Abstract
1H NMR (nuclear magnetic resonance spectroscopy) has been used for metabolomic analysis of ‘Riesling’ and ‘Mueller-Thurgau’ white wines from the German Palatinate region. Diverse two-dimensional NMR techniques have been applied for the identification of metabolites, including phenolics. It is shown that sensory analysis correlates with NMR-based metabolic profiles of wine. 1H NMR data in combination with multivariate data analysis methods, like principal component analysis (PCA), partial least squares projections to latent structures (PLS), and bidirectional orthogonal projections to latent structures (O2PLS) analysis, were employed in an attempt to identify the metabolites responsible for the taste of wine, using a non-targeted approach. The high quality wines were characterized by elevated levels of compounds like proline, 2,3-butanediol, malate, quercetin, and catechin. Characterization of wine based on type and vintage was also done using orthogonal projections to latent structures (OPLS) analysis. ‘Riesling’ wines were characterized by higher levels of catechin, caftarate, valine, proline, malate, and citrate whereas compounds like quercetin, resveratrol, gallate, leucine, threonine, succinate, and lactate, were found discriminating for ‘Mueller-Thurgau’. The wines from 2006 vintage were dominated by leucine, phenylalanine, citrate, malate, and phenolics, while valine, proline, alanine, and succinate were predominantly present in the 2007 vintage. Based on these results, it can be postulated the NMR-based metabolomics offers an easy and comprehensive analysis of wine and in combination with multivariate data analyses can be used to investigate the source of the wines and to predict certain sensory aspects of wine.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-011-9487-3) contains supplementary material, which is available to authorized users.
Keywords: Nuclear magnetic resonance metabolomics, Wine sensory attributes, Multivariate data analysis, Wine types, Vintages, Phenolics
Introduction
Wine, a beverage of long tradition and high value, contains a complex mixture of compounds at a wide range of concentrations. The compounds in wine cover a diverse range of metabolites including primary (e.g., sugars, organic acids, amino acids) and secondary metabolites (e.g., flavonoids, hydroxycinnamates, hydroxybenzoates, anthocyanins) (Ali et al. 2010). Unlike ethanol and glycerol, generally low concentrations of other components are present in wine (Amaral and Caro 2005; Kosir and Kidric 2002). Many compounds influence sensory attributes of wine including color, mouth-feel, flavor, and aroma (Pickering and Pour-Nikfardjam 2010).
Different factors like grapevine variety, harvest time, vineyard environment, yeast strain, winemaking technologies, storage time, and human practices, have an affect on wine quality which is determined by the concentration and composition of the compounds present. Thus on this chemical basis, classification and characterization of wine is feasible through qualitative and quantitative information of the wine constituents (Pérez-Magariño and González-San José 2006; Castillo-Sánchez et al. 2006; Matejícek et al. 2005; Guasch-Jane et al. 2004). The most important and basic factor for making wine of good quality is the grape variety and because of this many articles on grapes and wine quality relationships have been published (Gergaud and Ginsburgh 2008).
Cell regulatory processes result in several intermediates and end products which are generally regarded as metabolites. Out of these ‘secondary metabolites’ are produced due to the interaction of the organism with its environment (Pereira et al. 2005). Metabolomics, generally defined as metabolic snap shots of living systems, is a holistic approach aimed at better understanding of primary and secondary metabolism of the biological systems (Clayton et al. 2006). With the advancement in the field of analytical chemistry, more powerful and sophisticated tools (like mass spectrometry and nuclear magnetic resonance) for such chemical analyses have been introduced. The development of methods and algorithms for the multivariate statistical modeling have contributed much to metabolomics as they opened the way for handling the huge datasets of large-scale metabolic analyses (Crockford et al. 2006).
The chemical analysis of complex mixtures like wine can be of great importance as it can be used for the differentiation of wines based on the grapevine variety, yeast strain, geographical origin, terroir, and vintage. Many reports have been published using methods like mass spectrometry (MS) (Giraudel et al. 2007), nuclear magnetic resonance (NMR) spectroscopy (Son et al. 2009), mid-infrared (MIR) spectroscopy (Bevin et al. 2008), gas-chromatography (GC) (Aznar and Arroyo 2007), liquid-chromatography (LC) (Matejícek et al. 2005), atomic emission spectroscopy (AES) (Iglesias et al. 2007), inductively coupled plasma emission spectroscopy (ICP) (Moreno et al. 2008), along with the hyphenation of these methods (Guasch-Jane et al. 2004) for the wine analysis.
Nuclear magnetic resonance has an exceptional place in the chemical analyses of food products and is broadly renowned as one of the most influential modern instrumental analysis techniques. Apart from its routine use in the identification, characterization, and structure elucidation of molecules, NMR is now increasingly popular in the area of metabolome analysis (Son et al. 2009). Although criticized for its low sensitivity, the most promising features of NMR are its non destructive nature, simple sample preparation, in relative short time. A major advantage of NMR is that materials such as wine, beverages, and body fluids can be measured as such, and quantification is easy as for all compounds it is only dependent on the mole concentration. That means with a single internal standard compounds can be quantified without the need for calibration curves for each single compound as needed for all other analytical methods. Moreover sensitivity is not a problem in case of abundant materials such as wine.
Since nearly no sample pretreatment is required in NMR spectroscopy, the inherent properties of the sample are well kept. Since NMR is non selective so it is an ideal tool for the profiling of broad range metabolites in the field of agriculture (Dixon et al. 2006). NMR has been already demonstrated to be a robust method and unaffected by instrumental and experimental factors as it is the case in other analytical methods. Some recent publications reported the use of NMR for the studies like quality control, authenticity or geographical characterization of different food stuffs like coffee, olive oil, tomato and orange juices, wine and beer (Nord et al. 2004; Charlton et al. 2002; D’Imperio et al. 2007; Sobolev et al. 2003; Le Gall et al. 2001).
The aim of the present work was to identify the wine phenolics using 1H NMR spectroscopy with some two-dimensional NMR techniques without any chromatographic fractionation, along with the correlation of sensory data (quality score rating) of the different wine samples with the identified metabolites using different multivariate data analysis methods. The metabolic characterization of wines based on grape cultivar and vintage was also a target.
Materials and method
Wine samples and extraction
Two types of German white wines from Palatinate region (‘Riesling’ and ‘Mueller-Thurgau’) were used in this study. The experimental design included 40 samples of ‘Riesling’ and 19 samples of ‘Mueller-Thurgau’ from the vintages 2006 and 2007. Each sample has been tasted by a panel of experts and scored/rated accordingly. On the basis of scores, samples are classified into four classes. The class number actually indicates the quality grade of wine samples i.e., wines from class 1 were low quality wines whereas wines from class 4 were high quality wines (Table S1).
To generate reference data wines were directly analyzed for their content of ethanol, glucose, fructose, tartaric acid, malic acid, lactic acid, and glycerol by a standard procedure of FTIR (Fourier Transform Infrared Spectroscopy) using WineScanTM 120 FT (FossGmbH, Rellingen, Germany). For NMR spectroscopy each wine sample (20 ml) has been extracted by an equal volume of ethyl acetate. The extraction was repeated two more times and all the three extracts were pooled together. The pooled ethyl acetate fraction was treated with Na2SO4 to remove residual water. Then the Ethyl Acetate was evaporated using a rotary evaporator and stored at −20°C until further use.
NMR spectroscopy
The evaporated ethyl acetate extract was redissolved in 1 ml of deutrated methanol (CH3OH-d 4). An aliquot of 800 μl of sample was transferred to the 5 mm NMR tube and used for the NMR analysis.
