Abstract
The molecular and cellular mechanisms underlying the pathogenesis of chronic obstructive pulmonary disease (COPD) remain incompletely understood. We have investigated the potential role of macro-autophagy, a cellular homeostatic mechanism, in COPD and cigarette smoke-induced lung-cell injury. Autophagy is a dynamic process for the turnover of organelles and proteins, which regenerates metabolic precursors through the lysosomal-dependent catabolism of cellular macromolecules. It is typically associated with survival pathways, especially in nutrient deficiency states. The role of autophagy in human diseases is less clear, and has been associated with both protective and detrimental consequences, depending on the disease model. While autophagy is considered cytoprotective, this process is often found in association with cell death, and the relationships between autophagy and cell death remain ambiguous. We have found elevated autophagy in COPD lung specimens, as well as in response to cigarette smoke exposure in vitro and in vivo. In our studies, the activation of autophagic proteins was associated with epithelial cell apoptosis in response to cigarette smoke, with pathogenic implications in COPD. Further studies are needed to determine the functional significance of autophagy in COPD and other diseases of the lung.
Keywords: apoptosis, autophagy, chronic obstructive pulmonary disease, cigarette smoke, emphysema
Chronic obstructive pulmonary disease (COPD), a leading cause of morbidity and mortality worldwide, is characterized by progressive airflow limitation and destructive alveolar loss (emphysema) [1]. The pathogenesis of COPD involves exaggerated inflammatory responses of the lung to chronic particle and irritant exposure in the airways, which can be amplified by infections, protease/antiprotease imbalance and oxidative stress [2–4]. Cigarette smoke (CS) exposure is the major risk factor for COPD, which progresses in approximately 15% of smokers [1]. CS contains approximately 4700 chemical constituents, and can increase endogenous reactive oxygen species (ROS) production in target cell populations (i.e., inflammatory cells and epithelial cells) [5–7]. Pro-oxidant states occur when the production of ROS exceeds endogenous antioxidant defense and repair mechanisms, resulting in the impaired function of critical cellular macromolecules [5]. Proteases (i.e., elastases and matrix metalloproteinases [MMPs]), when unchecked by anti-protease activities, can induce tissue damage, leading to emphysema. The protease/anti-protease imbalance arises from inflammatory cell activity and oxidative stress triggered by CS or, in rare cases, directly from the genetic disorder α-1 antitrypsin (α-1 AT) deficiency [2–4].
Programmed cell death mechanisms (i.e., apoptosis) can potentially contribute to epithelial cell loss, leading to the pathogenesis of emphysema [8–11]. Several studies report increased numbers of apoptotic cells in the lungs of COPD patients [12–15]. Studies from our laboratory and others demonstrate activation of apoptosis signaling pathways in inflammatory, epithelial and fibroblast cells subjected to CS exposure [16–19]. The factors that regulate apoptotic pathways and their significance to the etiology of COPD remain partially understood.
Conversely, the potential role of (macro)-autophagy, a fundamental cellular homeostatic process, has not been previously studied in COPD. Our recent studies demonstrate the occurrence and elevation of autophagic markers in human clinical samples from patients with COPD [20]. This article will focus on the functional significance of autophagy and its relationships to apoptosis, in the context of CS-induced lung cell injury and emphysema pathogenesis (Figure 1).
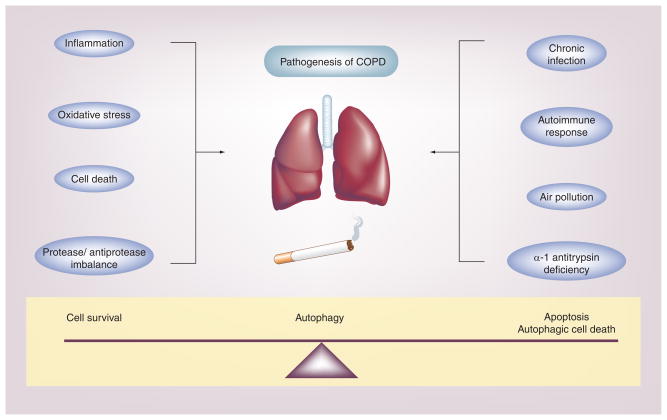
Figure 1. Chronic obstructive pulmonary disease is a multifactorial disease involving airflow limitation, bronchitis and emphysema, induced primarily by cigarette smoking.
The molecular mechanisms governing the pathogenesis of COPD remain unclear, but may involve several factors. Of these, oxidative stress, inflammation, apoptosis and protease/antiprotease imbalance may play critical roles. COPD pathogenesis can also be influenced by additional factors, such as chronic exposure to air pollution of microbial pathogens, genetic factors, such as α-1 antitrypsin deficiency, and autoimmune responses. Autophagy, a fundamental homeostatic process, may play a critical role in COPD pathogenesis by influencing epithelial cell fate in response to environmental stress (i.e., cigarette smoke) [19].
COPD: Chronic obstructive pulmonary disease.
Autophagy: general mechanism
Autophagy is a regulated process for the turnover of cytoplasmic proteins and organelles through a lysosome-dependent degradation pathway. The autophagic pathway progresses through a series of distinct steps that include the formation of an isolation membrane or phagophore, the formation of a double membrane-bound autophagosome that engulfs cytoplasmic material targeted for digestion, the fusion of the autophagosome to the lysosome and, finally, the digestion of sequestered cargo by proteolytic activity in the resulting autolysosome (Figure 2) [21]. The molecular machinery of autophagic regulation has been extensively studied and recently summarized in greater detail elsewhere [21–23]. The autophagy-related genes, designated Atg, which are critical in the regulation of autophagy in yeast, and their homologues in higher mammals, now include over 30 identified genes and gene products [23].
