Abstract
We explored whether breast cancer outcomes are associated with endoxifen and other metabolites of tamoxifen, and to examine potential correlates of endoxifen concentrations including CYP2D6 metabolizer phenotype and body mass index (BMI). Tamoxifen, endoxifen, 4-hydroxytamoxifen and N-desmethyltamoxifen concentrations were measured from 1370 estrogen receptor positive breast cancer patients participating in the Women’s Healthy Eating and Living (WHEL) Study, and tested for associations with breast cancer outcomes. Breast cancer outcomes were not associated with tamoxifen, 4-hydroxytamoxifen or N-desmethyltamoxifen concentrations. For endoxifen, a threshold was identified suggesting that women in the upper four quintiles of endoxifen had a 26% lower recurrence rate than women in the bottom quintile. (HR=0.74; 95% CI, [0.55, 1.00]). Predictors of membership in this higher risk bottom quintile were poor/intermediate metabolizer genotype, higher BMI, and low tamoxifen concentrations. This study suggests a minimal threshold at which endoxifen is effective against breast cancer recurrence, which 80% of tamoxifen-takers achieve.
Keywords: Cancers, CYP, Epidemiology, Pharmacogenetics, Genotype
INTRODUCTION
Tamoxifen reduces breast cancer recurrence(1) by blocking estrogen receptors;(2) an effect produced mainly through its metabolites, particularly endoxifen (4-hydroxy-N-desmethyltamoxifen) (3–5). Polymorphisms in the metabolizing enzyme, cytochrome P450 2D6 (CYP2D6) can lead to lower circulating endoxifen concentrations, and possibly tamoxifen effectiveness. (6–8) However, studies of an association between CYP2D6 functionality and breast cancer-specific outcomes have produced inconsistent results (9), highlighting the need for more research examining the association between tamoxifen metabolite concentrations, CYP2D6 functionality, and breast cancer survival.(10)
While CYP2D6 genotype is associated with tamoxifen metabolite concentrations, a functional categorization of the genotype does not explain all of the variance observed.(11) Compliance in taking tamoxifen may influence metabolite concentrations. Furthermore, drug-gene interactions, particularly some of the serotonin reuptake inhibitors (SSRIs) – which physicians may prescribe for depression or for vasomotor symptoms in breast cancer patients (12–14) – are potent inhibitors of CYP2D6 (8, 15–16) and can thus potentially reduce the effectiveness of tamoxifen.
The Women’s Healthy Eating and Living (WHEL) Study offers a unique opportunity to study the relationship between CYP2D6 genotype, tamoxifen metabolites, and breast cancer outcomes.(17–19) This study enrolled a large sample of women with early stage breast cancer diagnosed between 1991 and early 2000, when tamoxifen was the endocrine therapy of choice in the adjuvant setting and before SSRIs were reported to be a treatment for vasomotor symptoms in this population. The study collected extensive treatment data, stored blood samples, and followed women over an average of 7.3 years, medically verifying all breast cancer outcomes. Participants were enrolled up to four years after their initial diagnosis.
Given the complexity of tamoxifen metabolism and the inconsistent results in the literature regarding CYP2D6 genotype and breast cancer outcomes, the aim of this study was to explore the relationship between concentrations of tamoxifen metabolites and long-term breast cancer outcomes in a subset of 1370 estrogen receptor positive breast cancer patients. While CYP2D6 genotype, co-medications, compliance and other factors are all potential factors that may lead to reduced effectiveness of tamoxifen therapy, using the actual metabolite concentrations to predict breast cancer outcomes can provide a more direct measurement of this putative association.
RESULTS
Demographic and clinical characteristics of the 1370 participants selected for this study are described in Table 1. Consistent with the results from our entire WHEL cohort, breast cancer events (n=178) were significantly associated with breast cancer stage and grade.(19) In our sample of 1370 participants, radiation, chemotherapy and BMI were not associated with breast cancer events when added to a Cox model of disease free survival that included stage and grade. Quintiles of tamoxifen and metabolites, and raw event rates for each quintile, are presented in Table 2. Tamoxifen concentrations did not differ by CYP2D6 phenotype, but concentrations of endoxifen, 4-hydroxytamoxifen and N-desmethyltamoxifen were strongly associated with CYP2D6 phenotype (Table 3). The Chi-squared test statistic for testing Hardy-Weinberg equilibrium was 23.92 (df= 22; p=0.35) indicating that the genotypes were in equilibrium.
Table 1.
Demographic and clinical characteristics of 1370 WHEL Study participants
| Demographic or Clinical Characteristic | No. of Participants (N = 1370) | % |
|---|---|---|
| Age at baseline, years | ||
| <50 | 577 | 42 |
| 50–59 | 519 | 38 |
| 60 and older | 274 | 20 |
| Race/ethnicity | ||
| White, Non-Hispanic | 1191 | 87 |
| Hispanic | 68 | 5 |
| Asian/Pacific Islander | 57 | 4 |
| Black/African American | 31 | 2 |
| Other | 23 | 2 |
| Breast cancer treatments | ||
| Mastectomy | 707 | 52 |
| Received radiation | 867 | 63 |
| Received chemotherapy | 846 | 62 |
| Stage | ||
| I | 512 | 37 |
| IIA | 451 | 33 |
| IIB | 175 | 13 |
| IIIA | 180 | 13 |
| IIIC | 52 | 4 |
| Tumor Grade | ||
| Well differentiated | 283 | 21 |
| Moderately differentiated | 651 | 48 |
| Poorly differentiated | 307 | 22 |
| Not reported | 129 | 9 |
| Outcomes | ||
| No breast event | 1192 | 87 |
| Breast cancer recurrence | 160 | 12 |
| New primary breast cancer | 18 | 1 |
| Time from Diagnosis to study entry (months) | ||
| 1–12 | 229 | 17 |
| 13–24 | 485 | 35 |
| 25–36 | 373 | 27 |
| 37–48 | 283 | 21 |
Table 2.
