Abstract
Objectives:
Mitoxantrone is an anticancer drug widely used in the treatment of various cancers. In the present study the effect of mitoxantrone on chromatin proteins of intact hepatocytes nuclei was investigated and compared with soluble chromatin.
Materials and Methods:
UV/Vis spectroscopy, SDS polyacrylamide gel electrophoresis, and western bolting were used.
Results:
The results show that exposure of intact nuclei to various concentrations of mitoxantrone resulted in the release of histone H1 family proteins, H1 and H1°, in a dose-dependent manner but not core histones and high mobility group proteins. Western blot analysis using antiserum against histones H1 and H1° revealed cross-reactivity and confirmed the result. Spectroscopy results showed that mitoxantrone binds to nuclear components and reduces the absorbances at 608 and 400 nm. The binding isotherms revealed cooperative binding with one binding site.
Conclusion:
From the results it is suggested that mitoxantrone binds to intact nuclei and chromatin with different affinities and linker DNA can be considered as a main binding site for mitoxantrone at the nuclei level.
Keywords: Chromatin, hepatocytes, histone H1 family, Mitoxantrone
Introduction
Mitoxantrone is a synthetic antibiotic widely used as a chemotherapeutic drug for the treatment of solid tumors, leukemia, and lymphoma.[1–3] Numerous studies on the interaction of mitoxantrone with DNA have been undertaken and all indicate that the drug exerts its biological function via intercalation into DNA double strands.[4,5] Mitoxantrone also belongs to the class of topoisomerase II inhibitor.[6,7]
In eukaryotes, DNA is complexed with histones and other nuclear proteins producing a defined structure known as chromatin.[8] There are five main histones in the cell nucleus: the linker histones of the H1 family and four core or nucleosomal histones. The core histones are small, basic proteins with more than 50% of their amino acid composition is lysine and arginine. They are H2A, H2B, H3, and H4, which associate as H2A/H2B dimers and H3/H4 tetramer, arranged in an octamer form. Histone H1 is a very lysine-rich histone fraction of chromatin, which binds to linker DNA between adjacent nucleosomes to facilitate the folding of the chromatin fiber.[9,10] The high mobility group (HMG) proteins are a small set of non histone proteins with relatively low molecular weights and high solubility in water. They bind to DNA and chromatin and act as “architectural elements” that induce both short- and long-range changes in the structure of their binding sites and facilitate various DNA-related activities.[11,12]
In the cell, chromatin has been introduced as a tool for study of genome function in cancer.[13] We have previously shown that mitoxantrone binds to chromatin with higher affinity compared to DNA, implying that the histone proteins may play an important role in the chromatin- mitoxantrone interaction process.[14] In the present study, we have attempted to characterize the binding of anticancer drug mitoxantrone to intact nuclei and compared to soluble chromatin with especial emphasize on chromatin proteins. The results suggest release of proteins upon drug binding to nuclei and linker region are introduced as a preferable sites for mitoxantrone interaction.
Materials and Methods
Chemicals
Mitoxantrone hydrochloride (20 mg/ml in water) was purchased from Helale Ahmar, Tehran, Iran (manufactured by Ebewe Pharma Ges.m.b.H.Nfg.KG, Austria) and stored at 4 ºC in the dark. Before use, it was diluted to desired concentrations with 10 mM Tris--HCl (pH 7.4) and its concentration determined spectrophotometrically using a molar extinction coefficient of 19 200 M-1 cm-1 at 608 nm.
The antiserum against histones (H1 and core histones), H1°, and HMG proteins were raised in rabbits and after purification were used through the experiments.
Nuclei and Chromatin Preparation
The nuclei were prepared from rat liver (hepatocytes) as described.[15] All steps were carried out at 4 ºC in the presence of protease inhibitor phenylmethylsulfonylfluoride (PMSF) at a final concentration of 1 mM and cocktail protease inhibitor (1/100 V/V). The purified nuclei were suspended in reaction buffer (0.25 M sucrose, 25 mM NaCl and 10 mM Tris--HCl, pH 7.4) and its DNA content was determined by measuring the absorbance at 260 nm. Soluble chromatin was prepared from such nuclei as described before.[16] Briefly, the nuclear suspension at A260 = 100 was digested with 3 units of micrococcal nuclease/mg of DNA at 37 ºC for 10 min. The solution was then brought to 10 mM EDTA on ice and centrifuged at 8000 g for 5 min. The pellet was lysed in EDTA and after removing the insoluble material, the clear supernatant was designated as soluble chromatin.