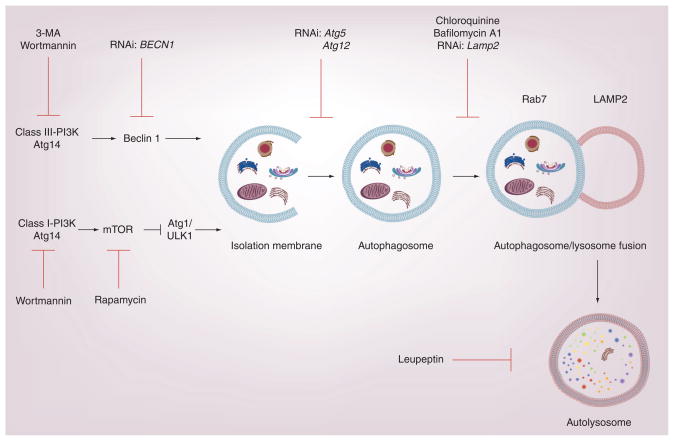
Figure 2. The activation of autophagy is a multistep process that involves the formation of an isolation membrane, the formation of an autophagosome with assimilation of organelles or proteins to be degraded, subsequent autophagosome–lysosome fusion, and finally the proteolytic degradation of the cargo.
Beclin 1 in complex with class III PI3K and Atg14 acts as a major positive regulator of autophagy. The rapamycin-sensitive mTOR/class I PI3K pathway, which is sensitive to cellular nutrient status, acts as a negative regulator of autophagy. Consequently, rapamycin acts as an activator of the autophagy pathway. Autophagy can be inhibited by molecules targeting the class III PI3K, including wortmannin, and 3-MA. Wortmannin can also inhibit the class-I PI3K associated with mTOR signaling. siRNA strategies targeting autophagic proteins can be used to inhibit autophagy at several steps, as indicated on the diagram. Autophagosome–lysosome fusion can be inhibited by bafilomycin A1. Lysosomal turnover of LC3B can be inhibited by protease inhibitors such as leupeptin.
3-MA: 3-methyladenine; LAMP2: Lysosomal-associated membrane protein 2; ULK1: Mammalian uncoordinated 51-like protein kinase-1.
The autophagic pathway is tightly regulated and highly inducible in response to metabolic signals, particularly those that relate to nutrient status, such as amino acid levels and energy charge [22,23]. Autophagy is negatively regulated under nutrient-rich conditions by a signaling pathway involving class I PI3K and mTOR. The antibiotic rapamycin acts as a potent inducer of autophagy, by inhibiting mTOR. Suppression of mTOR kinase activity by amino acid starvation or rapamycin results in activation of mammalian uncoordinated-51-like protein kinase ULK1 (yeast: Atg1), a key regulator of autophagic initiation [24]. ULK1, which forms a multiprotein complex with additional protein factors (i.e., the 200 kD focal adhesion kinase family-interacting protein, FIP200 and Atg13) is critical for starvation-induced autophagy, and initiates autophagosome formation [25,26].
The Bcl-2 interacting protein Beclin 1 (yeast: Atg6) also acts as key regulator of autophagy in mammalian cells [27]. Beclin 1 forms a multiprotein complex that includes class III PI3K (yeast: Vps34) and p150 (yeast: Vps15). Additional proteins that can interact with this complex include the mammalian homolog of Atg14, and the UV radiation resistance-associated tumor suppressor gene protein [28–30]. Upon cellular stimulation, the increased production of phosphatidylinositol-3-phosphate by class III PI3K/Vps34 regulates the formation of nascent autophagosomes [28–30]. The binding of Bcl-2 or Bcl-XL with Beclin 1 impairs autophagy by inhibiting the PI3K activity of this complex [31,32].
The elongation of the isolation membrane or phagophore also depends on the action of two ubiquitin-like conjugation systems. In the first of these pathways, Atg5 is conjugated to Atg12 by Atg7 (E1-like) and Atg10 (E2-like) enzymes. The resulting Atg5–Atg12 forms a complex with Atg16, which in turn assimilates with the isolation membrane. Another pathway involves the microtubule-associated protein-1 light chain (LC)3 (yeast: Atg8), which is cleaved by Atg4 and then conjugated with phosphatidylethanolamine by the sequential action of Atg7 (E1-like) and Atg3 (E2-like) activities [21–23]. In mammals, the conversion of LC3 from LC3-I (free form) to LC3-II (phosphatidylethanolamine-conjugated form) represents a key step in autophagosome formation [33]. During autophagosome–lysosome fusion, LC3 is degraded by lysosomal hydrolases within the autophagosome, or de-lipidated at the membrane surface by Atg4. The autophagosome–lysosome fusion step requires additional accessory factors (i.e., Rab7, LAMP2) [22,23]. In the final stages of the autophagic pathway, encapsulated cargo are degraded by lysosomal proteases and released. In this way, autophagy provides a mechanism not only for the turnover of cellular constituents, but also for the replenishment of metabolic precursors during nutrient-poor conditions.
Observations of autophagy in COPD
In the recent paper by Chen et al., we have shown marked increases of autophagic proteins in lung tissue derived from COPD patients [20]. The expression level of LC3B-II, a marker of autophagosome formation, as well as the expression of additional autophagy-associated proteins Atg4, Atg5–Atg12 and Atg7, was increased in COPD relative to control lung [20]. Electron microscopic analysis of lung tissue from COPD patients revealed enhanced production of autophagosome formation in COPD lung compared with control tissues [20].