NMR parameters
1H NMR spectra were recorded at 25°C on a 500 MHz Bruker DMX-500 spectrometer (Bruker, Karlsruhe, Germany) operating at a proton NMR frequency of 500.13 MHz. CH3OH-d 4 was used as the internal lock. Each 1H NMR spectrum consisted of 128 scans requiring 10 min and 26 s acquisition time with the following parameters: 0.16 Hz/point, pulse width (PW) = 30° (11.3 μs), and relaxation delay (RD) = 1.5 s. A pre-saturation sequence was used to suppress the residual H2O signal with low power selective irradiation at the H2O frequency during the recycle delay. FIDs were Fourier transformed with LB = 0.3 Hz. The resulting spectra were manually phased and baseline corrected, and calibrated to methanol at 3.3 ppm, using XWIN NMR (version 3.5, Bruker). 2D J-resolved NMR spectra were acquired using 600 MHz (Bruker DMX-600) 8 scans per 128 increments for F1 and 8 k for F2 using spectral widths of 5,000 Hz in F2 (chemical shift axis) and 66 Hz in F1 (spin–spin coupling constant axis). A 1.5 s relaxation delay was employed, giving a total acquisition time of 56 min. Datasets were zero-filled to 512 points in F1 and both dimensions were multiplied by sine-bell functions (SSB = 0) prior to double complex FT. J-Resolved spectra tilted by 45°, were symmetrized about F1, and then calibrated, using XWIN NMR (version 3.5, Bruker). 1H–1H correlated spectroscopy (COSY) and heteronuclear multiple bonds coherence (HMBC) spectra were recorded on a 600 MHz Bruker DMX-600 spectrometer (Bruker). The COSY spectra were acquired with 1.0 s relaxation delay, 6,361 Hz spectral width in both dimensions. Window function for COSY spectra was sine-bell (SSB = 0). The HSQC spectra were obtained with 1.0 s relaxation delay, 6,361 Hz spectral width in F2 and 27,164 Hz in F1. Qsine (SSB = 2.0) was used for the window function of the HSQC. The HMBC spectra were recorded with the same parameters as the HSQC spectra except for 30,183 Hz of spectral width in F2. The optimized coupling constants for HSQC and HMBC were 145 and 8 Hz, respectively.
Statistical analyses
The 1H NMR spectra were automatically reduced to ASCII files. Spectral intensities were scaled to total intensity and reduced to integrated regions of equal width (0.04) corresponding to the region of δ 0.0–10.0. The regions of δ 4.85–4.95 and δ 3.20–3.40 were excluded from the analysis because of the residual signal of D2O and CD3OD, respectively. Bucketing was performed by AMIX software (Bruker). Principal component analysis (PCA) based on Pareto scaling while partial least squares projections to latent structures (PLS), orthogonal projections to latent structures (OPLS), and bidirectional orthogonal projections to latent structures (O2PLS) analysis based on UV scaling were performed with the SIMCA-P+ software (v. 12.0, Umetrics, Umeå, Sweden).
Results and discussion
FTIR analysis for wine samples
The reference data for all the wines regarding their ethanol, glucose, fructose, sucrose, tartaric acid, malic acid, lactic acid, and glycerol generated by standard procedure of FTIR is shown in supplementary table. Several other parameters like density, alcohol content, fermented sugar, pH, acidity, and volatile acidity for each wine sample were also measured and represented (Supplementary Table S1).
Extraction and visual analysis of 1H NMR spectra
A total of fifty-nine different wine samples belonging to two wine types from two vintages were analyzed. The extraction procedure followed in this study was found quite effective as compared to most common methods in NMR based wine analyses. Methods like direct analysis of wine (Lopez-Rituerto et al. 2009), lyophilization (Baderschneider and Winterhalter 2001), and the use of nitrogen-flow (Amaral and Caro 2005), are associated with low reproducibility, time consuming, and signal overlapping by dominant compounds in wine, e.g., ethanol, and shifting of signals due to differences in pH of wine, respectively. The ethyl acetate extraction used in this study was found to be fast, does not require a pH control, and has less signal intensity for wine dominating metabolites like ethanol and lactic acid. Around fifty metabolites were identified in the different wine samples using different 2D NMR techniques like J-resolved, COSY (correlated spectroscopy), HMBC (heteronuclear multiple bonds coherence), and HSQC (heteronuclear single quantum coherence) spectra. The metabolites identified cover a wide diversity including amino acids, organic acids, carbohydrates, hydroxycinnamates, hydroxybenzoates, stilbenes, and flavonoids. All assignments were done by comparing the spectra with previous reports (Ali et al. 2009; Liang et al. 2006), and with 1D and 2D NMR spectra of more than 500 common metabolites in our in-house library.
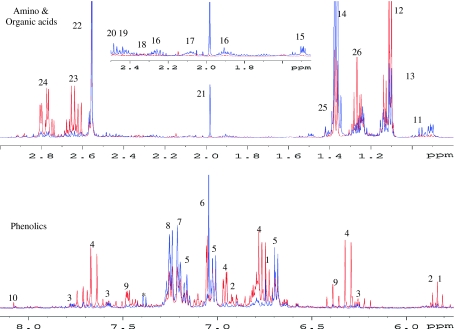
Conventionally a 1H NMR spectrum is divided into three regions. The area between δ 0.8 and 4.0 corresponds to amino acids with few organic acids signals as well. The region of δ 4.0–5.5 is considered to be the region for anomeric protons of carbohydrate and the remaining part, i.e., δ 5.5–8.5, is known as the phenolic region. Figure 1 presents a comparison of 1H NMR spectra of two different wines, ‘Riesling’ and ‘Mueller-Thurgau’ (from the 2007 vintage), showing differences not only in the phenolic contents but also in the organic and amino acids profile of the wines. Different vintages of ‘Riesling’ wine were also compared and the spectral differences among the wine types and vintages are clearly observed and are discussed below.
Fig. 1.
1H NMR spectra of ‘Riesling’ (red) and ‘Mueller-Thurgau’ (blue) from 2007 vintage. 1: (+)-catechin, 2: (−)-epicatechin, 3: quercetin, 4: caffeoyl moiety, 5: cis-resveratrol, 6: gallic acid, 7: tyrosine, 8: phenylalanine, 9: coumaroyl moiety, 10: kaempferol, 11: leucine, 12: valine, 13: 2,3-butanediol, 14: threonine, 15: alanine, 16: GABA, 17: methionine, 18: proline, 19: glutamate, 20: glutamic acid, 21: acetate, 22: succinate, 23: citrate, 24: malate, 25: lactate, 26: ethanol
Compound identification
The use of NMR in metabolomics studies offers many advantages and is considered as a powerful analytical tool for such studies. A problem associated with NMR based metabolomics is the congestion of signals in the spectra especially in case of complex mixtures like wine. This signal overlapping represents the major difficulty in compound identification. This problem is overcome by the use of different 2D techniques like J-resolved, 1H–1H COSY, 1H–13C HMBC, and 1H–13C HSQC, which provide additional information regarding the structure of metabolites. Among the 2D NMR techniques used in this study, the J-resolved is characterized by short measuring time and good quantitative features as compared to other 2D NMR experiments (Viant 2003). The 1H–1H COSY was found very effective in terms of confirming some characteristic correlations among the protons, especially for phenylpropanoids and flavonoids as shown in many metabolomics studies (Ali et al. 2009; Liang et al. 2006). The 13C-related NMR techniques, like 1H–13C HMBC, and 1H–13C HSQC, are not very common in metabolomics studies due to their long measuring time but recently the potential of these techniques in metabolomics has been discussed (Hyberts et al. 2007; Lewis et al. 2007). The assignments of signals to various metabolites identified in the present study are based on our in-house library of NMR data of common metabolites.
