Abstract
Genes expressed in the somatopleuric mesoderm, the embryonic domain giving rise to the vertebrate pelvis, appear important for pelvic girdle formation. Among such genes, Pbx family members and Emx2 were found to genetically interact in hindlimb and pectoral girdle formation. Here, we generated compound mutant embryos carrying combinations of mutated alleles for Pbx1, Pbx2, and Pbx3, as well as Pbx1 and Emx2, to examine potential genetic interactions during pelvic development. Indeed, Pbx genes share overlapping functions and Pbx1 and Emx2 genetically interact in pelvic formation. We show that in compound Pbx1;Pbx2 and Pbx1;Emx2 mutants, pelvic mesenchymal condensation is markedly perturbed, indicative of an upstream control by these homeoproteins. We establish that expression of Tbx15, Prrx1, and Pax1, among other genes involved in the specification and development of select pelvic structures, is altered in our compound mutants. Lastly, we identify potential Pbx1-Emx2-regulated enhancers for Tbx15, Prrx1, and Pax1, using bioinformatics analyses.
Introduction
In most mammalian tetrapods, the pectoral and pelvic girdles are crucial proximal limb structures involved in weight bearing and propulsion during locomotion (Romer and Parsons, 1986). The pelvic girdle consists of three segments. Ventrally, there are two pelvic bones, each of which comprises of a rostrally facing ilium, a ventrally oriented pubis, and a dorso-laterally positioned ischium (Fig. 1, representation of the hindlimb (HL) axes within red inset; Fig. 2). Dorsally, there is the sacrum, which in different mammal species bears a variable number of vertebrae. In the pelvic girdle, the two pubic elements articulate ventrally at the pubic symphysis, while each ilium (or iliac blade) articulates dorsally at the sacroiliac joint with the sacral vertebrae. The three main bones of the pelvis also form a laterally facing socket called the acetabulum, whereby the head of the femur articulates (Romer and Parsons, 1986).
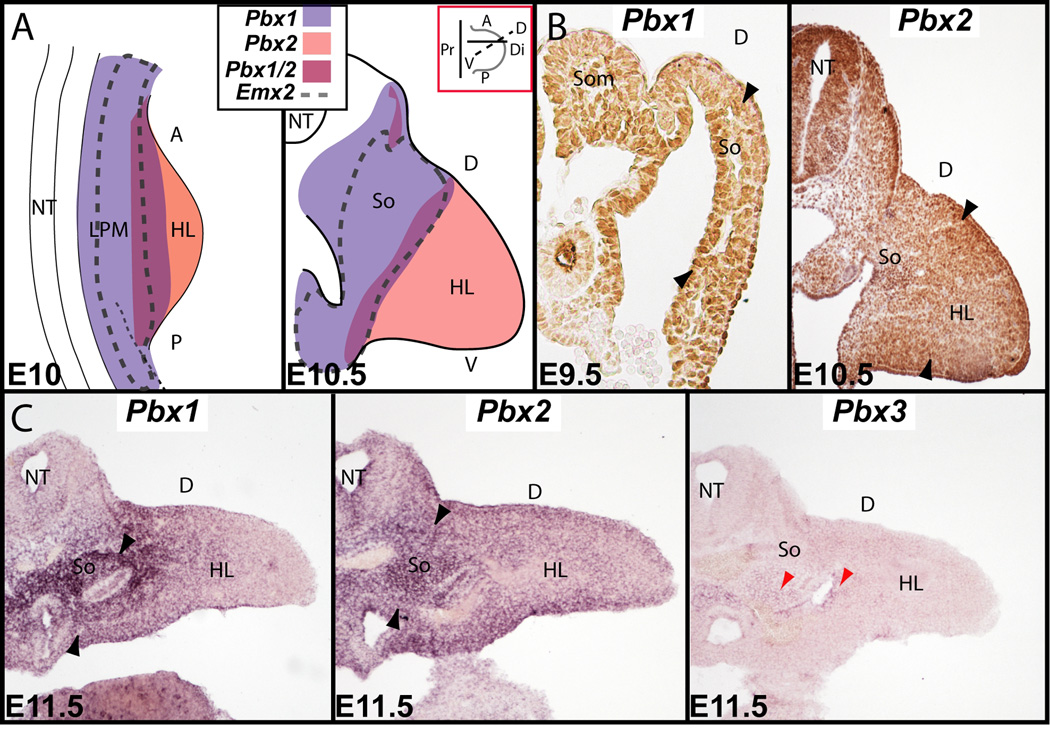
Fig. 1.
Pbx family member and Emx2 expression during pelvic development via in situ hybridization and immunohistochemistry. A: Summary illustration of Pbx1 and Pbx2 co-expression with Emx2 during early HL and pelvic development (see also Capellini et al., 2006; Di Giacomo et al., 2006). At E10 (left) to E10.5 (right), all three genes overlap within the LPM and mesodermal portion of the somatopleure of the proximal HL bud. Red inset shows orientations along plane of section used in the larger illustration below and the Pbx2 immunohistochemistry results in (B). B: Immunohistochemistry of Pbx1 (left) at E9.5 and Pbx2 (right) at E10.5 in the HL mesodermal portion of the somatopleure (black arrowheads). C: At E11.5, Pbx1 (left, black arrowheads) and Pbx2 (middle, black arrowheads) are strongly co-expressed in the mesodermal portion of the somatopleure, while Pbx3 is only weakly expressed (right, red arrowheads). Abbreviations: A, anterior; D, dorsal; Di, distal; HL, hindlimb; LPM, lateral plate mesoderm; NT, neural tube; P, posterior; Pr, proximal; So, somatopleure; Som, somite; V, ventral.
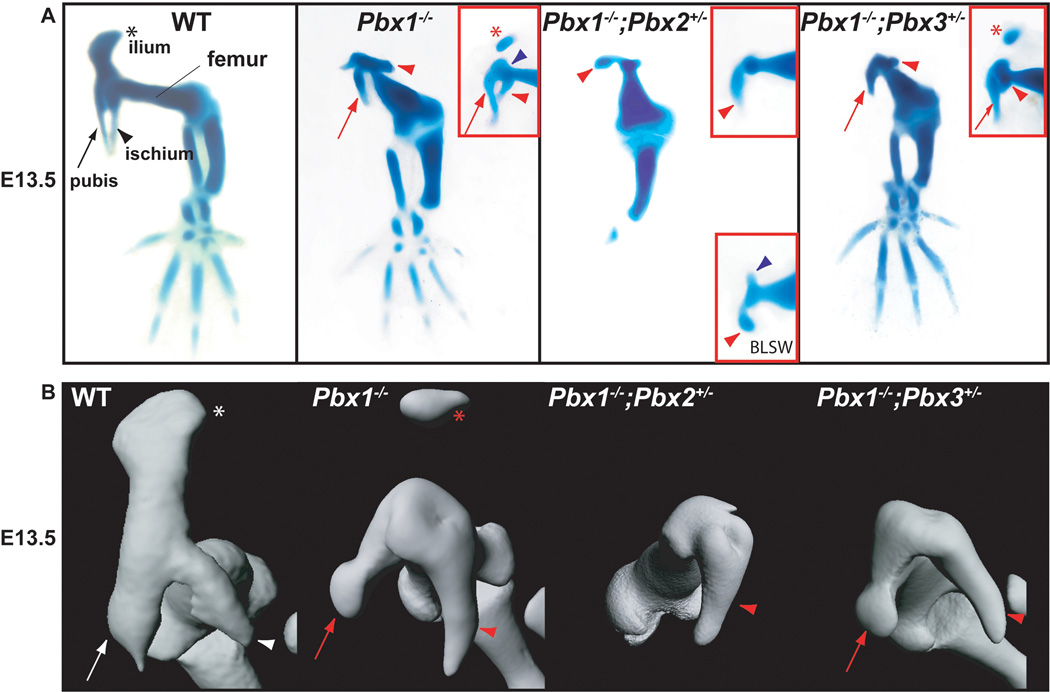
Fig. 2.
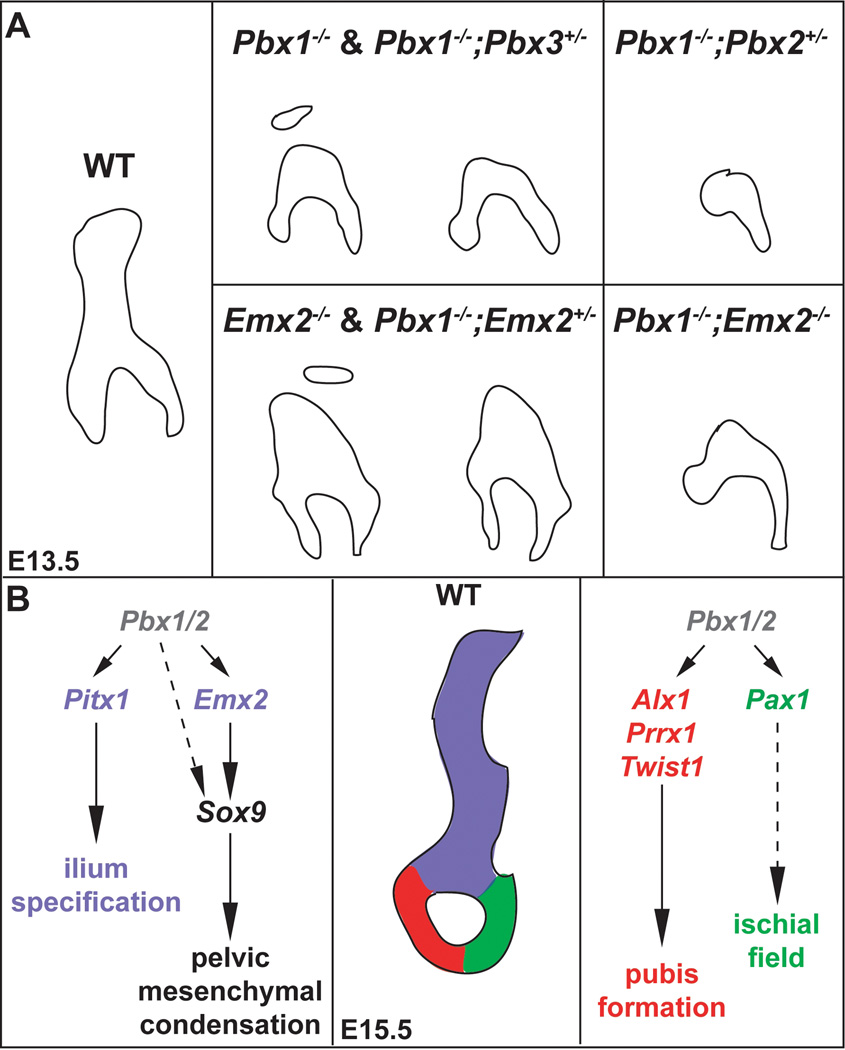
Pelvic skeletal phenotypes of compound Pbx family member mutants at E13.5 as assessed using Alcian blue and Alizarin red staining and OPT. A: Compared to WT HL (left panel): Pbx1−/− embryos exhibit either absent or detached ilia (red inset, red asterisk), and reduced pubis (red arrow) and ischia (red arrowhead); Pbx1−/−;Pbx2+/− mutants lack ilia and pubis and display rudimentary ischia (red arrowheads in main panel and insets). Insets show additional embryos acquired via breeding schemes described in Materials and Methods and Results. Lower red inset depicts the presence of a small tissue rudiment (blue arrowhead), often found in mutants repeated out-crossed to Black Swiss (BLSW); Pbx1−/−;Pbx3+/− mutants phenocopy single Pbx1 homozygotes in all defects (red asterisk, arrows, and arrowheads), and reveal that Pbx3, in this mutant model, is not required for distal HL development. B: OPT of WT (white symbols) and select Pbx family member mutants highlights defects illustrated above (red symbols), and that Pbx1−/−;Pbx2+/− mutants exhibit only truncated ischial rudiments (red arrowhead).