We have also observed increased markers of autophagy in the lungs of C57Bl/6 mice exposed to environmental CS for 12 weeks. CS-exposed mouse lung displayed increased autophagosome formation by electron microscopic analysis and increases in LC3B-II [20].
In addition, we have used an in vitro model of CS exposure, involving application of aqueous CS extract (CSE) to cultured cells. Our in vitro experiments demonstrate a dose-dependent induction of LC3B-II by CSE treatment in primary human bronchial epithelial (HBE) cells at the air–liquid interface (ALI), and in human small airway epithelial cells or bronchial epithelial cells (Beas-2B) [20]. CSE induced a marked induction of autophagosome formation in primary epithelial cells as determined by electron microscopy [19,20]. The appearance of punctate staining in green fluorescent protein (GFP)-LC3-expressing cells and tissues is regarded as an indicator of autophagosome formation [21,33]. CSE exposure was found to cause marked autophagosome formation in Beas-2B cells, as determined by the formation of GFP-LC3-labeled puncta [19,20]. The induction of LC3B-II levels by CSE in primary human bronchial cells was enhanced by bafilomycin A1, a compound that prevents the maturation of autophagic vacuoles by inhibiting autophagosome–lysosome fusion, and by lysosomal protease inhibitors pepstatin A and E64d, indicating that CS-induced LC3B expression reflected increased autophagic flux [19,20]. We have also recently observed CS-induced autophagy using a physiological in vitro model of fully differentiated respiratory epithelial cells grown at ALI. Similar to the human lung epithelial cells, autophagosomes and autolysosomes were also significantly increased by mainstream CS in ALI-cultured cells relative to air-treated cultures [Lam H, Choi AMK, Unpublished Data]. Taken together, our studies demonstrate elevated autophagy in COPD lung tissue, and as a response to CS exposure in lung cells and tissue, suggestive of a potential role for autophagy in lung epithelial cell injury [20].
Molecular mechanisms for autophagic regulation by CS
The mechanisms by which CS induces autophagy in epithelial cells and mouse lung tissue are not well understood. Previous studies have suggested links between oxidative stress, elevated ROS (i.e., H2O2 and O2−) production and activation of autophagy [34–37]. Since CS exposure is known to promote pro-oxidant states in epithelial and other cell types [5–7,19], it can be hypothesized that oxidative stress secondary to CS exposure is responsible, at least in part, for autophagic activation during CS stimulation. This hypothesis is supported by observations that CS-induced autophagy can be inhibited by administration of antioxidant compounds such as N-acetyl-L-cysteine. Furthermore, chemical inhibitors of NADPH oxidases, a membrane-dependent source of ROS, also inhibited CSE-inducible activation of LC3B [20].
Experiments in other chemical exposure models have further characterized the role of ROS in autophagic activation. For example, starvation-induced autophagosome formation was associated with increased intracellular ROS levels in cultured cells, and was reversible by treatment with the antioxidants N-acetyl-L-cysteine or catalase [35]. Direct administration of the oxidant H2O2 induced a caspase-independent cell death associated with autophagic activation in transformed or cancer cell lines. The siRNA-dependent knockdown of autophagy genes (i.e., Beclin 1, Atg5 and Atg7) prevented H2O2-induced autophagic cell death [36]. Similarly, treatment of transformed cell lines with inhibitors of mitochondrial respiration (i.e., rotenone) increased intracellular ROS production, induced autophagy and produced a caspase-independent form of cell death [37]. The autophagic protein Atg4 has been identified as a redox-regulated protein in the autophagic pathway. Mutations in a specific cysteine residue occurring in Atg4 protein(s) abolished the regulation of autophagy by H2O2 [35]. The precise redox-dependent mechanisms that regulate autophagy during CS exposure currently remain unknown and warrant further examination.
The mechanisms governing the transcriptional regulation of specific autophagy genes remain unclear. Sequence analysis of the LC3B promoter revealed consensus binding sites for the transcription factor early growth response-1 (Egr-1). Egr-1 is the product of an immediate early response gene that is induced rapidly following various cellular stresses in vitro, which plays multifunctional roles in the regulation of apoptotic and inflammatory signaling cascades [38]. Egr-1 binding to the LC3B promoter increased in response to stimulation by CSE in epithelial cells. Knockdown of Egr-1 inhibited the activation of LC3B in response to CS exposure in epithelial cells [20]. Interestingly, Egr-1−/− mice were resistant to the pro-apoptotic and pro-autophagic effects of chronic CS exposure in vivo, as well as to CS-induced emphysematous changes. However, Egr-1−/− mice also exhibited significant basal airspace enlargement, indicating a potential role for Egr-1 in lung developmental processes [20].
Epigenetic factors may also play an important role in gene regulation following CS exposure. The studies of Ito et al. demonstrate that histone deacetylases (HDACs), which are important regulators of transcription and chromatin function, are generally downregulated in response to CS exposure [39]. HDAC activities were also downregulated in the COPD lung as a function of disease severity [39]. In CSE-induced stress, the activation of autophagy as well as of Egr-1 were also linked to the downregulation of HDAC activities [20]. General inhibition of HDACs with tricostatin A resulted in increased autophagosome formation, LC3B conversion and Egr-1 activation in epithelial cells.
