-catenin signaling in the limb bud. Axin2, a negative feedback regulator, is a target of Wnt/ß-catenin signaling (Jho et al., 2002; Leung et al., 2002; Lustig et al., 2002). Expression of Axin2 was strongly detected in the AER of the control embryo (Fig. 5G); however, its expression domain in the posterior region was significantly smaller in the Hmgb1-/-; Hmgb2+/- forelimb bud (Fig. 5H). Since we detected normal expression of Fgf8 (Fig. 4A-D), the reduction in Axin2 expression is probably not due to a loss of posterior AER structure. Rather, this result suggests that Wnt/ß-catenin signaling is low in the posterior AER. DKK1 is a negative regulator of Wnt/ß-catenin signaling, and is also a target of Lef1/TCF-ß-catenin signaling (Gonzalez-Sancho et al., 2004; Niida et al., 2004; Chamorro et al., 2005). As previously shown (Mukhopadhyay et al., 2001), Dkk1 expression is detected in the anterior and posterior margin of the limb mesenchyme (Fig. 5I). In the Hmgb1-/-; Hmgb2+/- forelimb bud, Dkk1 expression domain in the posterior region was reduced (Fig. 5J). Together with the smaller expression domain of Cyclin D1 (Fig. 3K, L), a target of Shh and Wnt/ß-catenin signaling, our results suggest that ß-catenin pathway activity is lower in the posterior region of the Hmgb1-/-; Hmgb2+/-forelimb bud.
Global downregulation of canonical Wnt signaling in hmgb1-hmgb2 knockdown zebrafish
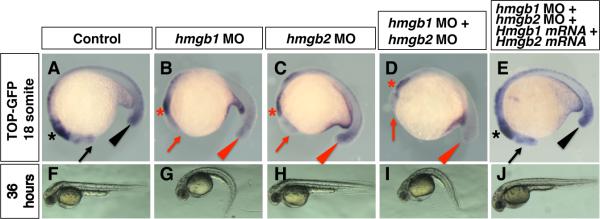
HMGB is a well-conserved nuclear protein found in multicellular organisms (Sessa and Bianchi, 2007), suggesting conserved functions in animal species. To further examine the function of hmgb genes in the enhancement of Wnt/ß-catenin pathway during embryonic development, we used Top-dGFP zebrafish embryos (Dorsky et al., 2002), in which the expression of dGFP is dependent on TCF/Lef1-ß-catenin activity. We knocked down hmgb1 and hmgb2 by injecting morpholinos (MOs) and monitored GFP expression by in situ hybridization. As previously shown (Dorsky et al., 2002), GFP expression is detected in known tissues with high Wnt/ß-catenin activities at the 18 somite-stage, such as forebrain, midbrain and tail bud (Fig. 6A). Both hmgb1-morphants and hmgb2-morphants show reduced GFP signal in all these tissues in a dose-dependent manner (Fig. 6B, C, Table 3) with more downregulation by hmgb1-MO. Furthermore, downregulating both hmgb1 and hmgb2 resulted in more severe downregulation of GFP expression at higher frequency (Fig. 6D, Table 3). This synergistic activity was also confirmed with a second set of morpholinos (Supplemental Table 1). While injection of synthetic mRNA for hmgb1 and hmgb2 did not affect Top-GFP expression, co-injection of mouse Hmgb1 mRNA and Hmgb2 mRNA could partially rescue the expression of Top-GFP (Fig. 6E, Table 4). These results demonstrate that enhancement of Wnt/ß-catenin signaling by hmgb1 and hmgb2 is a general feature in vertebrate embryos, and that this enhancement operates not only in the limb bud but also in other sites where Wnt/ß-catenin signaling is active.
Fig. 6. hmgb1 and hmgb2 are required for enhancement of Wnt/ß-catenin signaling in zebrafish embryos.
Lateral views of 18 somite stage embryos (A-E) and 36 hpf embryos (F-J) injected with control MO (A, F), hmgb1 MO (B, G), hmgb2 MO (C, H), both hmgb1 MO and hmgb2 MO (D, I) and hmgb1 MO, hmgb2 MO, Hmgb1 mRNA and Hmgb2 mRNA (E, J). (A-E) Top-GFP expression is visualized by in situ hybridization. Wnt/ß-catenin signaling in forebrain (arrows), midbrain (asterisks) and tail bud (arrowhead) is visualized in control morphants (A), but downregulated in hmgb-downregulated embryos (B-D). In mRNA co-injected double morphants, Top-GFP expression is partially restored (E). Normal body extension (F) is impaired in hmgb1-morphants (G). The phenotype is less severe in hmgb2-morphants (H), but becomes more severe in double morphants (I). This phenotype was partially restored by co-injection of Hmgb1 mRNA and Hmgb2 mRNA (J).