While the girdles have been the subjects of anatomical (Romer and Parsons, 1986), functional (Young, 2006), and evolutionary (Sanchez-Villagra and Maier, 2002) studies, their embryological derivation and underlying genetic networks have largely been neglected. To date, research conducted mainly in the chick (Huang et al., 2006) but also in the mouse (Pomikal and Streicher, 2010; Valasek et al., 2010), has partially elucidated the tissue origins for each girdle. Notably, the scapula has been shown to develop from multiple tissues, such as the dermomyotome of the somites, the mesodermal portion of the somatopleure (a domain of the lateral plate mesoderm or LPM) and neural crest cell-derived mesenchyme (Huang et al., 2000; Matsuoka et al., 2005; Wang et al., 2005). However, only the mesodermal portion of the somatopleure has been shown to give rise to each pelvic bone (Malashichev et al., 2008; Pomikal and Streicher, 2010), while sclerotome gives rise to sacral vertebrae (Christ et al., 2000).
Only a sparse number of genes have been identified so far that control girdle formation (Niswander, 2003; Huang et al., 2006). Specifically, more is known of the genetic pathways that govern the development of the scapular blade, head, and neck versus the pelvic girdle (Kuijper et al., 2005; Capellini et al., 2010). Interestingly, of the genes involved in scapular blade morphogenesis few have only minor roles in pelvic development. For example, Tbx15 regulates the murine scapular blade, through its genetic interactions with Gli3, but does not singularly or cooperatively control the formation of the pelvic girdle (Kuijper et al., 2005). However, in humans, mutations in TBX15 do produce pelvic iliac defects, suggesting potential differences in the genetic control of girdle development across species (Lausch et al., 2008). Notably, the genetic interactions identified between Aristaless family genes for the formation of the scapular blade were also identified in the pelvis. Specifically, compound Alx4;Alx1 mutants exhibit a reduction of the superior scapular blade and the loss of the anterior pubic ramus (also somewhat evident in single Alx4 homozygous mutants and Alx3;Alx4 double homozygous mice) (Kuijper et al., 2005). These pelvic defects, however, are not exacerbated by the compound loss of Tbx15 (Kuijper et al., 2005).
One emergent finding is that the somatopleuric mesoderm constitutes a domain that is important to both pectoral and pelvic girdle development (Malashichev et al., 2008; Pomikal and Streicher, 2010), as it is specifically involved in progenitor cell fate specification, patterning, and condensation formation (Kuijper et al., 2005; Capellini et al., 2010). Not surprisingly, of the genes required for pelvic development in the mouse, most are expressed in this specific domain. Compound null mutations in Prrx1 and Prrx2 (ten Berge et al., 1998), genes of the Aristaless family that are expressed in the mesodermal portion of the somatopleure, result in the loss of the pubic symphysis. Likewise, levels of Twist1 activity in the somatopleure and LPM are known to control pubis formation (Krawchuk et al., 2010). Furthermore, ilium reduction is observed when Pitx1 or Fgf10 are lost from the early HL bud LPM and somatopleure, as well as when Tbx4 is conditionally inactivated from the proximal HL LPM (Lanctot et al., 1999; Sekine et al., 1999; Naiche and Papaioannou, 2007).
Emx2−/− and Pbx1−/− embryos exhibit pelvic defects, as both have reduced ilia that are detached from the pubis and ischium (Pellegrini et al., 2001; Selleri et al., 2001). Previous studies have further implicated Pbx Three-Amino acid-Loop-Extension (TALE) homeoproteins as regulators of HL development (Selleri et al., 2001; Capellini et al., 2006). Lastly, Emx2 and Pbx1 were shown to interact genetically and molecularly in scapular blade formation (Capellini et al., 2010) within the mesodermal portion of the somatopleure. Here, we demonstrate that Pbx1 and Pbx2 share overlapping functions in the somatopleure and LPM during pelvic development. Compound Pbx1−/−;Pbx2+/− mutant embryos, but not Pbx1−/−;Pbx3+/− mutants, present exacerbations of the iliac defects of single Pbx1−/−, as well as novel caudal pelvic phenotypes absent from Pbx1−/− embryos (Capellini et al., 2006). Conversely, single loss of Pbx2 or Pbx3 does not cause pelvic phenotypes (Rhee et al., 2004; Selleri et al., 2004). We reveal that Pbx family members act as upstream regulators of multiple genes that are critical for ilium and pubis formation. Additionally, we demonstrate that in pelvic development Pbx1 interacts with Emx2, albeit less markedly than in the pectoral girdle. Lastly, we independently reveal potential regulatory elements that are putative targets of Pbx1/Emx2 in the proximity of genes expressed and involved in pelvis formation, such as Tbx15, Prrx1, and Pax1. Of note, these latter genes are down-regulated in Pbx compound mutant embryos. Our results delineate previously unknown genetic networks in pelvic girdle formation.
Materials and Methods
Mice
Intercrosses between Pbx1+/−(Selleri et al., 2001), Pbx2+/− (Selleri et al., 2004), Pbx3+/− (Rhee et al., 2004) and Emx2+/− (Pellegrini et al., 2001) mice were performed to obtain Pbx1+/−;Pbx2+/−, Pbx1+/−;Pbx3+/−, Pbx2+/−;Pbx3+/− and Pbx1+/−;Emx2+/− mutants. On a C57BL/6 background, the double heterozygous numbers obtained were below expected Mendelian ratios. To increase litter numbers and sizes, C57BL/6 double heterozygous males for each mutant line were crossed to a Black-Swiss strain [NIH-BL(S)]. Next, double heterozygous C57BL/6 females and mixed double heterozygous C57BL/6-Black-Swiss males were intercrossed and their progeny analyzed for skeletal phenotypes. On more out-crossed mixed genetic backgrounds, marked ameliorations of C57BL/6 pelvic skeletal and gene expression phenotypes were observed leading us to use only progeny from the first C57BL/6-to-Black-Swiss intercross in our in situ hybridization experiments. However, some mutants, obtained from parents that had been out-crossed to the Black Swiss strain more than once, were used in our analysis of skeletal phenotypes and are highlighted as such in the text. To generate more complex Pbx compound genotypes, Pbx1+/−;Pbx2+/− and Pbx1+/−;Pbx3+/- mutants were intercrossed to obtain triple Pbx1+/−;Pbx2+/−;Pbx3+/− mice. However, in utero lethality of Pbx triple heterozygotes prevented further intercrossing.
Skeletal preparations
Skeletons of E13.5–E14.5 mouse embryos were harvested according to standard protocols following IUACC guidelines, then dehydrated in ethanol, treated next with acetone, stained with Alcian Blue/Alizarin Red S and cleared using KOH and glycerol (McLeod, 1980; Depew et al., 1999). At least four embryos per genotype were analyzed.
Optical Projection Tomography
As described in detail (Sharpe et al., 2002; Sharpe, 2003), fixed specimens from skeletal preparation were washed in PBS and briefly rinsed with distilled H2O before being embedded in 1% LMP agarose in H2O. The agarose block was then trimmed to a pyramidal shape around the specimen, leaving about 5 mm on the base. The embedded specimens were dehydrated in 100% methanol (MetOH) (2×2–4 hours, 1× overnight) and cleared in BABB (1:2 mixture of Benzyl Alcohol and Benzyl Benzoate), overnight or until transparent, at room temperature and protected from light. The cleared specimens were glued by the base to a magnetic mount and placed into the OPT scanner. Light source and intensity, exposure, filters, position and focus were adjusted to maximize the dynamic range but avoiding saturation. Four hundred tiff images of each specimen were captured during a 360-degree single axis rotation.
Using the OPT Recon software (NRecon v1.6.2.0 ©Skyscan 2009) to correct alignment and signal intensity thresholds, the individual images were then reconstructed to produce a 3D voxel data-set of the specimen, which could be visualized with both OPT Viewer and Imaris software (BiOPTonics © Viewer V.1.6.1 and Imaris ×64 6.3.1 ©Bitplane AG). From these data, using the Imaris software, iso-surface 3D representations of the specimens were produced.
Section and whole mount in situ hybridization
For both section and whole mount in situ hybridization, harvested embryos were first somite-matched to control for developmental variations within each litter, then rinsed in PBS1X, fixed overnight at 4 C, and processed accordingly. As compound mutants from several distinct Pbx and Emx2 lines were acquired using multiple, different genetic crosses, each experiment was performed using specific littermate and somite-matched controls. For this reason, we have, for sake of scientific accuracy, displayed the proper controls for each experiment in the appropriate figures in this text. At least three mutant embryos per genotype per experimental assay were analyzed. For section in situ hybridization, embryos were embedded in Tissue Tek OCT compound, stored at –80 C, and sectioned at 10 µm. Section in situ hybridization was conducted as described (Di Giacomo et al., 2006). For whole mount in situ hybridization, embryos were passed through a methanol dehydration series, stored at – 20 C until their use in in situ protocols (see (Selleri et al., 2001). For both types of in situ hybridization experiments, digoxygenin-labeled RNA probes were generated as described (Di Giacomo et al., 2006) for the following probes: Alx1 (Kuijper et al., 2005), Alx4 (Beverdam and Meijlink, 2001), Col2a (Cheah et al., 1991), Emx2 (Pellegrini et al., 2001), Gli3 (Beverdam and Meijlink, 2001), Pax1 (Chalepakis et al., 1991), Pbx1 (Brendolan et al., 2005), Pbx2 (Selleri et al., 2004), Pbx3 (Di Giacomo et al., 2006), Pitx1 (Lanctot et al., 1999), Prrx1 (Beverdam and Meijlink, 2001), Scx (Brent and Tabin, 2004), Sox9 (Wright et al., 1995), Tbx15 (Kuijper et al., 2005) and Twist1 (Krawchuk et al., 2010).
Bioinformatic analysis
To assess the functionality of Emx2-Pbx1 binding events genome-wide by a bioinformatic approach, we utilized the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010). GREAT evaluates sets of cis-regulatory elements by assigning each element to its likely target gene(s). The annotations for each gene are then associated with each cis-element and statistical enrichment is assessed using two complementary statistical tests, which account for biases in the distribution of genes across the genome. We entered into GREAT 2,826 conserved instances of the Emx2-Pbx1 dimer motif in the mouse genome (mm9). We used default input parameters and default output display settings available at the GREAT website (url: http://great.stanford.edu/) to identify “hindlimb morphogenesis” and “cell fate specification” as the most significantly enriched Gene Ontology (GO) terms.
Results
Pbx1, Pbx2, and Emx2 are expressed in the mesodermal portion of the somatopleure and proximal hindlimb
Pelvic girdle formation begins approximately at the time of initial HL bud outgrowth (in mouse at E9.5–10), when cells of the mesodermal portion of the somatopleure are specified as pelvic progenitors (Malashichev et al., 2005; Malashichev et al., 2008). This process leads to the formation of a single mesenchymal mass (at around E11.5) that undergoes chondrogenesis (E12.5–E13.5) and osteogenesis (E14.5) to form each pelvic bone (Pomikal and Streicher, 2010). Previous studies established that Pbx1, Pbx2, and Emx2 are expressed in mesodermal tissues that undergo endochondral ossification (Pellegrini et al., 2001; Selleri et al., 2001; Capellini et al., 2006; Capellini et al., 2010). Additionally, Pbx1 and Pbx2, but not Pbx3, are co-expressed within the LPM of the posterior embryo together with Emx2 during early HL bud outgrowth (Fig. 1A; Pellegrini et al., 2001; Capellini et al., 2006). However, once HL buds elongate, Pbx1 remains localized proximally with Emx2 in the pelvic field within the mesodermal portion of the somatopleure and LPM, while Pbx2 becomes restricted distally (Fig. 1A; Capellini et al., 2006). Thus, at E9.5–10.5, Pbx1 is localized to the proximal HL and mesodermal portion of the somatopleure and Pbx2 is present in the distal HL bud, as revealed by immunohistochemistry (Fig. 1B; Capellini et al., 2006). At E11–11.5, while Pbx1, Pbx2, and Emx2 maintain the expression patterns described, Pbx3 transcripts emerge, albeit at low levels, in the mesodermal portion of the somatopleure (Fig. 1C). The expression patterns of Pbx1, Pbx2, and Emx2, which initially overlap in the mesodermal portion of the somatopleure and early HL bud, and then are partially co-expressed with Pbx3 in the proximal HL bud after E11, suggest roles in pelvic girdle formation.