Our recent studies also suggest a role for caveolin-1 in the regulation of CS-induced autophagy [40]. Caveolin-1 is a structural component of caveolae, which are cholesterol-and glycosphingolipid-rich domains of the plasma membrane. In addition to this function, caveolin-1 interacts with and regulates the activity of numerous membrane-associated signal transduction proteins [41]. We found that caveolin-1-deficient cells and Cav-1−/− mice exhibited higher levels of autophagy and apoptosis in response to CS exposure in vitro and in vivo, respectively. The specific mechanisms by which caveolin-1 can regulate autophagy are the subject of current investigation [40].
Autophagy in human diseases: comparative analysis
In addition to COPD as recently described [20,42], autophagy has been implicated in several other human diseases, including cancer [43–46], neurodegenerative diseases [47], inflammatory bowel disease [48,49] and cardiovascular diseases [50–53]. The regulation and functional significance of autophagy in these diseases are poorly understood. Dysfunctional autophagy resulting from either loss-of-function or excessive activation, has been related to disease pathogenesis (Figure 3) [43]. Many of these studies describe elevations of autophagy or autophagic markers in preclinical animal models of the diseases in question, rather than from the direct analysis of human clinical samples. Owing to inherent limitations in animal modeling, further translational and clinical studies are urgently needed to determine the significance of autophagy in human disease.
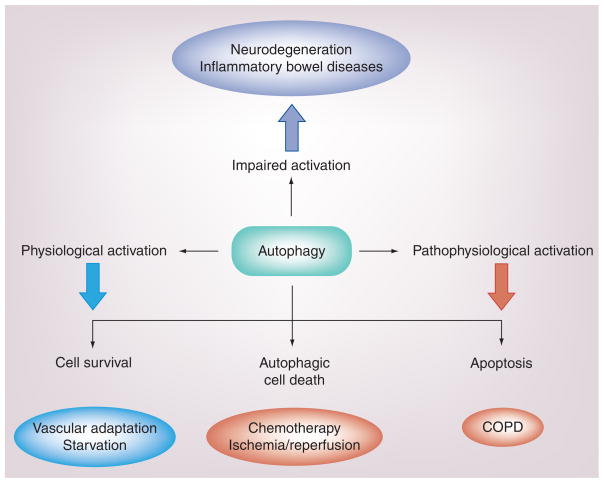
Figure 3. The activation of autophagy, as an endogenous inducible response to cellular stress, may have both adaptive and maladaptive consequences depending on the experimental model.
The protective functions of autophagy are associated with the homeostatic turnover of damaged cellular organelles and protein, thereby facilitating the recycling of essential metabolic precursors and the regeneration of energy equivalents. Thus, autophagy is generally thought of as a survival mechanism. However, excessive autophagy may be associated with degradation of intracellular constituents, leading to autophagic cell death or, alternatively, to apoptotic cell death, depending on the cell model, both of which may potentially play a contributory role in COPD pathogenesis.
COPD: Chronic obstructive pulmonary disease.
In cancer models, autophagy has been associated with tumor cell survival and tumor cell resistance to chemotherapeutic agents. In this context, autophagy is detrimental to the survival of the host. Paradoxically, a type of autophagic cell death has been observed in the action of certain chemotherapeutic agents [43–46].
Autophagy preserves neuronal cell integrity, whereas impaired autophagy promotes neurodegeneration in animal models [54,55]. Defects in autophagic processing have been associated with several neurodegenerative disorders, including Alzheimer’s and Parkinson’s diseases. On the other hand, excessive autophagic activity has also been implicated as a possible pathogenic factor in neurodegeneration caused by acute injury [47].
Defects in autophagic function have also been associated with the development of heart disease [56]. On the one hand, autophagy was found to be protective in the context of hemodynamic stress [57]. On the other hand, both protective and detrimental roles for autophagy were implicated in cardiac ischemia/reperfusion injury, depending on the phase [58]. Increased autophagy has been detected in arteriosclerotic plaques, and has been implicated in plaque stabilization [51,52]. Autophagy has also been implicated in the clearance of macrophages from arteriosclerotic plaques, and thus may play a beneficial role in the context of this disease [59].
Recent studies have suggested that genetic polymorphisms in autophagy genes may be linked to the progression of inflammatory bowel disease [48,49]. A small nucleotide polymorphism of the atg16L1 gene has been associated with increased susceptibility to Crohn’s disease [48,49]. Further studies are needed to determine whether or not genetic polymorphisms are relevant to the pathogenesis of other diseases, including chronic diseases of the lung.
Our own recent analysis of autophagy in lung diseases revealed elevated autophagy in human lung tissue from COPD patients at various stages of disease severity relative to normal lungs, although there was no clear correlation between the degree of apparent autophagy and disease severity. Our preliminary studies indicated no evidence for autophagy in lung tissues from patients with idiopathic pulmonary fibrosis, cystic fibrosis, sarcoidosis and systemic sclerosis [20]. However, we have observed increased autophagy in clinical samples of patients with various forms of pulmonary hypertension [60]. These observations suggest that autophagic proteins could represent potential biomarkers of distinct pulmonary disease conditions. Further research would be necessary to determine the specificity of these relationships.