Table 3.
Dose-dependency and synergistic function of hmgb1 morpholino and hmgb2 morpholino for the expression of Top-GFP in the zebrafish embryo
| Injection | Normal expression | Reduced expression | Affected embryos (%) |
|---|---|---|---|
| Control MO (2 ng) | 104 | 2 | 1.9 |
| hmgb1 MO (0.5 ng) | 35 | 5 | 12.5 |
| hmgb1 MO (1 ng) | 4 | 49 | 92.5 |
| hmgb2 MO (0.5 ng) | 34 | 7 | 17.1 |
| hmgb2 MO (1 ng) | 8 | 76 | 90.5 |
|
hmgb1 MO (0.5 ng) + hmgb2 MO (0.5 ng) |
20 | 31 | 60.8 |
|
hmgb1 MO (1 ng) + hmgb2 MO (1 ng) |
2 | 57 | 96.6 |
Table 4.
Rescue of Top-GFP expression in hmgb1/2-double morphants by hmgb1/2 mRNA at 18-somite stage
| Normal expression | Significantly reduced expression | Expression at an intermediate level* | |
|---|---|---|---|
| Control MO (2 ng) | 104 (98.1%) |
2 (1.9%) |
0 |
|
hmgb1 MO (1 ng) + hmgb2 MO (1 ng) |
2 (3.4%) |
55 (93%) |
2 (3.4%) |
|
Hmgb1 mRNA (100 pg) + Hmgb2 mRNA (100 pg) |
24 (100%) |
0 (0%) |
0 (0%) |
|
hmgb1 MO (1 ng) + hmgb2 MO (1 ng) + Hmgb1 mRNA (100 pg)+ Hmgb2 mRNA (100 pg) |
9 (13.2%) |
33 (48.5%) |
26 (38.2%) |
Weaker than normal expression but clearly stronger than expression with severe downregulation
At a later stage (36 hours post fertilization; hpf) hmgb1-morphants show a smaller head, an undulating notochord in small populations, and ventrally bent tail structures (Fig. 6G), similar to wnt8 knockdown embryos (Lekven et al., 2001; Shimizu et al., 2005). This phenotype was milder in hmgb2-morphants (Fig. 6H), but became more severe and frequent in hmgb1-hmgb2 double morphants (Fig. 6I, Table 5). This phenotype was also partially rescued by co-injecting Hmgb1 mRNA and Hmgb2 mRNA (Fig. 6J, Table 5). The correlation of the gross morphology phenotype and downregulation of Top-dGFP signal suggests that, similar to mouse mutant limb, hmgb1 function is more important than hmgb2 function in enhancing ß-catenin activity in the zebrafish embryo.
Table 5.
Synergistic function of hmgb1 and hmgb2 morpholinos and rescue by mRNA injection of the 36 hpf body morphology
| Injection | Normal | Short, curly tail |
|---|---|---|
| Control MO (2 ng) | 65 (100%) |
0 (0%) |
| hmgb1 MO (1 ng) | 8 (13.6%) |
51 (86.4%) |
| hmgb2 MO (1 ng) | 52 (92.9%) |
4 (7.1%) |
|
hmgb1 MO (1 ng) + hmgb2 MO(1 ng) |
13 (7.3%) |
166 (92.7%) |
|
Hmgb1 mRNA (100 pg) + Hmgb2 mRNA (100 pg) |
25 (100%) |
0 (0%) |
|
hmgb1 MO (1 ng) + hmgb2 MO (1 ng)+ Hmgb1 mRNA (100 pg) + Hmgb2 mRNA (100 pg) |
19 (22.1%) |
67 (77.9%) |
Downregulation of hmgb1 and hmgb2 causes downregulation of shh expression in the pectoral fin
The pectoral fin of zebrafish is derived from the lateral plate mesoderm and is homologous to the forelimb in tetrapod animals (Grandel and Schulte-Merker, 1998; Freitas et al., 2006). Remarkable similarities of developmental and genetic mechanisms between pectoral fin buds and forelimb buds have been found (Mercader, 2007). Thus, we examined the functional conservation of hmgb1 and hmgb2 during zebrafish pectoral fin development. The development of the pectoral fin bud in hmgb1-hmgb2 double morphants is not significantly affected at 36 hpf as visualized by prx1 expression (Fig. 7 A, B, Supplemental Table 2). However, the activation status of Wnt signaling, visualized by the expression of Top-dGFP, is significantly downregulated in hmgb1-hmgb2-knockdown fin buds (Fig. 7C, D, Table 6). Similar to the mutant mouse forelimb bud, expression of shh and ptc1 is significantly downregulated in hmgb1-hmgb2-knockdown fin buds (Fig. 7E-J, Table 6). These results demonstrate a conserved requirement of hmgb1 and hmgb2 for the enhancement of Wnt/ß-catenin and expression of shh in the vertebrate forelimb bud.
