Abstract
It is well established that natural killer (NK) cells confer resistance to many viral diseases, but in only a few instances the molecular mechanisms whereby NK cells recognize virus-infected cells are known. Here we show that CD94, a molecule preferentially expressed by NK cells, is essential for the resistance of C57BL/6 mice to mousepox, a disease caused by the Orthopoxvirus ectromelia virus. Ectromelia virus-infected cells expressing the major histocompatibility complex (MHC) class Ib molecule Qa-1b are specifically recognized by the activating receptor formed by CD94 and NKG2E. Because CD94-NKG2 receptors and their ligands are highly conserved in rodents and humans, a similar mechanism may exist during human infections with the smallpox and monkeypox viruses, which are highly homologous to ectromelia virus.
Graphical Abstract
Highlights
► Deficiency in CD94 results in susceptibility to lethal mousepox ► NK cells require CD94 to protect from mousepox ► CD94-NKG2E recognizes ECTV-infected cells in a Qa-1b-dependent manner
Introduction
Natural killer (NK) cells play important roles in protection from several, but not all, viral diseases (Lanier, 2008a, Lee and Biron, 2010). During some but not all viral infections, NK cells are activated and induced to proliferate by innate cytokines (Lee and Biron, 2010). Their ability to kill infected cells and protect from viral disease appears to require the interaction of specific activating receptors on the NK cells with ligands at the surface of infected cells (Lee and Biron, 2010). To date, only a few NK cell receptor-ligand interactions important for resistance to viral diseases have been characterized. The best studied involves the resistance to mouse cytomegalovirus (MCMV) infection by C57BL/6 (B6) mice where the activating receptor Ly49H specifically recognizes the viral protein m157 (Arase et al., 2002, Daniels et al., 2001, Smith et al., 2002) and by MA/My mice where the activating receptor Ly49P recognizes the major histocompatibility complex (MHC) class I molecule H2-Dk in complex with the viral protein m04 (Kielczewska et al., 2009). These mechanisms, however, are not conserved in humans because humans do not encode homologs of Ly49 molecules and because MCMV is fairly different from human cytomegalovirus (HCMV). Another less-well-characterized mechanism is the recognition of the influenza hemagglutinin by NKp46 on human and mouse NK cells (Gazit et al., 2006, Mandelboim et al., 2001).
Mousepox is a viral disease where an essential role for NK cells is well established (Delano and Brownstein, 1995, Fang et al., 2008, Jacoby et al., 1989, Parker et al., 2007). Mousepox is caused by the mouse Orthopoxvirus (OPV) ectromelia virus (ECTV), a close relative of the human pathogens variola virus (VARV, the agent of smallpox) and monkeypox virus (MPXV), which causes monkeypox, a grave endemic disease in central Africa that has recently caused an outbreak in the USA (Gross, 2003, Larkin, 2003). Young adult B6 mice are naturally resistant to mousepox when infected through the skin of the footpad, the natural route of infection. To understand the genetic mechanisms involved in mousepox resistance, Brownstein et al. (1991) infected a panel of B6×DBA2/J recombinant inbred strains of mice and determined virus titers in the liver after i.v. infection with ECTV. This resulted in the mapping of four genes involved in the B6 mice resistance to mousepox (Rmp1–Rmp4). Rmp1 resides in the natural killer (NK) complex (NKC) (Delano and Brownstein, 1995), a polymorphic region in the distal arm of chromosome 6 that varies widely among different strains of mice and encodes most of the inhibitory and activating receptors preferentially expressed by NK cells (Brown and Scalzo, 2008, Yokoyama et al., 1991). However, the identity of Rmp1 remains elusive. Moreover, the extent to which Rmp1 contributes to disease and lethality after footpad infection remains unknown.
NKG2D is an NK cell activating receptor that forms homodimers that bind host cell-encoded MHC class I-like proteins expressed by target cells. We have previously shown that NKG2D contributes to NK cell-mediated resistance to mousepox by providing optimal ability to kill targets expressing NKG2D ligands (Fang et al., 2008). However, our data also suggested that in addition to NKG2D, other activating receptors expressed by NK cells may also be essential for resistance (Fang et al., 2008).
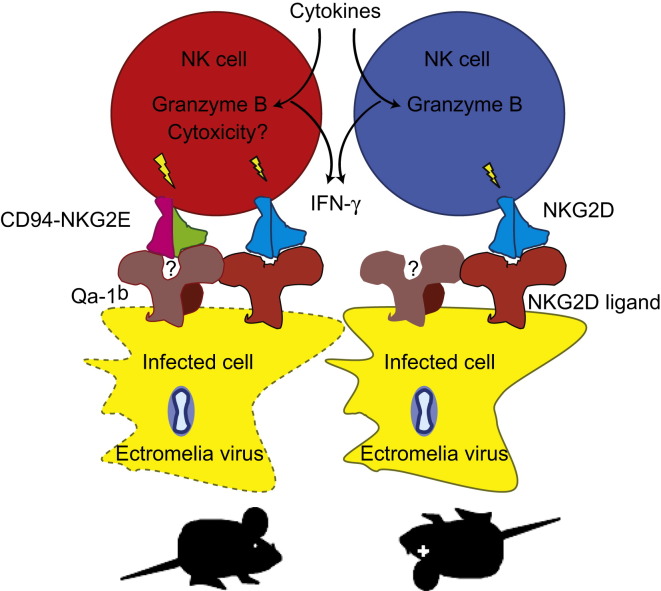
CD94 is a NKC-encoded molecule that forms heterodimeric receptors with NKC-encoded NKG2A, -C, and -E in the mouse and NKG2A, -C, and -E in humans. Despite their name and close linkage, these NKG2s are structurally and functionally unrelated to NKG2D and NKG2D does not pair with CD94 (Lanier, 2005). In rodents and primates, CD94-NKG2 heterodimers bind MHC class Ib molecules (Qa-1b in mice, HLA-E in humans) at the surface of cells. The CD94-NKG2A receptor is inhibitory and is the first MHC class I-binding receptor expressed during NK cell development, but its expression is dispensable for the normal differentiation of NK cells (Orr et al., 2010). CD94-NKG2C and CD94-NKG2E receptors are activating, but a specific role in NK cell biology remains to be described. The work presented here demonstrates that CD94 is essential for the resistance of B6 mice to mousepox. We also show that CD94 together with NKG2E recognizes ECTV-infected cells that express Qa-1b and that effective signaling requires NKG2D costimulation. This suggests that NK cells use a molecular mechanism reminiscent of T cell recognition to selectively target poxvirus-infected cells.
Results
The Natural Killer Complex from DBA2/J Mice Render B6 Mice Susceptible to Mousepox
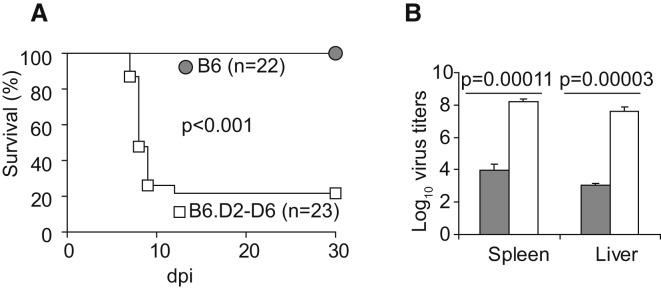
To characterize the role of the NKC in resistance to mousepox, we performed footpad infection of B6.D2-(D6Mit149-D6Mit15)/LusJ (herein B6.D2-D6) mice, a C57BL/6 congenic strain carrying the distal portion of chromosome 6, including the NKC, from DBA/2J mice (Davis et al., 2005). We found that B6.D2-D6 mice were highly susceptible to mousepox: the vast majority succumbed to infection (Figure 1A) and demonstrated very high virus loads in liver and spleen 7 days postinfection (dpi) (Figure 1B). These data showed that the DBA/2J NKC is sufficient to render B6 mice susceptible to mousepox after footpad inoculation but did not indicate which genes within the B6 NKC confer resistance. It is relevant to mention, however, that our data do not indicate that genes within the NKC are uniquely responsible for the susceptibility of DBA2/J mice or other susceptible strains to mousepox. In fact, three additional genes (Rmp2–4), not in the NKC, have been mapped in DBA2/J mice (Brownstein et al., 1991).
