Abstract
Objective
Recent genome-wide association studies (GWAS) identified a variant rs7575840 in the apolipoprotein B (APOB) gene region to be associated with LDL-C. However, the underlying functional mechanism of this variant that resides 6.5 kb upstream of APOB has remained unknown. Our objective was to investigate rs7575840 for association with refined apoB containing lipid particles; for replication in a non-Caucasian Mexican population; and for underlying functional mechanism.
Methods and Results
Our data show that rs7575840 is associated with serum apoB levels (P=4.85×10−10) and apoB containing lipid particles, very small VLDL, IDL and LDL particles (P=2×10−5 - 9×10−7) in the Finnish METSIM study sample (n=7,710). Fine mapping of the APOB region using 43 SNPs replicated the association of rs7575840 with apoB in a Mexican study sample (n=2,666, P=3.33×10−05). Furthermore, our transcript analyses of adipose RNA samples from 175 Finnish METSIM subjects indicate that rs7575840 alters expression of APOB (P=1.13×10−10) and a regional non-coding RNA (BU630349) (P=7.86×10−6) in adipose tissue.
Conclusions
It has been difficult to convert GWAS associations into mechanistic insights. Our data show that rs7575840 is associated with serum apoB levels and apoB containing lipid particles as well as influences expression of APOB and a regional transcript BU630349 in adipose tissue. We thus provide evidence how a common genome-wide significant SNP rs7575840 may affect serum apoB, LDL-C, and TC levels.
Keywords: Apolipoprotein B, association analysis, gene expression, adipose tissue, Mexicans
INTRODUCTION
Genome-wide association studies (GWAS) have been successful in discovering single nucleotide polymorphisms (SNPs) that affect LDL-C levels with low to moderate effect sizes1. However, many of the significantly associated SNPs are intergenic and belong to large linkage disequilibrium (LD) blocks, making it difficult to evaluate their functional relevance.
The SNP rs7575840 has been implicated in a recent GWAS for LDL-C1. As rs7575840 resides in the APOB gene region, 6.5 kb upstream of the gene, we tested it for association with refined lipid phenotypes and proton NMR spectroscopy measurements of lipid particles in a Caucasian population sample, the METabolic Syndrome in Men (METSIM) cohort (n=7,710). Furthermore, to also explore the role of this GWAS variant in a non-Caucasian population, we fine mapped the APOB region in a Mexican dyslipidemic case/control study sample and compared the regional LD between Caucasians and Mexicans. The Mexican population has a 44% prevalence of hypercholesterolemia defined by total cholesterol (TC) > 200mg/dL2. Other forms of dyslipidemias are highly common in this population as well. Yet to the best of our knowledge, no GWAS has been performed to date exploring lipid levels in Mexicans. Nor have the Caucasian GWAS signals for LDL-C been explored thoroughly in the Mexican population as of yet. Thus, Mexicans represent a population with a high susceptibility to dyslipidemia underinvestigated for the underlying genetic factors2.
ApoB is the sole protein component of the LDL-C particle and an essential core of triglyceride-rich lipoproteins. A mutation in APOB is known to cause a monogenic autosomal dominant disease called familial hypercholesterolemia3, further establishing the role of APOB in LDL metabolism. We hypothesized that rs7575840 and/or SNPs in LD with it may function as a regulatory cis-eQTL, and tested 175 adipose RNA samples from the Finnish METSIM study for allele-specific expression of transcripts within the APOB gene region.
METHODS
The study design was approved by the ethics committees of the participating centers and all subjects gave a written informed consent.
Study samples
METSIM cohort
The Finnish population-based cohort, METSIM (METabolic Syndrome In Men), was collected at the University of Kuopio, Kuopio, Finland as described previously4. The METSIM cohort consisted of 7,710 male subjects, age 50-70 years, randomly selected from the population of Kuopio in Eastern Finland. Each patient participated in an interview session to measure factors that may affect cardiovascular disease risk, including prescription drug use, weekly exercise, metabolites, cardiovascular family history, and other health questions. Phenotypic determinations were performed as described previously4. Plasma lipoproteins from the METSIM subjects were fractionated using proton NMR into HDL, LDL, intermediate lipoproteins (IDL), very low density lipoproteins, and chylomicron subclasses, as described previously5,6.