Pbx1 and Pbx2 genetically interact to regulate pelvic development
We tested for cooperative roles of genes of the Pbx family in the patterning and morphogenesis of the pelvic girdle by generating Pbx1, Pbx2, and Pbx3 single and compound mutant genotypes. Morphological analyses by skeletal preparation and optical projection tomography (OPT) of single Pbx1−/− embryos reveal: (1) absence of ilia, or presence of truncated and flattened ilia (Fig. 2A; red asterisk in red inset) that are separated from the remaining pelvis as single rudimentary elements (Fig. 2A, red inset); (2) presence of pubic and ischial structures that are slightly shortened rostral-caudally and detached from each other along their caudal rami (Fig. 2A); and (3) failure of the hip joint to form as the femoral head remains fused to the acetabulum (Fig. 2A, blue arrowhead in red inset). OPT of these skeletons at E13.5 (Fig 2B) shows hypoplastic and detached ilia and pubic and ischial elements with some vestige of normal morphological identity and position.
The severity of these defects becomes more pronounced when a single allele of Pbx2 is removed from a Pbx1 null background. Of note, due to the lethality of double homozygous Pbx1;Pbx2 embryos at E10 (Capellini et al., 2006), pelvic development could not be examined. In E13.5 Pbx1−/−;Pbx2+/− embryos, the only remaining pelvic element appears as a small rudiment that exhibits some apparent morphological and positional similarity to an ischium, as highlighted by OPT (Fig. 2B; Supp. Movie S1). When Pbx1;Pbx2 double heterozygous mice (on a C57BL/6 genetic background) were crossed to an outbred Black Swiss strain more than once, a marked amelioration of the phenotype was observed in Pbx1−/−;Pbx2+/− progeny with the appearance of a small skeletal element of uncertain identity, approximately 180 degrees opposite to the forming ischium (Fig. 2A, blue arrowhead in lower red inset). Pbx1−/−;Pbx2+/− mutants also exhibit a truncated femur that is fused to a single remaining pelvic element. The malformations observed in these compound mutants were not observed in other, even complementary genotypes, such as Pbx1+/−;Pbx2+/−, Pbx1+/−;Pbx2−/− and Pbx2−/− embryos (Selleri et al., 2004), all of which exhibit normal pelvic girdles.
Similar experiments were performed to evaluate potential genetic interactions between Pbx1 and Pbx3, as well as Pbx2 and Pbx3, in pelvic development. No significant genetic interactions were identified in any of the obtained compound genotypes at E13.5. In Pbx1−/−;Pbx3+/− embryos the abnormalities and hypoplasia of the pelvic structures and of the femur appeared to closely resemble the defects of single Pbx1−/− mutants (Fig. 2A and red inset), as clearly revealed by OPT (Fig. 2B; Supp. Movie S2). It was also apparent that in Pbx1−/−;Pbx3+/− embryos, HL skeletal elements distal to the femur (which is thicker, shorter, and dysmorphic as in single Pbx1−/− mutants) appear normal (Fig. 2A). Since double homozygous Pbx1;Pbx3 and Pbx2;Pbx3 embryos die early in gestation, prior to HL development, pelvic development could not be assessed. In all other double compound genotypes (i.e., Pbx1+/−;Pbx3+/−, Pbx1+/−;Pbx3−/−, Pbx2+/−;Pbx3+/−, Pbx2−/−;Pbx3+/−, and Pbx2+/−;Pbx3−/−) as well as triple (i.e., Pbx1+/−;Pbx2+/−;Pbx3+/−) mutants, no pelvic phenotype was detected (data not shown).
Pbx1;Emx2 compound mutants exhibit abnormal pelvic skeletal structures
Given the overlapping expression patterns of Pbx1 and Emx2 in the mesodermal portion of the somatopleure and proximal HL, as well as their reported genetic and molecular interactions in scapular development (Capellini et al., 2010), pelvic structures were assessed in single and compound Pbx1 and Emx2 mutants.
As for single Emx2 mutants, it was confirmed that at E14.5 Emx2+/− heterozygotes lack skeletal defects (Fig. 3A) (Pellegrini et al., 2001) and Emx2−/− homozygotes exhibit either absent or reduced iliac blades that are separated from the caudal pelvic elements (Fig. 3A). This pelvic phenotype only partially resembles that of single Pbx1−/− mutants in the iliac defects, because Pbx1−/− mutants have additional abnormalities caudally (Fig. 3A). Pbx1+/−;Emx2+/− compound mutants, which displayed modest alterations in the scapular blade (Capellini et al., 2010), did not exhibit pelvic phenotypes (data not shown). Additionally, in Pbx1+/−;Emx2−/− embryos, the loss of one allele of Pbx1 on an Emx2 null background did not lead to either exacerbated or novel pelvic phenotypes compared to those observed in single Emx2−/− mutants (Fig. 3A). Indeed, in these compound mutants the ilium is reduced and detached from the other pelvic elements, while the pubis and ischium remain intact.
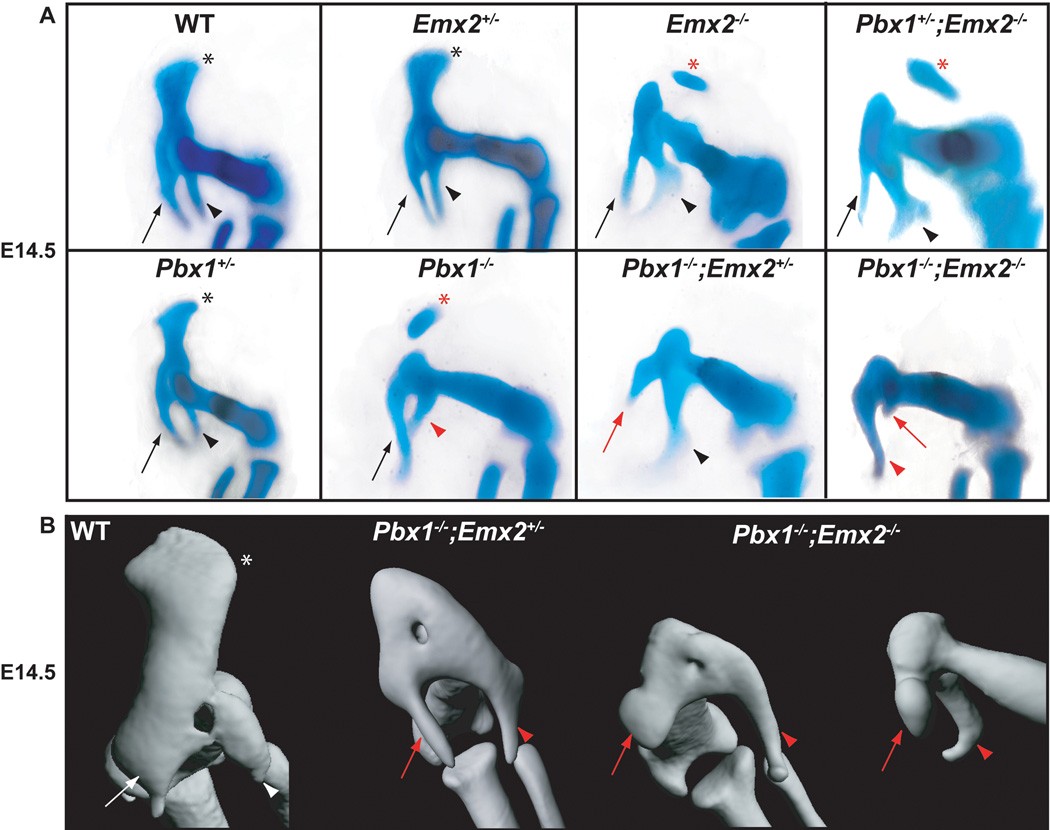
Fig. 3.
Pelvic skeletal phenotypes of single and compound Pbx1;Emx2 mutants at E14.5 as assessed using Alcian blue and Alizarin red staining and OPT. A: Compared to WT HL (left panel): single Pbx1+/− and Emx2+/− mutants display normal pelves (black symbols); single Emx2−/− and compound Pbx1+/−;Emx2−/− mutants phenocopy the rostral Pbx1−/− mutant pelvic morphologies with either absent or detached ilia (red asterisk); compound Pbx1−/−;Emx2+/− embryos additionally show modestly truncated pubis (red arrow); and compound Pbx1−/−;Emx2−/− embryos exhibit well formed ischia (red arrowhead), and truncated pubis (red arrow). B: OPT reveals that compared to WT (left panel; white symbols), and compound Pbx1−/−;Emx2+/− mutants, Pbx1−/−;Emx2−/− mutants show morphologically identifiable ischia and substantially reduced and rounded pubis (red arrows and arrowheads in both views, respectively).
Similar to the situation described for Pbx2, the reduction of functional Emx2 alleles on a Pbx1 null background leads to more severe pelvic malformations. At E14.5, Pbx1−/−;Emx2+/− mutants resemble single Pbx1−/− mutants in that all three pelvic elements are reduced; the ilium absent or detached; and the pubis and ischium partially truncated (Fig. 3A–B; Supp. Movie S3). However, compound Pbx1−/−;Emx2−/− mutants exhibit a more pronounced phenotype (Fig. 3A–B), with a single well-formed skeletal element, partly reminiscent of an ischium, and a smaller pubic rudiment (Fig. 3A, red arrow). OPT reveals a truncated, rounded pubis (Fig. 3B, red arrowheads; Supp. Movie S4). Also, the pelvis is fused to a truncated femur (Fig. 3B). Due to the lethality of compound Pbx1;Emx2 mutants and also Pbx1;Pbx2 mutants, it was not possible to produce triple Pbx1;Pbx2;Emx2 mutants to study their pelvic morphogenesis.
Genes of the Pbx family control Emx2 expression in the pelvic field
To understand the hierarchy in the control of pelvic development by genes of the Pbx family and Emx2, Pbx1, Pbx2, and Pbx3 expression was assessed in Emx2-deficient embryos, and Emx2 expression was evaluated in compound mutants deficient for different Pbx genes (Fig. 4). Gene expression was examined by whole mount in situ hybridization from E10 to E11.5, although only select gestational days are displayed here. In E10.5 Emx2−/− embryos, the only detectable down-regulation of Pbx genes was observed for Pbx1, whose mRNA was slightly reduced in the proximal-to-distal aspect of the HL bud (Fig. 4A). Pbx1 expression, however, persisted along the anterior-posterior boundary of the proximal flank (Fig. 4A). This modest spatial alteration of Pbx1 expression was not observed later at E11.5 (data not shown). In contrast, in both single Pbx1−/− mutants and in compound Pbx1−/−;Pbx3+/− mutants, Emx2 expression was reduced in the proximal flank and HL field (Fig. 4B). In Pbx1−/−;Pbx2+/− mutants, Emx2 expression was substantially down-regulated and nearly absent from domains of the proximal HL flank from E10 to E11.5 (Fig. 4B and data not shown), while being unperturbed in the more rostral flank domains (Fig. 4B).
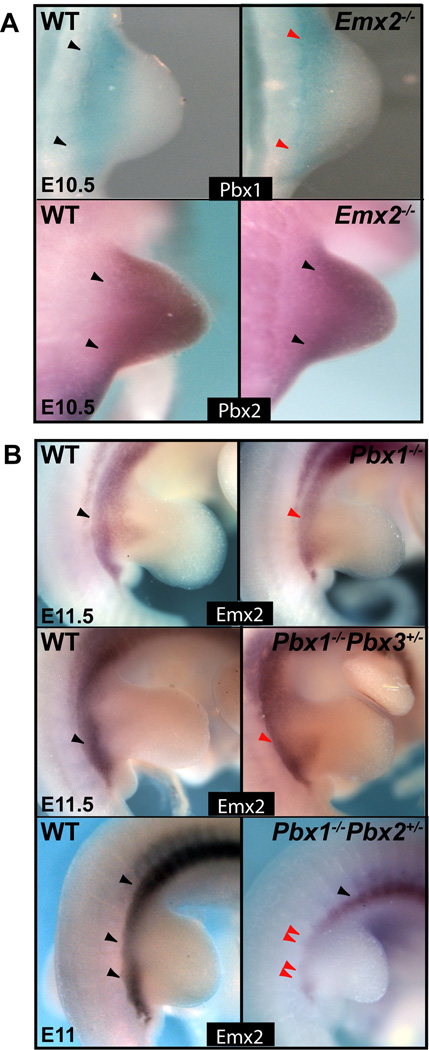
Fig. 4.