Amino acids, carbohydrates, and organic acids in wine
The amino acids not only contribute to wine taste and appearance (Hernandez-Orte et al. 2002) but also have an influence on aromas during the maturing process (Escudero et al. 2000). In wine, nearly all the sugars (from grapes) are consumed by the yeast during fermentation unless fermentation is arrested intentionally to make fortified, dry, late harvested or semisweet wines. Organic acids composition in wine and grape juice is of high importance as it influences the organoleptic properties, is involved in control of microbiological growth, and is a critical parameter in wine stabilization. These acids originated directly from the grapes or are formed in processes like alcoholic fermentation, oxidation of ethanol, and malolactic fermentation. Compounds like tartarate, malate, citrate (from grapes) along with succinate, oxalate, fumarate, citrate (from fermentation) are known to influence the pH of wine. Small amounts of other acids like galacturonate, citramalate, pyruvate, and ketoglutarate, are also present in wine (Castellari et al. 2000).
The high signal intensities in the amino acid region were helpful to elucidate a number of amino and organic acid signals. Amino acids like alanine, threonine, valine, proline, methionine, tyrosine, phenylalanine, glutamine, glutamate, arginine, and aspartate, were identified in wine by comparison with the reference spectra of these compounds. The signals in the carbohydrate regions were highly clustered and overlapped. This region showed the signals of the anomeric protons of β-glucose, α-glucose, and sucrose. Resonances for some other compounds like GABA, choline, glycerol, and 2,3-butanediol were also identified in the same region. A number of signals have been elucidated as organic acids like acetic acid, succinic acid, fumaric acid, formic acid, citric acid, malic acid, lactic acid, and tartaric acid. The 1H NMR chemical shifts for these metabolites are shown in Table 1.
Table 1.
1H NMR chemical shifts (δ) and coupling constants (Hz) of wine phenolics identified by references and using 1D and 2D NMR spectra
| Compounds | Chemical shifts (δ) |
|---|---|
| Alanine | 1.48 (d, J = 7.4) |
| Threonine | 1.32 (d, J = 6.5), 3.51 (d, J = 5.0), 4.27 (m) |
| Valine | 1.01 (d, J = 7.0), 1.06 (d, J = 7.0), 2.28 (m) |
| Proline | 2.35 (m), 3.37 (m) |
| Methionine | 2.15 (m), 2.65 (t, J = 8.0) |
| Tyrosine | 6.85 (d, J = 8.5), 7.19 (d, J = 8.5) |
| Phenylalanine | 3.15 (dd, J = 8.2, 14.5), 3.91 (t, J = 9.6) |
| Glutamine | 2.14 (m), 2.41 (td, J = 16.2, 7.5) |
| Glutamate | 2.13 (m), 2.42 (m), 3.71 (dd, J = 7.0, 1.9) |
| Arginine | 1.75 (m), 3.75 (t, J = 5.5) |
| Aspartate | 2.80 (m), 3.80 (m) |
| β-glucose | 4.58 (d, J = 7.8) |
| α-glucose | 5.17 (d, J = 3.7) |
| Sucrose | 5.39 (d, J = 3.9) |
| GABA | 1.90 (m), 2.31(t, J = 7.5), 3.01 (t, J = 7.5) |
| Choline | 3.20 (s) |
| Glycerol | 3.56 (m), 3.64 (m) |
| 2,3-butanediol | 1.14 (d, J = 6.5) |
| Acetic acid | 1.94 (s) |
| Succinic acid | 2.53 (s) |
| Fumaric acid | 6.52 (s) |
| Formic acid | 8.45 (s) |
| Citric acid | 2.56 (d, J = 17.6), 2.74 (d, J = 17.6) |
| Malic acid | 2.68 (dd, J = 16.6, 6.6), 2.78 (dd, J = 16.6, 4.7), 4.34 (dd, J = 6.6, 4.7) |
| Lactic acid | 1.40 (d, J = 7.0) |
| Tartaric acid | 4.35 (s) |
| cis-Resveratrol | 6.21 (t, J = 2.1), 6.31 (d, J = 2.1), 6.68 (d, J = 13.3), 6.71 (d, J = 8.5), 6.76 (d, J = 13.3), 7.18 (d, J = 8.5) |
| Gallic acid | 7.03 (s) |
| Syringic acid | 3.89 (s), 7.31 (s) |
| Vanillic acid | 3.90 (s), 6.77 (d, J = 8.2), 7.22 (m) |
| p-Benzoic acid | 6.83 (d, J = 8.7), 7.94 (d, J = 8.6) |
| p-Coumaric acid | 6.38 (d, J = 16.0), 6.84 (d, J = 8.8), 7.50 (d, J = 8.8), 7.59 (d, J = 16.0) |
| Caffeic acid | 6.24 (d, J = 16.0), 6.87 (d, J = 8.4), 7.02 (dd, J = 8.4, 2.0), 7.12 (d, J = 2.0), 7.52 (d, J = 16.0) |
| (+)-Catechin | 2.52 (dd, J = 16.1, 8.2), 2.83 (dd, J = 16.0, 5.4), 4.04 (m), 4.55 (d, J = 7.5), 5.89 (d, J = 2.2), 6.75 (d, J = 8.0), 6.80 (dd, J = 8.5, 2.0), 6.88 (d, J = 8.5), 6.9 (d, J = 2.0) |
| (−)-Epicatechin | 2.72 (dd, J = 16.8, 2.6), 2.89 (dd, J = 16.9, 4.6), 4.26 (m), 6.03 (d, J = 2.0), 6.06 (d, J = 2.0), 6.88 (brs), 6.96 (d, J = 2.2) |
| Quercetin | 6.27 (d, J = 2.0), 6.49 (d, J = 2.0), 6.99 (d, J = 8.6), 7.66 (dd, J = 8.5, 2.0), 7.71 (d, J = 2.0) |
| Kaempferol | 6.28 (d, J = 2.0), 6.52 (d, J = 2.0), 6.74 (d, J = 8.6), 8.04 (d, J = 8.6), |
| trans-Caftaric acid | 5.77 (s), 6.29 (d, J = 16.0), 6.88 (d, J = 8.4), 7.02 (dd, J = 8.4, 2.0), 7.12 (d, J = 2.0), 7.52 (d, J = 16.0) |
| trans-p-Coutaric acid | 5.84 (s), 6.36 (d, J = 16.0), 6.87 (d, J = 8.8), 7.51 (d, J = 8.8), 7.59 (d, J = 16.0) |
| cis-Caftaric acid | 5.34 (s), 5.92 (d, J = 13.0), 6.71 (d, J = 8.4), 6.81 (d, J = 13.0), 7.03 (dd, J = 8.4, 2.0), 7.44 (d, J = 2.0) |
| cis-p-Coutaric acid | 5.41 (s), 5.94 (d, J = 13.0), 6.73 (d, J = 9.2), 6.86 (d, J = 13.0), 6.93 (d, J = 9.2), 7.61 (d, J = 9.2) |
Phenolics in wine
Phenolics are a complex group of metabolites particularly contributing to the characteristics of grapes and wines. They are omnipresent and well known for their contribution in pigmentation of different organs along with their role against different biotic and abiotic stresses. In wine they usually are associated with appearance, taste, mouth-feel, to a certain extent fragrance, and antimicrobial activity (Kennedy 2008). Wine phenolics may arise from the fruit (skins and seeds), and/or may be the products of yeast metabolism. Wine phenolics can be classified into three categories which include flavonoids, stilbenoids, and simple phenolics.
Flavonoids are synthesized from the combination of the shikimic and polyketide pathway and are found primarily in the berry skins and seeds of the grapes. They can be further divided into several sub-groups. The most common flavonoids in wine are flavonols (quercetin, kaempferol, myricetin etc.), flavan-3-ols (catechin and epicatechin), and anthocyanins. Polymerization of polyhydroxy flavan 3-ol units, (+)-catechin and (−)-epicatechin, and their gallate esters produces oligomers and polymers called proanthocyanidins (often referred to as procyanidins). Even though procyanidins occur primarily as dimers in grapes (Kennedy 2008), they tend to be polymerized and predominate in wine as condensed tannins.