Medians, ranges, and raw recurrence rates for quintiles of tamoxifen and metabolites
| Median (ng/mL) | Range | Recurrence rate (%) | |
|---|---|---|---|
| Tamoxifen quintile | |||
| Q1 | 67.1 | 0–84.9 | 13.9 |
| Q2 | 100.0 | 84.9–114 | 12.8 |
| Q3 | 127.0 | 114–141 | 13.9 |
| Q4 | 160.0 | 141–179 | 11.2 |
| Q5 | 213.0 | 179–728 | 13.1 |
| Endoxifen quintile | |||
| Q1 | 4.2 | 0–5.9 | 16.0 |
| Q2 | 8.2 | 5.9–10.2 | 11.7 |
| Q3 | 12.4 | 10.2–14.6 | 14.7 |
| Q4 | 17.4 | 14.6–20.4 | 10.1 |
| Q5 | 26.2 | 20.4–73.7 | 12.4 |
| 4OH-tam quintile | |||
| Q1 | 1.0 | 0–1.28 | 14.7 |
| Q2 | 1.5 | 1.28–1.7 | 14.1 |
| Q3 | 1.9 | 1.7–2.21 | 8.8 |
| Q4 | 2.5 | 2.21–2.84 | 13.9 |
| Q5 | 3.8 | 2.84–10.9 | 13.4 |
| ND-tam quintile | |||
| Q1 | 135.0 | 0–168 | 14.0 |
| Q2 | 192.0 | 168–213.2 | 11.9 |
| Q3 | 234.5 | 213.2–258 | 15.9 |
| Q4 | 287.0 | 258–326 | 11.7 |
| Q5 | 381.5 | 326–869 | 11.3 |
Table 3.
Tamoxifen and tamoxifen metabolite concentrations (ng/mL) by CYP2D6 phenotype
| CYP2D6 Phenotype | Concentration (ng/mL) | ||||
|---|---|---|---|---|---|
| Endoxifen | Tamoxifen | 4OH-Tam | ND-Tam | ||
| Ultra-rapid metabolizer (UM) N=27 | Mean | 22.8 | 143.4 | 2.7 | 230.8 |
| Std. Deviation | 11.3 | 58.4 | 1.2 | 71.1 | |
| Median | 24.5 | 137.0 | 2.7 | 226.0 | |
| Extensive metabolizer (EM) N=1097 | Mean | 15.9 | 136.4 | 2.3 | 242.1 |
| Std. Deviation | 9.2 | 64.3 | 1.1 | 95.3 | |
| Median | 14.3 | 127.0 | 2.1 | 230.0 | |
| Intermediate metabolizer (IM) N=164 | Mean | 8.1 | 142.9 | 1.7 | 295.7 |
| Std. Deviation | 4.9 | 70.8 | 0.8 | 112.6 | |
| Median | 6.7 | 134.0 | 1.6 | 286.0 | |
| Poor metabolizer (PM) N=82 | Mean | 5.6 | 142.3 | 1.7 | 312.7 |
| Std. Deviation | 3.8 | 63.1 | 0.9 | 114.2 | |
| Median | 4.7 | 140.5 | 1.5 | 291.5 | |
| ANOVA F statistic | 77.1 | 0.69 | 20.6 | 25.3 | |
| p | <0.001 | 0.55 | <0.001 | <0.001 | |
Abbreviations: 4OH-Tam, 4-hydroxytamoxifen; ND-Tam, N-desmethyltamoxifen
The Cox linear trend analysis, controlling for breast cancer stage and grade, identified no evidence to support a linear relationship between tamoxifen and/or any of the measured metabolites and breast cancer outcomes: the hazard ratios (HR) for tamoxifen, endoxifen, 4-hydroxytamoxifen, and N-desmethyltamoxifen were 0.97 (95% CI, 0.92 to 1.03), 0.94 (95% CI, 0.84 to 1.06), 0.94 (95% CI, 0.62 to 1.44) and 0.97 (95% CI, 0.93 to1.02) change in HR per 1ng change in square-root of concentration respectively. We also examined the HR for the ratio of N-desmethyltamoxifen to endoxifen and the ratio was not associated with breast cancer outcomes (data not shown).
With no evidence of a linear association, we explored a potential threshold effect by dividing the continuous concentrations of tamoxifen and tamoxifen metabolites into quintiles and conducting separate Cox models, controlled for stage and grade (Figure 1). None of the upper four quintiles of tamoxifen had a significantly different risk than the lowest quintile. For endoxifen, two of the quintiles (2nd and 4th) were significantly different from the lowest quintile and all four upper quintiles had similar risk levels, suggesting a threshold effect. Three of the quintile cut-points for 4-hydroxytamoxifen had equivalent risk levels to the lowest quintile, with no suggestion of a threshold effect. Finally, for N-desmethyltamoxifen, only the second quintile had a significantly reduced risk compared to the lowest quintile. A Kaplan-Meier curve of unadjusted disease-free survival by endoxifen quintile is presented as Figure 2.
Figure 1.