Expression of Pbx family members and Emx2 in respective single Emx2 homozygous and compound Pbx1-3 mutants via in situ hybridization. A: Compared to WT (black arrowhead): in E10.5 Emx2−/− mutants, Pbx1 expression (top) is modestly spatially disrupted in its proximal-to-distal domain, but remains in the proximal HL (red arrowhead); and Pbx2 expression (bottom) is unaffected (black arrowhead). B: Compared to WT (black arrowheads): at E11.5, Emx2 expression is slightly reduced in single Pbx1−/− and compound Pbx1−/−;Pbx3+/− mutant proximal HL (red arrowheads), but is severely reduced-to-absent in compound Pbx1−/−;Pbx2+/− mutant proximal HL (double red arrowheads).
Genes of the Pbx family and Emx2 act upstream of pelvic mesenchymal condensation and early skeletogenesis
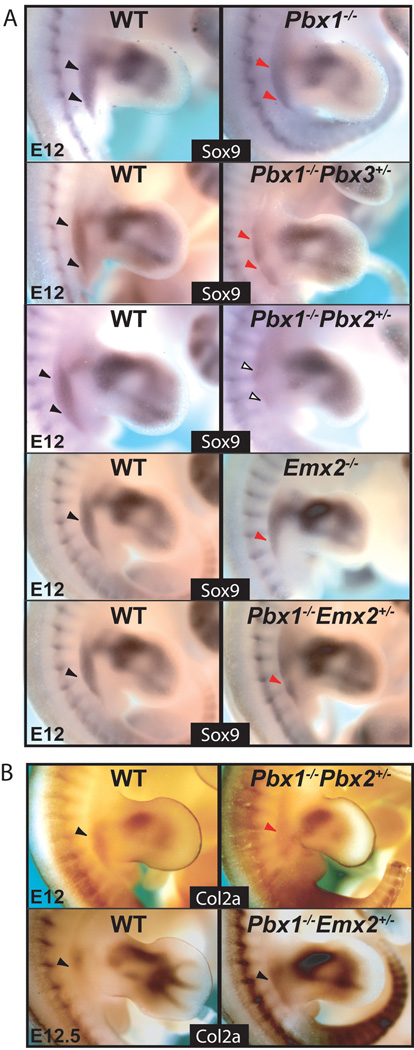
Given the observed skeletal defects in single Pbx1 and Emx2, as well as in select Pbx compound mutants and Pbx1;Emx2 mutants, gene expression analysis of mesenchymal markers was performed using whole mount in situ hybridization. For simplicity, hereafter, only those mutants that exhibit detectable changes in gene expression will be shown, while all other genotypes will be left for supplementary figures, or discussed as data not shown. Mesenchymal condensation and cartilage differentiation were evaluated by gene expression of Sox9 and Col2a, markers for both of these processes, respectively (Cheah et al., 1991; Wright et al., 1995). From E11–E12, Sox9 was reduced in single Pbx1 and Emx2 homozygous mutants (Fig. 5A). Likewise, Sox9 expression was down-regulated along the flank and proximal HL of Pbx1−/−;Pbx3+/−, Pbx1−/−;Emx2+/−, Pbx1+/−;Emx2−/−, and Pbx1−/−;Emx2−/− mutants (Fig. 5A; data not shown). Additionally, at E12, Col2a expression was unchanged or slightly reduced in most of these mutants compared to WT (data not shown). Intriguingly, in Pbx1−/−;Emx2+/− mutants, Col2a persisted along the flank and proximal HL, albeit slightly up-regulated. As expected, in complementary genotypes for these mutants, along with Pbx1+/−;Pbx2−/− embryos, Sox9 and Col2a were unchanged (data not shown). In E12 compound Pbx1−/−;Pbx2+/− mutants, which exhibit the most severe pelvic phenotypes, Sox9 was nearly absent from the pelvic field (Fig. 5A). A substantial down-regulation of Col2a at E12 was also detected in Pbx1−/−;Pbx2+/− mutants (Fig. 5B). These latter findings indicate that the pelvic abnormalities of Pbx1−/−;Pbx2+/− mutants may likely result from earlier perturbations of pelvic progenitor cell specification and patterning.
Fig. 5.
Expression of Sox9 and Col2a, markers of mesenchymal condensation and skeletal development, in single and compound Pbx family member and Pbx1;Emx2 mutants via in situ hybridization. A: Compared to WT (black arrowheads), at E12, in single Pbx1−/− and Emx2−/− mutants and compound Pbx1−/−;Pbx3+/− and Pbx1−/−;Emx2+/− mutants (red arrowheads), Sox9 expression is reduced, while in Pbx1−/−;Pbx2+/− mutants (empty arrowheads) it is nearly absent. B: Compared to WT (black arrowheads), in E12 Pbx1−/−;Pbx2+/− mutants (red arrowheads) Col2a expression is markedly reduced, while at E12.5, Col2a is modestly up-regulated in Pbx1−/−;Emx2+/− mutants (black arrowheads).
Pubis, ilium, and ischium marker gene expression is disrupted in Pbx compound mutants and in Pbx1;Emx2 mutants
Unlike for pectoral girdle development (Huang et al., 2006), it remains mostly unclear when and in which tissues the pelvic girdle is specified. For these reasons, we initially examined the expression of numerous genes whose individual or compound loss affects the formation of the entire pelvis or of one of its components. We performed these analyses at murine embryonic stages E10–11.5, when specification of the pelvic girdle is thought to occur based on comparable studies in chick (Malashichev et al., 2005; Malashichev et al., 2008). The genes analyzed by in situ hybridization comprised those whose loss most affects the pubis (Alx4, Alx1, Twist1) (Kuijper et al., 2005; Krawchuk et al., 2010); the pubic symphysis (Prrx1) (ten Berge et al., 1998); or the ilium (i.e., Pbx1, Emx2, Pitx1) (Lanctot et al., 1999; Pellegrini et al., 2001; Selleri et al., 2001). No known gene has yet been reported to specify uniquely the ischium (i.e., no single or compound homozygous mouse mutant exhibits a specific ischial reduction), although Pax1 is expressed in the early pelvic HL field and in the ischium cartilaginous rudiment (Timmons et al., 1994; LeClair et al., 1999).
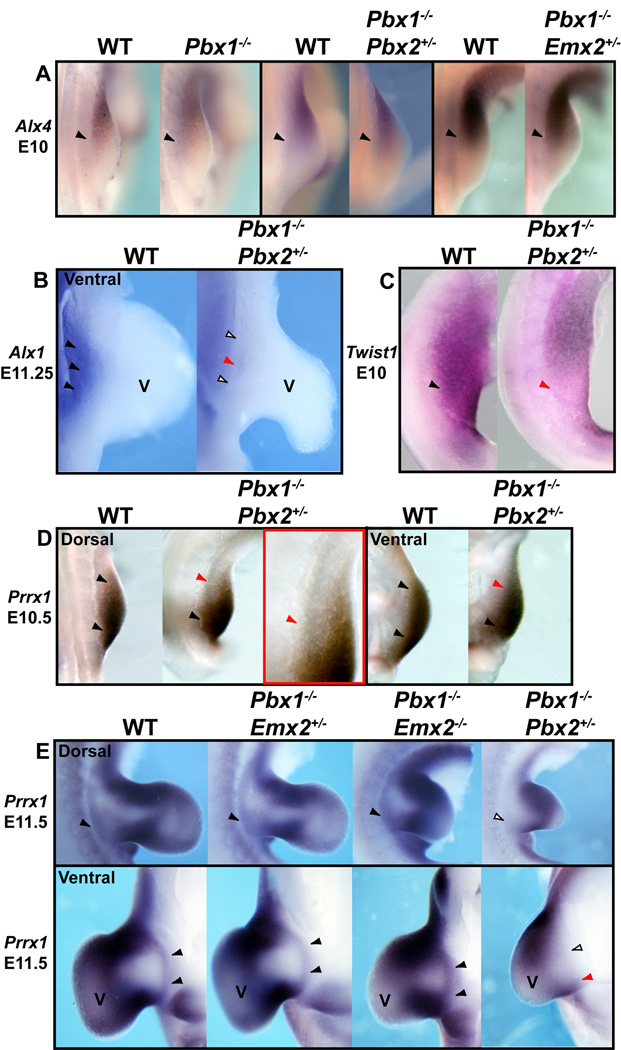
In E10 and E11.5 HL of all Pbx1;Pbx2, Pbx1;Pbx3, and Pbx1;Emx2 mutants, expression of Alx4, a gene whose loss results in pubic reduction (Kuijper et al., 2005), was unperturbed versus normal littermates (Fig. 6A, Supp. Fig. S1A, and data not shown). Next, expression of Alx1 (formerly Cart1), which aids in the specification of the pubis (Kuijper et al., 2005), was unperturbed in all compound mutants listed above, except in Pbx1−/−;Pbx2+/− embryos, in which it was lost from the ventral proximal somatopleure (Fig. 6B). Twist1, which contributes to pubic formation (Krawchuk et al., 2010), was found down-regulated only in the proximal posterior HL of E10 Pbx1−/−;Pbx2+/− embryos (Fig. 6C). Lastly, expression of Prrx1 (ten Berge et al., 1998) was reduced at E10.5 (Fig. 6D) and E11.5 (Fig. 6E; Supp. Fig. S1B) in both the dorsal and ventral domains of the proximal mesodermal portion of the somatopleure in Pbx1−/−;Pbx2+/− mutants, while it remained unperturbed in Pbx1−/−;Emx2−/− embryos (Fig. 6E; Supp. Fig. S1B).
Fig. 6.
Expression of genes involved in pubis development in single and compound Pbx family member and Pbx1;Emx2 mutants via in situ hybridization. A: Compared to WT (black arrowheads), at E10, expression of Alx4 remains unperturbed in single Pbx1−/− and compound Pbx1−/−;Pbx2+/− and Pbx1−/−;Emx2+/− mutants (black arrowheads). B: Compared to WT (black arrowhead), at E11.25, expression of Alx1 is absent or severely reduced from the ventral somatopleure of the proximal HL bud in Pbx1−/−;Pbx2+/− mutants (empty and red arrowheads). C: Compared to WT (black arrowhead), at E10, expression of Twist1 is reduced from the posterior margin of the HL bud in Pbx1−/−;Pbx2+/− mutants (red arrowhead). D: Compared to WT at E10.5 (black arrowheads), expression of Prrx1 is reduced from the anterior margin of the dorsal and ventral somatopleuric domains of the proximal HL bud in Pbx1−/−;Pbx2+/− mutants (red arrowheads). High magnification red inset shows the proximal anterior HL reduction in Prrx1 expression in Pbx1−/−;Pbx2+/− mutants (red arrowhead). E: Compared to WT and all other mutants shown (black arrowheads), at E11.5, expression of Prrx1 is absent or significantly reduced from the dorsal (top) and ventral (bottom) somatopleure of the proximal HL bud in Pbx1−/−;Pbx2+/− mutants (empty and red arrowheads). Abbreviation: V, ventral.