Fig. 7. Downregulation of Wnt signaling and shh signaling in hmgb-knockdown pectoral fin buds.
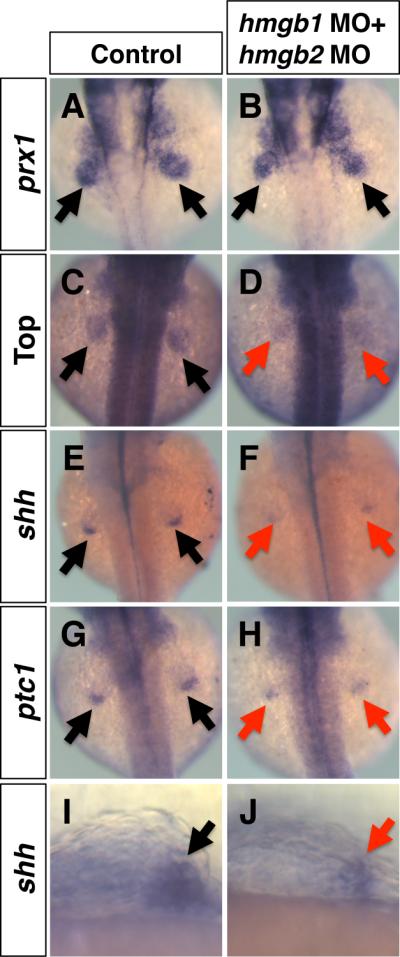
Dorsal views (A-H) and lateral views (I, J) of 36 hpf embryos injected with control MO (A, C, E, G, I) or hmgb1 MO + hmgb2 MO (B, D, F, H, J). (A, B) Normal expression of prx1 in the pectoral fin bud of control (A, arrows) and hmgb-knockdown embryos (B, arrows). (C-H) Expression of Top-GFP (D), shh (F) and ptc1 (H) in the pectoral fin bud of hmgb-knockdown embryos (D, F, H, red arrows) is downregulated, compared to control embryos (C, E, G, black arrows). (I, J) High magnification images of the pectoral fin bud stained with shh shows smaller expression domain of shh in the pectoral fin bud of hmgb-knockdown embryos (J, red arrow), compared to the control fin bud (I, black arrow).
Table 6.
Gene expression in the pectoral fin bud of embryos injected with hmgb1 MO and hmgb2 MO
| Analyzed Expression | Injection | Normal | Weak signal, small domain | Affected embryos (%) |
|---|---|---|---|---|
| Top-GFP | Control MO | 41 | 3 | 6.8 |
|
hmgb1 MO + hmgb2 MO |
4 | 31 | 88.6 | |
| shh | Control MO | 31 | 1 | 3.1 |
|
hmgb1 MO + hmgb2 MO |
4 | 46 | 92.0 | |
| ptc1 | Control MO | 32 | 0 | 0 |
|
hmgb1 MO + hmgb2 MO |
3 | 26 | 89.7 |
Injection amount is 2 ng for control MO, 1 ng for hmgb1 MO and 0.5 ng for hmgb2 MO.
DISCUSSION
Functional analyses of the Hmgb genes have identified their roles in adult animals, such as regulation of immune response, cancer, spermatogenesis and hematopoietic stem cell self-renewal (Ronfani et al., 2001; Dumitriu et al., 2005; Nemeth et al., 2006; Tang et al., 2010); however, their function in embryonic development was not well understood. In this report we examined the limb phenotype of Hmgb1 and Hmgb2 double mutants. Previous studies with individual mutant mice did not show any defects in the limb, except for the delay of endochondral ossification in late stage Hmgb1-/- mice (Taniguchi et al., 2007). Our attempt to obtain double null embryos revealed that Hmgb1-/-; Hmgb2-/- embryos arrested at E9.5 (Table 1), suggesting that the combined function of Hmgb1 and Hmgb2 is essential for the development of mouse embryos. Furthermore, our approach of double knockout of Hmgb1 and Hmgb2 found that these genes are genetically redundant, and that their combined function is required for proper forelimb development.
