Abstract
Inhibitor of κB (IκB) kinase (IKK) phosphorylates IκB proteins leading to their degradation and liberation of nuclear factor κB (NF-κB) for gene transcription. Here we report the crystal structure of IKKβ in complex with an inhibitor at 3.6 Å resolution. The structure reveals a tri-modular architecture with the kinase domain (KD), a ubiquitin-like domain (ULD) and an elongated, α-helical scaffold/dimerization domain (SDD). Surprisingly, the predicted leucine zipper and helix-loop-helix motifs do not form these structures but are part of SDD. The ULD and SDD mediate a critical interaction with IκBα that restricts substrate specificity, and the ULD is also required for catalytic activity. The SDD mediates IKKβ dimerization, but dimerization per se is not important for maintaining IKKβ activity, and instead is required for IKKβ activation. Other IKK family members IKKα, TBK1 and IKKi may share the similar tri-modular architecture and function.
NF-κB transcription factors are master regulators of inflammatory, immune and apoptotic responses 1,2. In the canonical pathway, NF-κB dimers are held in the cytoplasm via binding to IκB proteins, which mask their nuclear localization signals. When cells are stimulated by NF-κB inducers, IκBs are phosphorylated by the Ser/Thr-specific IKK, a modification that triggers their Lys48-linked polyubiquitination and subsequent proteasomal degradation 3. Freed NF-κB dimers enter the nucleus to regulate gene transcription 2. In the non-canonical pathway, activated IKK phosphorylates the IκB-like domain in the NF-κB family member p100/NF-κB2 3. NF-κB signaling pathways are associated with a vast number of human diseases including inflammation and cancer, which renders IKK a potentially important therapeutic target 4 (www-nf-kb.org).
IKK was originally purified from HeLa cells as a multi-protein complex that contains the kinase subunits IKKα and/or IKKβ, and the regulatory protein NEMO (also known as IKKγ or FIP-3) 5–11. IKKα and IKKβ both contain an N-terminal kinase domain (KD), predicted leucine zipper (LZ) and helix-loop-helix (HLH) domains, and a C-terminal NEMO binding domain (NBD) (Fig. 1a). IKKβ appears to have an additional ubiquitin-like domain (ULD) following the KD, which is not predicted in the corresponding region of IKKα 3. IKK-related kinases TBK1/NAK and IKKi/IKKε appear to share a similar domain organization 12. While IKKβ mediates activation of the canonical NF-κB pathway in response to pro-inflammatory stimuli, IKKα plays an indispensible role in non-canonical NF-κB signaling by phosphorylating p100/NF-κB2. Protein kinase assays suggested that IKKβ accounts for nearly all of the catalytic activity of the IKK holoenzyme towards IκBα 3,13.
Figure 1. Structure of xIKKβ.
a, Linear representation of IKKβ showing the boundaries for the kinase domain (KD), the ubiquitin-like domain (ULD) and the scaffold/dimerization domain (SDD). Sequences of hIKKβ and xIKKβ are of 756 and 758 residues, respectively, differing only at the most C-terminal region. The crystallized xIKKβ construct is shown. The previously designated leucine zipper (LZ) and helix-loop-helix (HLH) regions are shown in parentheses. b, Ribbon diagram of an xIKKβ protomer in the P1 crystal form. The N- and C-termini, KD N-lobe (orange) and C-lobe (yellow), ULD (magenta) and SDD (blue) are labeled. Secondary structural elements are labeled, with those in ULD followed by (‘) and those in SDD followed by (s). c, Ribbon diagram of an xIKKβ dimer. d, Superposition of ULD (magenta) with ubiquitin (gray). e, Ribbon diagram of an SDD dimer, showing locations of the previously designated LZ (red) and HLH (orange) regions.
The activation loop in both IKKα and IKKβ KDs contains the MEK consensus motif SxxxS (S177 and S181 in human IKKβ) 6–8,10. Some MEK kinase family members, such as TGF-β-activated kinase 1 (TAK1) and NF-κB-inducing kinase (NIK), were shown to phosphorylate IKKs 3,14,15. IKKα and IKKβ can also undergo autophosphorylation and activation as a result of overexpression or signal dependent NEMO clustering 10,16. Ala substitutions of the activation loop Ser residues prevent IKK activation whereas the phosphomimic, double Glu mutation S177E/S181E (EE) of IKKβ renders it constitutively active 7,13.
Tri-modular arrangement of IKKβ
We determined the crystal structure of Xenopus laevis IKKβ(xIKKβ) EE (residues 4–675) (Fig. 1a) in complex with either inhibitor Cmpd1 or Cmpd2 (Supplementary Fig. 1) at 4.0 and 3.6 Å resolutions in I4122 and P1 space groups, respectively (Supplementary Table 1 and 2, Supplementary Fig. 2). Eight IKKβ molecules in P1 and the one molecule in I4122 are highly similar to each other (Supplementary Fig. 3, Supplementary Table 3) and show conserved dimerization (see below). Structural description will use the first dimer (chains A and B) in P1. The hIKKβ and xIKKβ sequences share 74% identity with no gaps within the region of the structure; residue numbers designated for xIKKβ are also true for hIKKβ.
The IKKβ dimer structure resembles a pair of shears and has the overall dimensions of approximately 100Å x 130 Å x 60 Å (Fig. 1b, 1c). It contains KD (16–307), ULD (310–394), and a highly elongated domain we here refer to as the scaffold/dimerization domain (SDD, 410–666) (Fig. 1a, Supplementary Fig. 4). While KD and ULD form the handle of the shears, SDD is the blade. Both KD and ULD intimately associate with SDD, suggesting that KD does not function independently. Instead, the structure indicates that IKKβ is an integral tri-modular unit.