Mexican hypertriglyceridemia cases and controls
A total of 2,666 Mexican hypertriglyceridemic cases and controls were recruited at the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, as described previously7. Briefly, the inclusion criteria were fasting serum triglycerides > 2.3 mmol/L (200 mg/dL) for the cases and < 1.7 mmol/L (150 mg/dL) for the controls. Exclusion criteria were type 2 diabetes mellitus or morbid obesity (body mass index (BMI) > 40 kg/m2), and the use of lipid lowering drugs for the controls. Fasting lipid levels were measured using commercially available standardized methods as described previously7. Serum LDL-C levels were calculated using the Friedewald formula for subjects with TG < 400 mg/dL.
Genotyping and Real-time PCR
The SNP rs7575840 was genotyped in the METSIM study sample using the Sequenom genotyping platform. The 43 SNPs in the APOB region (+/−100 kb) were genotyped in the Mexican study sample using both Pyrosequencing and Illumina BeadArray technology platforms. The SNPs were in Hardy-Weinberg Equilibrium (p-value>0.05) in both study samples and had a genotyping call rate >90%. The real-time PCR of 175 METSIM RNAs for BU630349 was performed using Quantitect Reverse Transcription kit (Qiagen), as described in detail in the Supplementary methods.
Tagging of the APOB region in the Mexican study sample
We genotyped 43 SNPs for the APOB region (+/−100 kb) in 2,310 subjects of the Mexican dyslipidemic study sample. Using Haploview v4.28 and CEU HapMap data, we calculated that these 43 SNPs capture 90% of the variation with minor allele frequency (MAF) ≥5% and r2≥0.80. We utilized the CEU HapMap data for these calculations as the coverage of the Mexican American HapMap data is still incomplete.
Microarray data and gene expression analysis in METSIM fat biopsy samples
A total of 175 METSIM subjects underwent subcutaneous fat biopsies for adipose RNA isolation. The RNA isolation was performed according to the manufacturer’s instructions (Qiagen). The adipose RNA samples were hybridized to the Illumina HT-12 v3.0 expression chips. Genome Studio was used to perform quantile normalization and background subtraction. An absent call was made with the detection p-values>0.01. Probes with >50% absent calls were excluded from the analysis. All METSIM subjects were unrelated males. Microarray data for the transcripts in the APOB gene region (+/−500 kb) will be submitted to the NCBI’s Gene Expression Omnibus repository in MIAME compliant format (GSE27666).
To test whether the APOB or BU630349 expression in 175 METSIM fat biopsies are influenced by the rs7575840 genotypes, we log transformed the APOB and BU630349 expression values. Expression values more than 4 standard deviations from the mean were removed form the analysis. The two-sided Student’s T-test was performed to compare the means between carriers of the T rare allele and the homozygous common G group (i.e. dominant model). The ECR Vista browser was used to look at the conservation of BU630349 across 13 species.
Association analysis in METSIM
To test for association between the SNPs and lipid traits, and lipoprotein subclasses, we performed multivariate linear regression analysis for the additive genetic model using SPSS software. Trait values were adjusted for age and log transformed BMI. Subjects with BMI>40, lipid lowering medication, diabetes, or a trait value greater than 4 standard deviations from the mean were excluded from the analysis, leaving 5,054 METSIM individuals in the association analyses. Pearson correlation analyses were performed between the raw lipid particle measurements and gene expression (APOB and BU630349). The gene expression values were log transformed, and values greater than 4 standard deviations from the mean were removed.