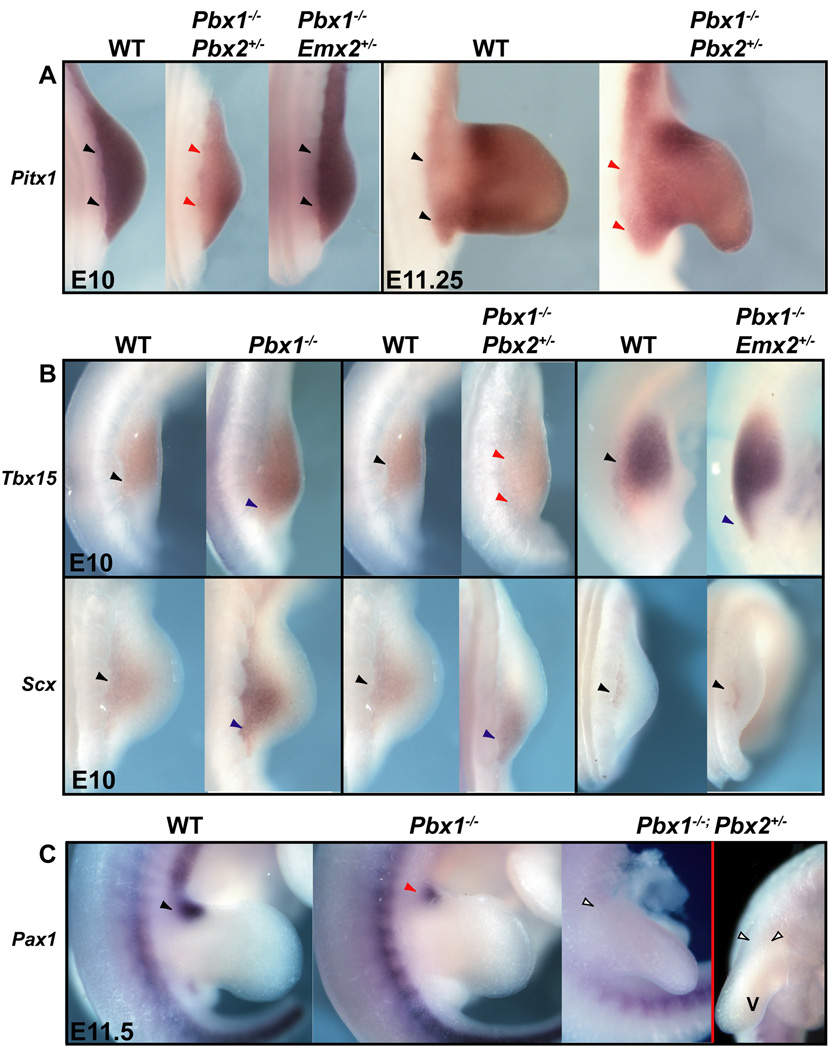
We also examined expression of ilium markers such as Pbx1, Emx2, and Pitx1 in our mutants (Lanctot et al., 1999; Pellegrini et al., 2001; Selleri et al., 2001). As mentioned, we observed a significant reduction of Emx2 expression in select Pbx compound mutants and only modest changes of Pbx1 expression in single Emx2 homozygous mutants (Fig. 4). In compound Pbx1−/−;Pbx2+/− mutants only, and not in Pbx1;Emx2 genotypes, Pitx1 expression was significantly reduced in the pre-pelvic and proximal HL field (Fig. 7A). We also detected Pitx1 down-regulation at later stages in Pbx1;Pbx2 mutants with the most severe pelvic phenotypes, but not in all other genotypes (Fig. 7A and Supp. Fig. S1C).
Fig. 7.
Expression of genes involved in ilium specification and hindlimb development in single and compound Pbx family member and Pbx1;Emx2 mutants via in situ hybridization. A: Compared to WT (black arrowheads), at E10, expression of Pitx1 is markedly reduced in its proximal domain but present in its distal limb domain in Pbx1−/−;Pbx2+/− mutants only (red arrowheads). At E11.25, Pitx1 expression is modestly reduced from the proximal HL of this same mutant genotype (red arrowheads). B: (top) Compared to WT (black arrowheads), at E10, expression of Tbx15 is posteriorly expanded in single Pbx1−/− and compound Pbx1−/−;Emx2+/− mutants (blue arrowheads) and greatly posteriorly expanded and reduced in Pbx1−/−;Pbx2+/− mutants (red arrowheads). (bottom) Compared to WT (black arrowheads), at E10, expression of Scx, a tendon marker, is slightly posteriorly expanded in single Pbx1−/− mutants (blue arrowhead), but markedly posteriorly shifted in Pbx1−/−;Pbx2+/− mutants (red arrowheads), and unchanged in Pbx1−/−;Emx2+/− mutants (black arrowheads). C: Compared to WT (black arrowhead), at E11.5, expression of Pax1, an ischium marker, is markedly reduced in single Pbx1−/− mutants (red arrowheads) and absent in Pbx1−/−;Pbx2+/− HL (empty arrowheads in left dorsal view and right superior-anterior view). Proximal is to the left in all panels. Abbreviation: V, ventral.
We next assessed whether overall posterior proximal HL development is markedly altered prior to limb outgrowth in compound mutants for different Pbx genes and in Pbx1;Emx2 mutants. Tbx15, a marker of pre-skeletogenic mesenchyme in the early mouse limb (Singh et al., 2005), with known roles in human pelvic defects (Lausch et al., 2008), was expanded posteriorly in E10.5 HL buds of all compound mutants bearing two Pbx1 mutant alleles (Fig. 7B). Additionally, in compound Pbx1−/−;Pbx2+/− HL buds, Tbx15 expression was substantially reduced (Fig. 7B). Later, at E11.5, a posterior expansion was also observed in Pbx1−/−;Pbx2+/− embryos, while no changes of Tbx15 expression were observed in other mutant genotypes compared to controls (Supp. Fig. S2A). The degree of abnormal specification and cellular disorganization in pelvic domains of single Pbx1−/− and compound Pbx1−/−;Pbx2+/− mutants was further highlighted by the posteriorization of transcripts for Scleraxis (Scx), a tendon marker (Perez et al., 2003) in E10–11.5 HL buds (Fig. 7B; Supp. Fig. S2B). Abnormal Scx expression was not detected in other mutants (Supp. Fig. S2B).
Lastly, Pax1, a marker for ischium condensation and pelvic domains (Timmons et al., 1994; LeClair et al., 1999), was down-regulated in E11.5 single Pbx1−/− embryos, as well as in other mutants lacking both Pbx1 alleles, such as Pbx1−/−;Pbx3+/−, Pbx1−/−;Emx2+/− and Pbx1−/−;Emx2−/− embryos (Fig. 7C; Supp. Fig. S3; and data not shown). In Pbx1−/−;Pbx2+/− mutants, Pax1 was completely absent from the proximal HL bud (Fig. 7C). Importantly, no detectable down-regulation of Pax1 was observed in any complementary genotypes, including single Emx2−/− mutants (Supp. Fig. S3).
Identification by bioinformatic GREAT analysis of Emx2-Pbx1 TF binding site enrichments in conserved, non-coding regions of the mouse genome
Given the remarkable phenotypes of Pbx compound mutants and Pbx1;Emx2 mutants, we analyzed whether Pbx1 and Emx2 can regulate, as heterodimers, genes involved in pelvic and proximal HL development. We previously determined the optimal DNA-binding consensus sequence of the Emx2-Pbx1 heterodimer (EP site; Capellini et al., 2010) and identified in vivo an Emx2-Pbx1 binding site within the Alx1 locus.
Here, based on those sequences, we generated a position weight matrix (PWM) (Supp. Fig. S4) to search for predicted Emx2-Pbx1 binding sites across a genome-wide multiple alignment of 32 eutherian mammals. By this approach, we identified 2,826 conserved Emx2-Pbx1 heterodimer binding sites in the mm9 version (July/2007) of the mouse genome (data not shown). We next used the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010) to assess functional enrichments of these predicted binding sites within the mouse genome (see Materials and Methods).
The top hits obtained from the analysis by GREAT were further examined for their Gene Ontology (GO) Biological Process annotations, revealing that our predicted Emx2-Pbx1 binding sites are enriched near genes involved in “HL morphogenesis” (Table 1). Indeed, of the 33 genes annotated by GO as “HL morphogenesis” genes, 16 have instances of the Emx2-Pbx1 binding motif in their regulatory domains. Significant enrichment was also observed for sites present near genes involved in cell-fate specification (Table 1), according to GO.
TABLE 1.
Results of the analysis by great*
| Term | Binomial P-value |
Fold enrichment |
Genes with annotation |
Genes hit |
Number of binding sites |
|---|---|---|---|---|---|
| Hindlimb morphogenesis | 7.3e-5 | 2.3 | 33 | 16 | 29 |
| Cell fate determination | 9.6e-5 | 2.1 | 30 | 15 | 33 |
| Embryonic hindlimb morphogenesis | 1.9e-4 | 2.3 | 25 | 12 | 24 |
In silico identification of potential Emx2-Pbx1 regulated enhancers near MGI-annotated genes involved in pelvic development
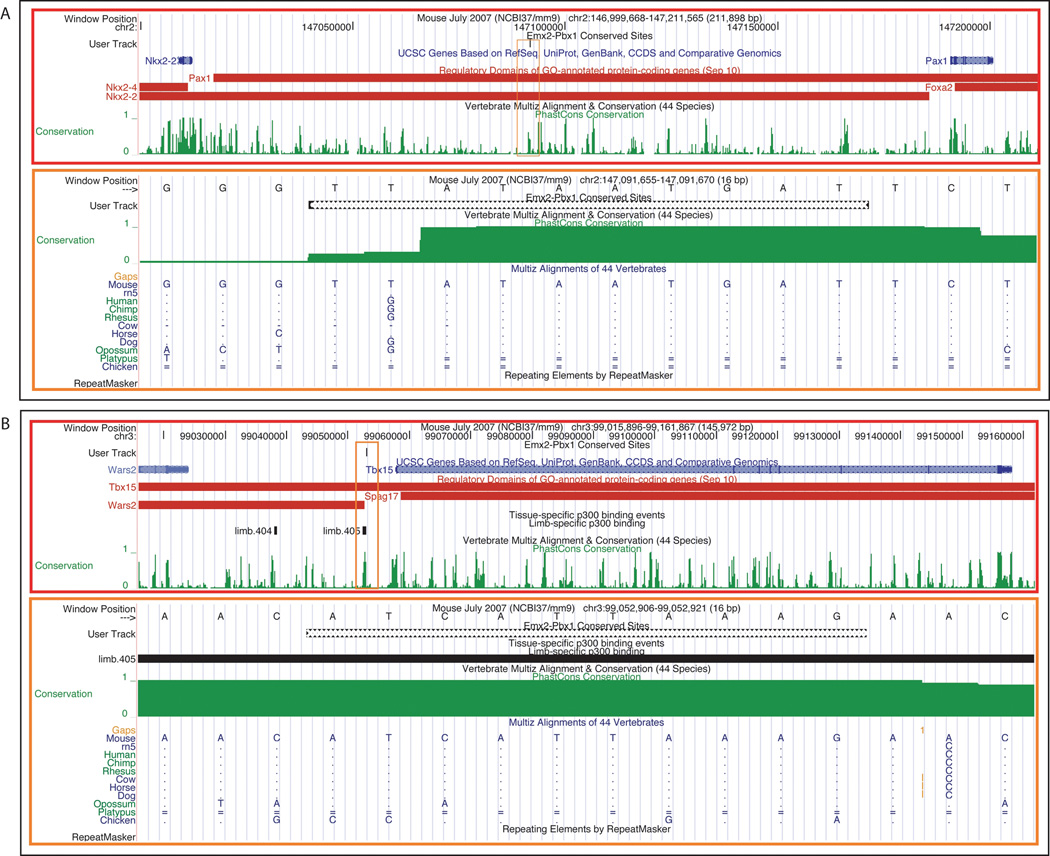
We next examined all of the conserved genome-wide Emx2-Pbx1 predictions to verify their occurrence near genes expressed and/or involved in pelvic development. To accomplish this goal, we extracted from the MGI database (http://www.informatics.jax.org/) all genes whose expression or phenotype matched the search term “pelvic girdle”. We then inspected approximately 200 Kb upstream and downstream of the transcriptional start site (TSS) of these genes for the presence of highly conserved Emx2-Pbx1 heterodimeric binding sites, using the UCSC genome browser (Kent et al., 2002). Of the 2,826 predicted sites, we found 53 sites located near 43 “pelvic girdle” genes (Table 2). These findings indicate that the identified conserved regions may indeed represent functional Emx2-Pbx1 regulatory elements for the nearby genes. Two specific examples are shown in Figure 8. It is of note that one site, approximately 5 Kb upstream of Tbx15, overlaps with a previously published binding site for the p300 co-activator identified through p300 ChIP-Seq analyses in limb tissue at E11.5 (Visel et al., 2009) (Fig. 8).
TABLE 2.