Spectroscopy
The nuclei suspension and soluble chromatin were diluted with the reaction buffer (10 mM Tris –HCl pH 7.4 or 0.25 M sucrose, 25 mM NaCl and 10 mM Tris--HCl, pH 7.4) to the final concentration of 100--200 μg/ml DNA and then incubated with various concentrations of mitoxantrone for 45 min at room temperature in the dark. The samples were then eppendorfed and the absorbance of the supernatants was measured at multi λ system using Shimadzo UV-160 spectrophotometer, equipped with quartz cells at constant temperature. Serial concentrations of mitoxantrone was made in the same buffer and used as a control.
The total and free drug concentrations (Ct and Cf , respectively) were determined using drug extinction coefficient of 19 200 M-1 cm-1 and the amount of bound drug (Cb) was obtained from Cb = Ct –Cf . Binding parameters were determined from the plot of r versus Cf according to the Scatchard method, where r is the molar concentration of bound drug.[17] In this representation, n denotes the apparent number of binding sites and Ka , apparent binding constant. The Hill coefficient nH was determined from the slope of the Ln (r/n–r) versus Ln Cf according to the Hill equation.[18]
Gel Electrophoresis and Western Blot Analysis
The nuclei and chromatin were treated with various concentrations of mitoxantrone and after removing the insoluble materials; the clear supernatants were precipitated with 12% trichloroacetic acid (TCA), and analyzed on 15% SDS-polyacrylamide gel electrophoresis as described by Laemmli.[19] The mini gels (10×10 cm) were run at 100 V for 2 h at room temperature. They were then stained with Coomassie brilliant blue, destained in methanol/acetic acid, and photographed.
For western blotting, the proteins were first run on SDS-PAGE and then transferred electrophoretically onto a nitrocellulose membrane (Hoffer) using 250 mA for 4 h at 4ºC, and then immunobloting.[20,21] The membranes with the immobilized proteins were incubated for 1 h at 37ºC with 1% (W/V) gelatin in buffer A (150 mM NaCl, 50 mM Tris-HCl, pH 7.4), and totally washed three times each for 15 min with buffer A. The reactions with the diluted antibodies in 1% gelatin in buffer A (W/V) were carried out at 4ºC overnight, following removal of excess antibody by washing with buffer A/Tween 20 (0.05%). The membranes were then incubated in peroxidase-conjugated goat anti-rabbit IgG antiserum for 2 h at room temperature. Another wash was performed and the membrane strips were incubated with the substrate solution for 30 min at 37ºC (1.2 ml of 0.3% 4-chloro-1-naphtol in methanol was mixed with 20 ml buffer A and 20 μl H2 O2) and developed.
The Effect of Salt on Mitoxantrone--Chromatin Interaction
A series of chromatin solution in10 mM Tris--HCl buffer pH 7.3 was prepared at a DNA concentration of 100 μg/ml. To one set, 40 μM of mitoxantrone was added and the other set kept as a control. Both sets (with and without drug) were incubated for 45 min at 23 °C in the dark and then various concentrations of NaCl in Tris buffer (0-0.8 M) was added with respect to 4M NaCl. The samples were vortexed and incubated for further 45 min in the same condition as above. Drug treated and the controls were eppendorfed and the supernatants were run on 15% SDS polyacrylamide gels.
Results
Comparison of the Effect of Mitoxantrone on Intact Nuclei and Chromatin
Nuclei and soluble chromatin were prepared from hepatocytes and after determination of their DNA content, they were individually treated with various concentrations of mitoxantrone in the same experimental condition. The absorbance was monitored at 608 and 400 nm in which absorbance at 608 nm is specifically related to the drug wavelength and nuclei or chromatin does not show any absorbance in this region. Also, absorbance at 400 nm defines the turbidity of the reaction mixture. The absorbance reduction at 608 nm was calculated, normalized with respect to the concentration of the reactants and the result is shown in Figure 1a. As is seen, at low concentrations of mitoxantrone (<50 μM), nuclei and chromatin exhibit a similar binding pattern but at higher concentrations the pattern is different. In intact nuclei, absorbance reduction at 608 nm is increased as mitoxantrone concentration is raised. This reduction in absorbance is continued up to 150 μM of drug and then levels off. However, in the case of chromatin, the absorbance reduction is completed at about 75 μM of mitoxantrone.
