Abstract
Hemes c are characterized by their covalent attachment to a polypeptide via a widely conserved CXXCH motif. There are multiple biological systems that facilitate heme c biogenesis. System I, the cytochrome c maturation (CCM) system, is found in many bacteria and is commonly employed in the maturation of bacterial cytochromes c in Escherichia coli-based expression systems. System III, cytochrome c heme lyase (CCHL), is an enzyme found in the mitochondria of many eukaryotes and is used for heterologous expression of mitochondrial holocytochromes c. To test CCM specificity, a series of Hydrogenobacter thermophilus cytochrome c552 variants was successfully expressed and matured by the CCM system with CXnCH motifs where n=1–4, further extending the known substrate flexibility of the CCM system by successful maturation of a bacterial cytochrome c with a novel CXCH motif. Horse cytochrome c variants with both expanded and contracted attachment motifs (n=1–3) were also tested for expression and maturation by both CCM and CCHL, allowing direct comparison of CCM and CCHL substrate specificities. Successful maturation of horse cytochrome c by CCHL with an extended CXXXCH motif was observed, demonstrating that CCHL shares the ability of CCM to mature hemes c with extended heme attachment motifs. In contrast, two single amino acid mutants were found in horse cytochrome c that severely limit maturation by CCHL, yet were efficiently matured with CCM. These results identify potentially important residues for the substrate recognition of CCHL.
Introduction
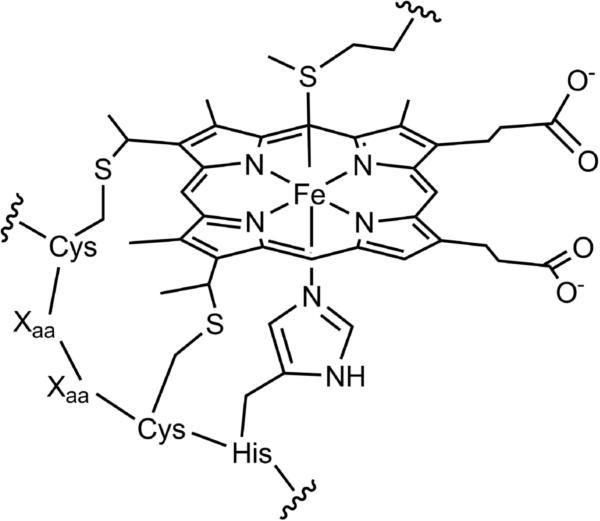
Heme c is a biological cofactor that consists of iron protoporphyrin IX covalently bound to a protein via two thioether linkages (Fig. 1). These thioether bonds are derived from two cysteine residues that are most commonly found in a Cys-Xaa-Xaa-Cys-His (CXXCH) heme attachment sequence. At least five maturation systems have been identified;1-3 of these, three systems (System I, II, and III) have been more well characterized.4 System I, the cytochrome c maturation (CCM) system, involves genes ccmA–H5 and is found in various bacteria and in plant mitochondria.6,7 System I matures cyts c in the periplasmic space of bacteria. System II, cytochrome c synthesis (CCS), consists of four modular components and is in various bacteria8 as well as the thylakoid membranes of plants and algae.9 System III, also called cytochrome c heme lyase (CCHL),10 is a single enzyme found in the mitochondria of fungi, vertebrates and invertebrates.6 Current knowledge of each of the mechanisms is incomplete; there are a number of reviews on the subject.6,11–13
Fig. 1.
Heme c and CXXCH heme attachment motif showing His and Met as the axial ligands.
CCM shows low substrate specificity, which appears to be simply a CXXCH polypeptide sequence with no dependence on flanking amino acids identified to date. Some exceptions exist, for example, the Eschericia coli periplasmic disulfide isomerase DsbC has a CXXCH motif but is not matured by CCM. In this case, it is likely that the target protein folds too fast and is too stable to allow heme attachment.14 However, this avoidance of heme c maturation of a CXXCH by CCM should be interpreted as a special case. Artificially engineered CXXCH sites as well as short peptides containing CXXCH motifs can be successfully matured.15 In nature the intervening (XX) residues have been found to be any amino acid except for cysteine. However, mutational analysis has shown that maturation by CCM can occur with cysteines in and around the CXXCH motif.16 The histidine, which in a mature heme c is the proximal axial ligand, is strictly necessary,17 as well as both ligating cysteines.18 The CCM system is unable to mature the CXXCK motif found in E. coli NrfA19 and a AKGCH2 motif found in W. succinogenes MccA;20 these cytochromes c (cyts c) have dedicated heme lyases.
For the majority of hemes c the heme attachment motif contains two residues between the cysteines. However, some CX3CH and CX4CH sequences exist, for example, in tetraheme cyts c3. These extended motifs still use CCM for maturation.21 In addition, similar extended motifs have been engineered into a cytochrome b with successful maturation by CCM.22 Attempts to mature heme c motifs with five or six intervening residues, however, resulted in heterogeneous products and aberrant heme attachment containing extra sulfurs.23 There is a known example of a CX15CH heme c motif, but this requires a dedicated heme lyase for maturation.3,24 Overall, despite these known limitations, CCM is a flexible system with extremely broad and robust substrate recognition.