Figure 1.
The Natural Killer Complex from DBA/2J Mice Renders B6 Mice Susceptible to Mousepox
(A) B6 and B6.D2-D6 mice were infected with ECTV and survival was monitored.
(B) Virus titers in spleen and liver 7 dpi (gray columns, B6 mice; white columns, B6.D2-D6 mice).
Data correspond to the average ± SD of three individual mice.
B6 Congenic Mice Deficient in CD94 Are Susceptible to Mousepox
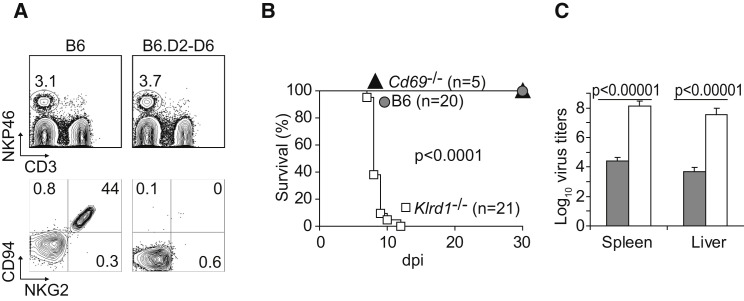
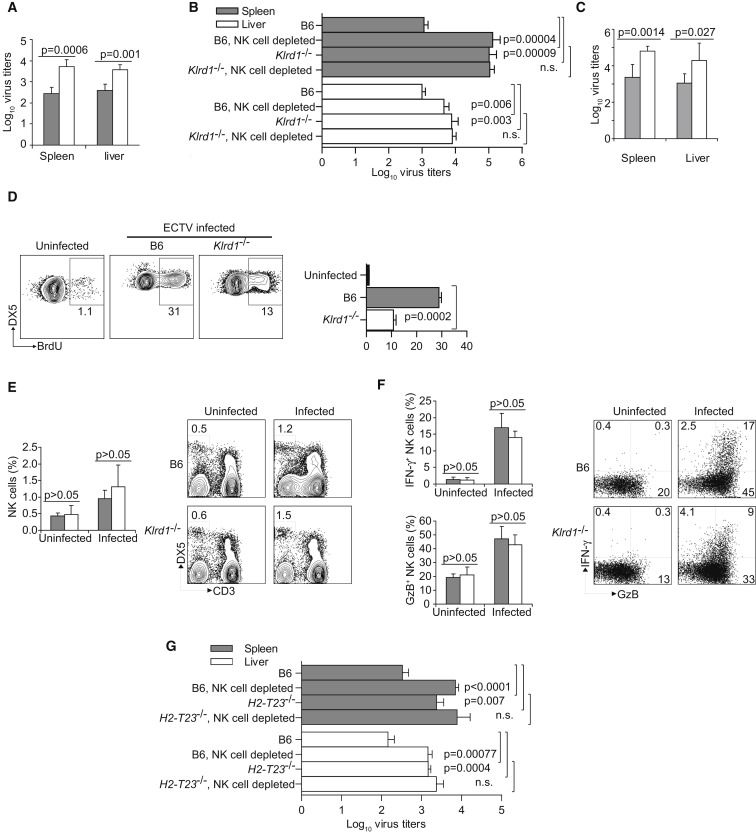
CD94 is a molecule expressed by NK cells and some activated T cells, NKT cells, and γδ T cells. It is encoded by Klrd1 within the NKC and forms inhibitory receptors with NKG2A and activating receptors with NKG2C and NKG2E, which are structurally and functionally unrelated to NKG2D (Lanier, 2005). Although it has been shown that fibroblasts infected with HCMV can drive preferential proliferation of human NK cells expressing CD94-NKG2C in vitro (Gumá et al., 2006), a specific role for CD94-NKG2 in controlling viral diseases has never been established. It has previously been reported that the parental DBA/2 mouse strain expresses CD94 on NK cells but that the DBA/2J substrain does not (Vance et al., 2002). Consistent with this observation, NK cells from B6.D2-D6 mice (with the NKC from DBA/2J) did not express CD94. Furthermore, because cell surface expression of NKG2A, NKG2C, and NKG2E requires CD94, B6.D2-D6 mice did not express any of these molecules at the surface of their NK cells (Figure 2A). This raised the enticing possibility that CD94-NKG2 receptors might be essential for resistance to mousepox. Arguing against this hypothesis, however, DBA/2 mice (the parental strain of DBA/2J) are also susceptible to mousepox even though they do express CD94. This could be due to Rmp2–Rmp4 or other unidentified genes in the DBA/2 background. To clarify this issue, we infected B6 mice that expressed the 129 strain NKC with a targeted deletion of Klrd1 (Klrd1 −/−, CD94-deficient) (Orr et al., 2010) and found that they were also very susceptible to mousepox, as indicated by high mortality (Figure 2B). Of note, the susceptibility of CD94-deficient mice to mousepox is not a general defect of the strain to resist viral infections; they control the highly related vaccinia virus (VACV) and lymphocytic choriomeningitis virus as efficiently as do wild-type (WT) B6 controls (Orr et al., 2010). It should be noted, however, that although CD94-deficient mice were backcrossed to the B6 background for at least nine generations, they still carry the NKC from the original 129 strain (Orr et al., 2010). Nonetheless, similarly to B6 mice, 129 mice are naturally resistant to mousepox and NK cells are essential for their resistance (Fang et al., 2008), strongly suggesting that the 129 NKC does not confer susceptibility to mousepox. In support of this, B6 mice that express the 129 strain NKC with a targeted deletion of Cd69 (Murata et al., 2003), but possessing a functional Klrd1 gene of 129 origin, were resistant to mousepox (Figure 2B). Consistent with their susceptibility to lethal mousepox infection, CD94-deficient mice had significantly higher viral loads in spleen and liver 7 dpi as compared with wild-type B6 mice (Figure 2C).
Figure 2.
B6 Congenic Mice Deficient in CD94 Are Susceptible to Mousepox
(A) Flow cytometry analysis of white blood cells in B6.D2-D6 mice. Top: Gated on lymphocytes according to forward angle (FSC) and side angle (SSC) light scattering properties. Staining for NKp46 and CD3 is shown. Bottom: gated on NKp46+CD3− (NK) cells as indicated in the upper plots. Staining for CD94 and NKG2 is shown.
(B) Wild-type B6, CD69-deficient, and CD94-deficient mice were infected with ECTV and survival was monitored. p value for wild-type B6 or CD69-deficient versus CD94-deficient mice.
(C) Virus titers in spleen and liver 7 dpi (gray columns, B6 mice; white columns, CD94-deficient mice).
Data correspond to the average ± SD of six individual mice from two independent experiments.
CD94 Is Essential for Resistance to Mousepox
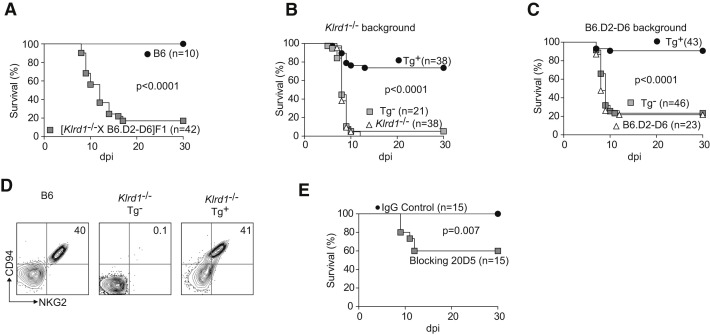
We performed additional experiments to conclusively establish an essential role for CD94 in resistance to mousepox. First, we found that [B6.D2-D6 × CD94-deficient] F1 mice were also susceptible to mousepox (Figure 3A), demonstrating that B6.D2-D6 and CD94-deficient genotypes did not complement each other. This strongly suggested that the reason for the susceptibility of both strains is the same, i.e., loss of CD94. Second, klrd1 expressed as a transgene (Tg+) (Orr et al., 2010) restored resistance to mousepox in CD94-deficient mice (Figure 3B) and B6.D2-D6 mice (Figure 3C), whereas Tg− littermates remained as susceptible as the original CD94-deficient strains. As expected, the klrd1 transgene rescued NKG2 expression in CD94-deficient mice (Figure 3D) and B6.D2-D6 mice (not shown). Third, treatment of mice with 20D5HCmIgG1-Q, which is a monoclonal antibody that blocks ligand binding by the CD94-NKG2 receptors but without depleting NK cells in vivo (Figure S1 available online; Orr et al., 2010), resulted in significant lethality in ECTV-infected wild-type B6 mice (Figure 3E). Although antibody treatment did not result in lethality in all of the infected mice, most probably because of incomplete in vivo blockade, the data reinforce the notion of a direct role for CD94-NKG2 receptors in mousepox resistance, rather than a developmental defect of NK cells in the B6.D2-D6 and CD94-deficient strains.