Apoptosis–autophagy relationship in COPD
Apoptosis has been implicated in the pathogenesis of COPD, although its precise role(s) remain controversial [8]. Several previous studies have identified increased alveolar cell apoptosis in human emphysematous lung tissue sections using in situ DNA end labeling techniques such as terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL) [13,14]. TUNEL-positive cells in emphysematous lung mainly represent endothelial cells from capillaries and arterioles but also alveolar epithelial cells, interstitial cells and inflammatory cells, whereas control lungs were negative [13–15]. The activated subunits of caspase-3 and the increased expression of pro-apoptotic proteins (i.e., Bax and Bad) were detected in emphysema lungs, while the anti-apoptotic protein Bcl-2 was not detected in either normal or emphysematous lung tissue [15]. Increases in apoptotic epithelial and endothelial alveolar septal cells were observed in emphysematous lungs compared with the lungs from nonsmokers, healthy smokers and patients with primary pulmonary hypertension, with no significant difference in apoptosis indicators between healthy nonsmokers and smokers without emphysema [61]. Recent data have demonstrated that apoptosis persists despite smoking cessation, suggesting that cigarette smoke itself is not the sole agent causing apoptosis once COPD is established, although COPD individuals are more likely to be susceptible to smoke-induced cell damage and apoptosis [12]. This finding indicates that additional factors including protease/antiprotease imbalance, inflammation and oxidative stress involved in the pathogenesis of emphysema may also contribute to increased apoptosis [12].
In our analysis of clinical specimens, we noted that caspase-3 activation was detectable in lung tissue from advanced-stage COPD (Global initiative for chronic Obstructive Lung Disease [GOLD]4) human lung specimens, but not detectable in specimens with less severe disease (i.e., GOLD0–2). By contrast, biochemical and morphological markers of autophagy were found to be elevated at all stages of disease progression (GOLD0–4) relative to healthy nonsmokers [20]. These findings imply that autophagy may precede apoptosis as a general response to chronic CS exposure in the lung [20]. The repetition of these studies with larger sample sets and more sensitive methods to detect lung apoptosis may shed further light on this issue.
Our recent in vitro studies in CS-induced lung cell injury models suggest a regulatory role for LC3B in epithelial cell apoptosis induced by CSE [19,20]. We have observed evidence for upregulation of both autophagy and apoptosis in lung epithelial cells exposed to CSE. CSE initiated the extrinsic apoptosis pathway involving assembly of the Fas-dependent death-inducing signaling complex (DISC) and activation of caspase-8, induced the expression and conversion of the autophagic regulator LC3B, increased autophagosome formation and, ultimately, increased caspase-3 activation in epithelial cells [19]. The role of autophagic proteins in mediating CSE-induced epithelial cell death was demonstrated in genetic interference experiments. The siRNA-directed knockdown of the autophagic proteins Beclin 1 or LC3B in epithelial cells inhibited the assembly of the Fas-dependent DISC. While initiating events in apoptosis signaling, such as DISC formation, appear to precede the activation of LC3B, the activation of executioner caspases occurs later than morphological signs of autophagy. Thus while autophagy and apoptosis were concurrently regulated after CS exposure in epithelial cells, the temporal and causal relationship of these processes remains unclear [19,20]. In our recent studies, we found that LC3B interacted with Fas under basal conditions in Beas-2B cells, whereas this interaction was rapidly disrupted after exposure to CSE. This molecular interaction localized to plasma membrane lipid rafts and was dependent on caveolin-1. Further experiments in progress will determine the significance of these findings [Choi AMK, Unpublished Data].
Relationship of autophagy to cell death mechanisms: unanswered questions
While recent consensus favors the hypothesis that autophagy primarily represents a cell survival mechanism, the functional relationships between autophagy and various forms of cell death, including apoptosis, remain the subject of extensive current debate [21,62].
A substantial body of literature has defined autophagy as a cellular process essential for normal homeostatsis, which provides a survival advantage under conditions of nutrient deprivation or metabolic stress [21,62–66]. In this regard, autophagy serves to replenish metabolic building blocks essential for survival by salvaging them from the degradation of damaged or dysfunctional cellular macromolecules. In this capacity, autophagy may prevent cell death by necrosis, as well as inhibit or delay the initiation of the apoptotic program. In support of this hypothesis, recent studies demonstrate that impaired autophagy, whether achieved by genetic or chemical means, promotes apoptosis under conditions of nutrient deprivation [67].
Since autophagy functions primarily as a degradative or catabolic process, it is conceivable that excessive or deregulated autophagy, leading to degradation of vital cellular components beyond the degree necessary to maintain homeostasis, may in some cases also trigger apoptosis [62–66].
Apoptosis (type I programmed cell death) has been functionally characterized by plasma membrane blebbing, chromatin condensation, DNA fragmentation and activation of caspases [66]. By contrast, the term ‘autophagic cell death’ (also type II programmed cell death) has been previously introduced to describe a form of cell death that occurs in association with apparent increases in autophagic vacuole (AV) formation, without evidence of caspase activation or other morphological signs of apoptosis [63,64]. Evidence for autophagic cell death relies in part on observation of increased vacuolization and cell death in apoptosis-compromised (caspase inhibitor-treated) cells [68,69]. The occurrence of AVs or autophagic markers in dying cells, however, does not establish a casual relationship between the autophagic process and cell death [67]. Furthermore, increases in AV formation do not necessarily indicate that an active autophagic process is occurring, as AV numbers can also increase as a result of inhibited autophagosome–lysosome fusion [21,67]. Boya et al. reported that cells displaying elevated AV formation resulting from impaired fusion were not necessarily committed to death [67]. Furthermore, these authors reported that biochemical indicators of apoptosis could be detected in cells that were undergoing active autophagy, thus questioning the classification of autophagy as a fully independent death pathway under physiological conditions [67]. By these arguments, the elevated occurrence of autophagy in dying cells may rather signify a homeostatic mechanism that only partially compensates but fails to counteract prevailing pro-death stimuli.