In the presented study, we successfully identified some flavonoids including both flavonols and flavan-3-ols. Flavonols like quercetin, kaempferol, and myricetin were also identified in the aromatic region. The quercetin signals at δ 6.49 of H-6 is correlated in the 1H–1H COSY spectrum with the signal at δ 6.27 of H-8 and a signal at δ 6.99 of H-5′ with one at δ 7.66 of H-6′. Similar correlations for the signals of kaempferol at δ 8.04 of H-2′ and H-6′ with δ 6.74 of H-3′ and H-5′, and at δ 6.52 of H-8 with δ 6.28 of H-6 were also observed. Likewise myricetin signals, δ 6.47 of H-8 with δ 6.25 of H-6, also showed 1H–1H COSY correlations. Compounds like (+)-catechin and (−)-epicatechin were also identified. For both the (+)-catechin and (−)-epicatechin, signals of H-6′ and H-5′ along with signals of H-6 and H-8 showed correlations in 1H–1H COSY spectra.
Stilbenoids is another class of phenolic compounds in grapes (non flavonoids) and attracted great interest because of their potential valuable health effects but occur in relatively low concentrations as compared to other phenolics (Guebaila et al. 2006). The stilbene contents in wines varied significantly due to factors like climate, grape variety, fungal infection (Perrone et al. 2007), UV light, metal ions (Pussa et al. 2006) and enological methods (Gambuti et al. 2004). Resveratrol (a monomeric stilbene) is the major stilbene of grapes, synthesized from phenylalanine and can exist in two isomeric forms. In wines, both isomeric forms were detected due to photochemical isomerization of trans-form into cis-form during wine making (Sun et al. 2003). Resonances like δ 6.21 (t, J = 2.1 Hz), δ 6.31 (d, J = 2.1 Hz), δ 6.68 (d, J = 13.3 Hz), δ 6.71 (d, J = 8.5 Hz), δ 6.76 (d, J = 13.3 Hz), and δ 7.18 (d, J = 8.5 Hz) are assigned to stilbenoid. The up-field shift of the olefinic protons signals, H-8: from δ 6.79 to δ 6.68, and H-7: from δ 6.89 to δ 6.76, with the reduced coupling constants (from 16.1 to 13.2 Hz), suggests that this compound is the cis-isomer of resveratrol. The 1H–1H COSY spectrum showed correlations among the signals of these olefinic protons along with other correlations like H-4 (δ 6.21) with H-2 and H-6 (δ 6.30), and also between H-6′ (δ 7.18) with H-3′ (δ 6.71).
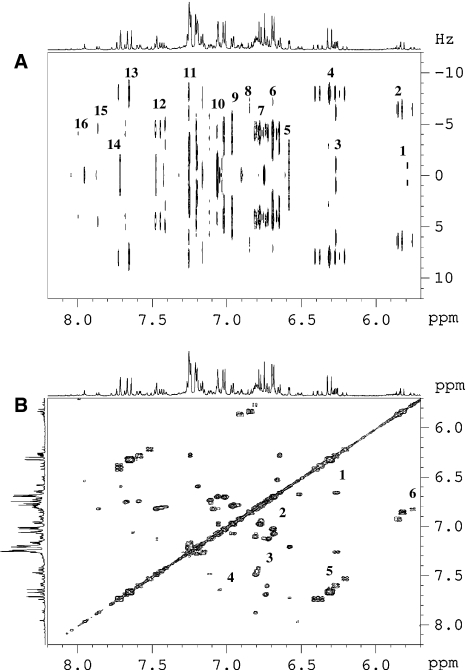
Simple phenolics of grape origin are initially biosynthesized from phenylalanine, whereas those of yeast origin are derived from acetic acid. Simple phenolics are derivatives of hydroxycinnamic acids (HCA) and hydroxybenzoic acids (HBA). Primarily, they are stored in cell vacuoles of grape cells and are released by crushing. In this study, we have identified major HBAs and HCAs along with their esterified forms. When compared with HCA, wines have lower levels of hydroxybenzoic acids (HBAs). The most common HBAs are gallic acid, gentisic acid, protocatechuic acid, and p-hydroxybenzoic acid, which are mainly found in their free form (Pozo-Bayon et al. 2003). In wine the presence of gallic acid has been shown by many studies (Sladkovsky et al. 2004) along with other compounds like protocatechuic, vanillic, and syringic acids (Baderschneider and Winterhalter 2001). The 2D-J resolved and 1H–1H COSY spectra are shown in Fig. 2a, b, respectively.
Fig. 2.
Two dimensional 1H–1H J-resolved (a) and 1H–1H COSY (b) spectra of ‘Riesling’ wine in the range of δ 5.5–δ 8.5. J-resolved (a) shows 1: H-6 of flavan-3-ols, 2: H-8 of cis-phenylpropanoids, 3: H-6 of flavonols, 4: H-8 of trans-phenylpropanoids, 5: H-2 & H-6 of cis-resveratrol, 6: H-8 of cis-resveratrol, 7: H-5 of phenylpropanoids, 8: H-7 of cis-phenylpropanoids, 9 & 10: H-6 of phenylpropanoids, 11: 1H of phenylalanine, 12: H-6 of p-coumaric acid, 13: H-7 trans-phenylpropanoids, 14: H-2 of flavonols, 15: H-2 & H-6 of p-benzoic acid, 16: H-2 of kaempferol. COSY (b) shows correlations between 1: H-6 and H-8 of quercetin, 2: H-5 and H-6 of phenylpropanoids, and H-7 and H-8 of resveratrol, 3: H-5 and H-6 of p-coumaric acid, 4: H-5 and H-6 of quercetin, 5: H-7 and H-8 of trans-phenylpropanoids, 6: H-7 and H-8 of cis-phenylpropanoids
The aromatic part of the 1H NMR spectra of wine show some signals of hydroxybenzoates like gallic acid, salicylic acid, syringic acid, vanillic acid, and p-hydroxybenzoic acid. Resonances for gallic acid, syringic acid, salicylic acid, vanillic acid, and p-hydroxybenzoic acid are also observed. 1H–1H COSY correlations for these compounds have been observed for the signals like δ 7.22 with δ 6.77 of vanillic acid, and δ 7.94 with δ 6.83 of p-hydroxybenzoic acid.
The concentrations of different HCAs depend on factors like grape variety, growing conditions, and climate. Most common hydroxycinnamic acids reported in wine are p-coumaric acid, caffeic acid, sinapic acid, and ferulic acid. The esterified form of HCA with tartaric acid is more common in wine than free HCA. The caftaric acid (caffeic acid conjugated with tartaric acid) is predominant (up to 50%) of the total HCA (Ricardo-Da-Silva et al. 1993). Evidence of the presence of glucosides like 4-O-glucosides of ferulic- and coumaric acid (both cis- and trans-forms), glucosides of caffeic-, coumaric-, ferulic-, and sinapic acid have been shown in some studies (Baderschneider and Winterhalter 2001; Monagas et al. 2005).
Characteristic doublets with a coupling constant of 16.0 Hz in the range of δ 6.20–6.41 and δ 7.51–7.75 represents the H-8′ and H-7′ (olefinic protons) of phenylpropanoids, respectively. The 1H–1H COSY spectra also confirmed the correlation between H-8′ and H-7′ of phenylpropanoids, by the coupling with the carbonyl carbon at δ 168.3 in the HMBC spectra. In 1H NMR spectra of wine samples, these resonances are assigned to four different hydroxycinnamic acids which include caffeic acid, coumaric acid, ferulic acid, and sinapic acid (Table 1). The 1H–1H COSY spectra show correlations among signals like δ 6.92 with δ 6.82 and δ 7.03 of caffeic acid; δ 7.50 with δ 6.84 of coumaric acid; δ 7.09 with δ 6.79 and δ 7.19 of ferulic acid.