Figure 3.
CD94 Is Essential for Resistance to Mousepox
(A) Wild-type B6 or [B6.D2-D6 × CD94-deficient] F1 mice were infected with ECTV and survival was monitored.
(B) CD94 transgenic (Tg+) and nontransgenic (Tg−) CD94-deficient × [CD94-deficient × B6-CD94Tg] F1 littermate mice and parental CD94-deficient mice were infected with ECTV and survival was monitored.
(C) Transgenic (Tg+) and nontransgenic (Tg−) B6.D2-D6 × [B6.D2-D6 × B6-CD94Tg] F1 littermate mice and parental B6.D2-D6 mice were infected with ECTV and survival was monitored.
(D) Flow cytometric analysis of white blood cells from the indicated mice. Plots are gated on NKp46+CD3− NK cells. Staining for CD94 and NKG2 is shown.
Data are representative of three similar experiments.
(E) B6 mice were treated with 150 μg 20D5HCmIgG1-Q mAb or an isotype-matched control mAb, infected with ECTV and survival monitored.
Data are cumulative from three independent experiments.
NK Cells Require CD94 to Protect from Mousepox but Not to Migrate to the Draining Lymph Nodes
We have previously shown that 2–3 days after ECTV infection in the footpad, activated NK cells are recruited to the draining popliteal lymph node (D-LN) where they produce IFN-γ and presumably kill infected cells (Fang et al., 2008). The recruitment of NK cells and the activation of their effector functions are essential to curb early systemic virus spread. Depletion of NK cells or their inability to produce IFN-γ or perforin (Fang et al., 2008) or their inefficient migration to the D-LN resulting from age (Fang et al., 2010) results in increased virus titers in the liver starting 3 dpi (a time point at which T cells are not yet involved in virus control) with the consequence of lethal mousepox infection (Fang et al., 2008). Similarly, absence of CD94 in the B6.D2-D6 mice (Figure 4A), CD94-deficient mice (Figure 4B), and antibody blockade of CD94-NKG2 in wild-type B6 mice (Figure 4C), all resulted in increased virus loads in the liver 3 dpi, indicating an NK cell defect. Moreover, depletion of NK cells in B6 mice with Asialo GM1 antibody increased virus loads in the liver and spleen 3 dpi to titers similar to those in CD94-deficient mice, whereas NK cell depletion in CD94-deficient mice did not further increase virus loads (Figure 4B). In addition, we have previously shown extensive proliferation of NK cells in the spleen of ECTV-infected B6 mice 5 dpi (Fang et al., 2008). Consistent with defective NK cell response in the absence of CD94, NK cell proliferation 5 dpi was significantly reduced in the spleens of CD94-deficient mice as compared with wild-type B6 mice (Figure 4D). However, CD94-deficient mice did not differ from wild-type B6 mice in the recruitment of NK cells to the D-LN (Figure 4E) or their activation, as determined by the expression of the effector molecules IFN-γ and granzyme B (GzB) (Figure 4F), which are essential for NK cells to protect from mousepox (Fang et al., 2008). Similar activation and recruitment of NK cells to the D-LN were also seen in B6.D2-D6 mice and in other mousepox-susceptible strains such as BALB/c and DBA2/J (not shown). Together these results suggest that CD94 deficiency does not prevent the migration of NK cells to the D-LN or their activation (which is probably due to cytokines) but might specifically affect their ability to kill virus-infected target cells. The ligand for CD94-NKG2 heterodimers is the class Ib MHC I molecule Qa-1b, which is encoded by the H2-T23 gene (Vance et al., 1999). Of interest, similar to CD94-deficient mice, Qa-1b-deficient B6 mice (H2-T23 −/−, which express CD94-NKG2 receptors and the entire B6 NKC) exhibited significantly higher virus titers in liver and spleen 3 dpi as compared to WT B6 mice and, different from WT B6 mice, NK cell depletion did not result in an additional increase in virus loads in Qa-1b mice (Figure 4G). This suggested that NK cells may use CD94-NKG2s to recognize Qa-1b on ECTV-infected cells. It should be mentioned, however, that only 20% of Qa-1b-deficient mice succumbed to mousepox (not shown). Still, it is very possible that other mechanisms can compensate the NK cell deficiency in Qa-1b-deficient mice, allowing substantial survival despite initial high virus loads. For example, it has been shown that Qa-1b-deficient mice lack CD8+ regulatory T cells and this may result in enhanced effector CD8+ T cell activity (Hu et al., 2004, Lu et al., 2006).
Figure 4.
Lack of CD94 Results in Defective NK Cell Control of ECTV Spread
(A) B6 mice (gray columns) and B6.D2-D6 mice (white columns) were infected with ECTV, and virus titers in spleen and liver were determined 3 dpi. Data correspond to the average ± SD of five individual mice and are representative of two similar experiments.
(B) Wild-type B6 mice or CD94-deficient mice were depleted or not of NK cells with 15 μl anti-Asialo GM antibody and infected with ECTV. Virus titers in spleen (gray bars) and liver (white bars) were determined 3 dpi. Data correspond to the average ± SD of five individual mice from two independent experiments.
(C) Wild-type B6 mice treated with blocking 20D5HCmIgG1-Q mAb (gray columns) or an isotype-matched control mAb (white columns) were infected with ECTV and virus titers determined 3 dpi. Data correspond to the average ± SD of six individual mice from two independent experiments.
(D) Wild-type and CD94-deficient B6 mice were infected with ECTV. NK cell proliferation was determined 5 dpi in the spleen by BrdU incorporation. Bar graph is the cumulative data for three similar experiments with three mice/group. Representative flow cytometry plots of gated DX5+CD3− NK cells are shown on the right.
(E) Wild-type B6 and CD94-deficient mice were infected with ECTV or not and the draining popliteal lymph node was analyzed by flow cytometry 2 dpi. Shown are the cumulative data for five independent experiments as a column graph and representative plots indicating the proportion of NK cells (DX5+, CD3−) gated on lymphocytes as determined by FSC and SSC.
(F) Column graphs show the cumulative data for GzB- and IFN-γ-positive cells in the DX5+CD3− NK cell gate (from D) for five independent experiments. Representative flow cytometry plots as used for the columns graphs are shown on the right.
(G) B6 Wild-type or Qa-1b-deficient B6 mice depleted or not of NK cells with NK1.1 mAb treatment were infected with ECTV and the virus titers in the liver and spleen were determined 3 dpi. Data correspond to the mean ± SD of five mice (bars as indicated).