CCHL, found in higher organisms, is a dedicated system that shows greater specificity than CCM. For example, while the CCM apparatus from E. coli can mature non-native mitochondrial cyts c, yeast CCHL cannot effectively mature bacterial cyts c.25 In yeast mitochondria, both cytochrome c (cyt c) and cyt c1 have specific heme lyases known as CCHL and CC1HL. However, higher organisms like humans have only a single heme lyase that matures both cyt c and cyt c1.26 CCHL and CC1HL share a common heme-binding motif (Cys-Pro-Val) yet have little sequence homology. The sequences of cyt c and cyt c1 likewise share little homology, but yeast CCHL has been shown to mature enough cyt c1 for cell survival in yeast. Human CCHL can likewise mature both yeast cyt c and cyt c1.26
The knowledge of heme c biogenesis has allowed high-yield recombinant expression of bacterial cyts c in E. coli. The genes ccmA–H were cloned into a plasmid (pEC86) that results in efficient maturation of bacterial monoheme27,28 and multiheme29 cyts c in the periplasmic space upon coexpression with the cyt c structural gene. A signal sequence for periplasmic delivery that is later cleaved in vivo is required for maturation. A mitochondrial cyt c from horse heart with an added signal sequence has also been properly matured by E. coli CCM in moderate yield using this method.30 However, for high-yield mitochondrial cyt c expression, coexpression of the gene for Saccharomyces cerevisiae CCHL (CYC3) along with the cyt c structural gene generally gives higher yields.31 This approach results in correct maturation of mitochondrial cyts c from various sources in the cytoplasm of E. coli.32 Although the factors determining CCHL substrate recognition are largely unknown, it has been found that the second X residue in the heme binding motif of cyt c1 (CXXCH) is important for its recognition as a substrate to CC1HL33 and CCHL.26
In this work, the yields in E. coli of a bacterial holo-cyt c552 from Hydrogenobacter thermophilus (Ht cyt c) are determined for variants with extended and contracted heme attachment motifs matured by CCM. Also, the maturation of variants of horse mitochondrial cyt c (h-cyt c) is explored using both periplasmic CCM maturation and cytoplasmic CCHL maturation. The use of both CCM and CCHL as heme attachment systems for h-cyt c variants allows direct testing and comparison of the factors influencing substrate recognition by CCM and CCHL.
Results
Extended heme attachment motifs
Variants containing extended heme attachment motifs (CXnCH where n = 3 or 4) were constructed for Ht cyt c. For h-cyt c, a variant with a CX3CH motif was constructed. In Ht cyt c the native CMACH motif was extended to CMAGCH (Ht 3X) and CAMAGCH (Ht 4X). In h-cyt c, the native CAQCH (Hh WT) was extended to CAQGCH (Hh 3X). The mutation of h-cyt c was performed in the pBTR(hCc) vector34 that also contains the gene for yeast CCHL for maturation in the cytoplasm of E. coli. The mutation was also performed for the h-cyt c gene in the pSHHC vector30 that includes a signal sequence on the 5’ side of the h-cyt c gene for translocation of the expressed polypeptide into the periplasmic space. The hcyt c gene within pSHHC was co-expressed with the ccmA–H genes within the pEC86 plasmid.28 The two h-cyt c constructs allow comparison between maturation by CCHL and CCM for a single protein CX3CH variant.
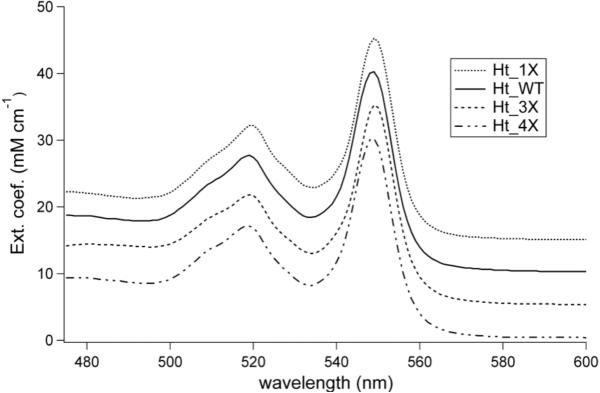
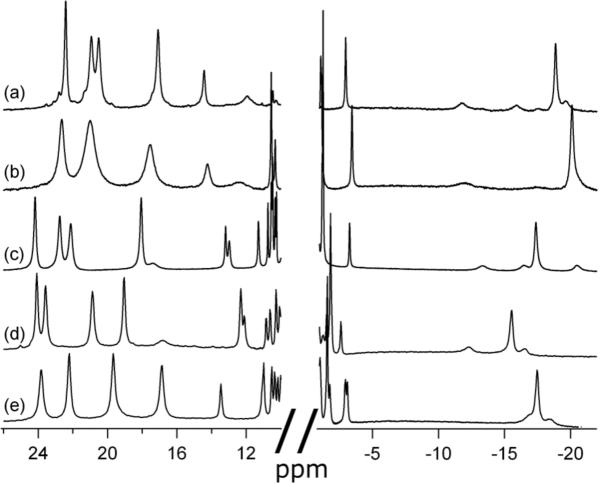
Purification of both Ht 3X and Ht 4X, coexpressed with CCM, yields a major fraction that elutes with the same profile as Ht wild-type (Ht WT), suggesting that major rearrangements in folding do not occur as a result of these mutations. All yields are reported in Table 1. The pyridine hemochrome spectra were measured by denaturation of heme protein by NaOH in the presence of excess pyridine, giving a bis-pyridine adduct. These spectra are indistinguishable from that of Ht WT under the same conditions with absorbance maxima at 549 nm (Fig. 2). The 1D 1H-NMR spectrum for each oxidized Ht cyt c mutant displays one set of four heme methyl resonances in the downfield region, as expected for homogeneous heme environment (Fig. 3c–e). The upfield (~ -20 to -25 ppm) resonance arising from the Met ε -CH 353 is observed, demonstrating methionine ligation in the variants.
Table 1.