Tartaric esters of hydroxycinnamic acid were also identified using 1D and 2D NMR spectra of wine samples. Three singlets for tartaric acid were observed in the region of δ 5.77–5.84 in 1H NMR spectrum. These singlets are downfield shifted from the typical tartaric acid signal at δ 4.80 due to their bonding to the carboxylic function of cinnamates which is confirmed by their correlation with the signal at δ 167.5–168.5 in the HMBC spectrum. The signals of olefinic protons (H-8′ and H-7′) were also slightly shifted. Based on these assignments, three compounds were identified which include caftaric acid (caffeic acid conjugated with tartaric acid), fertaric acid (ferulic acid conjugated with tartaric acid), and p-coutaric acid (coumaric acid conjugated with tartaric acid) (Table 1).
Along with the trans-forms, the cis-forms of these conjugated cinnamic acids, i.e., cis-caftaric acid and cis-coutaric acid, were also detected. When compared to their trans-configuration, the cis-forms showed the up-field shifting of the signals for their H-8′ and H-7′ along with the reduction in the coupling constants from 16.0 to 13.0 Hz. Two clear doublets of 13.0 Hz at δ 5.92 and δ 5.94 were detected for the H-8′ of cis-configuration. The 1H–1H COSY spectra also confirmed this by showing the correlation of these signals with their respective H-7′ protons at δ 6.81 and δ 6.86. It was also confirmed by the correlation of this signal with the carbonyl resonance at δ 167.2 in the HMBC spectra.
Multivariate data analyses (MvDA)
The 1H NMR data were further subjected to principal component analysis (PCA) but before that data were mean centered and then scaled using Pareto scaling. Unlike UV scaling, in which standard deviation is used as the scaling factor, Pareto scaling is characterized as a method in between UV scaling and no scaling and uses the square root of standard deviation as the scaling factor. 1H NMR data (δ 0.32–10.0) of both wines were first reduced by PCA and the results are projected in a score plot (data not shown). The PCA failed to highlight any difference based on quality scores and no grouping of samples was observed due to very high variability among the different wine samples.
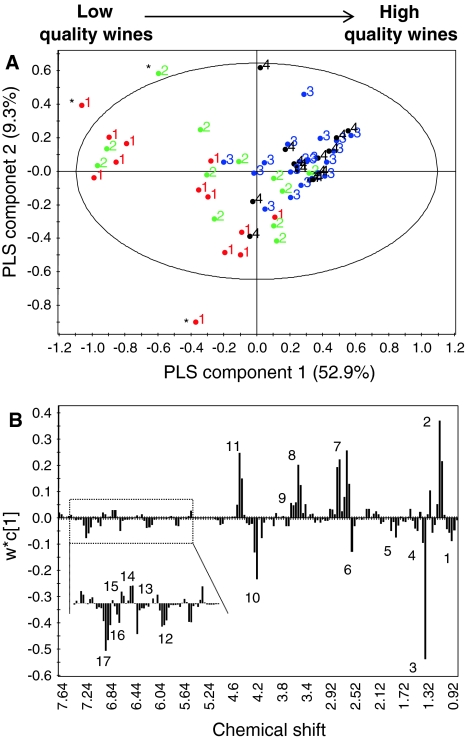
In order to emphasize the differences among the wine samples, we used another data analysis method known as PLS (partial least squares projections to latent structures) on the same NMR data. It is a projection method that, by using a linear multivariate model, can relate the two blocks of variables, i.e., X and Y. In our case X-variables are the values obtained after bucketing the 1H NMR spectra and the Y-variables are the quality scores corresponding to each wine sample. The regression has been validated using cross validation-analysis of variance (CV-ANOVA), with a P-value <0.001. The score plot showed (Fig. 3a) a pattern of arrangement of wine samples according to their quality scores, from inferior to superior tasting wines arranged from negative to positive side of component 1 (52.9%), respectively.
Fig. 3.
Partial least squares projections to latent structures (PLS) score (a) and loading line (b) plots. The score plot (a) shows arrangement of wine samples from low to high quality along component-1. Numbers (1–4) indicate the class to the wine sample belongs. Samples with ‘*’ are outliers. The column plot shows higher levels of compounds like 2: 2,3-butanediol, 7: malate, 8: proline, 9: arginine, 11: tartarate, 13, quercetin, 14: (+)-catechin, and 15: (−)-epicatechin, where as metabolites like 1: valine and leucine, 3: lactate, 4: alanine, 5: acetate, 6: succinate, 10: threonine, 12: caffeoyl moiety, 16: gallate, 17: vanillate, were found discriminating for low quality wines
By examining the loading column plot (Fig. 3b), several compounds were found participating in the discrimination of different wines. The superior wines (belonging to class 3 and 4) were found containing higher levels of amino acids like proline and arginine along with 2,3-butanediol. Organic acids like malic and tartaric acids were also found discriminating for the higher quality scores wine along with some phenolics like quercetin, (+)-catechin, and (−)-epicatechin. For the poor taste wines, resonances for the lactic, acetic, and succinic acid were found higher with some amino acids like threonine and alanine. Among the phenolics caffeic acid, gallic acid, and vanillic acid signals showed higher intensity in low taste wines.
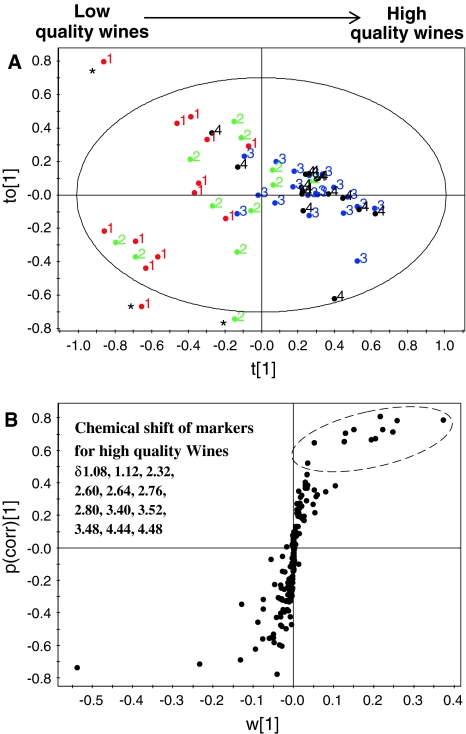
Finally for the identification of quality markers for high quality wine, we used another multivariate data analysis method known as bidirectional orthogonal projection to latent structures (O2PLS). It is a multivariate projection method that removes the structured noise by extracting linear relationships from X and Y data blocks (Trygg and Wold 2002, 2003). Analysis like PLS regression can cause systematic variation of any data block due to structured noise present in the data blocks. The structures noise can be removed from X and Y, in a bidirectional way, by O2PLS regression and results in the decomposition of systematic variation into two model parts: the predictive or parallel part and the orthogonal part. The regression has been validated using cross validation-analysis of variance (CV-ANOVA), with a P-value <0.001. As indicated by Fig. 4a (the score plot of O2PLS regression) the wine samples are arranged according to their quality grade (almost same as PLS). For the identification of markers for the high quality wines, we use S-plot (Fig. 4b). By analyzing the loadings, compounds like 2,3-butanediol, malate, proline, and GABA were found as markers for good taste. For low quality wines, signals of lactate and alanine were identified.
Fig. 4.
Bidirectional orthogonal projections to latent structures (O2PLS) score (a) and loading S-plot (b). The score plot (a) shows arrangement of wine samples from low to high quality. Samples with ‘*’ are outliers. The S-plot (b) shows markers (buckets) for the high quality wine
Proline is the most prominent amino acid in wine. The higher proline content is mainly because of the inability of yeast to use it as a nitrogen source and may be important to give wine the apparent mouth feel or body. This is because a salivary protein, which has a strong affinity for polyphenols, is rich in proline (Bennick 1982). 2,3-butanediol is the major dialcohol in wine, produced from pyruvate or acetoin, and contributes in wine sensory attributes with a slightly bitter taste and viscous body. Higher levels of 2,3-butanediol indicate reduction of acetoin which has a negative impact on wine taste with low taste and odor threshold (Cambon et al. 2006).