Deficient NK Cell Control of Virus Loads in the Absence of CD94 Results in a Defective CD8+ T Cell Response
Although expressed predominantly by NK cells, CD94 is also expressed by some activated CD8+ T cells. The death of B6.D2-D6 and CD94-deficient mice at approximately 10 dpi was consistent not only with defective NK cell function, but also with defective CD8+ T cell responses (Fang and Sigal, 2006). Thus, we analyzed the ECTV-specific CD8+ T cell responses in CD94-deficient mice (Figure S2A) and B6.D2-D6 mice (not shown). We found that they were highly deficient, as evidenced by decreased frequencies of CD8+ T cells expressing GzB and IFN-γ after ex vivo restimulation with infected cells and also after staining with H2-Kb-dimers loaded with TSYKFESV, the most immunodominant ECTV CD8 T cell determinant (Tscharke et al., 2005). However, as it occurs with aged B6 mice (Fang et al., 2010) or after NK cell depletion (Fang et al., 2008), the defective CD8+ T cell response was not CD8+ T cell intrinsic because the CD8+ T cell responses of wild-type B6 and CD94-deficient mice were similarly strong when infected with vaccinia virus, an OPV that is poorly pathogenic in mice but shares most of the CD8+ T cell determinants with ECTV, including TSYKFESV (Figure S2B). Moreover, CD94-deficient CD8+ T cells responded as efficiently as did CD94-sufficient CD8+ T cells in a CD94-sufficient environment (Figure S2C), while CD94-sufficient CD8+ T cells responded as poorly as did CD94-deficient CD8+ T cells in a CD94-deficient environment (Figure S2C) as determined by proliferation and H2-Kb-TSYKFESV dimer staining after adoptive transfer of T cells. Moreover, adoptive transfer of memory CD8+ T cells obtained from CD94-deficient mice immunized with VACV protected naive CD94-deficient mice from mousepox (Figure S2D). Together, our studies of the CD8+ T cell response show that the defective CD8+ T cell response and susceptibility of CD94-deficient mice to mousepox were due to a defect in the ability of their NK cells to control early virus spread and not to intrinsically defective CD8+ T cells.
CD94-NKG2E Recognizes ECTV-Infected Cells in the Presence of NKG2D and in a Qa-1b-Dependent Manner
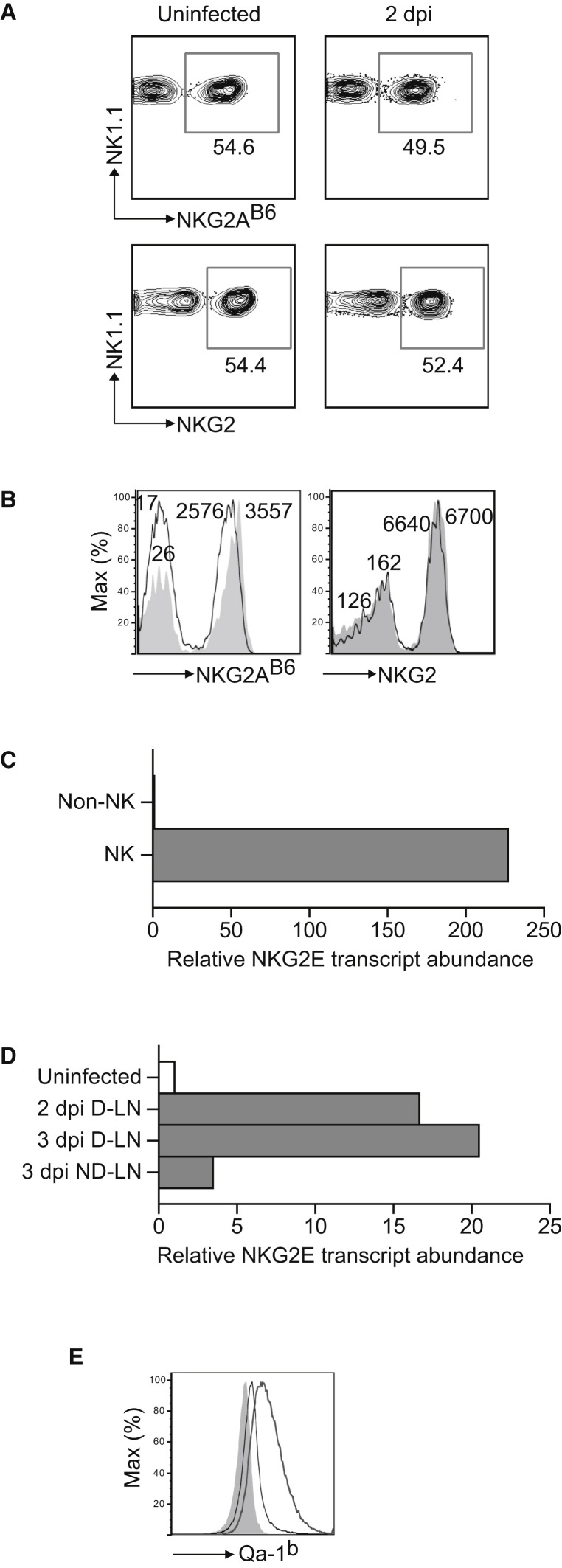
CD94-NKG2A receptors are inhibitory whereas CD94-NKG2C and CD94-NKG2E are activating (Lanier, 2008b). When we compared the D-LN of uninfected and infected (2 dpi) B6 mice, we found that in the infected mice there was a decrease in the proportion (Figure 5A) and mean fluorescence intensity (Figure 5B) of cells that stained with the NKG2A mAb 16A11 but not with the NKG2 (i.e., recognizing NKG2A, NKG2C, and NKG2E) mAb 20D5. This suggested an increase in the expression of activating NKG2s and a decrease of inhibitory NKG2A at the surface of NK cells in the D-LN. We were unable to directly test this possibility at the protein level because Abs specific for NKG2C and E are not available. This was important to confirm, however, because it is known that mouse splenic NK cells express very low amounts of activating NKG2s (Vance et al., 1999, Vance et al., 2002). Therefore, we performed quantitative polymerase chain reaction (qPCR) after reverse transcription for NKG2E. NKG2E transcripts were detected in the NK cell fraction but not in the NK cell-depleted fraction of LNs after magnetic sorting, confirming some expression in the NK cells of naive mice (Figure 5C). It should be noted, however, that the signal for NKG2E was weaker than that obtained with primers for NKG2A (not shown), supporting the notion that NKG2A is expressed at much higher levels than activating NKG2E. Still, 2 and 3 dpi NKG2E transcripts were increased 16- and 20-fold in the D-LN, respectively, as compared to the LN of uninfected mice while little change was observed in the ND-LN (Figure 5D). The qPCR assay was specific because it generated amplicons when using a plasmid encoding for NKG2E but not NKG2A or NKG2C as the template. Thus, we conclude that low levels of NKG2E were expressed in NK cells and that expression increased in the D-LN after ECTV infection. Because the fold increase in transcripts is higher than the fold increase of NK cells in the D-LN, the data suggest upregulation of NKG2E by NK cells after infection.
Figure 5.
Upregulation of NKG2E and Qa-1b during ECTV Infection
(A) Wild-type B6 mice were infected with ECTV in the footpad or not and the expression of NKG2A and NKG2 in the draining popliteal lymph node was determined. Flow cytometry plots correspond to pools of LNs from three mice and are representative of two similar experiments. Data show cells gated on NK1.1+CD3− NK cells.
(B) As in (A) but showing histograms to highlight changes in mean fluorescence intensities (shaded histogram, uninfected mice; black line, infected mice). The numbers indicate the mean fluorescence intensity of the closest histogram.
(C) LN cells were magnetically sorted into DX5+ (NK cells, 40% pure) and DX5− (non-NK, >99% pure). RNA was isolated, reverse transcribed, and the NKG2E message was quantified by qPCR with a specific primers/probe set. Data correspond to the mean ± SD of two wells and are representative of two similar experiments.
(D) RNA was obtained from the LNs of the indicated mice and analyzed by qPCR as in (C). Data correspond to the mean ± SD of two wells and are representative of two similar experiments.
(E) L cells were infected for 4 hr with 1 PFU/cell of sucrose-purified ECTV virus. Surface expression of Qa-1 was analyzed by flow cytometry. Data are representative from three similar experiments (shaded gray, isotype-matched control Ig; thin line, anti-Qa-1 staining of uninfected L cells; thick line, anti-Qa-1 staining of infected L cells).