Additional studies using non-physiological experimental conditions relying on the chemical or genetic impairment of apoptosis have implicated the activation of autophagy as an alternative pathway to cell death that can substitute for a dysfunctional apoptosis pathway [68–72]. Inhibition of caspase activation using chemicals such as Z-Val-Ala-Asp (OMe)-fluoromethylketone (z-VAD-fmk) simultaneously increases autophagy and induces cell death in several cell types [68–72]. The functional role of autophagy in this model of caspase inhibitor-induced cell death was variably attributed to both promotion [68–71] and inhibition [72] of cell death when chemical modulators of autophagy were used. Genetic knockdown of autophagic proteins typically inhibited caspase inhibitor-induced cell death [68–71]. In cells genetically deficient in apoptosis, for example Bax−/− Bak−/− murine embryonic fibroblasts, treatment with pro-apoptotic agents (i.e., etoposide) resulted in non-apoptotic cell death accompanied by excessive AV formation [73]. This caspase-independent cell death was blocked by chemical inhibitors of autophagy, or by genetic interference of autophagic proteins [73].
Literature reports on the functional consequences of autophagy (whether pro-survival or pro-death) appear to vary with cell type [74,75] or with the inducing conditions [76]. For example, one study demonstrated a differential functional outcome of autophagy in response to chemical stimulation, whereby the same stimuli promoted cell survival in transformed cell lines but caused cell death in corresponding untransformed lines [74]. Furthermore, chemical induction of autophagy with endoplasmic reticulum (ER) stress agents inhibited cell death in normal fibroblasts but promoted cell death in apoptosis-impaired (Bax−/− Bak−/−) fibroblasts [75]. The inhibition of autophagy by genetic interference was reported to have differential functional outcomes depending on the nature of the pro-death stimuli. For example, Atg5 knockdown enhanced fibroblast apoptosis in combination with pro-apoptotic stimuli (i.e., death receptor pathway agonists), but inhibited apoptosis in response to xenobiotics [76].
The relationships between the underlying molecular mechanisms that provide fine regulation of autophagy and those that regulate apoptosis also remain incompletely resolved [62]. Recent studies imply that cross-interaction can occur between the molecules that regulate autophagy and apoptosis (Figure 4) [63,64]. Autophagic proteins can interact with and modify the activity of apoptosis-associated factors, and vice versa. Beclin 1 interacts with anti-apoptotic Bcl-2 family members, including Bcl-2 and Bcl-XL [31,32,77]. Binding of Bcl-2 family proteins to Beclin 1 inhibits autophagy by preventing the association of Beclin 1 with the class III PI3K complex. Overexpression of Bcl-2 resulted in impaired autophagy. However, overexpression of a mutant Beclin 1 deleted for the Bcl-2 binding site resulted in increased autophagy relative to overexpression of the wild-type Beclin 1, and increased cell death [32]. In contrast to the aforementioned findings, overexpression of Bcl-2 during stimulation with pro-apoptotic agents such as etoposide was found to promote non-apoptotic cell death associated with increased AV formation [73]. More research is needed to deduce the function of Bcl-2-related proteins in autophagic regulation under physiological conditions. The autophagic protein Atg5 may affect death receptor-dependent apoptosis pathways through interactions with the Fas-associated death domain protein, a component of DISC [78]. A calpain fragment of autophagic protein Atg5 enhances apoptosis by binding to and inhibiting Bcl-XL [79]. In addition, several signal transduction proteins associated with apoptosis (i.e., p53, DRAM and c-Jun-NH2-terminal kinase) also regulate autophagy [80,81]. Our ongoing studies aim to elucidate these relationships in the context of CS-induced cell injury.
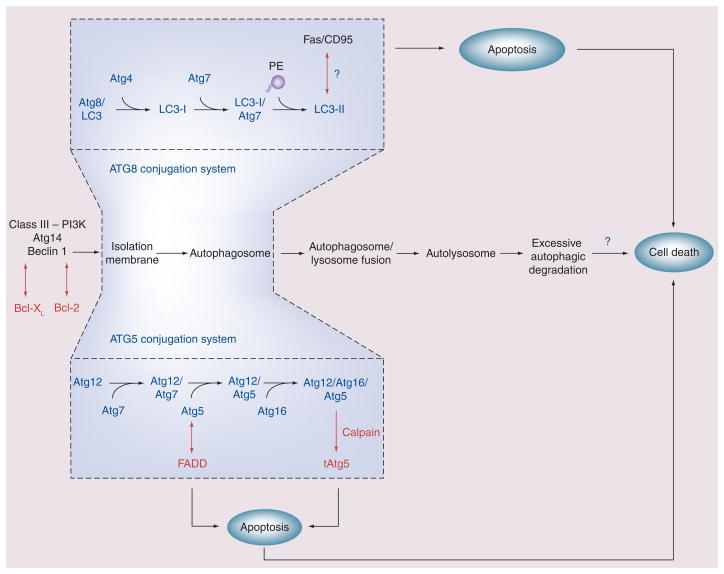
Figure 4. Several regulatory elements of autophagy have been shown to interact with apoptosis signaling pathways in mammalian cells.
Autophagosome formation requires two ubiquitin-like conjugation systems: the Atg8 (LC3) conjugation system and the Atg5–Atg12 conjugation system. Conjugation of LC3 with PE is a major indicator of autophagosome formation. Our recent studies indicate a possible pro-apoptotic interaction of LC3B with the Fas-dependent extrinsic apoptotic pathway in the CS exposure model [18]. Atg5 possibly links to cell death through interaction with FADD. A calpain-dependent truncation production of Atg5 (tAtg5) may act as a pro-apoptotic factor. Beclin 1 forms binding complexes with anti-apoptotic molecules Bcl-2 and Bcl-XL. These interactions generally inhibit autophagy by suppressing Beclin 1-dependent signaling and may influence the outcome of autophagy-associated cell death.