Cytochrome c yields in milligrams of protein per gram of wet cells. Maturation was accomplished using either yeast cytochrome c heme lyase (CCHL) or the cytochrome c maturation (CCM) system.
| Horse cyt c | Ht cyt c | ||
|---|---|---|---|
| Variant | CCHL | CCM | CCM |
| 1Xa | N.D. | N.D. | 0.23 |
| WT | 1.03 | 0.60 | 2.93 |
| 3Xb | 0.27 | 0.77 | 2.62 |
| 4Xc | - | - | 2.73 |
| G13ins | 0.3d | 0.68 | - |
| F10A | N.D. | 0.30 | - |
CGCH heme attachment sequence for both horse and Ht cyt c.
CAQGCH for horse and CMAGCH for Ht cyt c.
CAAQGCH for horse and CAMAGCH for Ht cyt c.
Heterogeneous product; presence of correctly matured protein is uncertain.
N.D. indicates that expressed cytochrome was not detected.
Fig. 2.
Pyridine hemochrome spectra for reduced Ht WT and the Ht heme attachment variants matured with CCM. The alpha-band positions at 549 nm indicates c-type heme attachment through two thioether bonds for all variants. Spectra are offset by 5 units on the vertical axis to aid comparison.
Fig. 3.
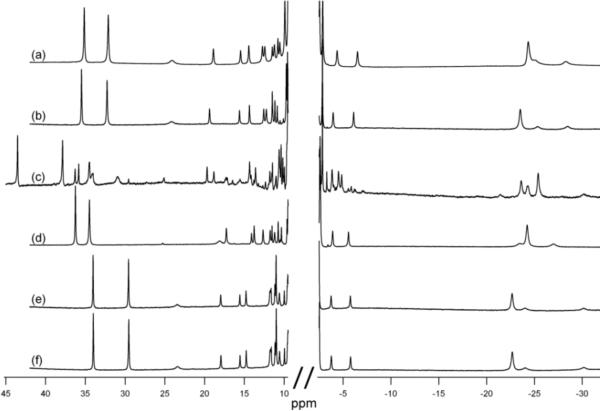
1H-NMR spectra for oxidized Ht cyt c variants matured with CCM. (a) Ht 1X at 40 °C (b) Ht 1X at 25 °C (c) Ht WT at 25 °C (d) Ht 3X at 25 °C (e) Ht 4X at 25 °C.
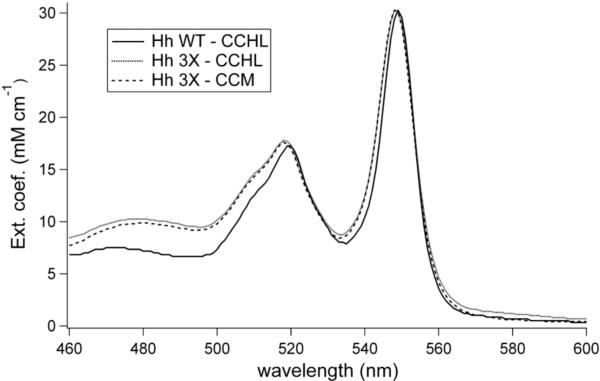
The Hh 3X mutant was expressed and purified with maturation by either CCHL or the CCM system. The FPLC elution profiles are the same as that of Hh WT, indicating no major perturbation of folding. For both samples, pyridine hemochrome spectra are similar to Hh WT (Fig. 4), consistent with the presence of heme c. The 1H-NMR spectrum displays downfield-shifted heme methyl resonances indicating a homogenous heme environment (Fig. 5e and f). The Met ε-CH3 resonance also is observed.
Fig. 4.
Pyridine hemochrome absorption spectra of the reduced Hh 3X variant matured with either the cytochrome c heme lyase (CCHL) or cytochrome c maturation (CCM) system. A pyridine hemochrome spectrum of reduced Hh WT is included for comparison. The spectra were normalized to a pyridine hemochrome alpha-band extinction coefficient of 30.27 mM-1 cm-1.
Fig. 5.
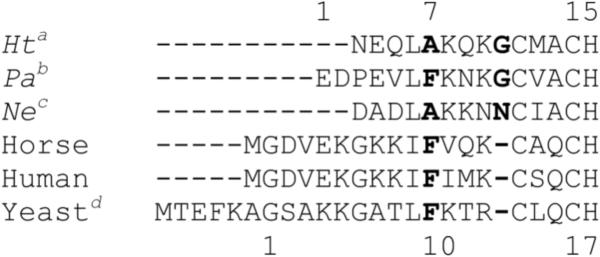
Sequence alignment of cyts c by Clustal showing the N-terminal helix region and the CXXCH heme attachment motifs. The numbering scheme for bacterial cyts c is included above the sequences, and the scheme for mitochondrial cyts c is included below the sequences. The position varied for Hh F10A is in bold type, as well as the position of insertion for the G13ins variant. Bacterial cyts c are the cyt c552 or cyt c551 proteins from (a) Hydrogenobacter thermophilus, (b) Pseudomonas aeruginosa and (c) Nitrosomonas europaea. Mitochondrial cyts c are from horse, human and (d) Saccharomyces cerevisiae.
Contracted heme attachment motifs
To further test the substrate flexibility of CCM, the Ht 1X mutant was made with the heme attachment sequence as CGCH. Glycine was chosen as the intervening residue for its conformational flexibility. The same CGCH sequence was used for h-cyt c. For Ht 1X, pink cell pellets were obtained indicating expression of holo-cytochrome c. The yield is decreased over 12-fold relative to Ht WT (Table 1). The UV-vis spectrum of reduced Ht 1X shows an alpha band slightly blue-shifted from 552 to 550 nm (data not shown), but the alpha band for the pyridine hemochrome remains at 549 nm (Fig. 2). 1H-NMR shows one set of heme methyl peaks (Fig. 3b). However, these are broadened at room temperature relative to Ht WT. Two of the methyl resonances overlap at 25 °C, but increasing the temperature to 40 °C resolves the two peaks (Fig. 3a).