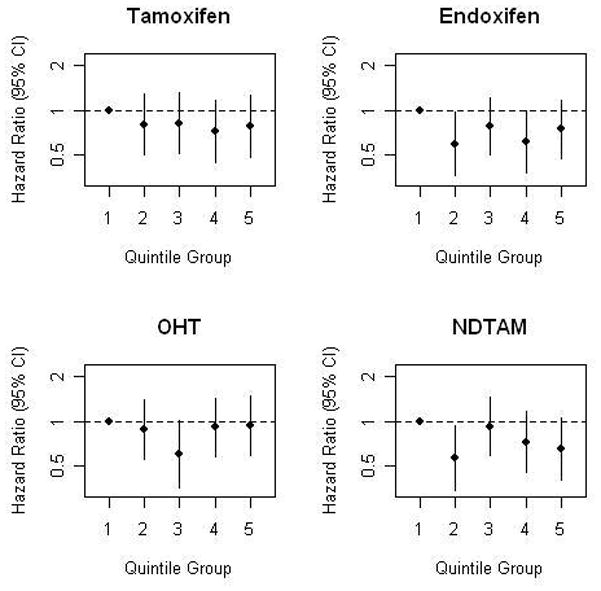
Plots of hazard ratios derived from delayed-entry Cox models, controlling for stage and grade.
Figure 2.
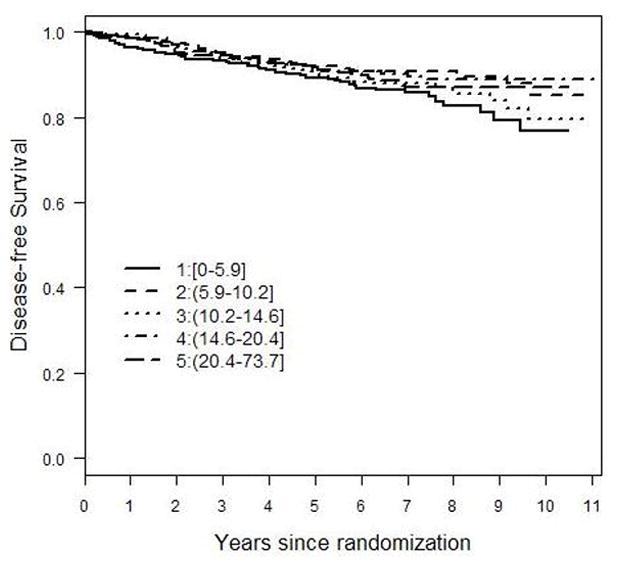
Kaplan-Meier plot of unadjusted recurrence rates for each quintile of endoxifen
We repeated the quintile Cox model analyses with CYP2D6 phenotype groups included in the model. For all four models, a likelihood ratio test indicated that the CYP2D6 phenotype was not statistically significant (p >= 0.6).
As endoxifen was the only metabolite that was suggestive of a threshold effect on breast cancer outcomes, we explored dichotomized optimal cut-points for a split regression analysis(20) of endoxifen concentrations with additional breast cancer events. After bootstrap resampling and cross-validation, we identified an at-risk subgroup as those with endoxifen below 5.97 ng/mL, a concentration that corresponds closely to the lowest quintile in our sample. In the Cox model, adjusted for both stage and grade, participants with endoxifen concentrations above 5.97ng/mL had a 30% lower risk of additional breast cancer events (HR=0.70, 95% CI, 0.52 to 0.94). The more conservative bias-corrected estimate(21) of reduced risk in this group of participants in the upper four quartiles of endoxifen concentrations was marginally significant (HR=0.74; 95% CI, 0.55 to 1.00). We repeated the Cox model stratifying by time since diagnosis, and the results were essentially identical to the unstratified model indicating that time from diagnosis to study entry did not affect our findings. We also repeated the model including BMI and found that BMI was not associated with breast cancer events, suggesting that the endoxifen effect is independent of BMI.
With a putative threshold for endoxifen concentrations identified (corresponding to the bottom quintile), we examined whether CYP2D6 phenotype, tamoxifen quintiles, race/ethnicity, age, or BMI were associated with membership in the bottom quintile of endoxifen (Table 4). Bivariately, CYP2D6 phenotype, tamoxifen concentrations, race/ethnicity, and BMI were all associated with membership in the bottom quintile of endoxifen. Using a logistic regression model, race/ethnicity and CYP2D6 genotype showed colinearity and so race/ethnicity was dropped from the model. (Table 5) The final model, controlling for age, indicated that CYP2D6 phenotype, BMI, and tamoxifen concentrations were all associated with membership in the bottom quintile of endoxifen.
Table 4.