IKKβ KD in complex with either inhibitor Cmpd1 or Cmpd2 exhibits the typical bilobal kinase fold 17. Although sharing only 21.1% sequence identity with human ubiquitin, ULD of IKKβ (310–394) indeed has the ubiquitin fold (Fig. 1d). A major difference is the presence of an eight-residue insertion (residues 373–380) that forms part of the loop connecting β4′ and β5′ in ULD, which is mostly disordered. The hydrophobic surface patch of ubiquitin 18, which is often recognized by ubiquitin binding proteins, is conserved in ULD, with residues V318, L354 and L389 equivalent to L8, I44 and V70 of ubiquitin (Supplementary Fig. 5).
SDD comprises six helices (α1s-α6s), among which α2s and α6s each contain 70 and 77 residues, respectively. They twist together with a stretch of three shorter helices between them to fold as an elongated structural ensemble. The KD C-lobe sits on the N-terminal end of SDD while ULD binds close to the middle of SDD. Surprisingly, formerly designated LZ (458–485) and HLH domains (605–644) 7,8,10 do not exist as such in the structure and are both parts of SDD (Fig. 1e). Most of the hydrophobic residues in the predicted LZ point inward and are not available for mediating dimerization as previously proposed.
Structure of inhibitor bound IKKβ KD
The inhibitor binds to IKKβ KD at the hinge loop connecting N- and C- lobes, a region that recognizes the adenine in ATP 19,20 and often used for inhibitor binding 21–23 (Fig. 2a, Supplementary Fig. 6). The KD conformation is incompatible with that of an active kinase 17,24,25 (Fig. 2b, 2c). The activation segment, which begins from D166-L167-G168 as the conserved DFG triad and extends to A190-P191-E192 in the conserved APE motif, is fully ordered, including phosphomimic residues E177 and E181 (Fig. 2b). However, the C-terminal anchor of the activation segment, including the APE motif itself, is completely out of place so that essential interactions with the catalytic center are lost (Fig. 2c). Gross conformation of the N-terminal anchor of the activation segment is preserved with β9 paired with the β6 strand that precedes the catalytic loop. Part of the activation loop (175–177) contains an additional β strand (β10) that inserts in between the lobes to form a three-stranded β sheet with β9 and β6. The αC helix is tilted up and outward (Fig. 2c) to make room for β10, which also disrupts its productive interactions with the DFG motif in active kinase structures.
Figure 2. Inhibitor bound xIKKβ kinase domain (KD).
a, Fo-Fc electron density map for Cmpd1 in the I4122 structure, contoured at 2.0 σ. Carbon, nitrogen and oxygen atoms are shown in green, blue and red, respectively. b, Structure of xIKKβ KD. Glycine-rich loop: cyan; activation segment: red except that the DLG and APE motifs are in black; Cmpd1: purple. Side chains of phosphomimic residues E177 and E181 are shown. c, Superposition between xIKKβ (orange and yellow) and PKA (cyan, PDB code 1ATP). The activation segments of xIKKβ and PKA are shown in red and black, respectively.
Interactions among KD, ULD and SDD
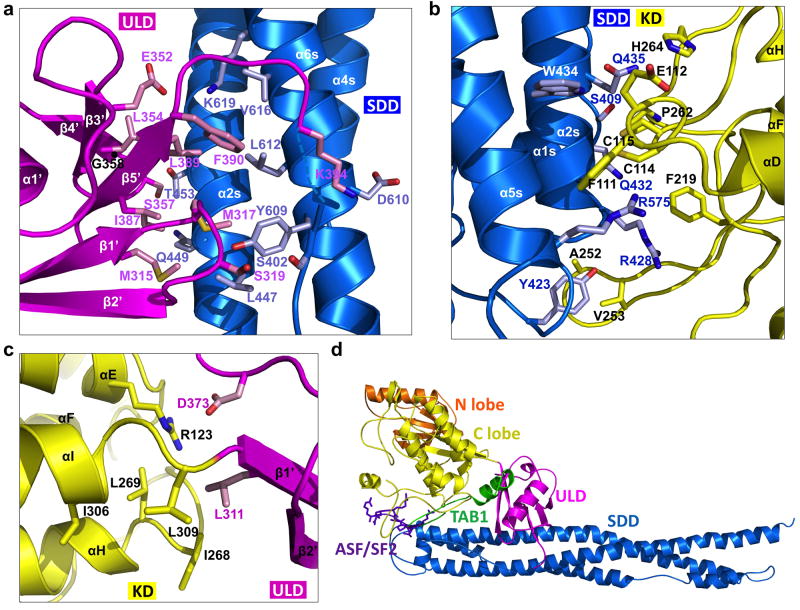
KD, ULD and SDD interact mutually, with the ULD: SDD interaction being most extensive, followed by the KD: SDD and the KD: ULD interactions. ULD binds close to the middle of SDD, contacting helices α2s and α6s (Fig. 3a). The interaction buries ~700 A2 surface area per partner. There are substantial hydrophobic contributions, supplemented by electrostatic interactions. Residues M315, M317, L354, I387, L389 and F390 on one side of ULD, pack against L612, Y609, L447 and main chain of α2s of SDD to form the central hydrophobic core of the interface. This hydrophobic patch of ULD is immediately adjacent to and overlaps with the conserved hydrophobic patch in the ubiquitin fold. Electrostatic interactions are observed between E352 and K619 and between K394 and D610. Additional interfacial residues include S319 and S357 of ULD and T453, Q449, and Y609 of SDD.