The Hh 1X variant matured by CCM, however, does not produce detectable expression of holo-cytochrome c. In fact, growth of E. coli transformed with both pEC86 and pSHHC containing the Hh 1X mutation is severely inhibited, indicating toxicity of the gene product. When the variant is expressed with yeast CCHL in the cytoplasm, the E. coli growth rate appears normal, but a green pellet is obtained in TB medium. A small but detectable amount of cytochrome is obtained after purification, yet this displays an abnormal pyridine hemochrome spectrum with a broad absorbance band at 575 nm that appears with the addition of dithionite, indicating improper heme attachment. In LB medium, the green color is absent and white cells are produced.
Mutations external to the heme attachment motif
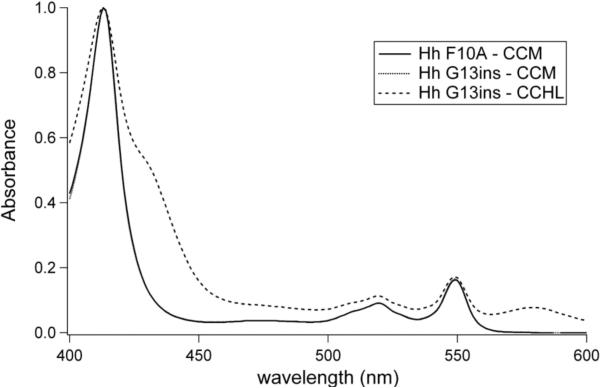
To further test the specificity of heme lyase, two mutations near the CXXCH motif were made in h-cyt c coexpressed with either CCHL or with the CCM genes. The mutations were modeled on the structure and sequence of Ht cyt c.. The Hh G13ins variant introduces a residue immediately before the CXXCH motif that is present in bacterial cyts c (Fig. 5). The residue in Ht WT is Gly11, although its identity is not conserved among bacterial cyts c. When expressed with heme lyase in the cytoplasm, dull pink cell pellets are obtained. However, the absorption spectrum of reduced protein after FPLC purification shows two unusual peaks at 564 and 622 nm (data not shown). The pyridine hemochrome spectrum also contains a large and broad absorbance at 579 nm which is abnormal for c-type cytochromes (Fig. 6). In addition, the NMR spectrum of oxidized Hh G13ins matured by CCHL (Fig. 7c) indicates heme environment heterogeneity, and displays heme methyl chemical shifts outside of the typical range.35 When matured by CCM in the periplasm, however, Hh G13ins has a typical absorption spectrum (Fig. 6) and an NMR spectrum like that of Hh WT, indicating a homogeneous heme environment (Fig. 7d).
Fig. 6.
Pyridine hemochrome spectra for reduced Hh G13ins matured with CCHL compared to both Hh G13ins and Hh F10A matured by CCM. The spectra of the proteins matured by CCM are indistinguishable. The anomalous spectrum of Hh G13ins matured by CCHL did not allow for typical pyridine hemochrome extinction coefficients to be used, so all spectra were normalized to a Soret absorbance of 1.0.
Fig. 7.
1H-NMR spectra of oxidized h-cyt c variants. (a) Hh WT matured by yeast CCHL, (b) Hh F10A matured by CCM, (c) Hh G13ins matured by CCHL, (d) Hh G13ins matured by CCM, (e) Hh 3X (CAQGCH attachment sequence) matured by CCHL, (f) Hh 3X matured by CCM.
Phe10 is conserved in mitochondrial cyts c.36 The corresponding residue in bacterial cyts c is in position 7 and is not strictly conserved (Fig. 5). It can be Phe as in cytochrome c551 from Pseudomonas aeruginosa (Pa), a protein closely related to Ht cyt c, but is more commonly Ala as it is in Ht cyt c.36 The mutation of Phe10 to Ala in h-cyt c abolishes maturation by heme lyase. Green cell pellets are obtained that give minimal heme absorbance following lysate clarification by ion exchange chromatography on CM Sepharose. However, no product is detected after FPLC purification. Upon growth in LB, a less rich medium, white cell pellets are obtained. In contrast, the F10A mutant of h-cyt c is able to be matured by expression in the periplasm by CCM (Table 1). In this case the purified protein has absorption (Fig. 6) and 1H-NMR spectra (Fig. 7b) similar to Hh WT.
Discussion
Heme attachment motif variants in Ht cyt c
Heme attachment motifs with three or four residues between the ligating cysteines are infrequent but do exist in nature and are matured with CCM.21 An artificial heme c protein, engineered from a heme b protein has already been successfully matured with CX3CH and CX4CH motifs.22 In addition, a functional cyt c2 has been matured by CCM with a CX3CH motif.37 The successful CCM-mediated maturation of Ht cyt c, a highly thermostable bacterial heme c protein, with extended heme attachment motifs is therefore not surprising. Although the yield of the CX3CH cyt c2 was reported to decrease two-fold upon extension of the motif,37 the comparable yields of homogenous Ht 3X and Ht 4X with Ht WT (Table 1) confirm the more recent results22 indicating that CCM-mediated maturation is not hindered by these extended motifs.
The pyridine hemochrome spectrum allows for determination of protein extinction coefficients, but is also an indicator of correct heme c attachment. A heme b or a heme attached by a single thioether bond would display an absorption maximum for the “alpha” (longest wavelength) band red-shifted by approximately 7 or 3 nm respectively.22 The Ht 3X and Ht 4X variants, however, displayed the alpha band absorption maximum at 549 nm, diagnostic of heme c bound by two thioethers (Fig. 2).