Factors associated with membership in the bottom quintile of endoxifen concentration
| N | % in Endoxifen Bottom Quintile | Chi-sq | p | |
|---|---|---|---|---|
| BMI | ||||
| <25 | 597 | 15.9 | 15.9 | 0.001 |
| 25 –29.9 | 429 | 21.2 | ||
| 30–34.9 | 199 | 22.6 | ||
| 35+ | 145 | 29.7 | ||
| CYP2D6 phenotype | ||||
| UM | 27 | 7.4 | 242.5 | <0.001 |
| EM | 1097 | 12.9 | ||
| IM | 164 | 41.5 | ||
| PM | 82 | 75.6 | ||
| Race/ethnicity | ||||
| White | 1191 | 21.2 | 12.04 | 0.017 |
| Hispanic | 68 | 7.4 | ||
| African-American | 31 | 22.6 | ||
| Asian/PI | 57 | 12.3 | ||
| Other | 23 | 8.7 | ||
| Age | ||||
| <45 | 137 | 17.5 | 3.00 | 0.22 |
| 45–54 | 548 | 22.3 | ||
| 55+ | 685 | 18.7 | ||
| Tamoxifen quintile | ||||
| Q 1 (low) | 274 | 39.8 | 108.6 | <0.001 |
| Q 2 | 273 | 23.7 | ||
| Q 3 | 279 | 16.1 | ||
| Q 4 | 269 | 13.5 | ||
| Q 5 (high) | 275 | 6.9 | ||
| Tamoxifen duration at time of blood draw | ||||
| ≤12 mos. | 581 | 19.4 | 2.43 | 0.489 |
| 13–24 mos | 432 | 22.0 | ||
| 25–36 mos | 266 | 19.5 | ||
| >36 mos | 91 | 15.4 | ||
Abbreviations: UM, Ultra-rapid Metabolizer; EM, Extensive Metabolizer; IM, Intermediate Metabolizer; PM, Poor Metabolizer
Table 5.
Logistic regression* predictors of bottom quintile of endoxifen vs. other
| p | OR | 95.0% C.I. | ||
|---|---|---|---|---|
| Lower | Upper | |||
| BMI | ||||
| <25 (ref) | ||||
| 25 –29.9 | 0.006 | 1.70 | 1.16 | 2.48 |
| 30–34.9 | 0.002 | 2.10 | 1.31 | 3.38 |
| 35+ | <0.001 | 3.21 | 1.93 | 5.33 |
| CYP2D6 phenotype | ||||
| UM | 0.359 | 0.49 | 0.10 | 2.27 |
| EM (ref) | ||||
| IM | <0.001 | 7.08 | 4.67 | 10.73 |
| PM | <0.001 | 43.73 | 23.37 | 81.81 |
| Tamoxifen quintile | ||||
| Q 1 (ref) | ||||
| Q 2 | <0.001 | 0.38 | 0.25 | 0.57 |
| Q 3 | <0.001 | 0.18 | 0.11 | 0.29 |
| Q 4 | <0.001 | 0.12 | 0.07 | 0.20 |
| Q 5 (high) | <0.001 | 0.05 | 0.02 | 0.09 |
Controlled for age at study entry
Abbreviations: OR, odds ratio; EM, Extensive Metabolizer; IM, Intermediate Metabolizer; PM, Poor Metabolizer, UM, Ultra-rapid Metabolizer
Finally, in an effort to describe the variance in endoxifen that is explained by CYP2D6 phenotype, we also looked at R-squared for predicting continuous endoxifen concentrations. Our first model included age, BMI, race/ethnicity and tamoxifen concentrations (square root). The second was the same but also included CYP2D6 phenotype. The R-squared for Model 1 was 0.28 and the R-squared for Model 2 was 0.46 (likelihood ratio test comparing model 1 to model 2 p < 0.00001), suggesting an absolute 18% increase in R-squared when the model includes CYP2D6 phenotype.
DISCUSSION
In a large cohort of early stage ER-positive breast cancer survivors using tamoxifen, the concentration of endoxifen, but not tamoxifen or other metabolites, was associated with risk of breast cancer recurrence or second breast cancer. However, this increased risk was confined to those with an endoxifen concentration in the bottom quintile of the distribution. Women with an impaired CYP2D6 metabolizer phenotype were more likely to be in this bottom quintile. Importantly, however, while the majority (76%) of poor metabolizers had low endoxifen levels, this leaves 24% of PMs who may be able to obtain therapeutic levels of endoxifen despite their PM status. Metabolizer phenotype alone may not be enough to determine whether tamoxifen is of potential benefit to any individual patient. Excess weight was also associated with lower endoxifen concentrations, as has previously been reported.(22) In addition, tamoxifen concentrations, a potential indicator of compliance, were associated with endoxifen concentrations. Overall, we have identified several variables (metabolizer phenotype, BMI and tamoxifen levels) which are associated with low endoxifen levels but are not independently associated with breast cancer outcomes in our sample. Taken together, these findings suggest that the relationships between these variables, metabolite levels and breast cancer outcomes is complex, which may help to explain the inconsistency in the published studies to date.
One recent study modeled the expected amount of tamoxifen (or its metabolites) required to affect estrogen receptors, and postulated that even women with a poor metabolizer CYP2D6 genotype might be able to bind over 99.6% of receptors and hence not be at increased risk for recurrence.(23) Biomarker studies have also suggested that there may be a case for reducing the current dose of tamoxifen to reduce side effects while maintaining efficacy.(24–25) In our study, the majority of participants who took tamoxifen appeared to have sufficient endoxifen concentrations to achieve its protective effect. However, our study suggests that the proportion of women on tamoxifen therapy who do not have a sufficient dose may be as high as 20%. Our results may help explain some of the varying results among studies of CYP2D6 genotype and breast cancer outcomes if an endoxifen threshold does exist. Individuals with poor metabolizer genotype may be much less likely to achieve endoxifen concentrations above the potential therapeutic threshold, while individuals with an intermediate metabolizer genotype may be more susceptible to other factors such as SSRI use or tamoxifen compliance that could decrease their endoxifen concentrations below the threshold. Importantly, however, only 46% of the variance in endoxifen concentrations was explained by known covariates (i.e., age, BMI, CYP2D6 genotype), indicating that other (unmeasured) factors are contributing to the observed variability in metabolite concentrations. This may include variables related to tamoxifen dose or compliance with taking the drug.