Figure 1.
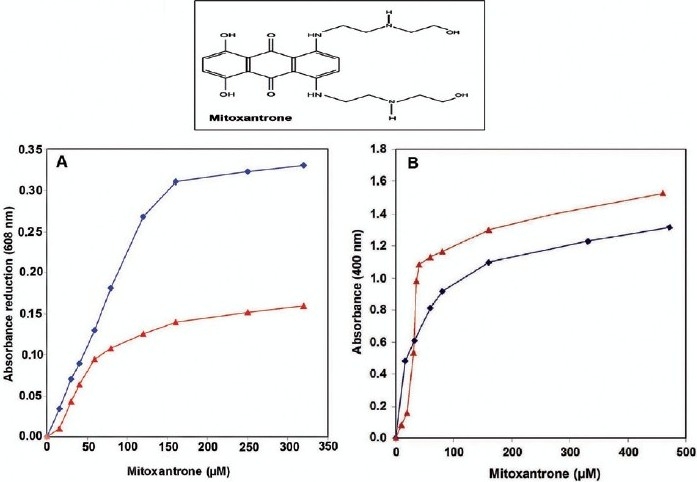
Absorbance changes at 608 nm (a) and turbidity measurements at 400 nm (b) of the interaction of various concentrations of mitoxantrone with hepatocytes nuclei (♦) and soluble chromatin (▲). Interaction was carried out at 23 °C for 40 min in 0.25 M sucrose, 25 mM NaCl and 10 mM Tris--HCl, pH 7.4 (n=4).
A gradual increase in drug concentration produced insoluble material which could be monitored at 400 nm. Figure 1b illustrates the amount of turbidity occurred by mitoxantrone action. In chromatin, turbidity of reaction mixture is continuously increased as mitoxantrone concentration is raised up to 40 μM and then remains unchanged. But in the case of a nuclei solution, turbidity is increased slower than in chromatin and continues up to 150 μM of drug.
Selective Release of Nuclear Proteins from Nuclei by Mitoxantrone
In the next step, we attempted to expose intact nuclei and also soluble chromatin to various concentrations of mitoxantrone to define whether such treatment can release a specific protein. To this end, after treatment of the nuclei and chromatin with mitoxantrone, the proteins were extracted and analyzed on SDS polyacrylamide gel electrophoresis and the result is shown in Figure 2. Calf thymus histones and HMG proteins were also run in parallel for comparison (lanes 1 and 2, respectively). Intact nuclei and chromatin in the absence of drug exhibit the release of numerous proteins (Figure 2a lane 3 and Figure 2b lane 1). Figure 2b shows the proteins pattern of soluble chromatin in the absence and presence of mitoxantrone. In contrast to nuclei, in the case of chromatin in the absence of drug (lane 1) exhibits core histones and a trace of histone H1 as analyzed parallel to the thymus histones (marked as lane 2 in Figure 2a) which correspond to the background of the experiment as described above. In the presence of various concentrations of mitoxantrone, the protein bands start to be retained and the histones tend to be unextractable therefore no band is visible on the gel (Figure 2b lanes 2--4). In other words, as drug concentration is increased the amount of core histone remained in the supernatants is decreased thus at 40 μM of drug all histones are lost on the gel.
Figure 2.