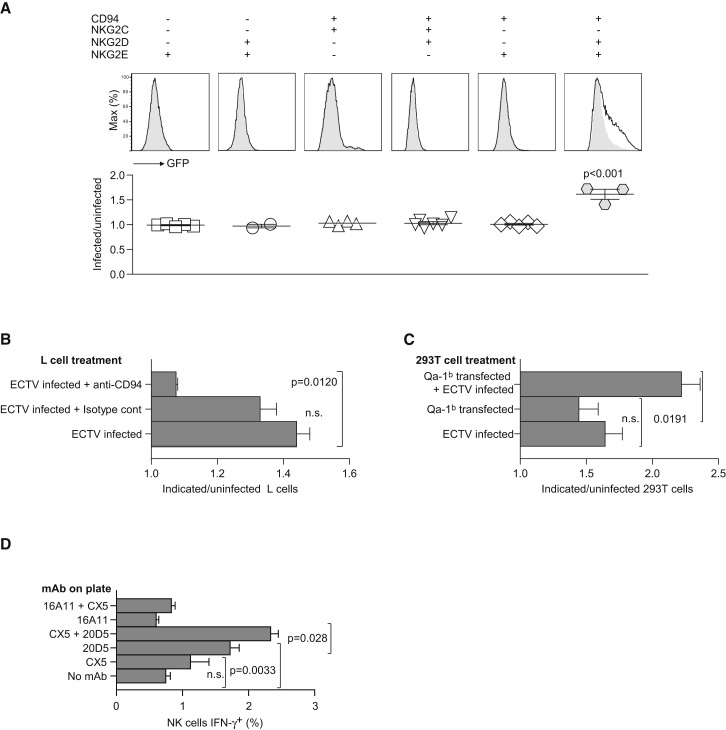
In vitro infection of mouse L cells with ECTV upregulated the expression of the Qa-1b glycoprotein (Figure 5E), further suggesting that NK cells might recognize ECTV-infected cells via activating NKG2s in a Qa-1b-dependent manner. To test for these possibilities, we generated reporter cell lines expressing green fluorescence protein (GFP) controlled by an NFAT promoter (NFAT-GFP) the signaling adaptor DAP12, CD94, and either activating NKG2C or NKG2E. Based on our prior studies implicating NKG2D in resistance to mousepox (Fang et al., 2008), we also cotransduced these reporter cells with the short form of NKG2D (NKG2D-s) that can pair and signal with DAP12. Expression of CD94, NKG2D, NKG2C, and NKG2E was verified on the cell surface of the reporter cell lines by flow cytometry (Figure S3). We found that the reporter cells expressing CD94, NKG2E, and NKG2D-s significantly increased GFP expression when coincubated with ECTV-infected as compared to uninfected L cells. None of the other reporters were significantly activated by ECTV-infected L cells, including those that expressed CD94 and NKG2E, with DAP12, in the absence of NKG2D-s (Figure 6A). The activation of the CD94+NKG2E+NKG2D+ reporter by ECTV-infected L cells was CD94 dependent because it was inhibited by a blocking CD94 mAb but not an isotype-matched Ig control (Figure 6B). Further, the CD94+NKG2E+NKG2D+ reporters responded to human 293T cells transfected with mouse Qa-1b and infected with ECTV, but much less to uninfected 293T cell transfected with Qa-1b or to 293T cells infected with ECTV but not transfected with Qa-1b (Figure 6C). Together, these results strongly suggest that NKG2D and CD94-NKG2E (but not CD94-NKG2C) synergize to recognize ECTV-infected cells in a Qa-1b-restricted manner.
Figure 6.
CD94-NKG2E, but Not CD94-NKG2C, Recognizes ECTV-Infected Cells in the Presence of NKG2D and in a Qa-1b-Dependent Manner
(A) L cells were infected with 1 PFU/cell purified ECTV virus for 2 hr, washed, and coincubated with the indicated reporters for 18 hr at 37°C. The expression of GFP by the reporters was then analyzed by flow cytometry, gating on the reporter cells as distinguished by forward angle light scatter and side angle light scatter properties. Top histograms are representative data (shaded gray, reporter cells with uninfected L cells; dotted thick line, reporter cells with ECTV-infected L cells). The bottom panel displays the results for 2–5 individual experiments where the y axis is the ratio of the geometric mean of the GFP fluorescence for the indicated reporter cells incubated with infected or uninfected L cells.
(B and C) L cells (B) and 293T cells (B) were treated as indicated or kept uninfected and incubated with CD94+NKG2E+NKG2D+ reporters for 16 hr, and the reporters' GFP fluorescence was determined by flow cytometry. Data represent the ratio of the geometric mean of the reporter cells incubated with L cells (B) or 293T cells (C) treated as indicated/GFP fluorescence of the reporter cells incubated with uninfected L cells. Data are representative of three similar experiments.
(D) Splenocytes from wild-type B6 mice were incubated for 5 hr with the indicated bound mAbs in the presence of 1000 U IL-2. Cells were analyzed by flow cytometry. Data show the proportion of NK cells (NK1.1+CD3−) that expressed intracellular IFN-γ. Data correspond to the mean ± SD of three replicate samples and are representative of two similar experiments.
The synergy between the NKG2D and activating CD94-NKG2 receptors in NK cell activation was further explored in assays where splenocytes from naive mice were incubated for 5 hr with plate-immobilized mAbs and the NK cells analyzed by flow cytometry for intracellular IFN-γ production. The results showed that CX5 (anti-NKG2D) and 20D5 (anti-NKG2) induced IFN-γ production in NK cells and that the proportion of NK cells producing IFN-γ was further increased when CX5 and 20D5 were combined. On the other hand, 16A11 (anti-NKG2AB6) did not induce IFN-γ production and the induction of IFN-γ production by CX5 was reduced to background amounts when combined with 16A11 (Figure 6D). Together, these experiments demonstrate a synergy between NKG2D and the activating CD94-NKG2 receptors in primary splenic NK cells and functionally demonstrate activating CD94-NKG2 receptor expression at the surface of primary NK cells. Further, because 20D5 binds activating NKG2s as well as inhibitory NKG2A, these results may indicate the existence of a subset of NK cells expressing activating CD94-NKG2C or E receptors in the absence of NKG2A expression.
Discussion
Here we demonstrated that the NK cell receptor CD94 has an essential role in the resistance of B6 mice to mousepox, adding to the still very limited number of NK cell receptors with known function in protecting from viral diseases. The susceptibility of CD94-deficient mice to mousepox was observed in B6 strains carrying the NKC haplotypes from the mousepox-susceptible DBA2/J strain with a spontaneous mutation in Klrd1 or from the resistant 129 strain with a targeted deletion of the Klrg1 gene. This strongly suggests that CD94 cannot be replaced by other NK cell receptors and that it is absolutely essential for the resistance of wild-type mice B6 to mousepox.
Consistent with a role for CD94 in the recognition of ECTV infection by NK cells, mice deficient in CD94 succumbed at times similar to those of NK cell-depleted mice (Fang et al., 2008, Parker et al., 2007) and, when compared with wild-type B6 mice, had significantly increased virus loads in liver 3 dpi, a time point when NK cells, but not T cells, play an important role in controlling virus spread from the D-LN (Fang et al., 2008). Thus, although CD94-deficient mice had a very diminished CD8+ T cell response, this was the consequence, rather than the primary reason, for the lack of virus control as we previously demonstrated for NK cell-depleted mice (Fang et al., 2008) and more recently for aged B6 mice (Fang et al., 2010). It remains possible, however, that CD94 on cells other than NK cells might also have contributed to protection.
Our study also revealed that ECTV-infected cells are specifically recognized by the activating receptor formed by CD94 and NKG2E and that this recognition requires the expression of the nonclassical MHC class I molecule Qa-1b at the surface of ECTV-infected cells. Moreover, at the peak of the NK cell response in the D-LN, the proportion of NK cells expressing CD94 paired to activating NKG2s increased at the expense of those expressing inhibitory CD94-NKG2A receptors. In addition, ECTV infection upregulated Qa-1b expression in cells. An important remaining question will be to understand how NK cells switch from the inhibitory CD94-NKG2A to the activating CD94-NKG2E receptor. NK cells do not proliferate in the D-LN 3 dpi (Fang et al., 2008). Thus, the most likely explanations are a change in the cellular expression of the different NKG2s as a consequence of activation or the preferential recruitment of cells expressing activating NKG2s into the D-LN.