Association analysis in Mexican hypertriglyceridemic cases and controls
To unify the association analysis with the METSIM analysis, we performed the multivariate linear regression analysis using SPSS software by adjusting the trait values for age, sex, log transformed BMI, and hypertriglyceridemia case-control status. Subjects with BMI>40, lipid lowering medication, diabetes, or a trait value greater than 4 standard deviations from the mean were excluded from the statistical analysis. Bonferroni correction (43 tested SNPs; P<0.001) was employed to evaluate significance. We did not correct for three tested traits, as apoB, LDL-C and TC are known to be highly correlated.
Individual ancestry (IA) analyses
We had genotype data for 82 evenly distributed ancestry informative markers (AIMs) for 2,310 of the Mexican hypertriglyceridemia case-control samples7. These AIMs were selected based on a published list of European/Amerindian AIMs9 and were used to calculate individual ancestry (IA) estimates using the STRUCTURE 2.2 software10, as described in the Supplementary methods.
RESULTS
The SNP rs7575840 resides in the APOB region, 6.5 kb upstream of APOB. To further investigate the association signal of rs7575840, we first tested the SNP for association with refined lipid levels and proton NMR spectroscopy measurements of lipid particles in the Finnish population sample METSIM. Table 1 shows that the rare allele T (MAF=28%) of rs7575840 is significantly associated with elevated apoB, LDL-C and TC levels in METSIM (n=7,710), apoB providing the most significant evidence of association (P=4.85×10−10). The proportion of 1 SD change in standardized apoB residual values for each copy of the risk allele was 0.14, and the contribution of this genetic effect can explain 0.8% of the total variance of apoB levels in METSIM (Table 1). The most associated lipid particles were the very small VLDL, IDL as well as all of the LDL subclasses (P=2×10−5-9×10−7) (Supplementary Table 1). Among the tested lipoprotein subclasses, rs7575840 explains the most variance for these same subclasses (Supplementary Figure 1). Furthermore, the strongest signals classified by positive beta were also observed for the VLDL, IDL and LDL subclasses (Beta=8×10−3-3×10−2) (Supplementary Table 1).
Table 1.
Association results of rs7575840 in the METSIM cohort
| SNP | Trait | p-value | n | Standardized Effect size |
Variance explained |
Effect Allele |
Effect Allele Freq‡ |
|
|---|---|---|---|---|---|---|---|---|
| B* | SE† | |||||||
| rs7575840 | ApoB | 4.85×10−10 | 5026 | 0.138 | 0.022 | 0.77% | T | 0.28 |
| LDL-C | 3.88×10−09 | 5054 | 0.131 | 0.022 | 0.68% | |||
| TC | 8.05×10−09 | 5052 | 0.128 | 0.022 | 0.66% | |||
Linear regression was performed on the METSIM cohort using an additive model. The effect allele is the allele associated with an increased trait value. Variance explained represents the percent of the overall variation in the standardized residuals that can be attributed to the additive model.
B (beta) represents the proportion of 1 SD change in standardized trait values for each copy of the rare allele
standard error of beta
the frequency of the effect allele.
Different populations may have different signals due to the underlying differences in LD. To investigate whether the effect of rs7575840 on serum apoB levels also extends to a non-Caucasian population, we first fine mapped the APOB gene region (±100 kb) by genotyping 43 SNPs in the Mexican dyslipidemic study sample (n=2,310). By utilizing the Caucasian HapMap CEU samples and the Mexican apoB controls (apoB levels <50th age/sex specific Mexican population percentile), we first observed that the LD pattern in Caucasians closely resembles the LD in Mexicans (Supplementary figure 2). When testing the 43 SNPs for association with apoB, we observed 4 non-redundant independent signals (r2<0.6) passing the Bonferroni correction (P<0.001) (Supplementary Table 2, Supplementary figure 2). Among the 43 tested SNPs, rs693, rs1367117, and rs7575840, that have been implicated in previous GWAS for LDL-C12,13, provided strong evidence of association with apoB, though rs1367117 and rs7575840 represent redundant signals, as described below. The p-values for these three SNPs were ranked in the top 5 for all of the three tested traits, apoB, LDL-C, and TC (Supplementary Tables 2-4). To further validate these associations, we extended the genotyping of rs693, rs1367117, and rs7575840 to 356 additional Mexican dyslipidemic samples available for study (total n=2,666, Table 2), which further strengthened the association signals for apoB (p-values of 2.90×10−06, 6.83×10−06, and 3.33×10−05; explaining 0.74%, 0.90%, and 0.76% of the apoB levels, respectively). The LDL-C and TC traits provided somewhat less significant p-values (Table 2). Interestingly, rs1367117, is a nonsynonymous SNP (T71I) in APOB. However, according to VISTA browser the amino acid T71 is only conserved in mammals, and furthermore, the amino acid change T71I was previously predicted to be benign using the PolyPhen, SIFT, and PANTHER algorithms11. The SNP rs693 is a synonymous variant. It is worth noting that the association signals were all for the same risk allele as in Caucasians. Hence, our results suggest that these Caucasian GWAS signals extend to the Mexican population.