Our results should be considered in the context of several limitations. First, the WHEL Study was designed as a randomized dietary trial, and our analyses are clearly secondary to the original hypotheses. However, the WHEL Study enrolled women when tamoxifen was the treatment of choice and is one of the few cohorts with adequate long-term endpoint data, stored biospecimens, and patient-reported covariates that allow for a secondary analysis of endoxifen and breast cancer endpoints. However, because WHEL participants were enrolled up to 4 years after their initial diagnosis, the study does not have an adequate representation of breast cancer patients who recur shortly after diagnosis. Rather, its strength is to predict risk for breast cancer events occurring 3 to 10 years post diagnosis. Importantly, considerable evidence suggests that early stage breast cancer survivors who recur quickly are more likely to have had ER-negative tumors.(26–29) A strength of our study is its large sample size of early stage breast cancer patients who were taking tamoxifen before aromatase inhibitors were available and before SSRI’s were prescribed to reduce vasomotor symptoms in this population. A weakness is that the WHEL Study did not assess SSRI use at baseline (which were used for the treatment of depression at that time), and retrospective recall of SSRI use indicated that at least 2% of participants who completed the WHEL Study may have been taking an SSRI at baseline. Similarly, we do not have detailed data on tamoxifen prescriptions (e.g. dose) or compliance. We note that, currently, there is no quality controlled standardized method for measuring endoxifen or other tamoxifen metabolites that is approved for clinical use, and so we caution against a clinical interpretation of the absolute metabolite values described in our study. There appears to be considerable variability in estimated metabolite concentrations across studies, though this could relate to variation in sample handling, storage, and measurement methods.
In summary, this study is the first to report on the association between endoxifen concentrations and breast cancer outcomes in tamoxifen-treated breast cancer patients. It is likely that a threshold of endoxifen must be achieved for a therapeutic effect of tamoxifen, rather than a linear dose-response effect. We have identified that CYP2D6 metabolizer phenotype, BMI, and tamoxifen concentrations contribute to having endoxifen concentrations below this threshold. Future studies of tamoxifen metabolites and breast cancer outcomes are needed to replicate these findings, and determine the extent to which other factors, such as BMI and CYP2D6 metabolizer phenotype, influence tamoxifen metabolism in breast cancer patients.
METHODS
This study used data and archived blood samples from the WHEL Study, which has been extensively described elsewhere.(18–19) Briefly, 3088 breast cancer patients with a verified diagnosis of invasive breast cancer (AJCC IV (30) stage: I ≥ 1 cm, II or IIIA) between 1991 and early 2000 were recruited from seven clinical sites in California, Arizona, Texas and Oregon. Internal review boards at each site approved the study and all participants provided written informed consent before enrolling.
At study entry, participants were within 4 years of diagnosis and had completed primary therapy, and had not experienced a recurrence or new breast primary cancer. At baseline (i.e., study entry) and subsequent clinic visits (1, 2 or 3, 4 and 6 years after study entry), participants were asked: “Have you ever taken an anti-estrogen? (e.g., tamoxifen, raloxifene)” Anti-estrogen users were asked to specify which drug they took, and the length of time that they had taken the drug. This study considers the 1370 participants who had estrogen receptor (ER)-positive tumors and had been taking tamoxifen for at least 4 months before the baseline survey (to ensure steady-state blood concentrations) and for whom we could successfully conduct genotyping of CYP2D6 (see below).
Height and weight were measured at the time of blood draw; and body mass index (BMI, weight [kg]/height [m2]) was computed.
Sample handling and DNA extraction
Blood (48 ml) was collected at a baseline clinic visit, placed on ice, and within the hour separated into plasma, serum, buffy coat and red blood cells using centrifugation at 2300 × g at 4° C for 10 minutes. Per the WHEL Operations Manual, all samples were immediately protected from light during sample processing (using aluminum foil, Red bags, or Amber bags) and were frozen promptly. Site laboratory procedures were audited during annual visits by the WHEL study director and principal investigator. To the best of our knowledge, WHEL samples were handled uniformly and were not thawed before analysis. Aliquots were stored at −80° C in cryogenic tubes until analysis. Genomic DNA was extracted from these archival samples using 200 uL of the buffy coat fraction (QIAcube robot with QIAamp DNA blood Mini Kit, Qiagen, Valencia, CA, USA). DNA was quantified with a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, according to manufacturers’ instructions).