The levels of organic acids like malate, lactate, tartarate, succinate, and citrate, mainly depend upon the grape cultivar and/or fermentation process. The malate, tartarate, and citrate, are the three major acids of grapes. The citrate and lactate is formed by malolactic fermentation while succinate is formed by alcoholic fermentation during the wine making process. Increased levels of malic or lactic acid have been associated with increased perception of sourness and astringency (pH dependent) and may be also with a milkier taste of wine (Son et al. 2008). Though the high molecular weight polyphenolics mainly account for color and astringency of wine but the presence of these organic acids apparently augments this (Hufnagel and Hofmann 2008).
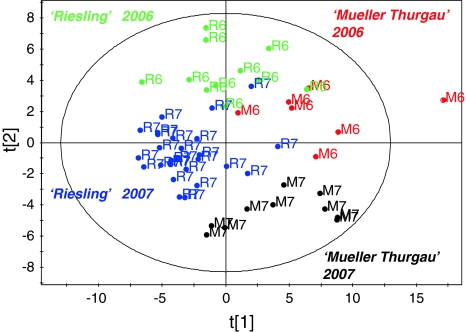
Wine phenolics and amino acids content can be used for the classification of wine (Bevin et al. 2008; Bennick 1982; Anastasiadi et al. 2009; Kosir and Kidric 2001). This study also resulted in characteristic phenolics and amino acids profiles of ‘Riesling’ and ‘Mueller-Thurgau’ wines, dominated by different sets of metabolites. As we have used two types of wine from two different vintages, another multivariate analysis method, i.e., orthogonal projections to latent structures (OPLS), was used to highlight differences among the samples based on grape cultivar and vintage. The samples were classified into four classes and each class was represented by samples from the same year of same cultivar. The regression has been validated using cross validation-analysis of variance (CV-ANOVA), with a P-value <0.001. By examining the score plot (Fig. 5), it can be easily observed that not only ‘Riesling’ was separated from ‘Mueller-Thurgau’, but also the samples from 2006 were distinct from the samples of 2007, in case of both the wine types. Figure 1 also highlights the different metabolic profiles of ‘Riesling’ and ‘Mueller-Thurgau’ wines. The loading plot reveals that among the phenolics compounds like (+)-catechin, (−)-epicatechin, caftarate, and coutarate are present in higher levels in ‘Riesling’ while resonances for quercetin, kaempferol, resveratrol, and gallate were more intense in ‘Mueller-Thurgau’. The amino and organic acid profiles of both wines were also found quite distinctive. Compounds like leucine, alanine, threonine, glutamine, and GABA, were more concentrated in ‘Mueller-Thurgau’ with some organic acids like lactate, acetate, and succinate. ‘Riesling’ was characterized by higher resonances for the amino acids like valine, proline, 2,3 butanediol, along with malate and citrate.
Fig. 5.
Orthogonal projections to latent structures (OPLS) score plot. The score plot clearly indicates differentiation among the samples based on wine types and vintage. The samples M6 with ‘*’ is an outlier
The vintage effect on metabolic profile of grapes and ultimately on wine has been extensively studied (Pereira et al. 2005; Lee et al. 2009) and the importance of the vintage is widely accepted. The amino acids and polyphenols contents are highly affected by the climatic conditions of a grape production area. It has been shown that a hot and dry climate results in a higher proline and phenolic contents in wine (Lee et al. 2009). Our findings also support the effects of vintage on the wine metabolome as samples from different vintages show different metabolic constituents. In the case of both ‘Riesling’ and ‘Mueller-Thurgau’, the vintage effect was also evident as indicated by the Fig. 5. By examining the loading plot, resonances responsible for the differentiation were identified and annotated. For the vintage of 2006, both these wines have higher levels of metabolites like leucine, phenylalanine, malate, acetate, citrate, tartarate, choline, GABA, along with higher levels of phenolics. The 2007 vintage was characterized mostly by higher levels of amino acids like valine, proline, alanine, arginine, threonine, together with organic acids like succinate and lactate.
In general, the enormous potential of NMR for the chemical analyses of complex food items has attracted the interests of many research groups around the world. One- and two-dimensional NMR experiments have been already used for wine (Son et al. 2009) and grape berries (Pereira et al. 2005) classification and characterization but very few reports are available for the quantification of wine components using intensities from 1H NMR spectra (Larsen et al. 2006; Viggiani and Morelli 2008). Most of the papers regarding the NMR analysis of wine either include some chromatography to identify the phenolic compounds or simply ignore the phenolics identification mainly because of the complexity of the spectra. In this study, 1H NMR has been combined with different 2D NMR techniques, like J-resolved, COSY, HMBC, and HSQC, and resulted in successful identification of nearly all the major classes of phenolics, along with amino acids, carbohydrates, and organic acids. This approach has proved quite powerful in identification of cinnamic acids, benzoic acid, stilbenoids, flavonols, and flavane-3-ols, along with differentiating in trans- and cis-forms.
Wine is considered as one of the most complex foods regarding the metabolic contents. The chemistry of wine is known to be highly influenced by many factors which include grape variety, climate, cultural practices, yeast strain, and the fermentation process as well (Rodríguez-Delgado et al. 2002). Several wine characteristics, like mouth feel, aroma, and color, are directly affected by the grape cultivars and many compounds can serve as a marker for wine discrimination (Stines et al. 2000). The present study is an attempt to underline those metabolic differences and link them with sensory analysis. The NMR spectroscopy in conjugation with multivariate data analysis was not only proven to be effective in identification of different compounds in wine but also in highlighting the differences among the wines belonging to different wine types and vintages and which have different quality grades.