Emx2-Pbx1 dimerization motifs within 200 Kb of pelvic genes
| Gene | Chr. | Motif Start | Motif End | Strand | To TSS (bp) |
MGI Term: TS: Description |
|---|---|---|---|---|---|---|
| Alx4 | chr2 | 93642898 | 93642908 | + | +160308 | Phenotype: small pubis |
| C77370 | chrX | 101551207 | 101551217 | − | −154752 | Expression: TS23: embryo;skeleton;pelvic girdle |
| C77370 | chrX | 101552028 | 101552038 | − | −155573 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Cd59a | chr2 | 103962170 | 103962180 | − | +26161 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Cdkn3 | chr14 | 47220880 | 47220890 | + | −159363 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Chrdl1 | chrX | 139677009 | 139677019 | − | +151708 | Expression: TS21: embryo;skeleton;pelvic girdle |
| Chrdl1 | chrX | 139837753 | 139837763 | − | −9027 | Expression: TS21: embryo;skeleton;pelvic girdle |
| Chst8 | chr7 | 35475321 | 35475331 | + | +122364 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Col11a1 | chr3 | 113834894 | 113834904 | − | +101437 | Expression: TS23: vertebral muscle; pelvic girdle |
| Col9a1 | chr1 | 24209022 | 24209032 | + | +24484 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Copb1 | chr7 | 121280770 | 121280780 | + | +117414 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Ctgf | chr10 | 24132107 | 24132117 | − | −183196 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Ctgf | chr10 | 24156973 | 24156983 | − | −158330 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Dpyd | chr3 | 118088581 | 118088591 | + | −176505 | Expression: TS23: vertebral muscle; pelvic girdle |
| Ext1 | chr15 | 53028807 | 53028817 | − | +148921 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Fat3 | chr9 | 16135872 | 16135882 | − | +46793 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Fat3 | chr9 | 16268418 | 16268428 | + | −85744 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Foxi1 | chr11 | 34014004 | 34014014 | − | +94075 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Foxi1 | chr11 | 34032581 | 34032591 | − | +75498 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Foxp1 | chr6 | 98976797 | 98976807 | + | +136216 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Gli3 | chr13 | 15508916 | 15508926 | − | −46630 | Phenotype: abnormal pubis morphology |
| Gli3 | chr13 | 15708609 | 15708619 | − | +153054 | Phenotype: abnormal pubis morphology |
| Hhip | chr8 | 82763655 | 82763665 | + | −181764 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Hmga2 | chr10 | 119881607 | 119881617 | + | +32374 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Hpse | chr5 | 101180712 | 101180722 | − | −32011 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Irx5 | chr8 | 94916857 | 94916867 | − | +35163 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Itpr3 | chr17 | 27275740 | 27275750 | + | +81260 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Kcnd2 | chr6 | 21194367 | 21194377 | + | +28259 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Lmx1b | chr2 | 33653044 | 33653054 | − | −157014 | Phenotype: abnormal ilium morphology |
| Magi1 | chr6 | 94286552 | 94286562 | + | −52655 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Ndufv2 | chr17 | 66388461 | 66388471 | − | +62360 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Ndufv2 | chr17 | 66392826 | 66392836 | − | +57995 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Nrf1 | chr6 | 29954775 | 29954785 | − | −43203 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Pax1 | chr2 | 147091657 | 147091667 | + | −99086 | Expression: TS21: embryo;skeleton;pelvic girdle |
| Pbx1 | chr1 | 170333299 | 170333309 | − | +29080 | Phenotype: abnormal pelvic girdle morphology |
| Pik3c3 | chr18 | 30314187 | 30314197 | − | −118353 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Pik3c3 | chr18 | 30542253 | 30542263 | + | +109704 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Plekhb1 | chr7 | 107780622 | 107780632 | + | +30247 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Prrx1 | chr1 | 165245123 | 165245133 | − | −1343 | Phenotype: abnormal pubis morphology |
| Ptch1 | chr13 | 63555761 | 63555771 | − | +111057 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Rasl11b | chr5 | 74572329 | 74572339 | + | −19012 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Rg9mtd2 | chr3 | 137794428 | 137794438 | + | −12064 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Runx2 | chr17 | 45008461 | 45008471 | + | −56886 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Satb2 | chr1 | 56955591 | 56955601 | + | +72577 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Tbx15 | chr3 | 98992619 | 98992629 | − | −65054 | Phenotype: abnormal pelvic girdle morphology |
| Tbx15* | chr3 | 99052908 | 99052918 | − | −4765 | Phenotype: abnormal pelvic girdle morphology |
| Tbx4 | chr11 | 85760419 | 85760429 | + | +56855 | Phenotype: abnormal pelvic girdle morphology |
| Tnmd | chrX | 130290990 | 130291000 | − | −94547 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Trp63 | chr16 | 25783048 | 25783058 | + | −18970 | Phenotype: abnormal pelvic girdle morphology |
| Trps1 | chr15 | 50674966 | 50674976 | − | +46611 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Unc5c | chr3 | 141241263 | 141241273 | − | +112736 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Unc5c | chr3 | 141303308 | 141303318 | − | +174781 | Expression: TS23: embryo;skeleton;pelvic girdle |
| Unc5cl | chr17 | 48752190 | 48752200 | − | +157943 | Expression: TS23: embryo;skeleton;pelvic girdle |
This specific motif overlaps a previously identified p300 limb element at E11.5 in mouse (Visel et al., 2009).
Fig. 8.
Bioinformatic analysis of two putative enhancers near Pax1 and Tbx15. A: (top – red box) UCSC browser window depicting the genomic location of Pax1 (right) in the mouse genome (mm9) relative to known sequence conservation across forty-four vertebrates (green peaks). (bottom – orange box) Zoom-in UCSC browser view of the specific location of the Emx2-Pbx1 heterodimeric TF prediction, showing sequence conservation to opossum. A dot in the alignment denotes an identical base to the reference species. B: (top – red box) UCSC browser window depicting the genomic location of Tbx15 in the mouse (mm9) genome relative to known sequence conservation across forty-four vertebrates (green peaks). (bottom – orange box) Zoom-in UCSC browser view of the specific location of the Emx2-Pbx1 heterodimeric TF prediction, showing conservation to opossum, and incomplete sequence conservation to chicken. This specific motif overlaps with a predicted P300 limb enhancer identified at E11.5 in mouse embryos (limb.405 line, above; horizontal black line, below) (see text for details). Red bars represent GREAT calculated regulatory domains for the genes listed to the left.
Discussion
Among Pbx genes, Pbx1 plays a primary role in pelvic development
In this study, we examined pelvic development using compound Pbx and Pbx1;Emx2 mutant mice. In all compound Pbx mutants bearing two Pbx1 null alleles, save compound Pbx1−/−;Pbx2+/− mutants whose phenotype is strikingly severe, the ilium is absent or reduced and only minor malformations of the pubis and ischium are present (Fig. 9). In compound Pbx1−/−;Emx2+/− mutants, ilium hypoplasia is likely due to the loss of Pbx1 function, since single Emx2+/− and compound Pbx1+/−;Emx2+/− mutants are normal. In complementary Pbx compound mutant genotypes, such as Pbx1+/−;Pbx2−/−, Pbx1+/−;Pbx3−/−, and triple Pbx1+/−;Pbx2+/−;Pbx3+/−, pelvic skeletal phenotypes are absent. All of these findings not only establish that Pbx1 plays a paramount role in pelvic formation, but also that, overall, Pbx1-2-3 gene dosage does not additively regulate pelvic girdle development. Importantly, Pbx1 dominant role over its family members has also been reported for the development of limb (Capellini et al., 2006), scapula (Capellini et al., 2010), and axial (Capellini et al., 2008) skeletons.
Fig. 9.
Summary and model of Pbx family member and Emx2 genetic functions during pelvic development. A: Illustrations depicting the phenotypes of the compound Pbx family member and Pbx1;Emx2 mutants analyzed in this study (see text for details). B: Model of Pbx family member and Emx2 genetic functions during pelvic girdle formation. (center panel) An E15.5 WT pelvis demonstrating the three distinct bony elements in color (blue – ilium, red – pubis, green – ischium). These colors reflect the genes involved in the formation of each structure, as depicted in the left and right panels. (left panel) Pbx1/Pbx2 hierarchically regulate Pitx1 and Emx2 during the specification and condensation of the ilium (blue), respectively. Dashed arrow indicates a potential control of Sox9 directly by Pbx family members. (right panel) Pbx1/Pbx2 hierarchically regulate genes involved in pubis development (red), such as Alx1, Prrx1, and Twist1, and ischial field expression (dashed green arrow), such as Pax1.
Pbx family members control Emx2 expression in the mesodermal portion of the somatopleure. Collaboratively, Pbx genes and Emx2 direct ilium formation by modulating Sox9 expression and mesenchymal cell condensation
Analyses performed in chick and mouse have illustrated that the skeletal elements of each pelvic bone form only from the somatopleure (Chevallier et al., 1977; Malashichev et al., 2008). Additional studies have demonstrated that the three adjoining pelvic elements (i.e., ilium, pubis, and ischium) arise from one single mesenchymal mass, which then undergoes chondrogenesis and osteogenesis in separate centers (Pomikal and Streicher, 2010). In this context, it has also been reported that: (1) Sox9 serves as a marker for mesenchymal condensations of the pectoral and pelvic girdles (Malashichev et al., 2005; Wang et al., 2005; Malashichev et al., 2008); and (2) Sox9 haploinsufficient mutant mice exhibit defects in most skeletal elements, including the pelvis, which is mostly affected in the ilium (Bi et al., 2001). Lastly, Emx2 expression has been shown prior to Sox9 activation in the developing girdles (Malashichev et al., 2005; Malashichev et al., 2008; Capellini et al., 2010), and may instruct programs for the establishment of the iliac segment of the pelvic mesenchymal condensation (Malashichev et al., 2008).
In single Pbx1 and Emx2 homozygous mutants, the marked down-regulation of Sox9 in the pelvic field (Fig. 5) precedes the reduction in ilium size (Fig. 9). Additionally, other mutants such as Pbx1−/−;Pbx3+/−, Pbx1−/−;Emx2+/−, and Pbx1+/−;Emx2−/−, exhibit corresponding reductions in Sox9 expression and ilium size. Strikingly, Sox9 expression is virtually absent in Pbx1−/−;Pbx2+/− mutants that lack most of the pelvis. Thus, Pbx1-2-3 and Emx2 act upstream of Sox9 in the posterior mesodermal portion of the somatopleure (Fig. 9B) and likely govern ilium formation, at least in part, through control of Sox9.
Importantly, we also observed Emx2 down-regulation in single Pbx1−/− and compound Pbx1−/−;Pbx3+/− mutants and a more drastic reduction of Emx2 expression in Pbx1−/−;Pbx2+/− mutants, indicating that Pbx family members control Emx2 in pelvic formation (Fig. 9B). Therefore, ilium hypoplasia in Pbx compound mutants is mediated, at least in part, through their genetic control of Emx2 in the pelvic field, although concomitant regulation of Sox9 expression by Pbx family members cannot be excluded. Likewise, the reduction of the ilium in single Emx2 homozygotes, as well as in compound Pbx1+/−;Emx2−/− and Pbx1−/−;Emx2−/− mutants, is likely due to down-regulation of Sox9 as a result of Emx2 loss (Fig. 9B). Similar hierarchical relationships among Pbx1-2-3, Emx2, and Sox9 genes were observed in pectoral somatopleuric mesenchyme (Capellini et al., 2010).
Pbx family members control ilium patterning genes Pitx1 and Tbx15 in early proximal HL bud, supporting early specification for pelvic girdle progenitors
While it has been shown that the pelvic girdle forms from a single mesenchymal condensation (Pomikal and Streicher, 2010), little is known about the primary cues that specify progenitor somatopleure cells to form one single mass and then separate into three distinct pelvic bones. In chick, signals from the paraxial mesoderm (Malashichev et al., 2008) and overlying ectoderm (Malashichev et al., 2005) are important in triggering mesodermal portion of the somatopleure gene expression and specifying the pelvic rudiments during initial HL outgrowth (Malashichev et al., 2008). Additionally, genes expressed in the mesodermal portion of the somatopleure that likely specify all three pelvic bones respond differentially to signals provided by the ectoderm (Malashichev et al., 2005). If the findings in the chick can be applied to murine pelvic girdle development, then these patterning and morphogenetic events likely occur at early stages of mouse embryogenesis during initial HL bud development (Malashichev et al., 2005).