Extraction and measurement of tamoxifen and its metabolites
Tamoxifen and its metabolites were measured in archived serum samples. First, 80x stock of deuterated internal standards, containing D5-Endoxifen (200ng/mL), D5-Tamoxifen (1ug/mL), and D5-N-Desmethyltamoxifen (4ug/mL) (Toronto Research Chemicals) was prepared in methanol and stored at −70*C. Before sample extraction, the internal standard mix was diluted with 0.5mM Ammonium Formate buffer, pH 3.0. Serum samples (200uL) were spiked with 800uL of diluted internal standard mix and extracted with Waters MCX 1cc solid phase extraction cartridges according to the manufacturer’s instructions. We used a 1200 series Agilent High Performance Liquid Chromatography (HPLC) system to resolve analytes by reverse phase chromatography with gradient elution. This gradient separates endoxifen and 4-hydroxyendoxifen from the corresponding 4′ and 4′hydroxy isomers. These latter two isomers are not measured in this assay. A Waters xTerra MS C18, 3.5μm (2.1×100mm) analytical column was used for the assay preceded by a Waters C18, 5μm (2.1×10m C18) guard column and column saver (MacMod). HPLC peaks were detected using a 3200 tandem/ion trap mass spectrometer (Applied Biosystems Sciex). We used Analyst Software (Applied Biosystems) to determine analyte concentration by comparing the peak area ratio of the analyte to the peak area ratio of the internal standard. D5-tamoxifen was used as the internal standard for tamoxifen, D5-N-desmethyltamoxifen was used as the internal standard for N-desmethyltamoxifen, and D5-endoxifen was used as the internal standard for endoxifen and 4-hydroxytamoxifen (separate standards were used for calibration purposes). The MRM transitions used to track analytes and internal standards were: tamoxifen m/z 372/72; N-desmethyltamoxifen 358/58, endoxifen 374/58, 4-hydroxytamoxefin 387/105, D5-tamoxifen 377/72, D5-N-desmethyltamoxifen 363/58, and D5-endoxifen 379/58. The total imprecision of this assay using quality control materials (n=17) was 7.0% and 6.6% for tamoxifen at 11.4 and 188 ng/mL, respectively, 7.8% and 4.8% for enxoxifen at 1.7 and 33 ng/mL, 5.3% and 4.5% for N-desmethyltamoxifen at 18.7 and 336 ng/mL, and 12.9% and 7.1% for 4-hydroxytamoxifen at 0.41 and 7.2 ng/mL. The linearity was up to 250 ng/mL for tamoxifen and endoxifen, 500 ng/mL for N-desmethyltamoxifen, and 6 ng/mL for 4-hydroxytamoxifen. Using the same serum samples, we compared results of this assay against an LC/MS assay from Mayo Medical Laboratories and observed concordance for tamoxifen, endoxifen, and N-desmethyltamoxifen, with less concordance for 4-hydroxytamoxifen (unpublished observations).
CYP2D6 genotyping and phenotyping
To determine CYP2D6 genotype, DNA samples were analyzed using the AmpliChip™ CYP450 Test (Roche Molecular Systems, Inc., Pleasanton, CA) by operators blinded to clinical data. This test combines a long multiplex polymerase chain reaction amplification with an oligonucleotide microarray manufactured by Affymetrix (Santa Clara, CA). After fragmentation and labeling of the PCR products, amplicons are hybridized to the microarray and stained using a streptavidin-phycoerythrin conjugate, and fluorescence associated with each probe feature is detected using a laser-illuminated, confocal scanner. Data analysis software interprets the hybridization pattern to a series of probes that are specifically complementary to either wild-type or variant sequences for each polymorphic site. From each site call, an allele call is inferred relative to an allele reference table.
The AmpliChip™ CYP450 Test queries 29 polymorphisms found in the CYP2D6 gene to identify 28 different alleles, including a variety of gene duplications. Each allele was assigned to one of four phenotypic categories according to its associated enzyme function:
Non-functional (poor metabolizer or PM) alleles include: CYP2D6*3, *4, *5, *6, *7, *8, *11, *14A, *15, *19, *20, and *40, and the *4XN gene duplication.
Reduced function (intermediate metabolizer or IM) alleles include: CYP2D6*9, *10, *17, *29, *36 and *41, and gene duplications *10XN, *17XN, and *41XN.
Fully functional (extensive metabolizer or EM) alleles include: CYP2D6*1, *2 and *35.
Increased function (ultrarapid metabolizer or UM) phenotype alleles include gene duplications such as CYP2D6*1XN, *2XN and *35XN.
Individuals are assigned a phenotype depending on the combination of alleles they carry (for assignments in our study population, see the online supplemental table)
Breast cancer outcomes
Disease-free survival was defined as the time from diagnosis of the original breast cancer toa second breast cancer event (including local and distant recurrences, metastatic disease, or new invasive primary breast cancer). Follow-up time was censored at the minimum time to a participant’s death (if not from breast cancer), the lastdocumented staff contact date, or study completion (June 1, 2006). Information on breast cancer recurrences and new primary breast cancers was obtained during semi-annual telephone contacts with participants during the WHEL Study. Any report of a study outcome triggered a request for medical records by study clinical co-investigators. Two independent oncologists reviewed medical records for all reported outcome events. Date of recurrence was coded as first confirmed incidence, clinical or pathological, of new or recurrent invasive breast cancer. Ductal carcinoma in situ (DCIS) was not considered a study outcome. Vital status and breast cancer outcome status (including date of recurrence or death) was available on 1328 of the 1370 (96.9%) WHEL participants in this analysis as of the study end date of June 2006.
Statistical methods
Descriptive statistics (means, proportions) were used to describe the clinical characteristics of the study population., Mean concentrations of tamoxifen and its metabolites were calculated for the four functional categories of the CYP2D6 phenotype (EM, IM, PM and UM). The Chi-squared test of Hardy-Weinberg equilibrium was conducted using the methods of Thomson et al for lumping genotype classes where expected values are <5 (31). Delayed-entry Cox models (32) were fit to examine whether there was a linear association between the concentration of tamoxifen and its metabolites (e.g. different models for (a) tamoxifen, (b) endoxifen, (c) ND-tamoxifen and (d) 4OH-tamoxifen) and time to a new breast cancer event. These models account for the variable time-interval between diagnosis and blood draw. In each of the models, we included cancer stage and grade as covariates as the WHEL cohort showed that these variables were strong predictors of outcome.(19) A square root transformation was applied to the tamoxifen and metabolite concentrations as this transformation indicated better fit of the Cox models, examined via residual plots.