We have recently demonstrated Hmgb2-mediated enhancement of Wnt/ß-catenin signaling in adult tissue. Reduction of Hmgb2 expression during aging correlates with reduction of Wnt/ß-catenin signaling in the adult diarthrodial joint, and HMGB2 enhances expression of Wnt/ß-catenin target genes in vitro (Taniguchi et al., 2009a; Taniguchi et al., 2009b). Moreover, the HMG domain within HMGB2 is responsible for interaction with Lef-1, suggesting that HMGB1 is also involved in enhancing Wnt/ß-catenin signaling (Taniguchi et al., 2009a). Our present data shows that hmgb2-dependent enhancement of the Wnt/ß-catenin signaling is also present in a variety of embryonic tissues in developing zebrafish embryos (Fig. 6). Furthermore, hmgb1 functions to enhance the Wnt/ß-catenin signaling in zebrafish embryos. The Wnt/ß-catenin reporter expression in zebrafish embryos demonstrates that hmgb1 and hmgb2 are functionally redundant, and suggests that the function of hmgb1 is more important than that of hmgb2 (Fig. 6). While exact mechanisms by which Hmgb genes function during embryonic development are still not well understood, enhancing Wnt/ß-catenin signaling is, at least in part, a mechanism for Hmgb1 and Hmgb2 to control organogenesis. Interestingly, Hmgb3 is also known to regulate Wnt/ß-catenin signaling activity. In contrast to Hmgb1 and Hmgb2, a study has demonstrated that abrogating Hmgb3 results in constitutive activation of Wnt/ß-catenin signaling in hematopoietic stem cells (Nemeth et al., 2006). Thus, although it has a high degree of structural similarity with Hmgb1 and Hmgb2, Hmgb3 seems to have an opposite function, and the enhancement of Wnt/ß-catenin signaling might be a specific function of Hmgb1 and Hmgb2.
Our analysis strongly suggested that the function of Hmgb1 and Hmgb2 affects not only Wnt/ß-catenin signaling but also other signaling pathways in developing limb buds. The skeletal phenotype of the Hmgb1-/-; Hmgb2+/- forelimb is the selective loss of digit 5, which is a reminiscent of early downregulation of Shh signaling, while other elements were intact. Because the development of digit 5 requires high level of Shh signaling (Harfe et al., 2004), a disruption of normal Shh signaling would contribute to the loss of digit 5 in the Hmgb1-/-; Hmgb2+/- forelimb bud. Indeed, a late onset and lower activation of Shh expression as well as reduced level of Shh target genes (Fig. 3) support this idea. However, reduction of Shh signaling alone does not simply account for the phenotype. For instance, a study utilizing an inducible Shh knockout has demonstrated that expansion of digit 3 primordia is the most sensitive to lowered Shh signaling, and that digit 3 is the first digit to be lost upon eliminating Shh during digit progenitor expansion period in the developing limb bud (Zhu et al., 2008). This is in contrast to the loss of digit 5 in the Hmgb1-/-; Hmgb2+/- forelimb bud. Furthermore, the anterior domain where Hoxd13 is not expressed, which is correlated with Gli3 processing (Wang et al., 2007a; Wang et al., 2007b), showed a slight expansion (Fig. 2G, H). Gli3 processing is counteracted by Shh signaling (Litingtung et al., 2002; te Welscher et al., 2002), suggesting that Shh-dependent counteraction of Gli3 processing is impaired, but not as severely in Shh mutant limb buds. Taken together, we favor the idea that Shh signaling is reduced in the Hmgb1-/-; Hmgb2+/- forelimb bud, however, the disruption of Shh signaling alone is not sufficient to cause the loss of digit 5.
The late onset and reduction of Shh expression (Fig. 3) suggest that Hox function is reduced. By eliminating multiple Hoxa and Hoxd genes in mice, it has been shown that 5’ Hox genes induce Shh expression in a gene-specific and dosage-dependent manner (Tarchini et al., 2006). Interestingly, HMGB1 is shown to interact with Hoxd proteins and enhance their transcriptional activity (Zappavigna et al., 1996). Thus, 5’-Hox gene function could be reduced in the Hmgb1-/-; Hmgb2+/- forelimb bud, which would contribute to the late onset and lower activation of Shh expression. Among four Hox clusters, the Hoxd genes are expressed with a posteriorly biased manner and have redundant function in the developing limb (Tarchini et al., 2006; Zakany and Duboule, 2007), therefore, Hoxd activity might also be reduced in the Hmgb1-/-; Hmgb2+/- forelimb bud, which may also contribute to the loss of digit 5 phenotype in the Hmgb1-/-; Hmgb2+/- forelimb bud.