The oxidized form of cytochrome c is paramagnetic (S = ½), and consequently has an 1H-NMR spectrum in which heme and axial ligand proton resonances are shifted outside of the crowded diamagnetic region, facilitating their detection. The downfield heme methyl peaks are most notable, as well as the upfield axial methionine peaks. These resonances provide a sensitive measure of the homogeneity and integrity of the heme environment.38 The 1H-NMR spectra for the Ht 3X and Ht 4X variants both have one set of downfield-shifted 3-proton intensity peaks consistent with heme methyls as well as the expected upfield-shifted 3-proton intensity axial Met ε-CH3 resonance (Fig. 3d,e), indicating homogenous product after purification. The shifting of these resonances relative to Ht WT indicates some perturbation to the heme conformation39 and/or heme axial ligands,40 which are not unexpected consequences of perturbing the heme attachment site.
Contracting the heme attachment sequence to a CGCH motif is a novel demonstration of the substrate flexibility of the CCM system. Ht 1X was successfully matured into a c-type cytochrome by CCM as indicated by pyridine hemochrome absorption spectra diagnostic of attachment of heme by two thioether bonds (Fig. 2). The absorption spectrum of folded protein also is consistent with a low-spin, six-coordinate heme iron. The yield was approximately 12-fold lower than that of Ht WT, likely due to a lower efficiency of maturation. It must also be considered that Ht 1X, like Ht WT, may be able to mature without the help of CCM maturation factors.41,42 But this is unlikely the route taken here, since expression of Ht 1X in E. coli without the cotransformation of pEC86 produced no detectable heme protein (data not shown).
The side chain of the axial ligand Met61 of Ht WT displays fluxionality between an R and an S configuration at sulfur that is fast on the NMR timescale. This has the effect of narrowing the range of heme methyl chemical shifts observed.43 The narrow range observed in the Ht variants (Fig. 3) indicates that the fluxionality of the axial methionine is still intact, and might also explain the broadness of the heme methyl peaks in Ht 1X at 25 °C (Fig. 3b). This broadness could be a result of a slower exchange between the R and S configurations of the axial methionine in this variant. Despite the conformational exchange, the 1H-NMR and absorption spectra both confirm the presence of heme c and a homogeneous heme environment for a cytochrome with a novel CXCH motif. The broad flexibility of CCM substrate recongnition has therefore been further extended.
Heme attachment motif variants in horse cyt c
Yeast CCHL has just two native substrate cyts c,10 each of which contains a typical CXXCH heme attachment motif. Extension to a CX3CH motif in h-cyt c matured by CCHL decreases the yield of mature h-cyt c 4-fold (Table 1). This decrease in yield when matured with CCHL could be due to a number of factors including mRNA stability,44 protein stability,45 or heme attachment efficiency. But since the yield of the Hh 3X variant does not decrease from that of Hh WT when matured with CCM (Table 1), it is likely that the lower yield of Hh 3X observed with CCHL maturation is due to a limited maturation efficiency of CCHL for this variant. However, despite the 4-fold decrease in yield, a high level of cyt c with the expected absorption spectrum and hemochrome absorption spectrum is obtained, indicating heme c attached by two thioether bonds (Fig. 4). Importantly, the properties of Hh 3X matured by CCHL are indistinguishable from those of the variant matured by CCM. The absorption (Fig. 4) and 1H-NMR spectra (Fig. 7e,f) of products matured by the CCHL and by CCM are indistinguishable, indicating that the holo-cyt c obtained is independent of its maturation pathway. The 1H-NMR also indicates homogeneity of the heme environment by the expected downfield-shifted heme methyl resonances and upfield-shifted axial methionine methyl resonance (Fig. 7e,f). Yeast CCHL is therefore capable of correctly recognizing a CX3CH motif for maturation.
Contraction of the h-cyt c motif to CGCH does not allow for maturation by either CCHL or CCM. In this case, no conclusions about substrate recognition can be drawn. The apparant toxicity of the variant when targeted to the periplasm for CCM maturation could be due to a low level of maturation resulting in an abnormally ligated, or improperly folded, toxic species. Expressed with CCHL in the cytoplasm, E. coli growth remains normal but no detectable yield is obtained. This could be due either to a lack of recognition by CCHL, or the protein fold not tolerating heme attachment to the contracted motif, causing instability and therefore proteolytic degradation after the post-translational modification.
Yeast heme lyase substrate recognition sites
The fact that h-cyt c can be matured either by CCM or CCHL allows comparison of enzyme specificities. The correct maturation of both the Hh G13ins and Hh F10A variants by CCM indicates that they are stable in the periplasm of the cell and upon purification. Therefore, low yields when matured by CCHL cannot be attributed to low mRNA or low holoprotein stability. For both variants, 1H-NMR spectra are similar to that of Hh WT, with only small shifts in the paramagnetically shifted resonances (Fig. 7a,b,d), demonstrating that the variants both remain 6-coordinate, low spin species and have a homogeneous heme environment. The intensity-3 upfield peak between -20 and -25 ppm is characteristic of methionine coordination also found in Hh WT. These data suggest that Hh G13ins and Hh F10A are minimally perturbed in terms of structure relative to Hh WT.
The Hh F10A variant, however, is not matured above a detectable level in the cytoplasm by CCHL (Table 1). Aside from the possibility that the protein stability is dramatically different in the cytoplasm relative to the periplasm, it must be that CCHL is unable to mature this variant and F10 is a recognition site for the enzyme.