Figure 3. Interactions among the KD, the ULD and the SDD.
a, Interaction between ULD (magenta) and SDD (blue). Important interfacial side chains are shown with nitrogen atoms in blue, oxygen atoms in red, sulfur atoms in orange, and carbon atoms in either pink (for ULD) or light blue (for SDD). Location of G358 is marked with a black ball on the main chain. b, Interaction between KD (yellow) and SDD (blue). c, Interaction between KD (yellow) and ULD (magenta). d, Locations of the TAB1 peptide (green ribbon, PDB code 2EVA) and the ASF/SF2 peptide (purple stick model, PDB code 1WBP) relative to the IKKβ structure after superposition of the corresponding kinase domains.
Consistent with an important role of ULD in interacting with SDD, mutations in residues that are not at the interface, P347A, L361A and Q351A, had minimal effects on NF-κB activation 26. In contrast, mutation in a residue within a ULD surface loop that contacts the SDD, G358A (Fig. 3a), affected IKKβ-induced NF-κB activity 26. It was also shown that L353 is absolutely required for IKKβ activity 26; however, L353 is buried in the ULD core and the L353A mutation may have disrupted IKKβ structural integrity. Double substitutions of human IKKα and IKKβ that are equivalent to I608R/Y609P of SDD of xIKKβ did not affect their interaction with WT IKKβ, but negatively impacted kinase activity 10; I608 is buried in the SDD core and Y609 directly interacts with ULD (Fig. 3a).
Similar to ULD, KD also makes contacts with the N-terminal end of SDD (Fig. 3b), burying a surface area of ~650 Å2 from each interface. The binding forces are mainly van der Waals in nature. Limited hydrophobic interactions are observed, between residues A252 and V253 of KD and Y423 of SDD, and between F111 of KD and the hydrophobic portions of R572, R575 and E576 ofα5s of SDD. KD is linked to ULD through a two amino acid linker between αI of KD and β1′ of ULD (Fig. 3c), burying only ~350 Å2 surface area. Side chain contacts between L311 of ULD and I268 of KD are observed, which together form the small hydrophobic patch at the KD-ULD junction consisting also of L269 and I306 of KD and L309 of the linker. Together with the ionic interaction between D373 of ULD and R123 of αE of KD, these interactions may confer rigidity to the KD-ULD junction despite the small buried surface area.
Structural comparison with other kinase structures revealed a similarity of the locations of SDD and ULD to several known docking sites for substrates and regulatory proteins 27. In the crystal structure of the Ser/Thr kinase SRPK1 in complex with a docking motif in its substrate ASF/SF2 27, the peptide motif interacts with the kinase at the location of SDD (Fig. 3d). In the structure of the TAK1 kinase domain fused with the C-terminal region of its binding protein TAB1 28, TAB1 interacts with the C-lobe of the kinase at a position analogous to both SDD and ULD, presumably to activate the kinase (Fig. 3d).
ULD-SDD binds IκBα C-terminal region
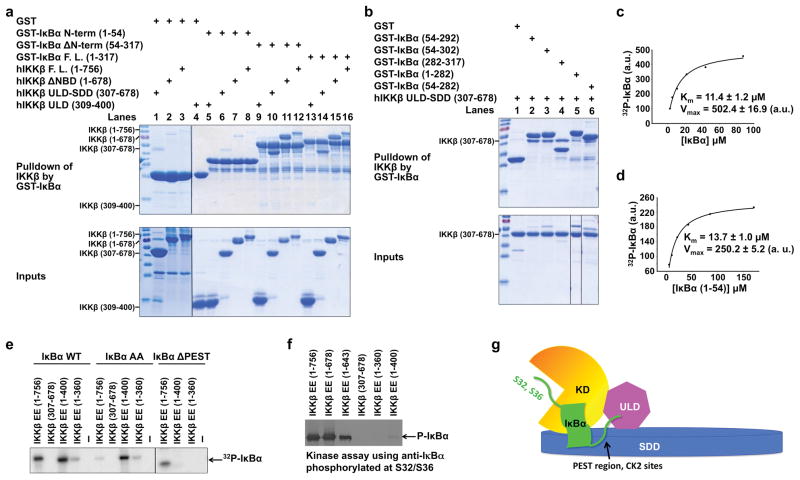
Previous studies have suggested that ULD in TBK1 and IKKi is involved in substrate recognition because its deletion impaired activity of the respective kinases and a GST-ULD fusion protein interacted with the specific substrates IRF3 or IRF7 29. Because ULD deletion in IKKβ also abolished its activity 26, we hypothesized that the ULD may recognize its specific substrate IκBα. However, we were surprised to find that GST-IκBα (1–317 and 54–317) pulled down only a minute amount of ULD (Fig. 4a, lanes 9 and 13), while GST alone did not pull down any (Fig. 4a, lane 4), suggesting that the interaction of IκBα with ULD is specific, but very weak. In contrast, GST-IκBα robustly pulled down full-length IKKβ or IKKβ lacking C-terminal NBD (Fig. 4a, lanes 15, 16).
Figure 4. ULD-SDD restricts IKKβ specificity while ULD is required for catalytic activity.
a, b,Pulldown of hIKKβ constructs using GST-IκBα constructs, showing the reciprocal interaction between ULD-SDD of IKKβ and C-terminal region of IκBα containing ankyrin repeats and PEST region. c, d, Measurement of Km and relative Vmax of IKKβ against full-length IκBα (c) and the N-terminal region of IκBα (1–54) (d). a.u.: arbitrary unit. e, Kinase assay of purified hIKKβ proteins against IκBα, its S32A/S36A mutant (AA), or its PEST-deletion construct (ΔPEST, 1–282) using [γ-32P]ATP. f, Kinase assay of purified hIKKβ proteins using antibody against IκBα phosphorylated at S32 and S36. g, A schematic model showing that the interaction between SDD of IKKβ and C-terminal region of IκBα may position the N-terminal cognate phosphorylation sites of IκBα to the active site of IKKβ.