In contrast to Shh signaling, Wnt/ß-catenin signaling seems to be affected both in the ectoderm and mesoderm in the forelimb bud. Downregulation of Axin2 in the AER, and Dkk1 and Msx2 in the mesenchyme suggest downregulation of Wnt/ß-catenin signaling in the posterior region of the AER and mesenchyme (Fig. 5). Several Wnt genes are known to function in mouse limb development, including Wnt3, Wnt5a, Wnt7a and Wnt9a. (Parr et al., 1993; Parr and McMahon, 1995; Yamaguchi et al., 1999; Barrow et al., 2003; Spater et al., 2006). Among them, normal expression of Lmx1b suggests that Wnt7a function is not affected in the Hmgb1-/-; Hmgb2+/- forelimb bud. Lmx1b is shown to be downregulated in a limb-mesenchyme-specific knockout of the Ctnnb1 gene (encoding ß-catenin) (Hill et al., 2006). Our data do not contradict with this report. ß-catenin signaling is clearly downregulated in hmgb1/2-knockdown zebrafish embryos, but not completely abolished (Fig. 6). Moreover, in the Hmgb1-/-; Hmgb2+/- forelimb bud, one copy of the Hmgb2 gene is still present. This condition might be sufficient for maintaining Lmx1b expression in the Hmgb1-/-; Hmgb2+/- forelimb bud. Similar to Shh signaling, we favor that Wnt/ß-catenin signaling is downregulated but it is not as severe as causing the skeletal phenotype by itself alone.
Downregulation of both Msx1 and Msx2 in the posterior mesenchyme suggests that BMP signaling is impaired in the Hmgb1-/-; Hmgb2+/- forelimb bud (Fig. 5A-F). However, compared to Wnt/ß-catenin signaling and Shh signaling, it is less clear how Hmgb1 and Hmgb2 regulate BMP signaling. Since HMGB proteins bind chromatin without sequence specificity and alter conformation to facilitate binding of other transcription factors (Pil and Lippard, 1992; Paull et al., 1993), it is conceivable to speculate the possibility that binding of HMGB proteins may facilitate Smad-dependent BMP signaling. Such mechanisms are left to be open for HMGB study in the future.
Our analysis of the Hmgb1-/-; Hmgb2+/- forelimb revealed a redundant function of Hmgb1 and Hmgb2 in mouse embryonic limb development. Gene expression analysis suggests that the combined function of Hmgb1 and Hmgb2 integrates Wnt/ß-catenin signaling, Shh signaling, BMP signaling and Hox activity (Fig. 8). Combined alteration of these signaling pathways at the posterior region in the Hmgb1-/-; Hmgb2+/- forelimb bud may have lead to the loss of digit 5. Furthermore, our study revealed redundant functions of hmgb1 and hmgb2 in the enhancement of Wnt signaling in a variety of embryonic structures in zebrafish. Since Hmgb1 and Hmgb2 are widely co-expressed in early stage mouse embryos (Supplemental Fig. 1), their combined function might be widely important for development of other organs. The data that double null embryos arrest at E9.5 supports importance of their function beyond this stage. Further study with conditional gene inactivation approach will identify combined function of Hmgb1 and Hmgb2 in organogenesis.
Fig. 8. A model of function of Hmgb1 and Hmgb2 during mouse forelimb development.
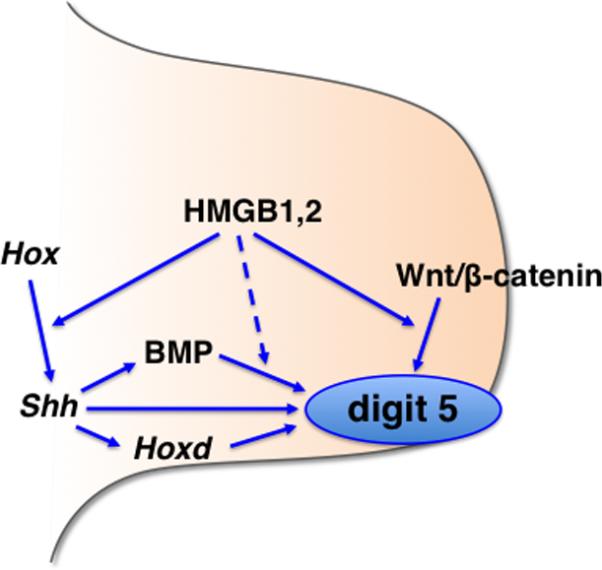
Combined function of Hmgb1 and Hmgb2 integrates the Hox-Shh pathway, Wnt/ß-catenin signaling and BMP signaling for proper development of digit 5 in the forelimb bud. See detail in the text.
EXPERIMENTAL PROCEDURES
Animals
Care and experimentation on mice and zebrafish were done in accordance with the Institutional Animal Care and Use Committee of the University of Minnesota, the Scripps Research Institute, and the National Institute of Advanced Industrial Science and Technology, Japan.
Both Hmgb1 mutant (Calogero et al., 1999) and Hmgb2 mutant (Ronfani et al., 2001) have been published previously. We generated Hmgb1+/-; Hmgb2+/- mice by breeding Hmgb1+/- and Hmgb2+/- mice, which were kept on Balb/c and C57BL/6 background, respectively. Mutant embryos were obtained from timed mating of the Hmgb1+/-; Hmgb2+/- mice. Embryo genotyping were done according to previous publications.