Conclusion
In conclusion, 1H NMR based metabolic profiling was shown to be effective in terms of identifying diverse metabolites, including amino and organic acids, carbohydrates, hydroxycinnamates, hydroxybenzoates, and flavonoids, in the complex mixtures present in wine. The extraction method used was found short, simple, and less time consuming with high reproducibility. In combination with 1H NMR, PLS, and O2PLS were found very effective in highlighting the differences among the wines based on quality scores. By using loadings of the score plot, many metabolites responsible for that discrimination have been identified. The effect of vintages on metabolic profiles of wine has also been studied (with OPLS) and clear differences were observed among the samples of different vintages, showing the effects of the climate. It should be mentioned that in terms of quality the interaction among the various components of wine are amazingly complex and knowledge regarding individual components might not be that much useful as such, increasing our insight in their interactions should be of interest. Metabolomics can not only offer to visualize the complexity of these specific interactions and provide better understanding of wines but more importantly how to make good quality wines.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors greatfully acknowledge Stefan Hilz from Landwirtschaftskammer Rheinland-Pfalz and his team for providing samples of wines for NMR analysis and their sensorial classification. This work was done in the frame of the EraNET Genomic Research-Assisted breeding for Sustainable Production of Quality GRAPEs and WINE (GRASP) in coordination with Dr. Eva Zyprian (http://urgi.versailles.inra.fr/projects/GRASP/). The authors thank Higher Education Commission (HEC) of Pakistan for the financial support of Kashif Ali.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ali K, Maltese F, Zyprian E, Rex M, Choi YH, Verpoorte R. NMR metabolic fingerprinting based identification of grapevine metabolites associated with downy mildew resistance. J Agric Food Chem. 2009;57:9599–9606. doi: 10.1021/jf902069f. [DOI] [PubMed] [Google Scholar]
- Ali K, Maltese F, Choi YH, Verpoorte R. Metabolic constituents of grapevine and grape derived products. Phytochem Rev. 2010;9:357–378. doi: 10.1007/s11101-009-9158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral FM, Caro MSB. Investigation of different pre-concentration methods for NMR analyses of Brazilian white wine. Food Chem. 2005;93:507–510. doi: 10.1016/j.foodchem.2004.09.039. [DOI] [Google Scholar]
- Anastasiadi M, Zira A, Magiatis P, Haroutounian SA, Skaltsounis AL, Mikros E. 1H NMR-based metabonomics for the classification of Greek wines according to variety, region, and vintage. Comaprison with HPLC data. J Agric Food Chem. 2009;57:11067–11074. doi: 10.1021/jf902137e. [DOI] [PubMed] [Google Scholar]
- Aznar M, Arroyo T. Analysis of wine volatile profile by purge-and-trap-gas chromatography-mass spectrometry. Application to the analysis of red and white wines from different Spanish regions. J Chromatogr A. 2007;1165:151–157. doi: 10.1016/j.chroma.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Baderschneider B, Winterhalter P. Isolation and characterization of noval Benzoates, Cinnamates, Flavanoids, and Liganans from Reisling wine and screening for antioxidant activity. J Agric Food Chem. 2001;49:2788–2798. doi: 10.1021/jf010396d. [DOI] [PubMed] [Google Scholar]
- Bennick A. Salivary proline-rich proteins. Mol Cell Biochem. 1982;45:83–99. doi: 10.1007/BF00223503. [DOI] [PubMed] [Google Scholar]
- Bevin CJ, Dambergs RG, Fergusson AJ, Cozzolino D. Varietal discrimination of Australian wines by means of mid-infrared spectroscopy and multivariate analysis. Anal Chim Acta. 2008;621:19–23. doi: 10.1016/j.aca.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Cambon B, Monteil V, Remize F, Camarasa C, Dequin S. Effects of GDP1 overexpression in S. cerevisiae commerical wine yeast strain lacking ALD6 genes. Appl Environ Microb. 2006;72:4688–4694. doi: 10.1128/AEM.02975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellari M, Versari A, Spinabelli U, Galassi S, Amati A. An improved HPLC method for the analysis of organic acids, carbohydrates and alcohols in grape musts and wines. J Liq Chromatogr Rel Technol. 2000;23:2047–2056. doi: 10.1081/JLC-100100472. [DOI] [Google Scholar]
- Castillo-Sánchez JJ, Mejuto JC, Garrido J, García-Falcón S. Influence of wine-making protocol and fining agents on the evolution of anthocyanin content, colour and general organoleptic quality of Vinhão wines. Food Chem. 2006;97:130–136. doi: 10.1016/j.foodchem.2005.03.030. [DOI] [Google Scholar]
- Charlton AJ, Farrington WHH, Brereton P. Application of 1H NMR and multivariate statistics for screening complex mixtures: quality control and authenticity of instant coffee. J Agric Food Chem. 2002;50:3098–3103. doi: 10.1021/jf011539z. [DOI] [PubMed] [Google Scholar]
- Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost J-P, Le-Net J-L, Baker D, Walley RJ, Everett JR, Nicholson JK. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK. Statistical heterospectroscopy, and approach to the integerated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- D’Imperio M, Mannina L, Capitani D, Bidet O, Rossi E, Bucarelli FM, Quaglia GB, Segre A. NMR and statistical study of olive oils from Lazio: a geographical, ecological and agronomic characterization. Food Chem. 2007;105:1256–1267. doi: 10.1016/j.foodchem.2007.02.045. [DOI] [Google Scholar]
- Dixon RA, Gang DR, Charlton AJ, Fiehn O, Kuiper HA, Reynolds TL, Tjeerdema RS, Jeffery EH, German JB, Ridley WP, Seiber JN. Applications of metabolomics in agriculture. J Agric Food Chem. 2006;54:8984–8994. doi: 10.1021/jf061218t. [DOI] [PubMed] [Google Scholar]
- Escudero A, Hernandez-Orte P, Cacho J, Ferreira V. Clues about the role of methional as character impact odorant of some oxidized wines. J Agric Food Chem. 2000;48:4268–4272. doi: 10.1021/jf991177j. [DOI] [PubMed] [Google Scholar]
- Gambuti A, Strollo D, Ugliano M, Lecce L, Moio L. trans-Resveratrol, quercitin, (+)-catechin, and (−)-epicatechin content in South Italian monovarietal wines: relationship with maceration time and marc pressing during winemaking. J Agric Food Chem. 2004;52:5747–5751. doi: 10.1021/jf0354895. [DOI] [PubMed] [Google Scholar]
- Gergaud O, Ginsburgh V. Natural endowments, production technologies and the quality of wines in Bordeaux. Does terrior matter? Econ J. 2008;118:F142–F157. doi: 10.1111/j.1468-0297.2008.02146.x. [DOI] [Google Scholar]
- Giraudel JL, Setkova L, Pawliszyn J, Montury M. Rapid headspace solid-phase microextraction-gas chromatographic-time-of-flight mass spectrometric method for qualitative profiling of ice wine volatile fraction. III. Relative characterization of Canadian and Czech ice wines using self-organizing maps. J Chromatogr A. 2007;1147:241–253. doi: 10.1016/j.chroma.2007.02.050. [DOI] [PubMed] [Google Scholar]
- Guasch-Jane MR, Ibern-Gomez M, Andres-Lacueva C, Juregui O, Lamuela-Ravents RM. Liquid chromatography with mass spectrometry in Tandem mode applied for the identification of wine markers in residues from ancient Egyptian vessels. Anal Chem. 2004;76:1672–1677. doi: 10.1021/ac035082z. [DOI] [PubMed] [Google Scholar]
- Guebaila HA, Chira K, Richard T, Mabrouk T, Furiga A, Vitrac X, Monti JP, Delaunay JC, Merillon JM. Hopeaphenol: the first resveratrol tetramer in wines from North Africa. J Agric Food Chem. 2006;54:9559–9564. doi: 10.1021/jf062024g. [DOI] [PubMed] [Google Scholar]
- Hernandez-Orte P, Cacho J, Ferreira V. Relationship between varietal amino acid profile of grapes and wine aromatic composition. Experiments with model solutions and chemometric study. J Agric Food Chem. 2002;50:2891–2899. doi: 10.1021/jf011395o. [DOI] [PubMed] [Google Scholar]