With lack of evidence for a linear association, we explored a potential nonlinear effect by dividing the population into quintiles based on the concentration of tamoxifen and its metabolites. Cox models including breast cancer stage and grade were fit with quintiles of each of tamoxifen and the other three metabolites. We also included CYP2D6 phenotype to determine if this variable contributed independently to breast cancer outcomes, after adjusting for metabolite concentration.
With evidence of a potential therapeutic threshold, we used methods developed by Tableman and Kim (20) to explore different cut-points for a split regression analysis. This analysis identifies an optimal cut-point above which tamoxifen metabolite concentrations might protect against additional breast cancer events. We used 1000 bootstrapped resamples to cross-validate the so-called optimal cut-point for identifying an at-risk subgroup. We then used a Cox model to assess the risk in this identified subgroup. As such an approach can lead to overfitting a model, to be conservative, we adjusted the estimate to account for this bias.(21)
Finally, we used logistic regression to identify whether having endoxifen concentrations below our putative threshold was associated with the CYP2D6 functional phenotype and/or BMI, both of which have been previously associated with concentrations of tamoxifen metabolites. The reference group for the logistic regression comprises the participants who are above the putative endoxifen threshold. Statistical analyses were conducted using the R statistical software package (http://www.r-project.org/) and SPSS (v. 14.0).
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT: The Women’s Healthy Eating and Living (WHEL) Study was initiated with the support of the Walton Family Foundation and continued with funding from NCI grant CA 69375. Some of the data were collected from General Clinical Research Centers, NIH grants M01-RR00070, M01-RR00079, and M01-RR00827. CYP2D6 blinded genotyping and partial funding for CYP2D6 analyses and tamoxifen metabolite measurements were provided by Roche Molecular Systems, Inc. Support for assessment of tamoxifen metabolites was also provided by philanthropic donations from Lynn and Bert Epsten and the McKenzie family (in remembrance of Muriel Harold).
The authors wish to thank the WHEL study participants. We thank Dennis Heath for the metabolite extractions; and Christine Hayes, Aimee Humphrey and Sheila Kealey for editorial assistance.
Footnotes
WHEL Study Group
WHEL Study Coordinating Center: University of California, San Diego, Cancer Prevention and Control Program, Moores UCSD Cancer Center, San Diego, CA (John P. Pierce, PhD; Susan Faerber, BA; Barbara A. Parker, MD; Loki Natarajan, PhD, Cheryl L. Rock, PhD, RD; Vicky A. Newman, MS, RD; Shirley W. Flatt, MS; Sheila Kealey, MPH; Linda Wasserman, MD, PhD; Wayne A. Bardwell, PhD; Lisa Madlensky, PhD.; Wael Al-Delaimy MD, PhD
WHEL Study Clinical Sites: Center for Health Research-Portland, Portland, OR (Njeri Karanja, PhD, Mark U. Rarick, MD); Kaiser Permanente Northern California, Oakland, CA (Bette J. Caan, DrPH, Lou Fehrenbacher, MD); Stanford Prevention Research Center, Stanford University, CA (Marcia L. Stefanick, PhD, Robert Carlson, MD); University of Arizona, Tucson & Phoenix, AZ (Cynthia Thomson, PhD, RD, James Warneke, MD, Cheryl Ritenbaugh, PhD, MPH); University of California, Davis, Davis, CA (Ellen B. Gold, PhD, Sidney Scudder, MD); University of California, San Diego, Moores UCSD Cancer Center, San Diego, CA (Kathryn A. Hollenbach, PhD, Vicky Jones, MD); University of Texas M.D. Anderson Cancer Center, Houston, TX (Lovell A. Jones, PhD, Richard Hajek, PhD, Richard Theriault, DO)
CONFLICT OF INTEREST/DISCLOSURE
D. Michele Nikoloff, Grantland Hillman, Marcel R. Fontecha, and H. Jeffrey Lawrence are employees of Roche Molecular Systems.
References
- 1.Fisher B, Dignam J, Bryant J, et al. Five versus more than five years of tamoxifen therapy for breast cancer patients with negative lymph nodes and estrogen receptor-positive tumors. J Natl Cancer Inst. 1996 Nov 6;88(21):1529–42. doi: 10.1093/jnci/88.21.1529. [DOI] [PubMed] [Google Scholar]
- 2.Jordan VC. Metabolites of tamoxifen in animals and man: identification, pharmacology, and significance. Breast Cancer Res Treat. 1982;2(2):123–38. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 3.Lien EA, Solheim E, Kvinnsland S, Ueland PM. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988 Apr 15;48(8):2304–8. [PubMed] [Google Scholar]
- 4.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004 May;85(2):151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 5.Lim YC, Li L, Desta Z, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006 Aug;318(2):503–12. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 6.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006 Jul;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005 Jan 5;97(1):30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 8.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003 Dec;95(23):1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 9.Ferraldeschi R, Newman WG. The Impact of CYP2D6 Genotyping on Tamoxifen Treatment. Pharmaceuticals. 2010;3(4):1122–38. doi: 10.3390/ph3041122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lash TL, Rosenberg CL. Evidence and practice regarding the role for CYP2D6 inhibition in decisions about tamoxifen therapy. J Clin Oncol. 2010 Mar 10;28(8):1273–5. doi: 10.1200/JCO.2009.26.7906. [DOI] [PubMed] [Google Scholar]