Table 2.
Association results of rs1367117, rs7575840, and rs693 in the Mexican study sample
| SNP | Trait | p-value | n | Standardized Effect Size |
Variance explained |
Effect Allele |
Effect Allele Freq |
|
|---|---|---|---|---|---|---|---|---|
| B | SE | |||||||
| rs1367117 | ApoB | 6.83×10−06 | 2250 | 0.146 | 0.032 | 0.90% | A | 0.27 |
| LDL-C* | 6.06×10−03 | 1551 | 0.107 | 0.039 | 0.49% | |||
| TC | 2.07×10−05 | 2384 | 0.135 | 0.032 | 0.76% | |||
| rs7575840 | ApoB | 3.33×10−05 | 2272 | 0.134 | 0.032 | 0.76% | T | 0.28 |
| LDL-C | 2.2×10−02 | 1563 | 0.089 | 0.039 | 0.34% | |||
| TC | 9.65×10−05 | 2406 | 0.123 | 0.031 | 0.63% | |||
| rs693 | ApoB | 2.90×10−06 | 2312 | 0.140 | 0.030 | 0.94% | A | 0.37 |
| LDL-C | 3.86×10−03 | 1598 | 0.103 | 0.036 | 0.52% | |||
| TC | 6.41×10−05 | 2446 | 0.116 | 0.029 | 0.65% | |||
Linear regression was performed in the Mexican study sample using an additive model, as described in the footnote to Table 1.
Serum LDL-C levels were calculated using the Friedewald formula only for subjects with TGs<400 mg/dL.
We next investigated the pairwise LD of rs7575840 with all other SNPs in the APOB gene region (±100 kb) using the Caucasian HapMap CEU sample and Mexican apoB controls (Supplementary figure 3). We observed that the regional differences in pairwise LD between the two populations are not large (Supplementary figure 3). The SNP rs693 was not in strong LD with rs7575840 or rs1367117 (r2 <0.55 both in HapMap CEU and Mexican apoB controls). The pairwise LD (using r2) between rs7575840 and rs1367117 in the Mexican apoB controls was 0.80 and in the CEU HapMap sample 0.91, indicating that in both populations rs7575840 and rs1367117 are proxies, likely reflecting the same functional signal.
Mexicans are an admixed population that descends from a recent mix of Amerindian and European ancestry with a small proportion of African ancestry14,15. Population admixture may confound allelic association if both the trait distribution and the allele frequency differ between ancestries. We previously demonstrated that the apoB distribution did not differ with individual ancestry (IA) estimates in 2,310 subjects of the Mexican hypertriglyceridemia cases and controls5. However, to further eliminate the possibility of spurious associations due to admixture, we also performed the association analyses of rs7575840, rs1367117, and rs693 while including IA estimates as a covariate in the regression model. In these adjusted analyses, we obtained P-values of 3.76×10−05, 6.55×10−06 and 1.30×10−06 for residual apoB levels, respectively, suggesting that the associations are not confounded by population admixture.