The Hh G13ins variant, unlike F10A, is produced in high yield upon maturation by CCHL. The product, however, even after FPLC purification, is heterogenous as shown by 1H-NMR (Fig. 7c). It is possible that appropriately matured cyt c is a minor fraction of the hetergenous mixture, but this is not certain. The pyridine hemochrome absorption spectrum is likewise abnormal, with significant differences relative to the same variant matured by CCM (Fig. 6). Since Hh G13ins was properly matured by CCM without any decrease in yield relative to Hh WT, the insertion does not interfere structurally with heme attachment or with holoprotein stability. Thus the poor result from attachment by CCHL is likely due to an altered substrate recognition and/or heme attachment process caused by the mutation.
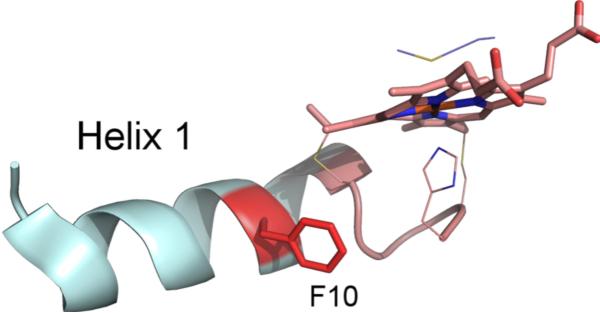
Based on the yields of both the Hh F10A and G13ins variants, the recognition of cyt c by CCHL involves the relationship between the N-terminal helix-1 and the CXXCH motif (Fig. 8), consistent with an early study that suggested that the N-terminal region of mitochondrial cyt c contains the necessary recognition elements for CCHL.46 The abolished maturation of h-cyt c F10A by CCHL strongly suggests that the reason for the conservation of F10 is for recognition by CCHL.
Fig. 8.
Partial structure of horse cyt c showing the N-terminal helix-1 (cyan) and the heme attachment motif with heme (pink). The side chain of the axial ligand Met80 is also included. F10 is highlighted in red. Coordinates from PDB: 1HRC.52 Figure generated with Pymol.
The surprising flexibility of CCHL to mature a CX3CH motif into heme c demonstrates that, despite its limited substrate diversity biologically, the flexibility of this system has not been fully explored. The points of substrate recognition discovered here by comparing the maturation of variants by both CCM and CCHL beg further study as well. Comparison of results of maturation of other mitochondrial cyt c variants by CCM and CCHL may provide additional information on the residues controlling substrate recognizion by CCHL. A better understanding of CCHL could lead to higher yielding recombinant cyt c expression, increasing the practicality of applications utilizing engineered cyts c.47 Also, expansion of the known substrate flexibility of the CCM system to include a CGCH motif allows for the study of the effect of this variation on heme properties and reactivity, with potential implications for the design of heme-containing proteins.
Conclusions
It has been demonstrated that the CCM system machinery for heme c maturation can tolerate a novel CXCH motif with correct, homogenous heme attachment. The known substrate specificity of yeast CCHL has also been broadened to include a CX3CH motif within h-cyt c. By maturation of h-cyt c position 10 and 13 mutants with both CCM and CCHL, two possible points of the recognition of mitochondrial cyt c by CCHL have been discovered, both involving helix-1 of h-cyt c. In this way, some sequence differences between bacterial cyts c relative to mitochondrial cyts c that hinder CCHL recognition of bacterial cyts c have been identified. Finally, a practical consideration for recombinant protein expression of mitochondrial cyt c variants is that low or absent holoprotein expression could be caused by diminished recognition by CCHL for the variant of interest, in which case maturation by the less specific CCM system may be a better route to high yields of pure cyt c variants.
Experimental
The plasmid containing a synthetic gene for Ht cyt c552 with an N-terminal signal sequence from Thiobacillus versutus cyt c550 (pSHC552, Ampr) for maturation by CCM has been previously described.42 The plasmid containing the CYC1 gene encoding h-cyt c and the CYC3 gene encoding yeast CCHL, pBTR(hCc) Ampr, for maturation of h-cyt c by CC was generously provided by Gary Pielak.34 A previously reported plasmid containing the gene for h-cyt c with a signal sequence from Thiobacillus versutus cyt c550 was used for maturation of h-cyt c by CCM (pSHHC, Ampr).30 Mutagenesis was performed using QuikChange (Stratagene) for all variants of Ht cyt c552. For mutants of h-cyt c, MEGAWHOP48 was performed using a promoter primer of the relevant plasmid and reverse complement mutagenic primers. DNA sequencing was performed to confirm successful mutagenesis.
Expression was carried out in BL21 (DE3) E. coli cells (Invitrogen) in a similar manner to that previously described.34 For maturation by the CCM system, E. coli were co-transformed with plasmids encoding a variant of either h-cyt c or Ht cyt c552 and with the pEC86 vector (Cmr)28 encoding genes ccmA–H. All media were supplemented with 50 μg/mL of the appropriate antibiotics. A 30-mL culture grown at 37°C in Luria Bertani (LB) medium was grown for 8 hours and used to inoculate 1 L of terrific broth (TB) medium for all variants except Hh F10A matured with CCM. For this variant, higher yields were obtained using 1-L cultures in LB medium. These large-scale cultures were grown in 4-L flasks at 37°C for 18 hours while shaking at 120 rpm. The cells were harvested by centrifugation at 10000 x g for 15 minutes. The wet cell paste was frozen and stored at -20°C.