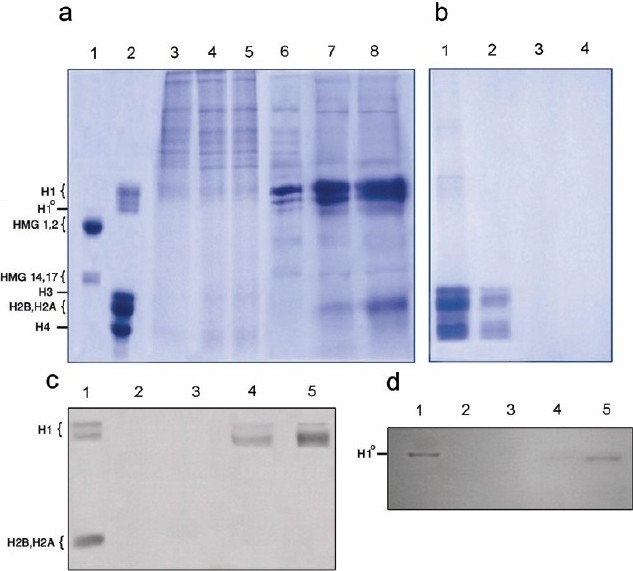
15% SDS-polyacrylamide gel electrophoresis of the proteins released from the chromatin (a) and nuclei (b) upon titration with mitoxantrone. (b), Lanes 1--4 are 0, 10, 20, and 40 μM of mitoxantrone, respectively. (b) Lanes 3-8 are 0, 10, 20, 40, 80, 160, and 220 μM of drug, respectively. Lanes 1and 2 are liver histone and HMG proteins as a standard proteins (n=3). The gels were blotted with histones antibody (c) and H1º antibody (d). Lane 1 is liver histone proteins and lanes 2-5 are nuclei treated with 0, 40, 80, and 160 μM of mitoxantrone. Number of experiments was 3
To identify the nature of the released proteins from the nuclei, we employed the western blot technique, using antiserum against core histone, histone H1, H1º, and HMG proteins in individual experiments and the results are shown in Figure 2c and d. As is seen, only histones H1 and H1º cross react with their antiserum (Figure 2c and d respectively), but core histones and HMG proteins dose not respond to thymus-related proteins antiserum (data not shown).
Considering the higher affinity of mitoxantrone to isolated chromatin compared to intact nuclei, the experiment was designed in the presence of monovalent cation, sodium chloride. Figure 3a represents the SDS gel electrophoresis of the proteins released to the supernatant after treating chromatin with various concentrations of NaCl (0--0.8 M) in the absence of mitoxantrone. As is shown, no significant changes in the proteins pattern of the chromatin as a function of salt is observed. Whereas in the presence of 40 μM (17.7 μg /ml) of mitoxantrone (drug concentration that had a considerable effect on chromatin as shown in Figure 1 and 2b) a significant change in the protein pattern is obtained Figure 3b. The release is started at 0.4 M NaCl, and at 0.8 M salt all five histones are seen on the gel; however, the quantity of the released proteins is still lower than the control in Figure 3a (lane 0).
Figure 3.
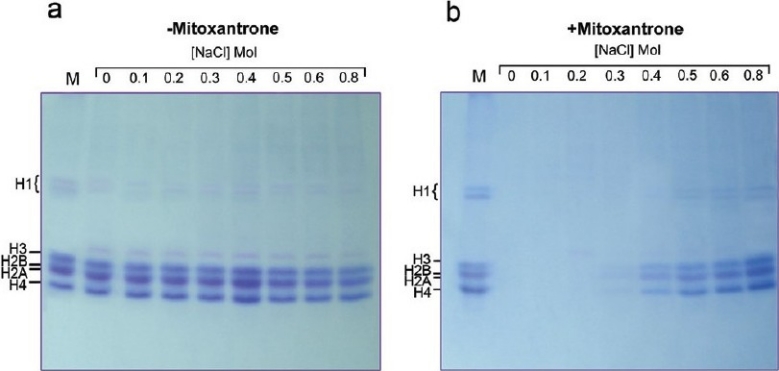
The effect of various concentrations of sodium chloride on the interaction of mitoxantrone with chromatin in the absence (– mitoxantrone) and presence (+mitoxantrone) of 40 μM of drug. Lanes are 0+0.8 M NaCl concentrations. M indicates liver histone proteins as a marker
Comparison of the Binding Isotherms
The binding isotherms obtained for the interaction of mitoxantrone with the intact nuclei and soluble chromatin is shown in Figure 4. Scatchard plots for the nuclei and chromatin shown in Figure 4a represent positive cooperative binding of drug to both nuclei and chromatin but with different extents. Drawing r against Cf Figure 4b indicates that as Cf is increased, r rises rapidly at first, and then levels off, indicating that the system approaches to an equilibrium or a saturating state. In the case of nuclei, the curve reaches a maximum at r =1.2, however in chromatin r =0.6. The number of binding sites (n), Hill coefficient value (nH), and binding constant (Ka) were calculated according to the equations given in the method section. As nH values show, in both cases, the Hill coefficients are >1 indicating that the binding is cooperative. Moreover, the Ka values of 1.4×105 and 7.1×106, and using the equation of ∆Go = –RT ln Ka, total macroscopic negative free energy values of –7.0 K and –9.3 K cal/mol were obtained for the binding of mitoxantrone to nuclei and chromatin, respectively.
Figure 4.