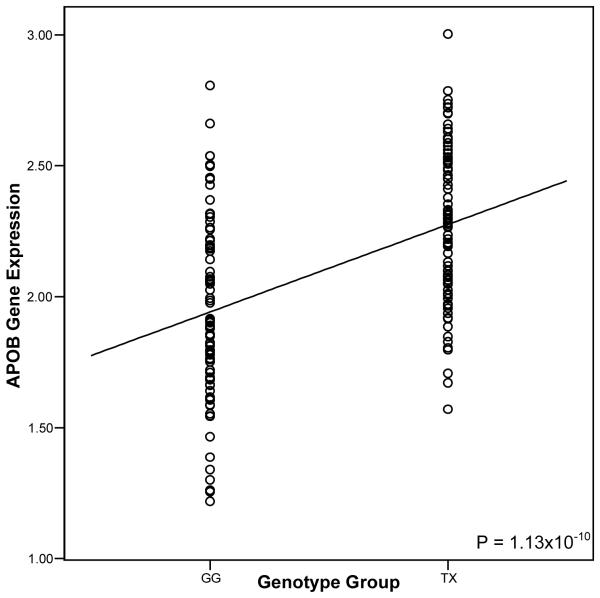
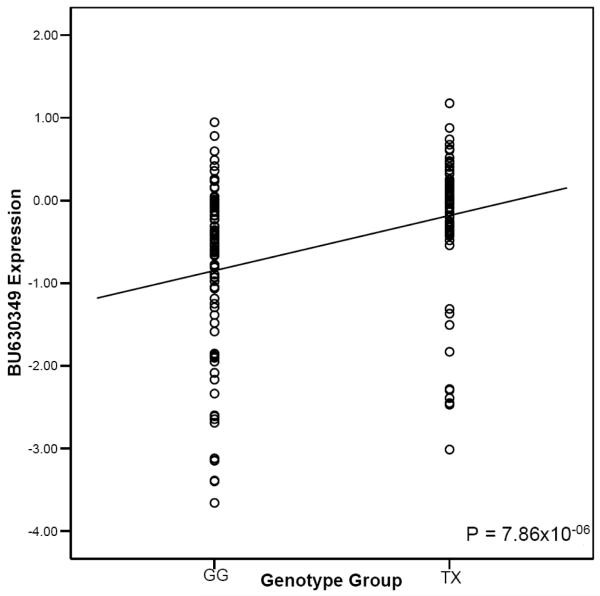
As rs7575840 resides 6.5 kb upstream of APOB, it may modify regulatory elements. To investigate whether rs7575840 affects the expression of APOB and/or transcripts in the APOB gene region, we searched for gene expression probes within 500 kb of APOB. The APOB probe ILMN_1664024, which corresponds with the APOB refseq ID NM_000384, was expressed in the 175 METSIM samples. No other expressed probes were observed within 500 kb of APOB on the microarray. We also tested a local transcript BU630349 for differential expression between the rs7575840 genotypes by RT-PCR, because it was previously found to be significantly differentially expressed based on the rs7575840 genotypes (i.e. cis-eQTL) in liver (P=1.62×10−54), passing a multiple testing correction of FDR <10%16. No effect was observed between the rs7575840 genotypes and APOB expression in liver16. BU630349 is a 662-bp long predicted non-coding RNA located 872 bp upstream of APOB and conserved in primates. To ensure we did not obtain spurious associations due to a small number of homozygous individuals, we utilized a dominant model to test for cis-eQTLs. The METSIM subjects carrying at least one minor allele of rs7575840 had significantly more APOB expression (P=1.13×10−10) and BU630349 expression (P=7.86×10−06) in adipose tissue relative to the homozygous common group (figures 1-2). The fold change between the two groups was 1.17X for APOB and 1.78X for BU630349, rs7575840 explaining 21.5% and 11.2% of the APOB and BU630349 variance, respectively (figures 1-2). The associations were also significant using an additive model (P=5.37×10−09 for APOB, P=2.18×10−06 for BU630349). The detected APOB differential expression is in line with the observation that individuals with the rare allele of rs7575840 have higher serum apoB levels (Table 1).