IκBα has an N-terminal region (1–54) that contains cognate phosphoacceptor sites at S32 and S36, followed by a C-terminal region (55–317) that contains multiple ankyrin repeats and the PEST region 30,31. Strikingly, the N-terminal region of IκBα did not pull down any IKKβ constructs (Fig. 4a, lanes 5–8), while the C-terminal region of IκBα interacted robustly with full-length IKKβ as well as its ULD-SDD region (Fig. 4a, lanes 9–12), and very weakly with ULD alone (Fig. 4a, lane 9). Further mapping on IκBα showed that both ankyrin repeats (1–282, or 54–282) and the PEST region (282–317) interacted with IKKβ ULD-SDD (Fig. 4b, lanes 4, 5, 6). The extent of pulldown suggests that the PEST region contributes more to IKKβ interaction than the ankyrin repeats. Despite trying multiple constructs, we could not obtain soluble protein for IKKβ SDD to test its interaction with IκBα. However, the pull down data suggest a dominant role of SDD in IκBα interaction. In any case, it is clear that the mutual interaction between IKKβ and IκBα is mediated by their ULD-SDD and C-terminal domains, respectively.
ULD-SDD limits specificity and ULD aids catalysis
The specific interaction between ULD-SDD of IKKβ and IκBα suggests that the Km of phosphorylation by IKKβ might be lower for full length IκBα than for its N-terminal region (1–54) alone. We performed kinase kinetics analysis of IKKβ EE against the two different substrates. Surprisingly, the measured apparent Km values were 11.4 μM and 13.7 μM for full-length and the N-terminal region of IκBα, respectively (Fig. 4c, d), suggesting that binding at SDD, an exosite, does not alter the Km of the reaction dramatically. This could be due to opposing effects of the SDD: IκBα interaction, which increases substrate interaction but slows down product dissociation. The relative Vmax values were 502.4 and 250.2, respectively, suggesting that the SDD: IκBα interaction moderately enhances catalysis.
Casein kinase 2 (CK2) phosphorylates the C-terminal PEST region of IκBα around residues 283–299 30,31. To determine whether the SDD: IκBα interaction restricts substrate specificity, we compared the kinase activity of purified IKKβ EE proteins against either IκBα or its S32A/S36A mutant (AA) using [γ-32P]ATP (Fig. 4e). Although the KD-ULD (1–400) construct gave rise to a small amount of protein, it produced robust phosphorylation on WT IκBα, comparable to full-length IKKβ (1–756), suggesting that it is catalytically competent. Remarkably, KD-ULD effectively phosphorylated the AA mutant which in contrast was a poor substrate for full-length IKKβ. The C-terminal PEST region appeared to harbor the major sites of phosphorylation by KD-ULD because a construct lacking PEST (1–282) was not significantly phosphorylated by KD-ULD but was phosphorylated by full-length IKKβ (Fig. 4e). In addition, when IκBα phosphorylation was detected by antibody against IκBα phosphorylated at S32/S36, only very weak phosphorylation was seen for the KD-ULD construct in comparison with full-length IKKβ (Fig. 4f). These experiments suggest that while the KD-ULD construct is active, it possesses an altered substrate specificity, causing preferential phosphorylation of the C-terminal PEST sequence, consistent with a previous observation 32. Hence, ULD-SDD appears to specifically position IκBα such that only its N-terminal region is accessible to the IKKβ catalytic pocket (Fig. 4g).
We expressed three KD constructs, 1–301, 1–310 and 1–360, in insect cells and only obtained small amount of protein for the 1–360 construct. Kinase assay showed that IKKβ EE (1–360) exhibited very low residual activity against IκBα or its AA mutant (Fig. 4e), suggesting that KD activity is compromised in the absence of ULD. Without an isolated KD structure, we cannot deduce the molecular mechanism by which ULD acts. However, in analogy to TAK1 activation by TAB1 28 (Fig. 3d), it may be speculated that this KD-ULD interaction allosterically potentiates kinase activity. Further kinase assay using antibody against IκBα phosphorylated at S32/S36 showed no detectable activity of KD alone (Fig. 4f), confirming that the low residual activity may also be directed towards the C-terminal PEST region, similar to KD-ULD. Therefore, while ULD-SDD is critical for IKKβ specificity, ULD is required for its catalytic activity.
SDD mediates IKKβ dimerization
Full-length hIKKβ (1–756) and its KD-ULD-SDD region (1–678) are dimers in solution as determined by gel filtration chromatography (Fig. 5a). In the P1 and I4122 crystals, two conserved dimerization interfaces exist, one mediated by SDD and the other mediated by KD. Because hIKKβ (1–643) that truncates into SDD is a monomer in solution (Fig. 5a), SDD-mediated dimerization (Fig. 5b) is likely of greater importance than KD-mediated dimerization.
Figure 5. Dimerization is critical for IKKβ activation but not for its activity.
a, Gel filtration profiles of various hIKKβ constructs showing that those containing the KD-ULD-SDD region (1–756, 1–678) are dimeric while a further truncated construct (1–643) is monomeric. b, The dimerization interface of xIKKβ. c, Structure-based mutations disrupt hIKKβ dimerization as shown by gel filtration and analytical ultracentrifugation. d, Kinase activity of HEK293T cell transfected or insect cell purified hIKKβ EE and dimerization defective mutants L654D/W655D (654–5), W655D/L658D (655–8) and L654D/W655D/L658D (654-5-8). e, Autoactivation of HEK293T cell transfected hIKKβ WT and its dimerization defective mutants. f, Transfection of hIKKβ into WT and NEMO−/− MEFs, showing reduced IKKβ activation in the absence of NEMO. g, Dimerization mutants of IKKβ showed reduced interaction with NEMO. h, A tetramer of xIKKβ in the P1 structure. i, A close-up view of the tetramer interface, showing that the activation loops of neighboring protomers (black) face each other.