Similar to class Ia MHC molecules, Qa-1b binds short peptides; however, most Qa-1b molecules are normally occupied by AMAPRTLLL (Aldrich et al., 1994), a peptide derived from the signal peptide of class Ia MHC I molecules (Aldrich et al., 1994, DeCloux et al., 1997). It appears that recognition of Qa1b by CD94-NKG2E is insufficient to trigger activation because the NFAT-GFP reporter cells expressing CD94-NKG2E receptors were not stimulated unless the target cells were infected with ECTV. It seems more probable that infection also results in the presentation of novel peptide(s) by Qa-1b (either a viral peptide or a peptide derived from an infection-induced cellular protein), resulting in Qa-1b-peptide complexes with higher affinity for CD94-NKG2E.
We previously reported that NKG2D is required for the ability of NK cells to protect from mousepox infection (Fang et al., 2008). We have also shown that ECTV upregulates expression of NKG2D ligands on infected cells and that resistance to mousepox infection is diminished by blocking the NKG2D receptor in vivo (Fang et al., 2008). Our present findings with reporter assays, as well as in vitro stimulation of primary NK cells with mAbs, suggest that CD94-NKG2E and NKG2D synergize to achieve optimal NK cell activation to mount a protective response against ECTV, which is consistent with the dual requirement for NKG2D and CD94 for resistance to mousepox. Although the molecular mechanism for this synergy remains unexplained, we speculate that stimulation through both receptors is required to overcome the inhibitory effects of NKG2A or other inhibitory receptors or, most likely, that the presence of both receptors at the immune synapse favor a stronger activating signal through the recruitment of DAP12. There is strong precedent for synergy between NKG2D and other activating receptors such as with KIR in human NK cells (Wu et al., 2000) or with the T cell receptor in humans (Groh et al., 2001) and mice (Jamieson et al., 2002).
Many of the activating NK receptors in the Ly49 and KIR families, as well as human CD94-NKG2C, have low to undetectable affinity for MHC class I. We speculate that the mouse CD94-NKG2E might have a low affinity for Qa-1b expressed on normal, healthy cells to avoid autoimmunity, but that peptides derived from ECTV-infected cells might generate higher-affinity Qa-1b ligands. Further, we hypothesize that because of the low affinity of the CD94-NKG2E receptor, productive activation might require synergistic signals derived from the NKG2D receptors. In light of our findings, we propose a model whereby NK cells, probably activated through cytokines, migrate to the D-LN and kill ECTV-infected cells via CD94-NKG2E for specific recognition of targets expressing a specific peptide(s) bound to Qa-1b, and NKG2D as a costimulator (Fang et al., 2008). This model is consistent with the current view that activating receptors are not essential for the cytokine-induced activation of the NK cells during a viral infection but are required for the cytolysis of the virus-infected target cells (Lee and Biron, 2010, Vance et al., 1998).
Importantly, different from the recognition of MCMV by Ly49-activating receptors, the CD94-NKG2 system is exquisitely conserved between rodents and primates (Vance et al., 1999) and the sequences of all OPVs are also highly conserved. This suggests that the recognition of OPVs by NK cells might be conserved between humans and mice and relevant in protection against smallpox and monkeypox infection.
Experimental Procedures
Tissue Culture
The dendritic cell line DC2.4 (Shen et al., 1997) was a gift from K. Rock (University of Massachusetts Medical Center, Worcester, MA). As L cells, we used A9-T7 cells (Elroy-Stein and Moss, 1990), which are A9 cells (ATCC #CCL-1.4), from the C3H/An mouse strain stably transfected with T7 polymerase (a kind gift from B. Moss, NIAID, MD). BS-C-1 cells (ATCC #CCL-26) were a gift from G. Cohen (University of Pennsylvania, Philadelphia, PA). 293T cells (ATCC #CRL-11268) were purchased from ATCC. Reporter cells were Ba/F3 cells transfected with a NFAT-inducible GFP construct (Hooijberg et al., 2000), Flag-tagged mouse DAP12 for signal transduction, HA-tagged mouse CD94, myc-tagged mouse NKG2C or NKG2E, and/or mouse NKG2D-S.
For cell culture, we used RPMI 10, which consisted of RPMI-1640 tissue culture medium (Invitrogen Life Technologies) supplemented with 10% FCS (Sigma-Aldrich), 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen Life Technologies), 10 mM HEPES buffer (Invitrogen Life Technologies), and 0.05 mM 2-ME (Sigma-Aldrich). For virus growth and titers, RPMI 2.5 (as above but with 2.5% FCS) was used. When required, 10 U/ml recombinant mouse interleukin-2 (IL-2) (BD) was added to RPMI 10 (RPMI 10-IL2). All cells were grown at 37°C with 5% CO2.
Viruses
Initial stocks of the WT ECTV Moscow were obtained from ATCC (#VR-1374). New stocks of ECTV were expanded in BSC-1 cells infected with 0.1 plaque-forming units/cell (PFU/cell). Titers in stocks were determined by plaque assays on confluent BSC-1 cells via 10-fold serial dilutions of the stocks in 0.5 ml RPMI 2.5 in 6-well plates (2 wells/dilution) for 1 hr. Two ml fresh RPMI 2.5 was added and the cells incubated at 37°C for 5 days. Next, the media was aspirated and the cells fixed and stained for 10 min with 0.1% crystal violet in 20% ethanol. The fix and stain solution was subsequently aspirated, the cells air-dried, the plaques counted, and the PFU/ml in stocks were calculated accordingly.
For the determination of virus titers in spleens or LNs, the organs were removed from experimental mice at indicated days after footpad infection, made into a single-cell suspension between two frosted slides, and resuspended in 10 ml RPMI 10 medium. One ml of the cell suspensions were frozen and thawed three times and titers determined in 10-fold serial dilutions of the cell lysates as above. Virus titers were calculated as PFU/spleen or PFU/LN. To determine the virus titers in liver, a portion of the liver was weighed and homogenized in medium by means of a Tissue Lyser (QIAGEN). The virus titers were calculated as PFU/100 mg liver.
Mice
The Fox Chase Cancer Center Institutional Animal Care and Use Committee approved the experimental protocols involving animals. B6 mice were purchased from Taconic when they were 8–10 weeks of age and rested at least a week before use in experiments. The B6.D2-D6 mice were initially purchased from Jackson Laboratory and bred in the Fox Chase Cancer Center Laboratory Animal Facility. The CD69-deficient mice (Murata et al., 2003), which were generated via 129 strain ES cells and backcrossed to C57BL/6, were a kind gift from J. Cyster (University of California San Francisco, San Francisco, CA). CD94-deficient and CD94-transgenic mice were made as described (Orr et al., 2010) and bred in the Fox Chase Cancer Center Laboratory Animal Facility. The CD94 transgenic mice were crossed with CD94-deficient mice or B6.D2-D6 mice. The F2 generation was genotyped by PCR with tail DNA with a forward primer for exon 3 (CTGAATGCTGTGTTTGCCTGGACAA) and a reverse primer for exon 6 (TCACAGGATTCAGCAGAAACGCTTTTG) that amplify a 334 bp fragment from cDNA and a 3010 bp fragment from the genomic Klrd1 gene. The F2 mice were also analyzed by flow cytometry after staining with NK1.1 (PK136) and CD94 (18d3) mAbs. Pure lines were established by intercrossing NK1.1−CD94+ animals. B6129S6-H2T3 (Qa-1b-deficient) mice were purchased from Jackson Laboratory.
Infections
Mice were infected in the left footpad with 25 μl PBS containing 3 × 103 PFU ECTV. After infections, mice were observed daily for signs of disease (lethargy, ruffled hair, weight loss, skin rash, eye secretions). When death was considered imminent (unresponsiveness to touch, lack of voluntary movements) the mice were euthanized and recorded as dead. Adoptive transfer of memory CD8+ T cells was exactly as before, but with CD8+ T cells purified from VACV-immune CD94-deficient mice instead of BALB/c mice (Xu et al., 2007).