FADD: Fas-associated death domain; LC: Light chain; PE: Phosphatidylethanolamine.
Relationship of autophagy with COPD pathogenesis
Our human studies indicate that autophagic markers are elevated at early stages of COPD, and that their presence in GOLD0 smokers perhaps indicate a general response to chronic smoke exposure that precedes the disease process [20]. In our studies, increased autophagy was also observed in the genetic variant of emphysema, α1-AT, whose etiology is independent of smoke or particle inhalation. Unlike the COPD specimens, where comprehensive smoking histories were available, no smoking histories were available for the α1-AT patients. Nevertheless, these observations suggest that intrinsic factors, such as increased matrix proteolysis or inflammation, may contribute to the activation of autophagy in COPD, in addition to direct cellular responses to CS.
Current studies aiming to elucidate the role of autophagy in COPD pathogenesis, as described in this article, have thus far only considered the potential impact of autophagy on the process of lung cell apoptosis. Although not yet tested directly in models of CS-induced lung injury, it remains possible that autophagy may impact on other important processes that are potentially involved in COPD pathogenesis, including the regulation of inflammation, innate immune responses and matrix proteolysis.
Recently, several relationhips between autophagy, inflammation and the activation of Toll-like receptor (TLR) signaling pathways have been proposed [82], although the role of these relationships in CS-induced lung injury has not yet been examined. Activation of TLRs is potentially involved in autophagy induction and autophagosome assembly [83–86]. In macrophages, the induction of autophagy by proinflammatory stimuli was reported to involve the TLR4 pathway, the adaptor TRIF, receptor-interacting protein 1, the p38 MAPK signaling pathway and the phagocytic NADPH oxidase [83,87]. Autophagy was also suggested to be responsive to stimulation by TLR3 and TLR7 ligands [84]. Further research will determine whether autophagy and inflammation cross-talk in the context of CS exposure models.
The role of MMPs in the regulation of autophagy, or vice versa, remains unclear. Recent studies indicate that MMP inhibitors may impair the induction of autophagy in cancer cells elicited by anti-cancer drugs [88]. Mitochondrial MMP-9 activity in cardio-myocytes has been linked to a pathway mediating mitochondria-specific autophagy (mitophagy) [89]. Nothing is currently known of the relationship between autophagy and matrix proteolysis as it may apply to COPD, and further experiments along these lines may be interesting.
Expert commentary
Additional studies are clearly needed to define the relationships between autophagy, apoptosis and cell death in the context of disease pathogenesis. Autophagy is primarily regarded as a cell survival mechanism under nutrient-poor conditions. Our recent studies, however, imply a potentially deleterious function of this process during CS exposure. Starvation represents a physiologically relevant model in which autophagy as a catabolic process can provide survival advantage by conserving cellular resources. CS exposure, however, represents a dramatically different model involving cellular exposure to a complex mixture of toxic particles, chemicals and reactive species. CS exposure may lead to the deposition of cellular debris, including exogenous material and insoluble protein aggregates within the cell. It is conceivable that autophagy, although induced as a protective mechanism in this context, fails to function in the intended fashion. This may imply a state in which autophagy is excessively activated but cannot cope with the substrate load, which may include foreign particles or large-scale accumulation of denatured protein, and leads to aggravation rather than resolution of the non-homeostatic state. The engulfment of foreign matter along with cellular constituents could be regarded as a ‘xenophagy’ rather than strictly autophagy, although direct evidence for this in the CS-exposure model is lacking. Frustrated autophagy may resemble frustrated phagocytosis, whereby futile phagocytic activity towards indigestible material leads to cellular injury. Chemicals that cause ER disruption, leading to intracellular accumulation of denatured protein, also cause a potent activation of autophagy in cell culture [74]. To date, the relationships between ER stress and autophagy have not yet been studied in the CS exposure model.
In the autophagy field, recent emphasis has been placed on the measurement of autophagic activity or flux and its distinction from the measurement of protein markers of autophagy alone. In vitro, this issue is currently addressed by the use of chemical inhibitors of lysosomal turnover or autophagosome–lysosome fusion (i.e., leupeptin and bafilomycin A1; see [21]). Molecular inhibitor studies lend support to the conclusion that the increases in autophagic markers in response to CS are associated with increased autophagic activity or flux, at least in vitro [19,20]. This relationship currently remains unclear in the case of COPD tissues and in mouse lungs exposed to chronic CS, and warrants further investigation. We cannot yet exclude the possibility that autophagic processes may be impaired in vivo, leading to accumulation of AV and apoptosis secondary to autophagic impairment. Few studies so far have attempted to dissect these relationships in vivo, and such an approach is likely to present a considerable challenge, in particular in chronic exposure models such as CS inhalation. Nevertheless, more rigorous testing of autophagic flux relationships in vivo with chemical inhibitor strategies may clarify these issues.