SDD in an IKKβ dimer does not form extensive interactions along the entire length dimension of the helical bundle. Rather, interactions are mostly localized to the end of the bundle (Fig. 5b), distal from KD and ULD and burying ~1,000 Å2 surface area from each monomer. The interaction is mostly hydrophobic. Residues that contribute significantly to dimerization include Q478, K482, F485, I492, K496, I505, Q651, L654, W655, L658, and I660 with residues L654, W655 and L658 from helix α6s burying the most surface area (Fig. 5b). This dimerization interface is entirely unexpected as it was predicted earlier that LZ mediates IKKβ dimerization. The structure now explains that failed dimerization of the LZ-defective L462S/L469S mutant of IKKα 10 is due to the structural role of L469, whose equivalent residue M472 of IKKβ is buried in the SDD core. Superposition of four IKKβ dimers in P1 and the one IKKβ dimer in I4122 shows that IKKβ dimers are conserved (Supplementary Fig. 7). In I4122, the distal part of SDD is not visible due to lack of crystal packing at this region and dynamic disorder, not degradation (Supplementary Fig. 8).
To confirm the observed interface in IKKβ dimerization, we performed structure-based mutagenesis on residues that bury the most surface area, L654, W655 and L658. These residues are not within the predicted LZ region. Two purified double mutants, L654D/W655D and W655D/L658D, failed to dimerize as shown by the considerable shift in gel filtration positions (Fig. 5c). Furthermore, equilibrium analytical ultracentrifugation experiments showed that WT IKKβ is indeed a dimer while the IKKβ mutants are either a monomer or have a 167-fold weaker dimerization affinity (Fig. 5c).
Dimerization in IKKβ activation but not activity
The geometry of the IKKβ dimer with KDs facing away from each other suggests that each IKKβ protomer is independent in its kinase activity. To confirm this, we transfected HEK293T cells with hIKKβ EE mutants, L654D/W655D (654–5), W655D/L658D (655–8) and L654D/W655D/L658D (654-5-8), and found that all mutants exhibited robust kinase activity. (Fig. 5d). Insect cell expressed and purified dimerization mutants showed the same results. In addition, a purified IKKβ construct with truncation into SDD (1–643) that is monomeric in solution (Fig. 5a) was active in phosphorylating IκBα (Fig. 5d).
Previous studies have shown that IKKβ can undergo trans-autophosphorylation and activation upon transfection 13,16. This autophosphorylation and phosphorylation by TAK1 likely both contribute to IKKβ activation upon cell stimulation. To determine whether the observed dimerization interface is critical for this activation, we tested dimerization mutations in IKKβ activation upon overexpression in HEK293T cells (Fig. 5e). While WT IKKβ was robustly activated, the L654D/W655D and W655D/L658D mutants completely and partially lost this activation, respectively, in a manner that correlates with the degree of impairment in dimerization (Fig. 5c, e). Interestingly, overexpression of IKKβ in NEMO-deficient mouse embryonic fibroblasts (MEFs) resulted in its activation, but to a lesser extent than in WT MEFs (Fig. 5f). We found that IKKβ dimerization mutants failed to interact with NEMO efficiently (Fig. 5g), perhaps due to reduced avidity towards oligomeric NEMO. Therefore, although IKKβ kinase activity does not depend on dimerization once its activation loop is phosphorylated, IKKβ activation requires dimerization and is likely enhanced by interaction with NEMO.
DISCUSSION
The IKKβ structure presented here predicts that IKKα, TBK1 and IKKi all exhibit an overall structural organization that comprises KD, ULD, and SDD. Although an ULD was not predicted in IKKα, conservation of buried residues in this region and of the ULD-SDD interface suggests that IKKα also has this domain (Supplementary Fig. 4). ULD and SDD likely share the common structural role but may have evolved additional, differential functions in the individual kinases. SDD-mediated dimerization may also be conserved. In particular, residues at the observed IKKβ dimerization interface are highly conserved in IKKα (Supplementary Fig. 4), explaining that IKKα and IKKβ can form both homo- and hetero-dimers 8,13. Given our structure of IKKβ and the previously determined structures of NEMO fragments 33–37, we may speculate that the full IKK complex is also a dimer, or a dimer of dimers with a molecular mass of around 270 kD or 540 kD. The apparent 700–900 kD molecular mass of the IKK holoenzyme on gel filtration 5–10 may be due to the elongated nature of NEMO and the complex (Supplementary Fig. 9).
Because the conserved IKKβ dimer structure does not place KD close to each other for trans-autophosphorylation, we wondered whether higher order oligomerization, which is likely enhanced by NEMO and its ability to bind ubiquitin 33,34, is responsible for this autoactivation. In both P1 and I4122, IKKβ exist as dimers of dimers (Fig. 5h, Supplementary Fig. 10). In particular, active sites of two neighboring protomers in the tetramer face each other such that activation loop from one protomer might reach into the active site of the other (Fig. 5i), which may mimic an autophosphorylation state.
The ULD-SDD region of IKKβ directly interacts with the C-terminal portion of IκBα. This interaction may serve several purposes. First, it likely orients IκBα so that its N-terminal cognate phosphorylation sites are presented to the KD active site (Fig. 4g). Without this interaction, IKKβ preferentially phosphorylates the C-terminal PEST motif of IκBα. Second, the interaction appears to enhance IKKβ activity. Third, phosphorylation at the PEST by CK2 or other kinases may regulate IκBα interaction with IKKβ and hence affect phosphorylation at the cognate sites. Fourth, the interaction may allow concerted phosphorylation at both S32 and S36 of IκBα without intervening dissociation. In MAP kinase cascades that involve dual phosphorylations, specific docking interactions occur between the kinases and their substrates 27,38,39. The β-catenin protein in the Wnt signaling pathway contains the same dual phosphorylated destruction box motif as IκBα 40. Consistently, β-catenin is brought to the responsible kinase GSK-3 via the adaptor protein Axin, allowing both specificity and concerted phosphorylation 41,42. Therefore, in a general statement that structure serves the function, the IKKβ structure seems to fit perfectly its function as the IκBα kinase, poised to turn on the important NF-κB pathway specifically, efficiently and concertedly in response to cellular physiology.