A9T7 cells (5 × 106) were plated in 6-well plates and cultured overnight to allow cells to adhere. The cells were then infected with 1 PFU ECTV/cell for 4 hr, collected, washed, stained, and analyzed for surface expression of Qa-1b.
In Vivo Depletion of NK Cells and NKG2 Blockade
Depletion of NK cells in B6 mice was performed by intraperitoneal (i.p.) inoculation of 200 μg NK1.1 mAb PK136 or 15 μl Asialo GM1 antisera (Wako Chemicals) 1 day before infection, as previously described (Fang et al., 2008). For in vivo blockade of the CD94-NKG2 receptors, mice were inoculated i.p. with 150 μg of the neutralizing, nondepleting 20D5HCmIgG1-Q mAb 1 day before infection. The production of neutralizing, nondepleting 20D5HCmIgG1 mAb that binds to NKG2A, NKG2C, and NKG2E has been described previously (Orr et al., 2010).
Flow Cytometry
Detection of NK and T cell responses was performed as described previously (Fang et al., 2008, Fang and Sigal, 2005, Fang and Sigal, 2006, Fang and Sigal, 2010). In brief, for T cell responses, lymphocytes were obtained from mice at different dpi and made into single-cell suspensions. After osmotic lysis of red blood cells with 0.84% NH4Cl, cells were washed and 106 cells were cultured at 37°C in 96-well plates in the presence of 2 × 105 VACV-infected DC2.4 cells or uninfected DC2.4 cells as control (stimulation with VACV- or ECTV-infected DC2.4 cells produces similar results when measuring anti-ECTV responses). For simplicity, we restimulated with VACV in all experiments. After 5 hr, Brefeldin A (BFA, Sigma) was added to block the secretory pathway and allow for the accumulation of cytokines inside the cells. After an additional 1.5 hr incubation, 2.4G2 mAb (anti-CD16 + CD32, ATCC) was added to block nonspecific binding of fluorochrome-conjugated mAbs to Fc receptors. The cells were then stained for cell surface molecules, fixed, permeabilized, and stained for intracellular molecules with the Cytofix/Cytoperm kit (Becton Dickinson, BD) according to the manufacturer's instructions. To determine NK cell responses in LNs, intact organs were incubated at 37°C for 1 hr in media containing 10 μg/ml Brefeldin A, made into single-cell suspensions, stained, and analyzed as described above. For the determination of proliferation by BrdU incorporation 5 dpi, mice were injected i.p. with 2 mg BrdU in sterile PBS and euthanized 3 hr later. Staining was exactly as described previously (Fang et al., 2008). The following mAb and staining reagents were used: anti-CD8α (53-6.7, BD), anti-CD3e (145-2C11, BD), anti-NKp46 (29A1.4), anti-CD94 (18D3), anti-NKG2 (20D5, eBioscience), anti-NKG2AB6 (16A11, eBioscience), anti-DX5 (DX5), anti-NK1.1 (PK136, BD), anti-IFN-γ (XMG1.2, BD), anti-Qa-1b (6A8.6F10.1A6, BD), anti-CD11b (M1/70, eBioscience), anti-CD27 (LG 3A10, BD), anti-NKG2D (C7, BD), anti-BrdU (eBioscience), and anti-human GzB (GzB, Caltag) that cross-reacts with mouse GzB (48). For TSYKFESV specific CD8+ T cell H2-Kb:Ig recombinant fusion protein (Dimer-X, BD) were incubated with synthetic TSYKFESV peptide (genScript) and used as recommended by the manufacturer. Stained cells were analyzed by flow cytometry at the Fox Chase Cell Sorting Facility with a LSR II system (BD).
Reporter Cell Assays
When A9T7 cells were used for restimulation, 4 × 105 A9T7 cells were plated in 24-well plates and cultured overnight to allow cells to adhere. The cells were then infected with 1 PFU ECTV/cell or left uninfected for 2 hr and washed, and 2 × 105 of the indicated NFAT-GFP reporter cells were added to the well and incubated for 18 hr. The reporter cells were washed and analyzed for expression of GFP by flow cytometry. For mAb blockade, the reporter cells were added to the indicated well in the presence of 20 μg/ml CD94 mAb (clone 18D3).
When 293T cells were used for restimulation, 2.4 × 104 cells were plated in 96-well plates and cultured overnight to allow cells to adhere. The cells were then transfected with 0.1 μg pCDNA3.1-Qa-1 (a gift from J. Forman, University of Texas Southwest Medical Center, Dallas, TX) with TransIT-LT1 reagent (Mirus) according to the manufacturer's instructions. The next day, some wells were infected with 1 PFU ECTV/cell for 2 hr and washed, and 2 × 104 reporter cells were added to the well and then incubated for 18 hr. The reporter cells were washed and analyzed for expression of GFP by flow cytometry.
In Vitro NK Cell Stimulation and IFN-γ Assay
Flat-bottom, high protein-binding plates (Thermos Fisher Scientific) were coated with NKG2D mAb CX5 (1.0 μg/ml) and/or anti-NKG2 Ab 20D5 (2.5 μg/ml), NKG2AB6 mAb 16A11 (2.5 μg/ml) at 4°C overnight. After extensive washing, B6 splenocytes (106/well) in RPMI 10 containing 10 μg/ml brefeldin A and 1000 U/ml recombinant human IL-2 (Roche, Basel, Switzerland; from the NCI Biological Resources Branch, Frederick, MD) were added to triplicate wells. Cells were incubated for 5 hr at 37°C, stained for surface cell markers and intracellular IFN-γ, and analyzed by flow cytometry.
Quantitative Real-Time PCR
Primers and probes for NKG2E transcripts (Mm00650941_m1∗) and GAPDH were purchased from Applied Biosystems. Total RNA was extracted from popliteal LNs of naive or ECTV-infected mice (2 dpi). In some cases, pools of LN cells from uninfected mice were sorted into DX5+ (NK cells) and DX5− (non-NK cells) with a mini-Macs separation unit, MS columns, and anti-DX5 MACS beads according to manufacturer's instructions (Miltenyi Biotech). The first-strand cDNA was synthesized with random primers. qRT-PCR was carried out with the ABI 7500 (Applied Biosystems). The cycling conditions for real-time PCR were 50°C for 10 min, followed by 45 cycles of 95°C for 30 s and 60°C for 2 min. Data were analyzed with the Sequence Detection v1.2 Analysis Software (Applied Biosystems).
Statistical Analysis
Unless indicated, all displayed data correspond to one representative experiment of at least three similar experiments with groups of three to five mice. Statistical analysis was performed with Excel or Graph Pad Prism software. For survival studies, p values were obtained with the Log-rank (Mantel-Cox) Test. All other statistical analyses were performed with unpaired two-tailed t test. When applicable, data are displayed with mean ± standard error of the mean (SEM).
Acknowledgments
We thank the following FCCC core facilities: Laboratory Animal, Flow Cytometry, Sequencing, Cell Culture, Transgenic, and Hybridoma. We thank H. Gillin for secretarial support. This work was supported by NIAID grants R01AI048849, R21AI077021, and R01AI065544 to L.J.S., NIH grant AI066897 to L.L.L., and NCI grant CA006927 to FCCC. M.F. was partly supported by the William J. Avery Fellowship. M.T.O. was supported by a fellowship from the Cancer Research Institute. L.L.L. is an American Cancer Society Professor. Author contributions are as follows. L.J.S. and L.L.L. conceived the initial idea; M.F. performed most of the experiments; L.J.S. supervised the project; M.T.O. generated some of the reporter cells; P.S. and T.E. produced the CD94 blocking mAb; L.J.S. and L.L.L. contributed reagents and analytic tools; M.F., M.T.O., L.L.L., and L.J.S. discussed the general design and the analysis of experiments; M.F. and L.J.S. designed most of the experiments; L.J.S. wrote the manuscript with M.F. collaboration; and M.F. and L.J.S. prepared the figures. L.J.S. and L.L.L. should be considered senior authors. All the authors approved the final manuscript.