In homozygous Pitx1 mutants, as well as in compound Pitx1;Pitx2 mutants (Lanctot et al., 1999; Marcil et al., 2003), the ilium is either missing or detached from the remaining girdle. We observed strikingly similar defects in Pbx compound and Pbx1;Emx2 mutants. Importantly, in Pbx1−/−;Pbx2+/− mutants, we also detected down-regulation of Pitx1 (Fig. 7) in the early proximal HL bud (e.g. E10–10.5), while Pitx1 reduction was less severe distally and at later stages when condensations form (E11.25; Fig. 7; and Supp. Fig. S1C). These findings suggest that ilium loss in Pbx1−/−;Pbx2+/− mutants is at least partially mediated by down-regulation of Pitx1. Given the persistence of Pitx1 distal expression in the limb proper of E10.5–11.5 Pbx1−/−;Pbx2+/− mutants, we contemplate the possibility that Pbx family members may control Pitx1 in the proximal mesodermal portion of the somatopleure (i.e., the pelvic progenitor field only), wherein both Pbx1 and Pbx2 expression patterns overlap (Fig. 9). These data also suggest that the timing of specification for the pelvic girdle likely takes place prior to, or at, early HL bud outgrowth, as in the chick, and support the hypothesis of an early specification model for pelvic progenitors (Dudley et al., 2002; Malashichev et al., 2005).
While Tbx15 homozygous mutant mice do not exhibit pelvic skeletal defects, human patients suffering from Cousin Syndrome bear TBX15 mutations and have dysmorphic and reduced iliac blades (Lausch et al., 2008). Notably, we observed a substantial down-regulation and spatial perturbation of Tbx15 expression in E10.5 Pbx1−/−;Pbx2+/− mutant proximal HL fields (Fig. 7). These changes are also present, but not as severe, in genotypes with homozygous loss of Pbx1 alone, (Fig. 7) placing Pbx family genes upstream of Tbx15 and supporting a model whereby the early proximal HL field may be incapable of specifying ilium progenitors in Pbx mutants.
Pbx family members control pubic (Twist1, Alx1, and Prrx1) and ischial (Pax1) genes in early proximal HL bud
Only in Pbx1−/−;Pbx2+/− mutants, which exhibit complete pubic loss, besides perturbation of genes involved in specifying the ilium, we also observed down-regulation of Twist1, Alx1, and Prrx1 at E10.5. Twist1 functions during pelvic (and limb) development have been revealed (Bialek et al., 2004; Krawchuk et al., 2010), as reducing Twist1 activity yields pubis loss (Krawchuk et al., 2010). In Pbx1−/−;Pbx2+/− mutants, we observed that Twist1 expression is partially down-regulated in the posterior HL. In Pbx compound mutants (Fig. 6 and Supp. Fig. S1), we also detected marked reduction of expression of Alx1, but not Alx4, which participate in pubic development (Kuijper et al., 2005). Alx1 down-regulation is intriguing, since Twist1 is also known to act upstream of Aristaless family genes, including Alx4, during limb development (Loebel et al., 2002; O'Rourke et al., 2002). However, while reductions in Alx4 expression were not identified in Pbx1−/−;Pbx2+/− mutants, it has to be noted that Twist1 is still normally expressed in the proximal anterior domain wherein Alx4 is actively transcribed.
In Pbx1−/−;Pbx2+/− mutants, we also observed significant down-regulation of Prrx1, a gene involved in the development of the pubic symphysis (ten Berge et al., 1998)(Fig. 6D–E). These results suggest that either Pbx genes regulate Prrx1 expression directly (see below), or that perturbation of as yet unknown Pbx-dependent upstream pathways result in Prrx1 loss (Fig. 9). Since compound Pbx1−/−;Pbx2+/− mutants die at E13.5, prior to symphysis formation, conditional gene targeting will help clarify whether ablation of Pbx genes in the pubic field results in abnormal symphyseal development.
Lastly, in all mutants analyzed, including the Pbx1−/−;Pbx2+/− genotype that is most severely affected, a rudimentary ischium remains, although it is significantly truncated in the latter genotype. We have found that the expression of Pax1, which is expressed in the proximal HL field and later in the ischium condensation (Timmons et al., 1994; LeClair et al., 1999), is reduced early in all mutants lacking two copies of Pbx1 and is absent in compound Pbx1−/−;Pbx2+/− mutants. However, Pax1 absence cannot explain ischium reduction in these mutants, since single Pax1 homozygotes or compound Pax1;Pax9 homozygous mutant mice lack ischial abnormalities (Peters et al., 1999). Ischium reduction in Pbx1−/−;Pbx2+/− mice may alternatively result from Sox9 down-regulation and reduced mesenchymal formation in these mutants, or from disruption of earlier, as yet unknown, pathways (see below).
Pbx family members are prime regulators of pelvic development
Our results establish that Pbx family members are critical upstream regulators of multiple genes involved in the specification, patterning, and morphogenesis of the murine pelvic girdle. We have previously demonstrated that Pbx homeoproteins control cell fate specification and morphogenesis of the HL proper (Capellini et al., 2006), the axial skeleton (Capellini et al., 2008), the pectoral girdle (Capellini et al., 2010), and the splenic anlage (Brendolan et al., 2005), among other organs (Moens and Selleri, 2006). In the pectoral girdle, we presented evidence that Pbx1 and Emx2 are bound in vivo to a putative regulatory region of Alx1, a scapular blade effector gene, and drive transcriptional activation of a luciferase reporter in cultured cells. Likewise, it will be important to evaluate if cell fates of specific progenitors for the ilium, ischium, and pubis are also controlled directly by Pbx homeoproteins within the early mesodermal portion of the somatopleure or during mesenchymal condensation.
Pbx roles in developmental programs as cell fate specification have long been considered primarily as those of Hox cofactors (reviewed in Moens and Selleri, 2006). However, misexpression or loss of Hox genes result in mild alterations of the pelvic girdle, including defects in mice misexpressing Hoxd12 in lateral plate derivatives (Knezevic et al., 1997); modest malformations of pelvic bones and sacrum in Hoxc10 mutants (Hostikka et al., 2009); and lack of uterosacral ligaments in Hoxa11 mutant mice (Connell et al., 2008). Therefore, it is unlikely that Pbx homeoproteins effect their roles in pelvis formation solely as Hox cofactors, suggesting instead cooperation with other proteins, e.g. Emx2.
Using GREAT (McLean et al., 2010), a new bioinformatics tool, we found that potential Emx2-Pbx1 regulated enhancers, defined by the presence of strongly conserved Emx2-Pbx1 binding sites, are more often enriched in the vicinity of genes involved in HL morphogenesis and cell fate specification than predicted by chance. Furthermore, we identified specific Emx2-Pbx1 dimerization motifs in well conserved non-coding sequences near (at least) three genes involved in ischium, pubis, and ilium development (i.e., Pax1, Prrx1, and Tbx15, respectively). Based on their conservation and close proximity to nearby genes, these sequences may represent functional regulatory elements. For the ischium, we identified the presence of a strongly conserved Emx2-Pbx1 dimerization site approximately 99kb from the TSS of Pax1 (Fig. 8A), which is down-regulated in compound Pbx1;Emx2 mutants and lost in Pbx1;Pbx2 mutants (Fig. 7). For Prrx1, a gene patterning the pubis, we identified a motif conserved to Medaka, inside a block of approximately 230 bp of sequence homology within 1.5 kb of Prrx1 TSS (data not shown). Of note, Prrx1 expression is also strikingly reduced in E10.5–11.5 Pbx1;Pbx2 mutants (Fig. 6), which have no pubis. Finally, our prediction for a functional enhancer of Tbx15, a gene involved in ilium formation in humans, is of exceptional interest, not only considering its close proximity to the Tbx15 TSS (~5 kb) (Fig. 8B), but also given its overlap with a separately identified target of p300 in limb cells at E11.5 (Visel et al., 2009). Independently, we observed down-regulation and spatial alteration of Tbx15 in vivo, in E10.5–11.5 Pbx1;Pbx2 and Pbx1;Emx2 compound mutants (Fig. 7) that lack the ilium. While these in silico findings provide strong associations, future experiments will be required to verify transcriptional regulation by Pbx and Emx2 homeoproteins via these potential enhancers identified in silico (Table 2). Among these potential regulatory elements, some may control cell fate specification and patterning of specific pelvic structures such as the pubis, ilium, and ischium in development.
In summary, our studies reveal that Pbx family members genetically interact and that concomitantly Pbx1 interacts with Emx2 to control pelvic development in mammalian embryogenesis. To perform these functions, Pbx genes regulate effector genes involved in the specification and development of the ilium (Pitx1 and Emx2), ischium (Pax1), and pubis (Alx1, Twist1, Prrx1), as well as genes responsible for mesenchyme formation and condensation (Sox9 and Col2). In the context of this overarching control by Pbx homeoproteins, our work highlights multiple potential target genes either known for their involvement in pelvic development, or of unknown function. Thus, our studies open the path for new research in an under-studied area of vertebrate embryonic development.
Supplementary Material
Supp. Fig. S1. Expression of pubis and ilium markers in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. A: Compared to control HL (black arrowheads), expression of Alx4 remains unchanged in all studied mutants. B: Compared to control HL (black arrowheads), expression of Prrx1 remains unperturbed in all mutants, except for Pbx1−/−;Pbx2+/− mutants, in which it is reduced in the proximal-posterior flank-HL (red arrowheads). C: Compared to control HL (black arrowheads), Pitx1 remains normally expressed in all mutants analyzed, except for Pbx1−/−;Pbx2+/− mutants (red arrowhead), in which it is reduced in the proximal posterior HL.
Supp. Fig. S2. Expression of mesenchymal and tendon markers in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. A: Compared to WT (black arrowheads), the expression of Tbx15 is normal in all mutants analyzed, except for Pbx1−/−;Pbx2+/− mutants, where it is posteriorly expanded and decreased in its proximal domain (red arrowhead). B: Compared to WT (black arrowheads), the expression of Scx is modestly expanded in single Pbx1−/− and Pbx1−/−;Emx2+/− compound mutants (blue arrowheads), while it is drastically posteriorly shifted in Pbx1−/−;Pbx2+/− mutants (red arrowheads).
Supp. Fig. S3. Expression of Pax1 in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. Compared to WT and single Emx2−/− mutants (black arrowheads), Pax1 expression is down-regulated in most genotypes bearing two Pbx1 null alleles (red arrowheads), and completely absent in Pbx1−/−;Pbx2+/− mutants (empty arrowheads).
Supp. Fig. S4. Pairwise matrix motif for the Emx2-Pbx1 heterodimer used in the GREAT analysis. PWM generated as described in Materials and Methods, and Results.
Hard-tissue phenotypes of E13.5 compound Pbx1−/−;Pbx2+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Pbx2+/− pelvic girdle consists of only an ischial rudiment.
Hard-tissue phenotypes f E13.5 compound Pbx1−/−;Pbx3+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Pbx3+/− pelvic girdle consists of pubic and ischial rudiments.
Hard-tissue phenotypes of E14.5 compound Pbx1−/−;Emx2+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Emx2+/− pelvic girdle consists of a detached iliac blade and relatively normal pubic and ischial skeletal elements.
Hard-tissue phenotypes of E14.5 compound Pbx1−/−;Emx2−/− mutants by OPT of skeletal preparations. The Pbx1−/−;Emx2−/− pelvic girdle consists of a relatively normal ischial element, along with a significantly reduced and rounded pubic rudiment.
ACKNOWLEDGMENTS
We are grateful to Michael Cleary for anti-Pbx antibodies; researchers of the Mouse Genetics Community for in situ probes; and Liz Lacy, Kat Hadjantonakis, Ann Foley, and Xiao P. Peng for helpful discussions. T.C. was the recipient of the CUNY Carole and Morton Olshan Fellowship; K.H. was recipient of a fellowship from the “French Association against Myopathies”; E.F. was recipient of a Marie Curie fellowship (MOIF-CT-2005 022003). S.L.C. is a Howard Hughes Medical Institute Gilliam Fellow in the MSTP Program, Stanford University, Stanford, California, USA. Work supported by grants from: the NIH (2RO1HD043997, 1RO1HD061403, and R21DE018031-02S1 to L.S.; RO1HD059862 to G.B); March of Dimes and Birth Defects Foundation (#6- FY03-071 to L.S.); the Italian Association for Cancer Research and the ASM Foundation to V.Z. G.B. is a Packard Fellow, Searle Scholar, Microsoft Fellow, and Sloan Research Fellow. L.S. is an Irma T. Hirschl Scholar and recipient of research awards from The Alice Bohmfalk Trust and The Frueauff Foundation.