Figure 1.
APOB expression in 175 METSIM fat biopsies based on the rs7575840 genotypes. The TX genotype group (n=85) contains carriers of the minor allele (T), and the GG genotype group (n=90) are homozygous for the common (G) allele. We added a regression line to show the trend of the difference in means.
Figure 2.
BU630349 expression in 175 METSIM fat biopsies using the same rs7575840 genotype groups as described in legend to figure 1. We added a regression line to show the trend of the difference in means.
To determine whether expression of APOB or BU630349 may be related to lipoprotein subclasses, we performed Pearson correlations in the 175 METSIM fat biopsy samples. Both expression of APOB and BU630349 were positively correlated with HDL-C concentration/particles and negatively correlated with triglycerides and VLDL concentration/particles (Supplementary figure 4). We also observed that raw serum apoB levels were negatively correlated with APOB expression in adipose tissue (p=0.005, r =−0.21).
To search for a possible mechanism of how rs7575840 influences APOB and BU630349 expression, we performed in silico analysis using the PROMO program to search for transcription factor binding sites (TFBS) that differ based on rs7575840 genotype. We found that the major allele uniquely codes for a TFBS for CEBPA and CEBPB transcription factors, and the minor allele codes for a YY1 TFBS. CEBPA expression was positively correlated with APOB (r=0.50, P=1.57×10−12) and BU630349 expression (r=0.21, P=6.91×10−03) in the 175 METSIM adipose RNA samples. This is in line with the fact that APOB and BU630349 expression were also correlated in adipose (r=0.49, p=1.46×10−11). After using the rs7575840 genotype as a covariate, the correlations between CEBPA and APOB were stronger and about 1000 times more significant (r=0.56 and P=8.38×10−16 for additive model; r=0.58 and P=6.92×10−17 for dominant model). The correlation between CEBPA and BU630349 were slightly stronger when correcting for rs7575840 genotype (r=0.24 and P=1.97×10−03 for additive genotype; r=0.23 and P=2.42×10−03 for dominant genotype). SNP rs7575840 was not significantly associated with CEBPB or YY1 expression. CEPBA and CEBPB were positively correlated with each other (r=0.37, p=3.30×10−07). Taken together, our in silico and gene expression data suggest that rs7575840 may affect APOB and BU630349 expression in adipose by influencing a CEBPA binding site.
DISCUSSION
Our data in two different populations, Finns and Mexicans, implicate the rs7575840 variant near the APOB gene for apoB, LDL-C and TC levels, the most affected traits in our analyses being apoB and apoB containing lipid particles. Furthermore, our novel data also revealed a possible functional mechanism how rs7575840 influences apoB, LDL-C and TC levels by altering the expression level of the APOB gene itself and a regional non-coding RNA (BU630349) in adipose tissue. The latter has also been shown to be differentially expressed by rs7575840 in 427 liver samples16. A nonsynonymous SNP, rs1367117, that has also been identified in a recent GWAS meta-analysis for LDL-C1 is in high LD with rs7575840 both in Caucasians and Mexicans. Our functional findings suggest a mechanism how these redundant, genome-wide significant GWAS signals, rs7575840 and rs1367117, may alter apoB levels.
Variants in APOB have been investigated for associations with lipids in multiple previous candidate gene studies mostly based on Caucasian populations11,17,18. In a recent Caucasian mega meta-analysis GWAS of mainly population-based samples, the amino acid change rs1367117 in APOB resulted in a p-value of 4.48×10−114 for LDL-C in 95,402 subjects, whereas rs7575840 (LD=0.9 by r2) resulted in a p-value of 1.67×10−98 for LDL-C in 41,912 subjects1. It is important to confirm whether the known loci have a consistent effect across ethnic groups to determine which variants are to be used in cardiovascular risk assessment. Furthermore, despite the high prevalence of high LDL-C (46%) in Mexicans2, little is known about the genetic factors predisposing to high cholesterol levels in this non-Caucasian population. Our trans-ethnic fine mapping shows the first replication of these two GWAS variants and another previous GWAS variant, rs69312, in the Mexican population with a high susceptibility to dyslipidemia.