Top-dGFP zebrafish line (Dorsky et al., 2002) was used to detect the activation status of Wnt signaling. Morpholino and mRNA injections were done as previously described (Kawakami et al., 2004; Oishi et al., 2006). Morpholinos were designed by GeneTools. The sequences of the MOs are hmgb1 MO: 5’-GATCCTTCCCCATCTTTGCCTAAAT-3’ (translation blocking MO) hmgb2 MO: 5’-AGTCCTTCTGTTCAACTCACCCTCC-3’ (processing blocking MO) hmgb1 MO(2): 5’- AAATGAGAGGTGTGACTGACCTTCC-3’ (processing blocking MO) hmgb2 MO(2): 5’- CACATTTGCACGCTACTCACGGTGG-3’ (processing blocking MO) Standard control MO from GeneTools (targeting human beta globin intron mutation 5’-CCTCTTACCTCAGTTACAATTTATA-3’) was used as a control. mRNA of mouse Hmgb1 and Hmgb2 are synthesized by using the mMessage mMachine Kit (Ambion) according to the manufacturer's instruction.
In situ hybridization analysis and skeletal preparation
Whole mount in situ mRNA expression analysis in mouse and zebrafish embryos was done following standard protocols (Kawakami et al., 2004; Kawakami et al., 2005; Oishi et al., 2006). Skeletal preparation of mouse embryos was done as previously described (Kawakami et al., 2009).
Immunohistochemistry
Samples were fixed with 4% PFA/PBS+0.1% Tween20 (PBT) overnight at 4°C, washed with PBT and dehydrated by serially increasing methanol concentration. After rehydration, samples were treated with 30% sucrose/PBS, embedded in OCT and sectioned at 14 μm. Sections were washed with PBT, blocked with 10% goat serum/PBT, incubated with anti-phospho ERK1/2 (Cell Signaling, #9101, 1:1000 dilution), washed with PBT and incubated with Alexa594 anti rabbit IgG (Invitrogen, 1:1000 dilution). After washing and counter staining with DAPI, we detected pERK1/2 signal by using Zeiss LSM 710 Laser Scanning Microscopes. Images were analyzed by ZEN2009 software, and pERK1/2 positive cell layers were manually counted.
Supplementary Material
Supplemental Fig. 1 Expression pattern of Hmgb genes in the mouse embryos at E10.5
Lateral views of whole mount mouse embryos hybridized with Hmgb1 (A), Hmgb2 (B), Hmgb3 (C) and Hmgb4 (D) probes are shown. Forelimb buds and hindlimb buds are indicated by arrows and arrowheads, respectively. Hmgb1 and Hmgb2 are highly and widely expressed, while Hmgb3 expression is detected in restricted regions and Hmgb4 expression was undetectable.
Supplemental Table 1 Effect of second set morpholinos on the Top-GFP expression at 18-somite stage
Supplemental Table 2 prx1 expression in the pectoral fin bud of embryos injected with hmgb1 MO and hmgb2 MO
ACKNOWLEDGEMENTS
We are grateful to investigators for generously sharing materials; Dr. Marco E. Bianchi (Hmgb1 mutant mice and Hmgb2 mutant mice), Dr. Randall Moon (Top-dGFP fish line), Dr. Yasushi Nakagawa (Axin2 probe), and Dr. Juan Carlos Izpisua Belmonte (Shh, Fgf8 and Hoxd13 probes). We are also grateful to Dr. Michael O'Connor for allowing us to use Zeiss LSM710, Thu Quach, Austin Johnson, Emily Hsu and Miu Yamashita for their excellent technical support and Krista Bluske for editorial assistance. This study is supported by grants from Minnesota Medical Foundation (Y.K.), NIH grants AG007996 and AR056026 (M.L) and Japan Society for the Promotion of Science, Grant-in-Aid for Scientific Research (C), 22570197 (I.O.)
Grant Sponsors: Minnesota Medical Foundation, National Institutes of Health, and Japan Society for the Promotion of Science.