- 11.Borges S, Desta Z, Jin Y, et al. Composite Functional Genetic and Comedication CYP2D6 Activity Score in Predicting Tamoxifen Drug Exposure Among Breast Cancer Patients. J Clin Pharmacol. 2010 Apr;50(4):450–8. doi: 10.1177/0091270009359182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Sloover Koch Y, Ernst ME. Selective serotonin-reuptake inhibitors for the treatment of hot flashes. Ann Pharmacother. 2004 Jul-Aug;38(7–8):1293–6. doi: 10.1345/aph.1D512. [DOI] [PubMed] [Google Scholar]
- 13.Loprinzi CL, Kugler JW, Sloan JA, et al. Venlafaxine in management of hot flashes in survivors of breast cancer: a randomised controlled trial. Lancet. 2000 Dec 16;356(9247):2059–63. doi: 10.1016/S0140-6736(00)03403-6. [DOI] [PubMed] [Google Scholar]
- 14.Pandya KJ, Raubertas RF, Flynn PJ, et al. Oral clonidine in postmenopausal patients with breast cancer experiencing tamoxifen-induced hot flashes: a University of Rochester Cancer Center Community Clinical Oncology Program study. Ann Intern Med. 2000 May 16;132(10):788–93. doi: 10.7326/0003-4819-132-10-200005160-00004. [DOI] [PubMed] [Google Scholar]
- 15.Flockhart D. CYP2D6 genotyping and the pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol. 2008 Jul;6(7):493–4. [PubMed] [Google Scholar]
- 16.Jeppesen U, Gram LF, Vistisen K, Loft S, Poulsen HE, Brosen K. Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51(1):73–8. doi: 10.1007/s002280050163. [DOI] [PubMed] [Google Scholar]
- 17.Mortimer JE, Flatt SW, Parker BA, et al. Tamoxifen, hot flashes and recurrence in breast cancer. Breast Cancer Res Treat. 2008;108:421–6. doi: 10.1007/s10549-007-9612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce JP, Faerber S, Wright FA, et al. A randomized trial of the effect of a plant-based dietary pattern on additional breast cancer events and survival: the Women’s Healthy Eating and Living (WHEL) Study. Control Clin Trials. 2002 Dec;23(6):728–56. doi: 10.1016/s0197-2456(02)00241-6. [DOI] [PubMed] [Google Scholar]
- 19.Pierce JP, Natarajan L, Caan BJ, et al. Influence of a diet very high in vegetables, fruit, and fiber and low in fat on prognosis following treatment for breast cancer: the Women’s Healthy Eating and Living (WHEL) randomized trial. Jama. 2007 Jul;18;298(3):289–98. doi: 10.1001/jama.298.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tableman M, Kim JS. Survival Analysis Using S: Analysis of Time-to-Event Data. Boca Raton: Chapman & Hall/CRC; 2004. [Google Scholar]
- 21.Schumacher M, Hollnder N, Sauerbrei W. Resampling and cross-validation techniques: a tool to reduce bias caused by model building? Stat Med. 1997;16:2813–27. doi: 10.1002/(sici)1097-0258(19971230)16:24<2813::aid-sim701>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 22.Wu AH, Pike MC, Williams LD, et al. Tamoxifen, soy, and lifestyle factors in Asian American women with breast cancer. J Clin Oncol. 2007 Jul 20;25(21):3024–30. doi: 10.1200/JCO.2006.10.5023. [DOI] [PubMed] [Google Scholar]
- 23.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009 Aug;10(8):825–33. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Decensi A, Robertson C, Viale G, et al. A randomized trial of low-dose tamoxifen on breast cancer proliferation and blood estrogenic biomarkers. J Natl Cancer Inst. 2003 Jun 4;95(11):779–90. doi: 10.1093/jnci/95.11.779. [DOI] [PubMed] [Google Scholar]
- 25.Guerrieri-Gonzaga A, Baglietto L, Johansson H, et al. Correlation between tamoxifen elimination and biomarker recovery in a primary prevention trial. Cancer Epidemiol Biomarkers Prev. 2001 Sep;10(9):967–70. [PubMed] [Google Scholar]
- 26.Anderson WF, Chen BE, Jatoi I, Rosenberg PS. Effects of estrogen receptor expression and histopathology on annual hazard rates of death from breast cancer. Breast Cancer Res Treat. 2006 Nov;100(1):121–6. doi: 10.1007/s10549-006-9231-y. [DOI] [PubMed] [Google Scholar]
- 27.Dignam JJ, Dukic V, Anderson SJ, Mamounas EP, Wickerham DL, Wolmark N. Hazard of recurrence and adjuvant treatment effects over time in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008 Oct 2; doi: 10.1007/s10549-008-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess KR, Pusztai L, Buzdar AU, Hortobagyi GN. Estrogen receptors and distinct patterns of breast cancer relapse. Breast Cancer Res Treat. 2003 Mar;78(1):105–18. doi: 10.1023/a:1022166517963. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan L, Pu M, Parker BA, et al. Time-varying effects of prognostic factors associated with disease-free survival in breast cancer. Am J Epidemiol. 2009 Jun 15;169(12):1463–70. doi: 10.1093/aje/kwp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleming ID, editor. AJCC. American Joint Committee on Cancer: Manual for Staging of Cancer. 6. New York, NY: Springer-Verlag; 2002. [Google Scholar]
- 31.Thomson G, Torres HM, Lancaster AK, et al. Testing fit of genotype frequencies to Hardy-Weinberg proportions. 2009. Hardy-Weinberg Proportions Methods Manual Version 0.1.2. [Google Scholar]
- 32.Therneau TM, Grambsch PM. Statistics for Biology and Health. New York: Springer-Verlag; 2000. Modeling Survival Data. Extending the Cox Model. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