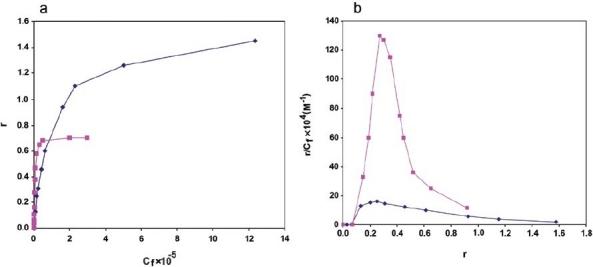
Binding isotherms obtained for the interaction of mitoxantrone with hepatocytes nuclei (♦) and chromatin (▲). The table represents the comparison of the binding parameters obtained for the two experimental states used (n =4)
Discussion
New drug design requires knowledge of the mechanism of the interaction of drugs with cellular targets, which is responsible for its pharmacological activity. Although the mechanism of the interaction is often implied from studies of the affinity of anticancer drugs to free DNA as a main target, the environment of nucleic acids in the cell, especially DNA--protein complexes, may significantly modulate these interactions. Mitoxantrone is an intercalating drug in which exerts its anticancer activity via interaction with a DNA double helix;[22–24] however, its mode of action at the nuclei and chromatin level is still poorly understood.
We have previously reported that mitoxantrone binds to soluble chromatin with higher affinity than to naked DNA.[14] In the present study, the effect of mitoxantrone on hepatocytes nuclei and selective release of nuclear proteins was investigated and compared with chromatin. Although a selective release of nuclear proteins by various antitumor drugs has been studied,[25,26] to date the nature of these proteins have not been identified. Hence, this is the first paper demonstrating the nature of released proteins from the nuclei as a function of an anticancer drug.
The SDS gels suggest that although mitoxantrone reduces the content of chromatin proteins remained in the supernatants, the situation is indeed different in the nuclei in which some proteins are released into environment. In the absence of drug, a spontaneous release of loosely bound proteins has also been observed by others and represents the background of the experiment.[25,27] As mitoxantrone concentration is increased, some proteins are released from the nuclei and some others are lost on the gel. Among them histone H1 and its subtype H1º (a subtype of histone H1 that accumulates in terminally differentiated nondividing cells such as hepatocytes[28,29] ) and the proteins with the same mobility as histone H2A, H2B, and possibly H4 are the most released proteins. Moreover, some of the proteins running ahead of histone H1 are stabilized in the presence of mitoxantrone and are lost on the gel as drug concentration is increased. Therefore, it is suggested that in chromatin, drug has direct access to chromatin components and increasing concentration of drug induces condensation of chromatin possibly via forming cross links between the chromatin components. Whereas in the case of nuclei, accessibility of drug to chromatin components is a different process because of nuclear envelope, hence the protein patterns are completely different. At a low concentration of mitoxantrone only some unknown proteins are released, but at higher concentrations, histone H1 and its subtype H1º are released with an unknown mechanism.
Histones of the H1 family are very lysine-rich protein fractions of chromatin with a very different structure and function from that of core histones. They bind to linker DNA between adjacent nucleosomes to facilitate the folding of the chromatin fiber,[10] and hence, they are also referred as linker histones. The release of these proteins from the chromatin by mitoxantrone action may suggest linker DNA as a main target for drug binding. However at this stage we cannot really speculate the mode of its interaction.
Moreover, in the presence of sodium chloride, chromatin treated with mitoxantrone exhibits a slight release of proteins compared to the control (no salt). This finding confirms our previous results indicating condensation of chromatin in the presence of mitoxantrone.[14]
Spectroscopic results indicated that mitoxantrone binds to chromatin with higher affinity than to intact nuclei representing EC50 values of 49 and 75 μM, respectively. Binding isotherms suggest a cooperative binding pattern for both nuclei and chromatin with a negative free energy indicating that the binding process is spontaneous (exergonic).
The results of the interaction of mitoxantrone with soluble chromatin is in agreement with the results reported previously[14] but the only explanation for slower binding of mitoxantrone to intact nuclei is the nuclear membrane barrier which slows down the accessibility of drug to the chromatin in the cell nucleus.