METHODS SUMMARY
Xenopus laevis IKKβ was expressed in insect cells and purified to homogeneity using Ni-affinity, ion exchange and gel filtration chromatography. It was crystallized at 4°C in polyethylene glycol (PEG) 6000 and K/Na phosphate for the P1 and I4122 crystals, respectively. The structure was determined by multi-wavelength anomalous diffraction (MAD) of the selenomethionyl protein.
Methods
Protein expression and purification
To elucidate the molecular basis of IKKβ function, we expressed IKKβ from a number of species using baculovirus mediated insect cell expression. The Xenopus laevis IKKβ (xIKKβ) sequence in the NCBI database starts at a Met residue that is equivalent to M17 of both the human and mouse IKKβ sequences (hIKKβ and mIKKβ). Translation of the DNA sequence preceding the ATG codon of M17 revealed sequences that are almost identical to residues 9–16 of hIKKβ and mIKKβ. These were taken as part of the xIKKβ sequence and residues 1–8 were taken from the corresponding mIKKβ sequence. This reconstructed xIKKβ sequence has the same residue numbering as the hIKKβ sequence until after the scaffold/dimerization domain.
Various constructs of IKKβ WT and the phosphomimic S177E/S181E mutant were designed with an N-terminal polyhistidine tag and a Tobacco Etch Virus (TEV) protease cutting site between the tag and the protein. Recombinant IKKβ baculoviruses were made in DH10BAC cells, amplified, and used to infect Hi5 insect cells in serum-free media (Invitrogen). The cells were cultured in suspension and harvested 48 hrs post-infection. The recombinant proteins were purified by Ni-affinity chromatography, anion-exchange and gel-filtration chromatography. For crystallization, the polyhistidine tag was cleaved by the TEV protease during protein purification.
All IκBα proteins were expressed in E. coli using pET28a, pGEX4T3 and pET-SUMO vectors and purified by their respective affinity tags. For kinase assays, the SUMO tag was cleaved off IκBα proteins. His-ULD and His-ULD-SDD of hIKKβ were also expressed in E. coli using the pET28a vector.
Crystallization and data collection
Unlike many protein kinases, IKKβ KD cannot be recombinantly expressed as a well-behaved biochemical entity for structural studies. In addition, after mapping a compact region by limited proteolysis, IKKβ was still refractory to crystallization, both alone and in the presence of various ATP analogues. To overcome this obstacle, we used several IKKβ inhibitors including Cmpd1 and Cmpd2 (Supplementary Fig. 1), which were identified against the S177E/S181E (EE) mutant, in co-crystallization. A human IKKβ (hIKKβ) EE construct (residues 1–678) lacking only the C-terminal NBD did crystallize; however, these crystals only diffracted to ~7.5 Å resolution. Searching IKKβ orthologues that may give better crystals led to a success in crystallizing the analogous region of Xenopus laevis IKKβ (xIKKβ) EE (residues 4–675) (Fig. 1a).
The xIKKβ (S177E/S181E) protein containing residues 4–675 was concentrated by ultrafiltration (Amicon) to about 15 mg/ml in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl and 10 mM dithiothreitol (DTT). It was mixed with an inhibitor compound in 1:2 molar ratios before crystallization. Cmpd1 is (4-((4-(4-(chlorophenyl)pyrimidin-2-yl)amino)phenyl)(4-(2-hydroxyethyl)piperazin-1-yl)methanone and Cmpd2 is 1-(4-(4-((4-(4-(pyridin-4-ylsulfonyl)phenyl)pyrimidin-2-yl)amino)benzoyl)piperazin-1-yl)ethanone. The P1 crystals were grown using hanging drop vapor diffusion at 4°C by mixing equal volumes of the purified protein and the crystallization condition of 100 mM N-(2-acetamido) iminodiacetic acid at pH6.5, 10 % (w/v) polyethylene glycol (PEG) 6000, 50 mM Li2SO4, 300 mM NaCl and 10 mM DTT. The I4122 crystals were grown at 4°C with well solution containing 1.8 M K/Na phosphate at pH 5.6 and 10 mM DTT. For data collection, all crystals were flash frozen in the respective crystallization conditions supplemented with 25 % (v/v) ethylene glycol. Diffraction data were collected at the 24ID-C beam line of the Advanced Photon Source (APS). Multi-wavelength anomalous diffraction (MAD) data of heavy-atom derivative crystals or selenomethionyl (Se-Met) crystals were collected near the respective absorption edges. All diffraction data were processed using the HKL2000 suite 43 and their statistics are shown in Supplementary Table 1 and Supplementary Table 2.
Structure determination, refinement and analysis
The initial xIKKβ crystals grew in the P1 space group in the presence of the inhibitor Cmpd1 or Cmpd2 and diffracted to 3.6 Å resolution. Selenomethionyl crystals were obtained, but we failed to locate the large number of expected Se sites. Among the extensive heavy atom searches, a Yb-derivative was obtained, with 8 well-defined sites, which likely correspond to 8 IKKβ molecules in the asymmetric unit. However, the electron density map calculated from a 3-wavelength Yb-anomalous diffraction data set was insufficient for tracing and phase combination with the selenomethionyl data set could not be performed due to non-isomorphism.