The recent Caucasian mega GWAS study1 focused on TC and LDL-C, whereas apoB levels were not available for the study. Although we also obtained significant association evidence for TC and LDL-C, we observed the strongest association signal with apoB. ApoB is the sole protein component of LDL particles and present in most atherogenic lipoprotein particles. Previous prospective epidemiological studies have clearly shown that apoB levels predict the CAD risk better than LDL-C and TC levels19. Our study demonstrates that rs7575840 is associated with VLDL, IDL, and LDL subclasses. These data further implicate the role of rs7575840 and/or SNPs in LD with it in apoB metabolism.
Discovering the underlying mechanisms behind GWAS signals has been very difficult, largely due to large linkage disequilibrium blocks, spanning many genes, and the absence of functional evidence provided by the GWAS. To search for a possible function of rs7575840, we utilized one of the largest subcutaneous fat biopsy gene expression cohorts published to date (n=175). Because adipose tissue is an important regulator of lipid metabolism in humans, we decided to determine whether rs7575840 genotype alters gene expression in this tissue. We discovered that rs7575840 is associated with APOB and BU630349 expression in adipose tissue, providing possible regulatory mechanisms how this SNP may alter apoB, LDL-C and TC levels. Importantly, the same cis-eQTL between rs7575840 and BU630349 has previously been observed in liver, although the APOB expression did not differ between the rs7575840 genotypes16. Differences in the used microarray platforms may have contributed to these discrepancies. However, even though the hepatic APOB expression did not differ by the rs7575840 genotypes, the regional non-coding RNA BU630349 may still serve as a post-transcriptional regulator of APOB, because besides expression long non-coding RNAs may affect splicing, transport, translation, and epigenetic regulation of genes20. Interestingly, we also found that the binding site of a transcription factor, CEBPA, is predicted to differ based on the rs7575840 genotypes, and its expression was positively correlated with APOB and BU630349 expression in the 175 METSIM adipose RNA samples. CEBPA is highly expressed both in adipose tissue and liver; known to modulate leptin and adiponectin expression; and induces expression of genes involved in differentiation of granulocytes, monocytes, adipocytes and hepatocytes21,22. Future studies are warranted to determine the detailed molecular mechanisms how rs7575840 or SNPs in LD with it ultimately impact apoB levels. Nevertheless, taken together our data provide the first piece of evidence how rs7575840 influences apoB and apoB containing lipid particles.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Finnish and Mexican individuals who participated in this study. We also thank Cindy Montes, Maribel Rodríguez-Torres, and Salvador Ramírez for laboratory technical assistance.
Sources of funding
This research was supported by the grants HL095056 and HL-28481 from the National Institutes of Health. B.E.H. is supported by NHGRI grant T32 HG02536. This work has also been supported by the Academy of Finland’s Responding to Public Health Challenges Research Programme (grant 129429 to MAK), and the Finnish Cardiovascular Research Foundation (MAK), and the Jenny and Antti Wihuri Foundation (AJK).
Footnotes
Disclosures
None.