Taking all together, from the results presented above, it is concluded that apart from DNA, the nuclear proteins play an important role in the drug--chromatin interaction. Although we have extensively studied the interaction of anthracycline antibiotic antitumor drugs, daunomycin and Adriamycin, with the histone proteins[30,31] the release of chromatin proteins upon these drugs action is still unknown. Also the possible binding of mitoxantrone to histone proteins is unclear and demands further work to be elucidated. According to the results presented above, we can consider linker DNA, which is the histone H1 family DNA-binding site in chromatin, as a main binding site for such an anticancer drug.
Conclusion
In the present study the binding affinity of anticancer drug, mitoxantrone, to hepatocyte nuclei and soluble chromatin was investigated. From the results presented above it is concluded that mitoxantrone binds to soluble chromatin with higher affinity than to intact nuclei. Binding of mitoxantrone to both chromatin and nuclei is cooperative and the negative free energy suggests that the binding process is spontaneous. Although mitoxantrone binds to chromatin and produces a compact structure possibly through DNA--protein or protein--protein cross links, the effect of mitoxantrone on hepatocyte nuclei is accompanied by selective release of some proteins. In this case, linker-binding histones H1 and its subtype H1º are the most potent suggesting linker DNA as a main target for mitoxantrone binding at the chromatin level. It is suggested that the protein components of chromatin can also be considered as a target for the activity of this antitumor drug. Studies on the interaction of mitoxantrone with the histone protein directly in solution may reveal the real mechanism of action of mitoxantrone with histone proteins in the nuclei.
Acknowledgments
This work was supported by the grant (no. 6401017/6/07) from the research office of the University of Tehran to AR.
Footnotes
Source of Support: Grant (no. 6401017/6/07) from the research office of the University of Tehran to AR.
Conflict of Interest: None declared.
References
- 1.Holmes FA, Yap HY, Esparza L, Buzdar AU, Hortobaqui GN, Blumenschein GR. Mitoxantrone, cyclophosphamid and 5-fluorouracil in the treatment of hormonally unresponsive metastatic breast cancer. Semin Oncol. 1984;11:28–31. [PubMed] [Google Scholar]
- 2.Thomas X, Archimband E. Mitoxantrone in the treatment of acute myelogenous leukemia: A review. Hematol Cell Ther. 1979;39:163–74. doi: 10.1007/s00282-997-0163-8. [DOI] [PubMed] [Google Scholar]
- 3.Velasquez WS, Lew D, Grogan TM, Spiridonidis CH, Balcerzak SP, Dakhil SR, et al. Combination of fludarabine and mitoxantrone in untreated stages III and IV low-grade lymphoma: 5950. J Clinical Oncol. 2003;21:1996–2003. doi: 10.1200/JCO.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 4.Lown JW, Morgen AR, Yen SF, Wang YH, Wilson DW. Characteristics of the binding of the anticancer agents′ mitoxantrone and ametantrone and related structures to deoxyribonucleic acids. Biochemistry. 1985;24:4028–35. doi: 10.1021/bi00336a034. [DOI] [PubMed] [Google Scholar]
- 5.Krishnamoorthy CR, Sau-Fong Y, Smith JC, Lown JW, Wilson WD. Stoppedflow kinetic analysis of the interaction of anthraquinone anticancer drugs with calf thymus DNA, poly [d(G-C)].poly[d(G-C)], and poly[d(A-T)].poly[d(A-T)] Biochemistry. 1986;25:5933–40. doi: 10.1021/bi00368a015. [DOI] [PubMed] [Google Scholar]
- 6.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: When enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–53. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 7.Giles GI, Sharma RP. Topoisomerase enzymes as therapeutic targets for cancer chemotherapy. Med Chem. 2005;1:383–94. doi: 10.2174/1573406054368738. [DOI] [PubMed] [Google Scholar]
- 8.Bradbury EM, Van Holde KE. In: Chromatin structure and dynamics: A historical perspective in chromatin structure and dynamics: State of the art. Zlatanova J, Leuba SH, editors. The Netherlands Elsevier: Amesterdam; 2004. [Google Scholar]