The structure determination was eventually successful in the alternative crystal form of I4122, which contains 1 molecule of IKKβ in complex with Cmpd1 and diffracted to 4.0 Å resolution, using multi-wavelength anomalous diffraction (MAD) of the selenomethionyl crystals (Supplementary Table 1 and 2, Supplementary Fig. 2). Twelve Se-sites were determined using the program Shelx D 44 and refined with the program MLPHARE in the CCP4 suite 45. MAD phases were calculated at 4.0 A resolution with data from I4122 crystals using the program SHARP 46. A few cycles of model building and refinement were carried out with the program O 47 and REFMAC with TLS parameterization 48. The I4122 crystals contain one monomer in the asymmetric unit and 80% solvent when calculated with the entire IKKβ construct and 84% solvent when considering only the ordered part of the structure. The inhibitor Cmpd1 has density in the MAD experimental map and the Fo-Fc omit map. The Dundee PRODRG2 Server was used to generate topology and restraint files of the compound for refinement. The refined model contains residues 16–236, 243–286, 290–376, 384–394, 401–475 and 528–637 and Cmpd1.
The structure of the P1 form was determined by molecular replacement using the refined model of the I4122 crystal form as the search model, in which 8 molecules were located. Se-sites of the SAD data set of a selenomethionyl crystal in the P1 space group was calculated by difference Fourier and used to verify residue registration in the P1 structure. Refinement in the P1 structure was conducted at 3.6 Å resolution and incorporated tight non-crystallographic symmetry (NCS) restraints (0.02 Å rmsd in atom positions). Upon several rounds of refinement at 3.6 Å resolution, new electron densities appeared in the P1 crystal form to complete the model building. Although Cmpd2 was in the crystallization condition, it did not have clear density and was not included in the refinement. The refined model of the P1 crystal form contains four IKKβ dimers in the asymmetric unit. Three of the dimers encompass residues 16–236, 243–286, 290–376, 384–394, 401–551 and 559–666. One dimer contains the same residues as the structure in I4122. The structures were analyzed using the CCP4 suite 45 and the Dali server 49, and the figures were made using PyMOL 50.
GST-pulldown
The tagged proteins were first purified with glutathione or Ni-NTA beads and their expression levels were assessed by SDS-PAGE. Beads containing estimated equivalent quantities of the tagged proteins were mixed with the cell lysates or the purified versions of the interaction partners. The mixtures were incubated at room temperature for 1 hr with rotation. After centrifugation, the supernatants were removed. The beads were then washed twice, eluted and subjected to SDS-PAGE analysis. All pulldown experiments were repeated two to four times with consistency.
Kinase assays using anti-phospho-IκBα antibody
The hIKKβ proteins (0.1 μg/μl) were incubated with recombinant IκBα (1 μg/μl) in 50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10 mM MgCl2, and 2 mM DTT for 30 min at 30°C. SDS-PAGE sample buffer was used to terminate the reactions. The products were separated on 15 % SDS-PAGE and transferred to PVDF membranes. Anti-phospho-IκBα antibody (Cell Signaling Technology, Inc.) was used to detect phospho-IκBα.
Determination of Km of hIKKβ for IκBα (1–54) and full-length
To derive the Km of hIKKβ for full-length IκBα, kinase assays were performed at substrate concentrations of 2.8 μM, 5.3 μM, 10.6 μM, 21.3 μM, 42.5 μM, and 85 μM. Similarly, to derive the Km of hIKKβ for IκBα (1–54), kinase assays were performed at substrate concentrations of 5.3 μM, 10.6 μM, 21.3 μM, 42.5 μM, 85 μM, and 170 μM. A time course of the kinase reactions was first performed to select a hIKKβ amount and a time point within which the reactions are linear with time. The final selected reactions contain 10 ng baculovirus-expressed hIKKβ, 100 mM cold ATP and 1 μl [γ-32P]ATP (3000Ci/mmole, 1mCi/100 μl) in 25 μl reaction buffer containing 50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10 mM MgCl2 and 2 mM DTT. The phospho-transfer reaction was allowed to proceed for 10 minutes at 30 °C and quenched with SDS-PAGE sample buffer. The products were separated on 15 % SDS-PAGE and subjected to autoradiography. The relative amounts of phosphorylated IκBα were quantified using ImageJ, plotted against total IκBα concentrations and fitted using non-linear regression to the Michaelis-Menten equation to obtain the Km using SigmaPlot.
Transfection, immunoprecipitation and kinase assay
The constructs Flag-hIKKβ EE and its truncation mutants, HA-hIKKβ EE and its dimerization mutants L654D/W655D, W655D/L658D and L654D/W655D/L658D, Flag-hIKKβ and its dimerization mutants L654D/W655D and W655D/L658D were generated in the vector pcDNA3 by conventional PCR. All IKKβ constructs were transfected in HEK293T cells with Lipofectamine 2000 (Invitrogen). After 24 hrs, cell extracts were immunoprecipitated with anti-Flag antibodies bound to agarose beads (M2, Sigma) or anti-HA bound to agarose beads (Sigma). IKKβ kinase assays were essentially done as described 6,13. Briefly, immunoprecipitates were incubated with 2 μM full-length IκBα (1–317) in 20 mM HEPES at pH 7.5, 10 mM MgCl2, 20 mM β-glycerophosphate, 10 mM PNPP, 50 mM Na3VO4, 1 mM DTT, 20 mM ATP, and (1–10 mCi) [γ-32P]ATP at 30°C for 30 min, and subjected to SDS-PAGE and autoradiography. Immunoblotting was performed using anti-Flag (Sigma), anti-HA (Sigma) or anti-IKKβ antibodies (Upstate, 05–535).