REFERENCES
- 1.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Cecile JW, Janssens A, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar-Salinas CA, Gómez-Pérez FJ, Rull J, Villalpando S, Barquera S, Rojas R. Prevalence of dyslipidemias in the Mexican National Health and Nutrition Survey 2006. Salud Publica Mex. 2010;52:S44–53. doi: 10.1590/s0036-36342010000700008. [DOI] [PubMed] [Google Scholar]
- 3.Innerarity TL, Weisgraber KH, Arnold KS, Mahley RW, Krauss RM, Vega GL, Grundy SM. Familial defective apolipoprotein B-100: low density lipoproteins with abnormal receptor binding. Proc Natl Acad Sci U S A. 1987;84:6919–6923. doi: 10.1073/pnas.84.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JC, Weissglas-Volkov D, Kyttälä M, Dastani Z, Cantor RM, Sobel EM, Plaisier CL, Engert JC, van Greevenbroek MM, Kane JP, Malloy MJ, Pullinger CR, Huertas-Vazquez A, Aguilar-Salinas CA, Tusie-Luna T, de Bruin TW, Aouizerat BE, van der Kallen CC, Croce CM, Aqeilan RI, Marcil M, Viikari JS, Lehtimäki T, Raitakari OT, Kuusisto J, Laakso M, Taskinen MR, Genest J, Pajukanta P. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am J Hum Genet. 2008;83:180–192. doi: 10.1016/j.ajhg.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soininen P, Kangas AJ, Würtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Järvelin MR, Kähönen M, Lehtimäki T, Viikari J, Raitakari OT, Savolainen MJ, Ala-Korpela M. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 6.Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, Kumpula LS, Hämäläinen E, Jousilahti P, Kangas AJ, Männistö S, Savolainen MJ, Palotie A, Salomaa V, Perola M, Ala-Korpela M, Peltonen L. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol. 2010;6:441. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissglas-Volkov D, Plaisier CL, Huertas-Vazquez A, Cruz-Bautista I, Riaño-Barros D, Herrera-Hernandez M, Riba L, Cantor RM, Sinsheimer JS, Aguilar-Salinas CA, Tusie-Luna T, Pajukanta P. Identification of Two Common Variants Contributing to Serum Apolipoprotein B Levels in Mexicans. Arterioscler Thromb Vasc Biol. 2009;30:353–359. doi: 10.1161/ATVBAHA.109.196402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 9.Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, Silva G, Belmont JW, Hanson RL, Knowler WC, Gregersen PK, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benn M, Nordestgaard BG, Jensen JS, Grande P, Sillesen H, Tybjaerg-Hansen A. Polymorphism in APOB associated with increased low-density lipoprotein levels in both genders in the general population. J Clin Endocrinol Metab. 2005;90:5797–5803. doi: 10.1210/jc.2005-0974. [DOI] [PubMed] [Google Scholar]
- 12.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salari K, Burchard E. Latino populations: a unique opportunity for epidemiological research of asthma. Paediatr Perinat Epidemiol. 2007;21(Suppl 3):15–22. doi: 10.1111/j.1365-3016.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 15.Price A, Patterson N, Yu F, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 18.Ronald J, Rajagopalan R, Ranchalis JE, Marshall JK, Hatsukami TS, Heagerty PJ, Jarvik GP. Analysis of recently identified dyslipidemia alleles reveals two loci that contribute to risk for carotid artery disease. Lipids Health Dis. 2009;8:52. doi: 10.1186/1476-511X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barter PJ, Ballantyne CM, Carmena R, Castro Cabezas M, Chapman MJ, Couture P, de Graaf J, Durrington PN, Faergeman O, Frohlich J, Furberg CD, Gagne C, Haffner SM, Humphries SE, Jungner I, Krauss RM, Kwiterovich P, Marcovina S, Packard CJ, Pearson TA, Reddy KS, Rosenson R, Sarrafzadegan N, Sniderman AD, Stalenhoef AF, Stein E, Talmud PJ, Tonkin AM, Walldius G, Williams KM. Apo B versus cholesterol in estimating cardiovascular risk and in guiding therapy: report of the thirty-person/ten-country panel. J Intern Med. 2006;259:247–258. doi: 10.1111/j.1365-2796.2006.01616.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen L-C, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SG, De Vos P, Guerre-Millo M, Wong K, Hermann T, Staels B, Briggs MR, Auwerx J. The adipocyte specific transcription factor C/EBPalpha modulates human ob gene expression. Proc Natl Acad Sci U S A. 1996;93:5507–5511. doi: 10.1073/pnas.93.11.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segawa K, Matsuda M, Fukuhara A, Morita K, Okuno Y, Komuro R, Shimomura I. Identification of a novel distal enhancer in human adiponectin gene. J Endocrinol. 2009;200:107–116. doi: 10.1677/JOE-08-0376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.