- 9.Bradbury EM. Nucleosome and chromatin structure and function. J Cell Biochem. 1988;30-31:177–84. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<177::AID-JCB22>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–9. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 11.Agresti A, Bianchi ME. HMG B proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–8. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 12.Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem Sci. 2001;26:152–3. doi: 10.1016/s0968-0004(00)01777-1. [DOI] [PubMed] [Google Scholar]
- 13.Urnov FD. Chromatin as a tool for study of genome functions in cancer. Ann N Y Acad Sci. 2003;983:5–21. doi: 10.1111/j.1749-6632.2003.tb05958.x. [DOI] [PubMed] [Google Scholar]
- 14.Hajihassan Z, Rabbani-Chadegani A. Studies on the binding affinity of anticancer drug mitoxantrone to chromatin, DNA and histone protein. J Biomed Sci. 2009;16:31–7. doi: 10.1186/1423-0127-16-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgoyne LA, Waqar MA, Atkinson MR. Calcium dependent priming of DNA synthesis in isolated rat liver nuclei. Biochem Biophys Res Commun. 1970;39:254–9. doi: 10.1016/0006-291x(70)90786-2. [DOI] [PubMed] [Google Scholar]
- 16.Rabbani A, Iskandar M, Ausio J. Daunomycin-induced unfolding and aggregation of chromatin. J Biol Chem. 1999;274:18401–6. doi: 10.1074/jbc.274.26.18401. [DOI] [PubMed] [Google Scholar]
- 17.Scatchard G. The attraction of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660–72. [Google Scholar]
- 18.Freifelder DM. Physical biochemistry. 2nd ed. New York: 1982. Chapter 18. [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Twobin H, Staehelin T, Groden J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;79:4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demaio A. Protein blot and immunoblotting using nitrocellulose membranes. In: Dunbar BS, editor. Protein blotting. Oxford: Oxford University Press; 1996. pp. 11–32. [Google Scholar]
- 22.Parker BS, Buley T, Evison BJ, Cutts SM, Neuman GM, Iskander MN, et al. A molecular understanding of mitoxantrone-DNA adducts formation: Effect of cytosine methylation and flanking sequence. J Biol Chem. 2004;279:18814–23. doi: 10.1074/jbc.M400931200. [DOI] [PubMed] [Google Scholar]
- 23.Li N, Ma Y, Yang C, Guo L, Yang X. Interaction of anticancer drug, mitoxantrone with DNA analyzed by electrochemical and spectroscopic methods. Biophys Chem. 2005;116:199–205. doi: 10.1016/j.bpc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg LS, Carvlin MJ, Krugh TR. The antitumor agent mitoxantrone binds cooperatively to DNA: Evidence for heterogeneity in DNA conformation. Biochemistry. 1986;25:1002–8. doi: 10.1021/bi00353a008. [DOI] [PubMed] [Google Scholar]
- 25.Bartkowiak J, Kapuscinski J, Melamed MR, Darzynkiewicz Z. Selective displacement of nuclear proteins by antitumor drugs having affinity for nuclei acids. Proc Natl Acad Sci USA. 1989;86:5151–4. doi: 10.1073/pnas.86.13.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassota P, Melamed MR, Darzynkiewicz Z. Release of specific proteins from nuclei of HL-60 and MOLT-4 cells by antitumor drugs having affinity to nucleic acids. Biochm Pharmacol. 1991;41:1055–65. doi: 10.1016/0006-2952(91)90214-p. [DOI] [PubMed] [Google Scholar]
- 27.Santi P, Papa S, Del Coco R, Falcieri E, Zini N, Marinelli F, et al. Modification of the chromatin arrangement induced by ethidium bromide in isolated nuclei, analyzed by electrone microscopy and flow cytometry. Biol Cell. 1987;59:43–54. doi: 10.1111/j.1768-322x.1987.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 28.Zlatanova JS, Doenecke D. Histone H1º: A major player in cell differentiation. FASEB J. 1994;8:1260–8. doi: 10.1096/fasebj.8.15.8001738. [DOI] [PubMed] [Google Scholar]
- 29.Ghadam P, Rabbani A. Occurrence of histone H1º protein in rat alveolar macrophages. Mol Cell Biochem. 2000;236:45–51. doi: 10.1023/a:1016193909193. [DOI] [PubMed] [Google Scholar]
- 30.Rabbani A, Abdosamadi S, Sari-Saraf N. Affinity of anticancer drug, daunomycin, to core histones in solution: Comparison of free and cross-liked proteins. Acta Pharmacol Sin. 2007;28:731–7. doi: 10.1111/j.1745-7254.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 31.Zahraei Z, Rabbani-Chadegani A. A comparison of the effects of anticancer drugs, idarubicin and Adriamycin, on soluble chromatin. Eur J Pharmacol. 2007;575:28–33. doi: 10.1016/j.ejphar.2007.07.045. [DOI] [PubMed] [Google Scholar]