For all protein variants, thawed cell pellets were resuspended in 10 mM Tris (pH 8) supplemented with 2 mg/mL deoxycholate sodium salt and sonicated for 1 minute at a power of 7 and 50% duty cycle on a Branson Sonifier 450. After centrifugation, the lysate was decanted and centrifuged again before being loaded onto a CM Sepharose Fast Flow cation exchange column (GE Life Sciences). Protein was eluted with a NaCl gradient, concentrated in an Amicon (Millipore) with 3-kDa molecular weight cut-off regenerated cellulose membranes and exchanged with a desalting column (PD10, GE Life Sciences) into 5 mM NaPi, pH 7.0 for Ht cyts c, and 50 mM NaPi, pH 7.0 for h-cyts c. The protein was oxidized with K[Co(EDTA)] synthesized from a literature method49 before loading onto an FPLC system equipped with a Mono S column (GE Life Sciences). The protein was eluted with an NaCl gradient from 0–160 mM in 5 mM NaPi, pH 7.0 for Ht cyts c, and 140–200 mM in 50 mM NaPi, pH 7.0 for h-cyts c. Yields were calculated from the alpha-band of the pyridine hemochrome spectrum50 using the extinction coefficient of 30.27 mM-1 cm-1. Absorption spectra were taken with a UV-Vis spectrophotometer (Shimadzu 2401PC) at ambient temperature. The spectra were also used as an indicator of correct heme c attachment through two thioether bonds.22 A few crystals of sodium dithionite were added directly to the sample for reduction of the protein. The extinction coefficient of 106.1 mM-1 cm-1 was used for hcyt c and its variants,34 while 105 mM-1 cm-1 was used for Ht cyt c552 and its variants.42 The yields given (Table 1) are an average of two separate growth and purifications.
1H-NMR spectra were obtained using a Varian INOVA 500-MHz NMR spectrometer. All experiments were performed with samples in 50 mM NaPi, pH 7.0, 10% D2O at either 25°C or 40°C. Data were processed with SpinWorks and graphed with Igor Pro 6.1. The chemical shift scale was referenced indirectly via the water resonance.51
Acknowledgments
The authors thank Prof. Linda Thöny-Meyer for the gift of pEC86 and Prof. Gary Pielak for the gift of pBTR(hCc). This work was supported by the NIH (R01-GM63170 to K.L.B.)
Notes and references
- 1.Bowman SEJ, Bren KL. Nat. Prod. Rep. 2008;25:1118–1130. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen JWA, Ginger ML, Ferguson SJ. Biochem. J. 2004;383:537–542. doi: 10.1042/BJ20040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartshorne RS, Kern M, Meyer B, Clarke TA, Karas M, Richardson DJ, Simon J. Mol. Microbiol. 2007;64:1049–1060. doi: 10.1111/j.1365-2958.2007.05712.x. [DOI] [PubMed] [Google Scholar]
- 4.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Microbiol. Mol. Biol. Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thöny–Meyer L, Fischer F, Kunzler P, Ritz D, Hennecke H. J. Bacteriol. 1995;177:4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giege P, Grienenberger JM, Bonnard G. Mitochondrion. 2008;8:61–73. doi: 10.1016/j.mito.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Sanders C, Turkarslan S, Lee DW, Daldal F. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckett CS, Loughman JA, Karberg KA, Donato GM, Goldman WE, Kranz RG. Mol. Microbiol. 2000;38:465–481. doi: 10.1046/j.1365-2958.2000.02174.x. [DOI] [PubMed] [Google Scholar]
- 9.Xie ZY, Merchant S. Biochim. Biophys. Acta. 1998;1365:309–318. doi: 10.1016/s0005-2728(98)00085-1. [DOI] [PubMed] [Google Scholar]
- 10.Dumont ME, Ernst JF, Hampsey DM, Sherman F. EMBO J. 1987;6:235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson SJ, Stevens JM, Allen JWA, Robertson IB. Biochim. Biophys. Acta. 2008;1777:980–984. doi: 10.1016/j.bbabio.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Goddard AD, Stevens JM, Rao F, Mavridou DAI, Chan WL, Richardson DJ, Allen JWA, Ferguson SJ. J. Biol. Chem. 2010;285:22880–22887. doi: 10.1074/jbc.M110.133421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richard-Fogal CL, Frawley ER, Bonner ER, Zhu HF, Francisco BS, Kranz RG. Embo Journal. 2009;28:2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavridou DAL, Braun M, Thony-Meyer L, Stevens JM, Ferguson SJ. Biochem. Soc. Trans. 2008;36:1124–1128. doi: 10.1042/BST0361124. [DOI] [PubMed] [Google Scholar]
- 15.Allen JWA, Barker PD, Ferguson SJ. J. Biol. Chem. 2003;278:52075–52083. doi: 10.1074/jbc.M307196200. [DOI] [PubMed] [Google Scholar]; Braun M, Thony-Meyer L. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12830–12835. doi: 10.1073/pnas.0402435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen JWA, Ferguson SJ. Biochem. Soc. Trans. 2006;34:91–93. doi: 10.1042/BST0340091. [DOI] [PubMed] [Google Scholar]
- 17.Allen JWA, Leach N, Ferguson SJ. Biochem. J. 2005;389:587–592. doi: 10.1042/BJ20041894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen JWA, Tomlinson EJ, Hong L, Ferguson SJ. J. Biol. Chem. 2002;277:33559–33563. doi: 10.1074/jbc.M204963200. [DOI] [PubMed] [Google Scholar]