References
- Beverdam A, Meijlink FU. Expression patterns of group-I aristaless-related genes during craniofacial and limb development. Mech Dev. 2001;107:163–167. doi: 10.1016/s0925-4773(01)00450-6. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Brendolan A, Ferretti E, Salsi V, Moses K, Quaggin S, Blasi F, Cleary ML, Selleri L. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development. 2005;132:3113–3126. doi: 10.1242/dev.01884. [DOI] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131:3885–3896. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Capellini TD, Vaccari G, Ferretti E, Fantini S, He M, Pellegrini M, Quintana L, Di Giacomo G, Sharpe J, Selleri L, Zappavigna V. Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development. 2010;137:2559–2569. doi: 10.1242/dev.048819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Zewdu R, Di Giacomo G, Asciutti S, Kugler JE, Di Gregorio A, Selleri L. Pbx1/Pbx2 govern axial skeletal development by controlling Polycomb and Hox in mesoderm and Pax1/Pax9 in sclerotome. Dev Biol. 2008;321:500–514. doi: 10.1016/j.ydbio.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Fritsch R, Fickenscher H, Deutsch U, Goulding M, Gruss P. The molecular basis of the undulated/Pax-1 mutation. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- Cheah KS, Au PK, Lau ET, Little PF, Stubbs L. The mouse Col2a-1 gene is highly conserved and is linked to Int-1 on chromosome 15. Mamm Genome. 1991;1:171–183. doi: 10.1007/BF00351064. [DOI] [PubMed] [Google Scholar]
- Chevallier A, Kieny M, Mauger A. Limb-somite relationship: origin of the limb musculature. J Embryol Exp Morphol. 1977;41:245–258. [PubMed] [Google Scholar]
- Christ B, Huang R, Wilting J. The development of the avian vertebral column. Anat Embryol (Berl) 2000;202:179–194. doi: 10.1007/s004290000114. [DOI] [PubMed] [Google Scholar]
- Connell KA, Guess MK, Chen H, Andikyan V, Bercik R, Taylor HS. HOXA11 is critical for development and maintenance of uterosacral ligaments and deficient in pelvic prolapse. J Clin Invest. 2008;118(3):1050–1055. doi: 10.1172/JCI34193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Di Giacomo G, Koss M, Capellini TD, Brendolan A, Popperl H, Selleri L. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr Patterns. 2006;6:747–757. doi: 10.1016/j.modgep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Ros MA, Tabin CJ. A re-examination of proximodistal patterning during vertebrate limb development. Nature. 2002;418:539–544. doi: 10.1038/nature00945. [DOI] [PubMed] [Google Scholar]
- Hostikka SL, Gong J, Carpenter EM. Axial and appendicular skeletal transformations, ligament alterations, and motor neuron loss in Hoxc10 mutants. Int J Biol Sci. 2009;5(5):397–410. doi: 10.7150/ijbs.5.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Christ B, Patel K. Regulation of scapula development. Anat Embryol (Berl) 2006;211 Suppl 1:65–71. doi: 10.1007/s00429-006-0126-9. [DOI] [PubMed] [Google Scholar]
- Huang R, Zhi Q, Patel K, Wilting J, Christ B. Dual origin and segmental organisation of the avian scapula. Development. 2000;127:3789–3794. doi: 10.1242/dev.127.17.3789. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic V, De Santo R, Schughart K, Huffstadt U, Chiang C, Mahon KA, Mackem S. Hoxd-12 differentially affects preaxial and postaxial chondrogenic branches in the limb and regulates Sonic hedgehog in a positive feedback loop. Development. 1997;124(22):4523–4536. doi: 10.1242/dev.124.22.4523. [DOI] [PubMed] [Google Scholar]
- Krawchuk D, Weiner SJ, Chen YT, Lu BC, Costantini F, Behringer RR, Laufer E. Twist1 activity thresholds define multiple functions in limb development. Dev Biol. 2010;347(1):133–146. doi: 10.1016/j.ydbio.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper S, Beverdam A, Kroon C, Brouwer A, Candille S, Barsh G, Meijlink FU. Genetics of shoulder girdle formation: roles of Tbx15 and aristaless-like genes. Development. 2005;132:1601–1610. doi: 10.1242/dev.01735. [DOI] [PubMed] [Google Scholar]
- Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- Lausch E, Hermanns P, Farin HF, Alanay Y, Unger S, Nikkel S, Steinwender C, Scherer G, Spranger J, Zabel B, Kispert A, Superti-Furga A. TBX15 mutations cause craniofacial dysmorphism, hypoplasia of scapula and pelvis, and short stature in Cousin syndrome. Am J Hum Genet. 2008;83:649–655. doi: 10.1016/j.ajhg.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair EE, Bonfiglio L, Tuan RS. Expression of the paired-box genes Pax-1 and Pax-9 in limb skeleton development. Dev Dyn. 1999;214:101–115. doi: 10.1002/(SICI)1097-0177(199902)214:2<101::AID-AJA1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Loebel DA, O'Rourke MP, Steiner KA, Banyer J, Tam PP. Isolation of differentially expressed genes from wild-type and Twist mutant mouse limb buds. Genesis. 2002;33:103–113. doi: 10.1002/gene.10091. [DOI] [PubMed] [Google Scholar]
- Malashichev Y, Borkhvardt V, Christ B, Scaal M. Differential regulation of avian pelvic girdle development by the limb field ectoderm. Anat Embryol (Berl) 2005;210:187–197. doi: 10.1007/s00429-005-0014-8. [DOI] [PubMed] [Google Scholar]
- Malashichev Y, Christ B, Prols F. Avian pelvis originates from lateral plate mesoderm and its development requires signals from both ectoderm and paraxial mesoderm. Cell Tissue Res. 2008;331:595–604. doi: 10.1007/s00441-007-0556-6. [DOI] [PubMed] [Google Scholar]
- Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx1 and Pitx2 are required for development of hindlimb buds. Development. 2003;130:45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, Koentges G. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- Niswander L. Pattern formation: old models out on a limb. Nat Rev Genet. 2003;4:133–143. doi: 10.1038/nrg1001. [DOI] [PubMed] [Google Scholar]
- O'Rourke MP, Soo K, Behringer RR, Hui CC, Tam PP. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol. 2002;248:143–156. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Pantano S, Fumi MP, Lucchini F, Forabosco Agenesis of the Scapula in Emx2 Homozygous Mutants. Dev Biol. 2001;232:149–156. doi: 10.1006/dbio.2001.0159. [DOI] [PubMed] [Google Scholar]
- Perez AV, Perrine M, Brainard N, Vogel KG. Scleraxis (Scx) directs lacZ expression in tendon of transgenic mice. Mech Dev. 2003;120:1153–1163. doi: 10.1016/j.mod.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Peters H, Wilm B, Sakai N, Imai K, Maas R, Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- Pomikal C, Streicher J. 4D-analysis of early pelvic girdle development in the mouse (Mus musculus) J Morphol. 2010;271:116–126. doi: 10.1002/jmor.10785. [DOI] [PubMed] [Google Scholar]
- Rhee JW, Arata A, Selleri L, Jacobs Y, Arata S, Onimaru H, Cleary ML. Pbx3 deficiency results in central hypoventilation. Am J Pathol. 2004;165:1343–1350. doi: 10.1016/S0002-9440(10)63392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AS, Parsons TS. The Vertebrate Body. New York: Saunders College Publishing; 1986. iii-679. [Google Scholar]
- Sanchez-Villagra MR, Maier W. Ontogenetic data and the evolutionary origin of the mammalian scapula. Naturwissenschaften. 2002;89:459–461. doi: 10.1007/s00114-002-0362-7. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O_Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Selleri L, DiMartino J, van Deursen J, Brendolan A, Sanyal M, Boon E, Capellini T, Smith KS, Rhee J, Popperl H, Grosveld G, Cleary ML. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol Cell Biol. 2004;24:5324–5331. doi: 10.1128/MCB.24.12.5324-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J. Optical projection tomography as a new tool for studying embryo anatomy. J Anat. 2003;202:175–181. doi: 10.1046/j.1469-7580.2003.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert AU. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- Timmons PM, Wallin J, Rigby PW, Balling R. Expression and function of Pax 1 during development of the pectoral girdle. Development. 1994;120:2773–2785. doi: 10.1242/dev.120.10.2773. [DOI] [PubMed] [Google Scholar]
- Valasek P, Theis S, Krejci E, Grim M, Maina F, Shwartz Y, Otto A, Huang R, Patel K. Somitic origin of the medial border of the mammalian scapula and its homology to the avian scapula blade. J Anat. 2010;216:482–488. doi: 10.1111/j.1469-7580.2009.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, He L, Ehehalt F, Geetha-Loganathan P, Nimmagadda S, Christ B, Scaal M, Huang R. The formation of the avian scapula blade takes place in the hypaxial domain of the somites and requires somatopleure-derived BMP signals. Dev Biol. 2005;287:11–18. doi: 10.1016/j.ydbio.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Young NM. Function, ontogeny and canalization of shape variance in the primate scapula. J Anat. 2006;209:623–636. doi: 10.1111/j.1469-7580.2006.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Fig. S1. Expression of pubis and ilium markers in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. A: Compared to control HL (black arrowheads), expression of Alx4 remains unchanged in all studied mutants. B: Compared to control HL (black arrowheads), expression of Prrx1 remains unperturbed in all mutants, except for Pbx1−/−;Pbx2+/− mutants, in which it is reduced in the proximal-posterior flank-HL (red arrowheads). C: Compared to control HL (black arrowheads), Pitx1 remains normally expressed in all mutants analyzed, except for Pbx1−/−;Pbx2+/− mutants (red arrowhead), in which it is reduced in the proximal posterior HL.
Supp. Fig. S2. Expression of mesenchymal and tendon markers in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. A: Compared to WT (black arrowheads), the expression of Tbx15 is normal in all mutants analyzed, except for Pbx1−/−;Pbx2+/− mutants, where it is posteriorly expanded and decreased in its proximal domain (red arrowhead). B: Compared to WT (black arrowheads), the expression of Scx is modestly expanded in single Pbx1−/− and Pbx1−/−;Emx2+/− compound mutants (blue arrowheads), while it is drastically posteriorly shifted in Pbx1−/−;Pbx2+/− mutants (red arrowheads).
Supp. Fig. S3. Expression of Pax1 in compound Pbx family member and Pbx1;Emx2 mutants at E11.5 by in situ hybridization. Compared to WT and single Emx2−/− mutants (black arrowheads), Pax1 expression is down-regulated in most genotypes bearing two Pbx1 null alleles (red arrowheads), and completely absent in Pbx1−/−;Pbx2+/− mutants (empty arrowheads).
Supp. Fig. S4. Pairwise matrix motif for the Emx2-Pbx1 heterodimer used in the GREAT analysis. PWM generated as described in Materials and Methods, and Results.
Hard-tissue phenotypes of E13.5 compound Pbx1−/−;Pbx2+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Pbx2+/− pelvic girdle consists of only an ischial rudiment.
Hard-tissue phenotypes f E13.5 compound Pbx1−/−;Pbx3+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Pbx3+/− pelvic girdle consists of pubic and ischial rudiments.
Hard-tissue phenotypes of E14.5 compound Pbx1−/−;Emx2+/− mutants by OPT of skeletal preparations. The Pbx1−/−;Emx2+/− pelvic girdle consists of a detached iliac blade and relatively normal pubic and ischial skeletal elements.
Hard-tissue phenotypes of E14.5 compound Pbx1−/−;Emx2−/− mutants by OPT of skeletal preparations. The Pbx1−/−;Emx2−/− pelvic girdle consists of a relatively normal ischial element, along with a significantly reduced and rounded pubic rudiment.