REFERENCES
- Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- Barrow JR, Thomas KR, Boussadia-Zahui O, Moore R, Kemler R, Capecchi MR, McMahon AP. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 2003;17:394–409. doi: 10.1101/gad.1044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazet JD, Bischofberger M, Tiecke E, Goncalves A, Martin JF, Zuniga A, Naef F, Zeller R. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Bluske KK, Kawakami Y, Koyano-Nakagawa N, Nakagawa Y. Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev Dyn. 2009;238:3297–3309. doi: 10.1002/dvdy.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Izpisua Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Catena R, Escoffier E, Caron C, Khochbin S, Martianov I, Davidson I. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol Reprod. 2009;80:358–366. doi: 10.1095/biolreprod.108.070243. [DOI] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. Embo J. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL. Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet. 1998;19:51–55. doi: 10.1038/ng0598-51. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Duboc V, Logan MP. Building limb morphology through integration of signalling modules. Curr Opin Genet Dev. 2009;19:497–503. doi: 10.1016/j.gde.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P. HMGB1: guiding immunity from within. Trends Immunol. 2005;26:381–387. doi: 10.1016/j.it.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Lunn JS, Dickinson RJ, Munsterberg AE, Sanz-Ezquerro JJ, Farrell ER, Mathers J, Keyse SM, Storey K, Tickle C. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr Biol. 2003;13:1009–1018. doi: 10.1016/s0960-9822(03)00381-6. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Cohn MJ. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature. 2006;442:1033–1037. doi: 10.1038/nature04984. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sancho JM, Brennan KR, Castelo-Soccio LA, Brown AM. Wnt proteins induce dishevelled phosphorylation via an LRP5/6- independent mechanism, irrespective of their ability to stabilize beta-catenin. Mol Cell Biol. 2004;24:4757–4768. doi: 10.1128/MCB.24.11.4757-4768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandel H, Schulte-Merker S. The development of the paired fins in the zebrafish (Danio rerio). Mech Dev. 1998;79:99–120. doi: 10.1016/s0925-4773(98)00176-2. [DOI] [PubMed] [Google Scholar]
- Grasser KD, Launholt D, Grasser M. High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta. 2007;1769:346–357. doi: 10.1016/j.bbaexp.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hill TP, Taketo MM, Birchmeier W, Hartmann C. Multiple roles of mesenchymal beta-catenin during murine limb patterning. Development. 2006;133:1219–1229. doi: 10.1242/dev.02298. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Duff EK, Sirard C. Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J Biol Chem. 2003;278:48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RL, Tabin CJ. Molecular models for vertebrate limb development. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Capdevila J, Buscher D, Itoh T, Rodriguez Esteban C, Izpisua Belmonte JC. WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell. 2001;104:891–900. doi: 10.1016/s0092-8674(01)00285-9. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Esteban CR, Matsui T, Rodriguez-Leon J, Kato S, Belmonte JC. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development. 2004;131:4763–4774. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisua Belmonte JC. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Uchiyama Y, Rodriguez Esteban C, Inenaga T, Koyano-Nakagawa N, Kawakami H, Marti M, Kmita M, Monaghan-Nichols P, Nishinakamura R, Izpisua Belmonte JC. Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development. 2009;136:585–594. doi: 10.1242/dev.027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Wada N, Nishimatsu S, Nohno T. Involvement of frizzled-10 in Wnt-7a signaling during chick limb development. Dev Growth Differ. 2000;42:561–569. doi: 10.1046/j.1440-169x.2000.00545.x. [DOI] [PubMed] [Google Scholar]
- Kengaku M, Capdevila J, Rodriguez-Esteban C, De La Pena J, Johnson RL, Belmonte JC, Tabin CJ. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science. 1998;280:1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J Biol Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev Growth Differ. 2007;49:421–437. doi: 10.1111/j.1440-169X.2007.00942.x. [DOI] [PubMed] [Google Scholar]
- Michos O, Panman L, Vintersten K, Beier K, Zeller R, Zuniga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem. 1989;185:303–310. doi: 10.1111/j.1432-1033.1989.tb15116.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Izpisua Belmonte JC, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004;255:332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood. 2005;105:627–634. doi: 10.1182/blood-2004-07-2551. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, Cline AP, Bodine DM. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102:1298–1306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci U S A. 2006;103:13783–13788. doi: 10.1073/pnas.0604006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JK, Kawakami Y, Buscher D, Raya A, Itoh T, Koth CM, Rodriguez Esteban C, Rodriguez-Leon J, Garrity DM, Fishman MC, Izpisua Belmonte JC. The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development. 2002;129:5161–5170. doi: 10.1242/dev.129.22.5161. [DOI] [PubMed] [Google Scholar]
- Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S, Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- Nissim S, Hasso SM, Fallon JF, Tabin CJ. Regulation of Gremlin expression in the posterior limb bud. Dev Biol. 2006;299:12–21. doi: 10.1016/j.ydbio.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Niswander L. Pattern formation: old models out on a limb. Nat Rev Genet. 2003;4:133–143. doi: 10.1038/nrg1001. [DOI] [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, Itoh N, Noji S. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- Oishi I, Kawakami Y, Raya A, Callol-Massot C, Izpisua Belmonte JC. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat Genet. 2006;38:1316–1322. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- Parr BA, McMahon AP. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature. 1995;374:350–353. doi: 10.1038/374350a0. [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- Pauken CM, Nagle DL, Bucan M, Lo CW. Molecular cloning, expression analysis, and chromosomal localization of mouse Hmg1-containing sequences. Mamm Genome. 1994;5:91–99. doi: 10.1007/BF00292334. [DOI] [PubMed] [Google Scholar]
- Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- Pizette S, Niswander L. BMPs negatively regulate structure and function of the limb apical ectodermal ridge. Development. 1999;126:883–894. doi: 10.1242/dev.126.5.883. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, Tabin C. Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell. 1995;83:631–640. doi: 10.1016/0092-8674(95)90103-5. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Scholer HR, Consalez GG, Bianchi ME. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001;128:1265–1273. doi: 10.1242/dev.128.8.1265. [DOI] [PubMed] [Google Scholar]
- Sessa L, Bianchi ME. The evolution of High Mobility Group Box (HMGB) chromatin proteins in multicellular animals. Gene. 2007;387:133–140. doi: 10.1016/j.gene.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Zechner D, Huelsken J, Mishina Y, Behringer RR, Taketo MM, Crenshaw EB, 3rd, Birchmeier W. Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsalventral axis in the limb. Genes Dev. 2003;17:1963–1968. doi: 10.1101/gad.263003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spater D, Hill TP, O'Sullivan RJ, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Yonei-Tamura S, Yano T, Yokoyama H, Ide H. The autopod: its formation during limb development. Dev Growth Differ. 2008;50(Suppl 1):S177–187. doi: 10.1111/j.1440-169X.2008.01020.x. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi N, Carames B, Kawakami Y, Amendt BA, Komiya S, Lotz M. Chromatin protein HMGB2 regulates articular cartilage surface maintenance via beta-catenin pathway. Proc Natl Acad Sci U S A. 2009a;106:16817–16822. doi: 10.1073/pnas.0904414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi N, Carames B, Ronfani L, Ulmer U, Komiya S, Bianchi ME, Lotz M. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci U S A. 2009b;106:1181–1186. doi: 10.1073/pnas.0806062106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi N, Yoshida K, Ito T, Tsuda M, Mishima Y, Furumatsu T, Ronfani L, Abeyama K, Kawahara K, Komiya S, Maruyama I, Lotz M, Bianchi ME, Asahara H. Stage-specific secretion of HMGB1 in cartilage regulates endochondral ossification. Mol Cell Biol. 2007;27:5650–5663. doi: 10.1128/MCB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D, Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature. 2006;443:985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- Towers M, Tickle C. Growing models of vertebrate limb development. Development. 2009;136:179–190. doi: 10.1242/dev.024158. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Beltrame M, Ferrari S, Bianchi ME. Hmg4, a new member of the Hmg1/2 gene family. Genomics. 1998;49:247–252. doi: 10.1006/geno.1998.5214. [DOI] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Warnken W, Izpisua Belmonte JC. Dorsal cell fate specified by chick Lmx1 during vertebrate limb development. Nature. 1995;378:716–720. doi: 10.1038/378716a0. [DOI] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. A hypermorphic mouse Gli3 allele results in a polydactylous limb phenotype. Dev Dyn. 2007a;236:769–776. doi: 10.1002/dvdy.21082. [DOI] [PubMed] [Google Scholar]
- Wang C, Ruther U, Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev Biol. 2007b;305:460–469. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yang Y. Wnts and wing: Wnt signaling in vertebrate limb development and musculoskeletal morphogenesis. Birth Defects Res Part C Embryo Today. 2003;69:305–317. doi: 10.1002/bdrc.10026. [DOI] [PubMed] [Google Scholar]
- Yang Y, Niswander L. Interaction between the signaling molecules WNT7a and SHH during vertebrate limb development: dorsal signals regulate anteroposterior patterning. Cell. 1995;80:939–947. doi: 10.1016/0092-8674(95)90297-x. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi ME. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. Embo J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 Expression pattern of Hmgb genes in the mouse embryos at E10.5
Lateral views of whole mount mouse embryos hybridized with Hmgb1 (A), Hmgb2 (B), Hmgb3 (C) and Hmgb4 (D) probes are shown. Forelimb buds and hindlimb buds are indicated by arrows and arrowheads, respectively. Hmgb1 and Hmgb2 are highly and widely expressed, while Hmgb3 expression is detected in restricted regions and Hmgb4 expression was undetectable.
Supplemental Table 1 Effect of second set morpholinos on the Top-GFP expression at 18-somite stage
Supplemental Table 2 prx1 expression in the pectoral fin bud of embryos injected with hmgb1 MO and hmgb2 MO