- Hufnagel JC, Hofmann T. Quantitative reconstruction of the nonvolatile sensometabolome of a red wine. J Agric Food Chem. 2008;56:9190–9199. doi: 10.1021/jf801742w. [DOI] [PubMed] [Google Scholar]
- Hyberts SG, Heffron GJ, Tarragona NG, Solanky K, Edmonds KA, Luithardt H, Fejzo J, Chorev M, Aktas H, Colson K, Falchuk KH, Halperin JA, Wagner G. Ultrahigh-resolution 1H–13C HSQC spectra of metabolite mixtures using nonlinear sampling and forward maximum extropy reconstruction. J Agric Food Chem. 2007;129:5108–5116. doi: 10.1021/ja068541x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M, Besalú E, Anticó E. Internal standardization-atomic spectrometry and geographical pattern recognition techniques for the analysis and classification of catalonian red wines. J Agric Food Chem. 2007;55:219–225. doi: 10.1021/jf0629585. [DOI] [PubMed] [Google Scholar]
- Kennedy JA. Grape and wine phenolics: observations and recent findings. Cienc Investig Agrar. 2008;35:107–120. [Google Scholar]
- Kosir IJ, Kidric J. Identification of amino acids in wines by one- and two-dimensional nuclear magnetic resonance spectroscopy. J Agric Food Chem. 2001;49:50–56. doi: 10.1021/jf0008137. [DOI] [PubMed] [Google Scholar]
- Kosir IJ, Kidric J. Use of modern nuclear magnetic resonance spectrscopy in wine analysis: determination of minor compounds. Anal Chim Acta. 2002;458:77–84. doi: 10.1016/S0003-2670(01)01549-5. [DOI] [Google Scholar]
- Larsen FH, van den Berg F, Engelsen SB. An exploratory chemometric study of 1H NMR spectra of table wines. J Chemometr. 2006;20:198–208. doi: 10.1002/cem.991. [DOI] [Google Scholar]
- Le Gall G, Puaud M, Colquhoun IJ. Discrimination between orange juice and pulp wash by 1H nuclear magnetic resonance spectroscopy: identification of marker compounds. J Agric Food Chem. 2001;49:580–588. doi: 10.1021/jf001046e. [DOI] [PubMed] [Google Scholar]
- Lee J-E, Hwang G-S, Berg FVD, Lee C-H, Hong Y-S. Evidence of vintage effects on grape wines using 1H NMR-based metabolomic study. Anal Chim Acta. 2009;648:71–76. doi: 10.1016/j.aca.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Sussman MR, Markley JL. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional 1H–13C NMR spectra. Anal Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YS, Kim HK, Lefeber AWM, Erkelens C, Choi YH, Verpoorte R. Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J Chromatogr A. 2006;1112:148–155. doi: 10.1016/j.chroma.2005.11.114. [DOI] [PubMed] [Google Scholar]
- Lopez-Rituerto E, Cabredo S, Lopez M, Avenoza A, Busto JH, Peregrina JM. A thorough study on the use of quantitative 1H NMR in Roja red wine fermentation processes. J Agric Food Chem. 2009;57:2112–2118. doi: 10.1021/jf803245r. [DOI] [PubMed] [Google Scholar]
- Matejícek D, Mikes O, Klejdus B, Sterbová D, Kubán V. Changes in contents of phenolic compounds during maturing of barrique red wines. Food Chem. 2005;90:791–800. doi: 10.1016/j.foodchem.2004.05.057. [DOI] [Google Scholar]
- Monagas M, Batolome B, Gomez-Cordoves C. Update knowledge about the presence of phenolic compounds in wine. Crit Rev Food Sci Nutr. 2005;45:85–118. doi: 10.1080/10408690490911710. [DOI] [PubMed] [Google Scholar]
- Moreno IM, González-Weller D, Gutierrez V, Marino M, Cameán AM, González AG, Hardisson A. Determination of Al, Ba, Ca, Cu, Fe, K, Mg, Mn, Na, Sr, and Zn in red wine samples inductively coupled plasma optical emission spectroscopy: evaluation of preliminary sample treatments. Microchem J. 2008;88:56–61. doi: 10.1016/j.microc.2007.09.005. [DOI] [Google Scholar]
- Nord LI, Vaag P, Duus J. Quantification of organic and amino acids in beer by 1H NMR spectrscopy. Anal Chem. 2004;76:4790–4798. doi: 10.1021/ac0496852. [DOI] [PubMed] [Google Scholar]
- Pereira GE, Gaudillere J-P, Leeuwen CV, Hilbert G, Lavialle O, Maucourt M, Deborde C, Moing A, Rolin D. 1H NMR chemometrics to characterize mature grape berries in four wine-growing areas in Bordeaux, France. J Agric Food Chem. 2005;53:6382–6389. doi: 10.1021/jf058058q. [DOI] [PubMed] [Google Scholar]
- Pérez-Magariño S, González-San José ML. Polyphenols and colour variability of red wines made from grapes harvested at different ripeness grade. Food Chem. 2006;96:197–208. doi: 10.1016/j.foodchem.2005.02.021. [DOI] [Google Scholar]
- Perrone G, Nocoletti I, Pascale M, De Rossi A, De Girolamo A, Visconti A. Positive correlation between high levels of ochratoxin A and resveratrol-related compounds in red wines. J Agric Food Chem. 2007;55:6807–6812. doi: 10.1021/jf070993l. [DOI] [PubMed] [Google Scholar]
- Pickering GJ, Pour-Nikfardjam MS. Sensory attributes of wine influenced by variety and berry shading discriminated by NMR metabolomics. Food Chem. 2010;121:1296–1304. doi: 10.1016/j.foodchem.2010.01.067. [DOI] [Google Scholar]
- Pozo-Bayon MA, Hernandez MT, Martin-Alvarez PJ, Polo MC. Study of low molecular weight phenolic compounds during the aging of sparkling wines manufactured with red and white grape varieties. J Agric Food Chem. 2003;51:2089–2095. doi: 10.1021/jf021017z. [DOI] [PubMed] [Google Scholar]
- Pussa T, Floren J, Kuldepp P, Raal A. Survey of grapevine Vitis Vinifera stem polyphenols by liquid chromatography-diode array detection-tandem mass spectrometry. J Agric Food Chem. 2006;54:7488–7494. doi: 10.1021/jf061155e. [DOI] [PubMed] [Google Scholar]
- Ricardo-Da-Silva JM, Cheynier V, Samson A, Bourzeix M. Effect of pomace contact, carbonic maceration, and hyperoxidation on the procyanidin composition of Grenach Blanc wines. Am J Enol Vitic. 1993;44:168–172. [Google Scholar]
- Rodríguez-Delgado MA, González-Hernández G, Conde-González JE, Pérez-Trujillo JP. Principal component analysis of the polyphenol content in young red wines. Food Chem. 2002;78:523–532. doi: 10.1016/S0308-8146(02)00206-6. [DOI] [Google Scholar]
- Sladkovsky R, Solich P, Urbanek M. High performance liquid chromatography determination of phenolic compounds in wine using off-line isotachophoretic pre-treatment. J Chromatogr A. 2004;1040:179–184. doi: 10.1016/j.chroma.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Sobolev AP, Segre A, Lamanna R. Proton high-field NMR study of tomato juice. Magn Reson Chem. 2003;41:237–245. doi: 10.1002/mrc.1176. [DOI] [Google Scholar]
- Son H-S, Kim KM, van den Berg F, Hwang G-S, Park W-M, Lee C-H, Hong Y-S. 1H Nuclear magnetic resonsnce-based metabolomic characterization of wines by grape varieties and production areas. J Agric Food Chem. 2008;56:8007–8016. doi: 10.1021/jf801424u. [DOI] [PubMed] [Google Scholar]
- Son H-S, Hwang G-S, Kim KM, Ahn H-J, Park W-M, Berg FVD, Hong Y-S, Lee C-H. Metabolomic studies on geographical grapes and their wines using 1H NMR analysis coupled with multivariate statistics. J Agric Food Chem. 2009;57:1481–1490. doi: 10.1021/jf803388w. [DOI] [PubMed] [Google Scholar]
- Stines AP, Grubb J, Gokowiak H, Henschke P, Hoj PB, Heewijk RV. Proline and arginine accumulation in developing berries of Vitis vinifera L. in Australian vineyards: influence of vine cultivar, berry maturity and tissue type. Aust J Grape Wine Res. 2000;6:150–158. doi: 10.1111/j.1755-0238.2000.tb00174.x. [DOI] [Google Scholar]
- Sun BS, Ferrão C, Spranger MI. Effect of wine style and winemaking technology on resveratrol level in wines. Cienc Tec Vitivinic. 2003;18:77–91. [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16:119–128. doi: 10.1002/cem.695. [DOI] [Google Scholar]
- Trygg J, Wold S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integeral OSC filter. J Chemom. 2003;17:53–64. doi: 10.1002/cem.775. [DOI] [Google Scholar]
- Viant MR. Improved methods for acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Comm. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- Viggiani L, Morelli MAC. Characterization of wines by nuclear magnetic resonance: a work study on wines from the Basilicata region in Italy. J Agric Food Chem. 2008;56:8273–8279. doi: 10.1021/jf801513u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.