- 19.Eaves DJ, Grove J, Staudenmann W, James P, Poole RK, White SA, Griffiths I, Cole JA. Mol. Microbiol. 1998;28:205–216. doi: 10.1046/j.1365-2958.1998.00792.x. [DOI] [PubMed] [Google Scholar]
- 20.Hartshorne S, Richardson DJ, Simon J. Biochem. Soc. Trans. 2006;34:146–149. doi: 10.1042/BST0340146. [DOI] [PubMed] [Google Scholar]
- 21.Herbaud ML, Aubert C, Durand MC, Guerlesquin F, Thony-Meyer L, Dolla A. Biochim. Biophys. Acta. 2000;1481:18–24. doi: 10.1016/s0167-4838(00)00117-5. [DOI] [PubMed] [Google Scholar]
- 22.Allen JWA, Sawyer EB, Ginger ML, Barker PD, Ferguson SJ. Biochem. J. 2009;419:177–184. doi: 10.1042/BJ20081999. [DOI] [PubMed] [Google Scholar]
- 23.Sawyer EB, Stephens E, Ferguson SJ, Allen JWA, Barker PD. J. Am. Chem. Soc. 2010;132:4974–4975. doi: 10.1021/ja908241v. [DOI] [PubMed] [Google Scholar]
- 24.Kern M, Eisel F, Scheithauer J, Kranz RG, Simon J. Mol. Microbiol. 2010;75:122–137. doi: 10.1111/j.1365-2958.2009.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders C, Lill H. Biochim. Biophys. Acta. 2000;1459:131–138. doi: 10.1016/s0005-2728(00)00122-5. [DOI] [PubMed] [Google Scholar]
- 26.Bernard DG, Gabilly ST, Dujardin G, Merchant S, Hamel PP. J. Biol. Chem. 2003;278:49732–49742. doi: 10.1074/jbc.M308881200. [DOI] [PubMed] [Google Scholar]
- 27.Arslan E, Schulz H, Zufferey R, Kunzler P, Thöny-Meyer L. Biochem. Biophys. Res. Comm. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 28.Fee JA, Chen Y, Todaro TR, Bren KL, Patel KM, Hill MG, Gomez-Moran E, Loehr TM, Ai JY, Thöny-Meyer L, Williams PA, Stura E, Sridhar V, McRee DE. Prot. Sci. 2000;9:2074–2084. doi: 10.1110/ps.9.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Costa PN, Conte C, Saraiva LM. Biochem. Biophys. Res. Comm. 2000;268:688–691. doi: 10.1006/bbrc.2000.2198. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg JA, Bren KL. Biochim. Biophys. Acta. 2002;1601:215–221. doi: 10.1016/s1570-9639(02)00471-5. [DOI] [PubMed] [Google Scholar]
- 31.Rumbley JN, Hoang L, Englander SW. Biochemistry. 2002;41:13894–13901. doi: 10.1021/bi026543y. [DOI] [PubMed] [Google Scholar]
- 32.Pollock WBR, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 33.Corvest V, Murrey DA, Bernard DG, Knaff DB, Guiard B, Hamel PT. Genetics. 2010;186:561–571. doi: 10.1534/genetics.110.120022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel CN, Lind MC, Pielak GJ. Prot. Express. Purif. 2001;22:220–224. doi: 10.1006/prep.2001.1438. [DOI] [PubMed] [Google Scholar]
- 35.La Mar GN, Satterlee JD, De Ropp JS. In: The Porphyrin Handbook. Kadish KM, Smith KM, Ruilard R, editors. Vol. 5. Academic Press; NY: 2000. pp. 185–298. [Google Scholar]
- 36.Moore GR, Pettigrew GW. Cytochromes c. Evolutionary, Structural and Physicochemical Aspects. Springer-Verlag; Heidelberg; Berlin: New York: 1990. pp. 115–155. Ch. 3. [Google Scholar]
- 37.Rios-Velazquez C, Cox RL, Donohue TJ. Arch. Biochem. Biophys. 2001;389:234–244. doi: 10.1006/abbi.2001.2330. [DOI] [PubMed] [Google Scholar]
- 38.Bren KL. In: Applications of Physical Methods to Inorganic and Bioinorganic Chemistry. Scott RA, Lukehart CM, editors. John Wiley & Sons; Chichester, UK: 2007. pp. 357–384. [Google Scholar]
- 39.Liptak MD, Wen X, Bren KL. J. Am. Chem. Soc. 2010;132:9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shokhirev NV, Walker FA. J. Biol. Inorg. Chem. 1998;3:581–594. [Google Scholar]
- 41.Sambongi Y, Ferguson SJ. FEBS Lett. 1994;340:65–70. doi: 10.1016/0014-5793(94)80174-6. [DOI] [PubMed] [Google Scholar]
- 42.Karan EF, Russell BS, Bren KL. J. Biol. Inorg. Chem. 2002;7:260–272. doi: 10.1007/s007750100292. [DOI] [PubMed] [Google Scholar]
- 43.Zhong LH, Wen X, Rabinowitz TM, Russell BS, Karan EF, Bren KL. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8637–8642. doi: 10.1073/pnas.0402033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross J. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsell DA, Sauer RT. J. Biol. Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 46.Veloso D, Juillerat M, Taniuchi H. J. of Biol. Chem. 1984;259:6067–6073. [PubMed] [Google Scholar]
- 47.Asher WB, Bren KL. Prot. Sci. 2010;19:1830–1839. doi: 10.1002/pro.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyazaki K, Takenouchi M. Biotechniques. 2002;33:1033–1038. doi: 10.2144/02335st03. [DOI] [PubMed] [Google Scholar]
- 49.Dwyer FP, Gyarfas EC, Mellor DP. J. Phys. Chem. 1955;59:296–297. [Google Scholar]
- 50.Berry EA, Trumpower BL. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 51.Gottlieb HE, Kotlyar V, Nudelman A. J. Org. Chem. 1997;62:7512–7515. doi: 10.1021/jo971176v. [DOI] [PubMed] [Google Scholar]
- 52.Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]