Published online: April 21, 2011
Footnotes
Supplemental Information includes three figures and can be found with this article online at doi:10.1016/j.immuni.2011.02.015.
Supplemental Information
References
- Aldrich C.J., DeCloux A., Woods A.S., Cotter R.J., Soloski M.J., Forman J. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- Arase H., Mocarski E.S., Campbell A.E., Hill A.B., Lanier L.L. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- Brown M.G., Scalzo A.A. NK gene complex dynamics and selection for NK cell receptors. Semin. Immunol. 2008;20:361–368. doi: 10.1016/j.smim.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein D.G., Bhatt P.N., Gras L., Jacoby R.O. Chromosomal locations and gonadal dependence of genes that mediate resistance to ectromelia (mousepox) virus-induced mortality. J. Virol. 1991;65:1946–1951. doi: 10.1128/jvi.65.4.1946-1951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels K.A., Devora G., Lai W.C., O'Donnell C.L., Bennett M., Welsh R.M. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.C., Schadt E.E., Smith D.J., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Lusis A.J. A genome-wide set of congenic mouse strains derived from DBA/2J on a C57BL/6J background. Genomics. 2005;86:259–270. doi: 10.1016/j.ygeno.2005.05.010. [DOI] [PubMed] [Google Scholar]
- DeCloux A., Woods A.S., Cotter R.J., Soloski M.J., Forman J. Dominance of a single peptide bound to the class I(B) molecule, Qa-1b. J. Immunol. 1997;158:2183–2191. [PubMed] [Google Scholar]
- Delano M.L., Brownstein D.G. Innate resistance to lethal mousepox is genetically linked to the NK gene complex on chromosome 6 and correlates with early restriction of virus replication by cells with an NK phenotype. J. Virol. 1995;69:5875–5877. doi: 10.1128/jvi.69.9.5875-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elroy-Stein O., Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Sigal L.J. Antibodies and CD8+ T cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J. Immunol. 2005;175:6829–6836. doi: 10.4049/jimmunol.175.10.6829. [DOI] [PubMed] [Google Scholar]
- Fang M., Sigal L.J. Direct CD28 costimulation is required for CD8+ T cell-mediated resistance to an acute viral disease in a natural host. J. Immunol. 2006;177:8027–8036. doi: 10.4049/jimmunol.177.11.8027. [DOI] [PubMed] [Google Scholar]
- Fang M., Sigal L. Studying NK cell responses to ectromelia virus infections in mice. Methods Mol. Biol. 2010;612:411–428. doi: 10.1007/978-1-60761-362-6_28. [DOI] [PubMed] [Google Scholar]
- Fang M., Lanier L.L., Sigal L.J. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 2008;4:e30. doi: 10.1371/journal.ppat.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Roscoe F., Sigal L.J. Age-dependent susceptibility to a viral disease due to decreased natural killer cell numbers and trafficking. J. Exp. Med. 2010;207:2369–2381. doi: 10.1084/jem.20100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R., Gruda R., Elboim M., Arnon T.I., Katz G., Achdout H., Hanna J., Qimron U., Landau G., Greenbaum E., et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- Groh V., Rhinehart R., Randolph-Habecker J., Topp M.S., Riddell S.R., Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- Gross E. Update on emerging infections: News from the Centers for Disease Control and prevention. Update: Multistate outbreak of monkeypox—Illinois, Indiana, Kansas, Missouri, Ohio, and Wisconsin, 2003. Ann. Emerg. Med. 2003;42:660–662, discussion 662–664. doi: 10.1016/S0196-0644(03)00819-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumá M., Budt M., Sáez A., Brckalo T., Hengel H., Angulo A., López-Botet M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- Hooijberg E., Bakker A.Q., Ruizendaal J.J., Spits H. NFAT-controlled expression of GFP permits visualization and isolation of antigen-stimulated primary human T cells. Blood. 2000;96:459–466. [PubMed] [Google Scholar]
- Hu D., Ikizawa K., Lu L., Sanchirico M.E., Shinohara M.L., Cantor H. Analysis of regulatory CD8 T cells in Qa-1-deficient mice. Nat. Immunol. 2004;5:516–523. doi: 10.1038/ni1063. [DOI] [PubMed] [Google Scholar]
- Jacoby R.O., Bhatt P.N., Brownstein D.G. Evidence that NK cells and interferon are required for genetic resistance to lethal infection with ectromelia virus. Arch. Virol. 1989;108:49–58. doi: 10.1007/BF01313742. [DOI] [PubMed] [Google Scholar]
- Jamieson A.M., Diefenbach A., McMahon C.W., Xiong N., Carlyle J.R., Raulet D.H. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- Kielczewska A., Pyzik M., Sun T., Krmpotic A., Lodoen M.B., Munks M.W., Babic M., Hill A.B., Koszinowski U.H., Jonjic S., et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J. Exp. Med. 2009;206:515–523. doi: 10.1084/jem.20080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Lanier L.L. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. Monkeypox spreads as US public-health system plays catch-up. Lancet Infect. Dis. 2003;3:461. doi: 10.1016/S1473-3099(03)00713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Biron C.A. Here today—Not gone tomorrow: Roles for activating receptors in sustaining NK cells during viral infections. Eur. J. Immunol. 2010;40:923–932. doi: 10.1002/eji.201040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Werneck M.B., Cantor H. The immunoregulatory effects of Qa-1. Immunol. Rev. 2006;212:51–59. doi: 10.1111/j.0105-2896.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., Davis D.M., Strominger J.L., Yewdell J.W., Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- Murata K., Inami M., Hasegawa A., Kubo S., Kimura M., Yamashita M., Hosokawa H., Nagao T., Suzuki K., Hashimoto K., et al. CD69-null mice protected from arthritis induced with anti-type II collagen antibodies. Int. Immunol. 2003;15:987–992. doi: 10.1093/intimm/dxg102. [DOI] [PubMed] [Google Scholar]
- Orr M.T., Wu J., Fang M., Sigal L.J., Spee P., Egebjerg T., Dissen E., Fossum S., Phillips J.H., Lanier L.L. Development and function of CD94-deficient natural killer cells. PLoS ONE. 2010;5:e15184. doi: 10.1371/journal.pone.0015184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A.K., Parker S., Yokoyama W.M., Corbett J.A., Buller R.M. Induction of natural killer cell responses by ectromelia virus controls infection. J. Virol. 2007;81:4070–4079. doi: 10.1128/JVI.02061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Reznikoff G., Dranoff G., Rock K.L. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- Smith H.R., Heusel J.W., Mehta I.K., Kim S., Dorner B.G., Naidenko O.V., Iizuka K., Furukawa H., Beckman D.L., Pingel J.T., et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke D.C., Karupiah G., Zhou J., Palmore T., Irvine K.R., Haeryfar S.M., Williams S., Sidney J., Sette A., Bennink J.R., Yewdell J.W. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E., Kraft J.R., Altman J.D., Jensen P.E., Raulet D.H. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b) J. Exp. Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E., Jamieson A.M., Raulet D.H. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E., Jamieson A.M., Cado D., Raulet D.H. Implications of CD94 deficiency and monoallelic NKG2A expression for natural killer cell development and repertoire formation. Proc. Natl. Acad. Sci. USA. 2002;99:868–873. doi: 10.1073/pnas.022500599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Cherwinski H., Spies T., Phillips J.H., Lanier L.L. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J. Exp. Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., Fang M., Klein-Szanto A., Sigal L.J. Memory CD8+ T cells are gatekeepers of the lymph node draining the site of viral infection. Proc. Natl. Acad. Sci. USA. 2007;104:10992–10997. doi: 10.1073/pnas.0701822104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama W.M., Ryan J.C., Hunter J.J., Smith H.R., Stark M., Seaman W.E. cDNA cloning of mouse NKR-P1 and genetic linkage with LY-49. Identification of a natural killer cell gene complex on mouse chromosome 6. J. Immunol. 1991;147:3229–3236. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.