Equilibrium analytical ultracentrifugation measurements
Experiments were performed in a Beckman XL-A/I analytical ultracentrifuge (Beckman-Coulter, Palo Alto CA, USA), utilizing six-cell centerpieces with straight walls, 12 mm path length and sapphire windows. Samples were kept and diluted in 50 mM Tris-HCl at pH 8.0 and 300 mM NaCl. Samples from wild-type protein were diluted to 6.9, 4.5 and 2.4 μM, mutant L654D/W655D was diluted to 7.4, 4.8, and 2.6 μM and mutant W655D/L658D to 4.9, 3.2, and 1.7 μM for channels A, B and C, respectively. Dilution buffer was used as blank. All samples were run at 4°C at 9000 rpm (held for 20 h, then four scans with 1 h interval), 11000 rpm (held 10 h then four scans with 1 h interval), 14000 rpm (held 10 h then four scans with 1 h interval), 17000 rpm (held 10 h then four scans with 1 h interval). Detection was by UV at 280 nm. Solvent density and protein v-bar at both temperatures were determined using the program SednTerp (Alliance Protein Laboratories, Corte Cancion, Thousand Oaks, CA, USA). For calculation of KD and apparent molecular weight, all useful data were used in a global fit, using the program HeteroAnalysis, obtained from University of Connecticut (www.biotech.uconn.edu/auf).
Supplementary Material
Acknowledgments
We thank Drs. Kanagalaghatta Rajashankar and Narayanasami Sukumar for data collection at the NE-CAT of APS, Dr. Beate Schwer for help with the kinase assay, Dr. Pascale Gaillard for help with the chemistry, and Drs. Goran Ahlsen, Lawrence Shapiro and Barry Honig for the ultracentrifugation experiments. This work was supported by the National Institute of Health (H.W. and M.K.), the American Heart Association (G.X. and Y.C.L.) and the Cancer Research Institute (Y.C.L.). M. K. is an American Cancer Society Research Professor.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
G.X. cloned, expressed, purified, crystallized and determined the crystal structure of xIKKβ and performed Km determination experiments. Y.C.L. cloned, expressed, purified and crystallized hIKKβ and performed pulldown experiments and kinase assays using phospho-IκBα antibody. Q.L. expressed the hIKKβ mutants in insect cells. G.N. and X.W. vperformed transfection, immunoprecipitation and kinase assays and M.K. supervised these experiments. H.W. supervised the project. G.X and H.W. made the figures and wrote the manuscript.
Author Information The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3QA8 and 3QAD. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132 (3):344. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 3.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25 (51):6685. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 4.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441 (7092):431. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84 (6):853. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 6.DiDonato JA, et al. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388 (6642):548. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 7.Mercurio F, et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278 (5339):860. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 8.Woronicz JD, et al. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278 (5339):866. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 9.Yamaoka S, et al. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93 (7):1231. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 10.Zandi E, et al. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91 (2):243. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 11.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395 (6699):297. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 12.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 13.Zandi E, Chen Y, Karin M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF-kappaB-bound substrate. Science. 1998;281 (5381):1360. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]
- 14.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6 (11):1087. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 15.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103 (31):11677. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang ED, et al. Roles for homotypic interactions and transautophosphorylation in IkappaB kinase beta IKKbeta) activation [corrected] J Biol Chem. 2003;278 (40):38566. doi: 10.1074/jbc.M304374200. [DOI] [PubMed] [Google Scholar]
- 17.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253 (5018):407. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 18.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10 (10):659. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49 (Pt 3):362. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 20.Bossemeyer D, et al. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5–24) EMBO J. 1993;12 (3):849. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu RM, Carmel G, Kuret J, Cheng X. Structural basis for selectivity of the isoquinoline sulfonamide family of protein kinase inhibitors. Proc Natl Acad Sci U S A. 1996;93 (13):6308. doi: 10.1073/pnas.93.13.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 23.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303 (5665):1800. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 24.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15 (5):661. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 25.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376 (6538):313. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 26.May MJ, et al. A novel ubiquitin-like domain in IkappaB kinase beta is required for functional activity of the kinase. J Biol Chem. 2004;279 (44):45528. doi: 10.1074/jbc.M408579200. [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith EJ, et al. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107 (11):5065. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown K, et al. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J Mol Biol. 2005;354 (5):1013. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda F, et al. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 2007;26 (14):3451. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato T, Jr, Delhase M, Hoffmann A, Karin M. CK2 Is a C-Terminal IkappaB Kinase Responsible for NF-kappaB Activation during the UV Response. Mol Cell. 2003;12 (4):829. doi: 10.1016/s1097-2765(03)00358-7. [DOI] [PubMed] [Google Scholar]
- 31.Barroga CF, Stevenson JK, Schwarz EM, Verma IM. Constitutive phosphorylation of I kappa B alpha by casein kinase II. Proc Natl Acad Sci U S A. 1995;92 (17):7637. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaul JD, Farina A, Huxford T. The human IKKbeta subunit kinase domain displays CK2-like phosphorylation specificity. Biochem Biophys Res Commun. 2008;374 (3):592. doi: 10.1016/j.bbrc.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 33.Lo YC, et al. Structural Basis for Recognition of Diubiquitins by NEMO. Mol Cell. 2009;33:602. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136 (6):1098. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Rushe M, et al. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure. 2008;16 (5):798. doi: 10.1016/j.str.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Bagneris C, et al. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Mol Cell. 2008;30 (5):620. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 37.Cordier F, et al. Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J Mol Biol. 2008;377 (5):1419. doi: 10.1016/j.jmb.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 38.Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol. 2006;16 (6):676. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87 (5):929. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 40.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11 (6):1445. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda S, et al. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17 (5):1371. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart MJ, et al. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8 (10):573. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58 (Pt 10 Pt 2):1772. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Bricogne G, et al. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59 (Pt 11):2023. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 47.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building models in electron density maps and the location of errors in those models. Acta Crystallgr A. 1991;47:110. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 48.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 49.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 50.Delano WL. The PyMol Molecular Graphics System. 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.