Some of the major autophagic regulators such as LC3B appear to interact with other signaling networks not necessarily related to autophagic function. LC3B acts as a regulator of fibronectin (Fn1) mRNA translation by interacting with an AU-rich region of the Fn1 3′-untranslated region [90,91]. LC3B-dependent fibronectin expression and resulting increases in connective tissue growth factor expression were associated with tumor cell proliferation, adhesion and invasiveness in fibrosarcoma cells [92]. In addition, LC3B acts as a regulator of microtubule assembly [93] and microtubule-dependent mRNA transport [94]. Thus, it remains unclear whether the involvement of an autophagy-associated protein (i.e., LC3B) in a particular process, such as the regulation of cell death, implies the involvement of autophagic activity, or rather reflects an association of the autophagic protein with cellular signaling pathways independent of autophagic activation. Since LC3B knockdown results in impaired apoptosis in the CSE exposure model, it is possible that the LC3B directly interacts with signaling pathways involved in the upstream regulation of apoptosis, which do not necessarily require an active autophagic process. Further resolution of this issue would be needed when considering autophagic proteins as targets for therapy.
The experiments conducted to date in the CS exposure model have relied on the use of homozygous or heterozygous knockout mice for the major autophagic regulator proteins LC3B or Egr-1, and Beclin 1, respectively. Due to the potential impact of these gene deletions (e.g., Egr-1) on normal lung development, an inducible or conditional cell-type specific (i.e., epithelial cell) approach to generation of knockout animals may remove this caveat and provide more specific information.
Another limitation of the studies described in this paper is that they were performed in Beas-2B, or in differentiated respiratory epithelial cells cultured at air–liquid interface. However, COPD consists of both emphysema and chronic bronchitis, and therefore studies in airway epithelia alone may not completely or accurately model physiological responses in COPD. Emphysema primarily reflects the selective destruction of type II alveolar epithelial cells. The observed responses in airway epithelia to cigarette smoke do not necessarily prove their occurrence or lack thereof in type II cells. Further experimentation in type II epithelial cells could be warranted to resolve these issues. Unfortunately, type II epithelial cells are relatively more difficult to isolate and culture, and this has somewhat delayed progress in this area.
Five-year view
The specific role(s) of autophagy in many human diseases, including chronic lung disease, as discussed in this article, remains incompletely resolved. It is still unclear if autophagy represents a ‘bystander’ phenomenon that occurs as a general adaptive response in stressed tissue, or whether clear links to disease mechanisms will emerge. Furthermore, it remains unclear whether ‘non-autophagic’ functions of autophagic proteins (i.e., LC3B and Beclin 1) may be more important in a disease process than the process of autophagy itself, as they may impact on signal transduction pathways. Over the next 5 years, we hope to see a resolution of these fundamental questions for various human diseases, including chronic diseases of the lung. Analysis of autophagic function in stressed cells may further broaden the understanding of how environmental stress can translate to disease. If the role of autophagy can be confirmed as either adaptive or pathogenic in any specific disease context, then the pharmacological modulation of this process, either positively or negatively, could potentially offer clinical benefit. To date, there are no regulators of the autophagic process, with the exception of rapamycin, that are approved for human clinical use, and no approved drugs that specifically or selectively target this process. The further resolution of the molecular mediators and mechanisms of autophagy would potentially uncover novel therapeutic targets, which would in turn facilitate new drug discovery and development. Further evolution of this field will ultimately determine whether molecular medicines based on autophagic pathway manipulation would have an impact on clinical medicine as a whole, and on specific disorders such as COPD.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
This work was supported in part by NIH grants R01-HL60234, R01-HL55330 and R01-HL079904, awarded to Augustine MK Choi. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Rabe KF, Hurd S, Anzueto A, et al. Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2.Macnee W. Pathogenesis of chronic obstructive pulmonary disease. Clin Chest Med. 2007;28:479–513. doi: 10.1016/j.ccm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Tuder RM, Yoshida T, Arap W, Pasqualini R, Petrache I. State of the art. Cellular and molecular mechanisms of alveolar destruction in emphysema: an evolutionary perspective. Proc Am Thorac Soc. 2006;3:503–510. doi: 10.1513/pats.200603-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao H, Rahman I. Current concepts on the role of inflammation in COPD and lung cancer. Curr Opin Pharmacol. 2009;9:375–383. doi: 10.1016/j.coph.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahman I, Biswas SK, Kode A. Oxidant and antioxidant balance in the airways and airway diseases. Eur J Pharmacol. 2006;533:222–239. doi: 10.1016/j.ejphar.2005.12.087. [DOI] [PubMed] [Google Scholar]
- 6.van der Toorn M, Smit-de Vries MP, Slebos DJ, et al. Cigarette smoke irreversibly modifies glutathione in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1156–L1162. doi: 10.1152/ajplung.00081.2007. [DOI] [PubMed] [Google Scholar]
- 7•.van der Toorn M, Rezayat D, Kauffman HF, et al. Lipid-soluble components in cigarette smoke induce mitochondrial production of reactive oxygen species in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2009;297:L109–L114. doi: 10.1152/ajplung.90461.2008. Study demonstrating the involvement of mitochondria in the generation of reactive oxygen species induced by cigarette smoke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JW, Ryter SW, Choi AM. Functional significance of apoptosis in chronic obstructive pulmonary disease. COPD. 2007;4:347–353. doi: 10.1080/15412550701603775. [DOI] [PubMed] [Google Scholar]
- 9.Henson PM, Vandivier RW, Douglas IS. Cell death, remodeling, and repair in chronic obstructive pulmonary disease? Proc Am Thorac Soc. 2006;3:713–717. doi: 10.1513/pats.200605-104SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrache I, Natarajan V, Zhen L, et al. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat Med. 2005;11:491–498. doi: 10.1038/nm1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Yoshida T, Tuder RM. Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol Rev. 2007;87:1047–1082. doi: 10.1152/physrev.00048.2006. Recent comprehensive review on the molecular and cellular mechanisms of chronic obstructive pulmonary disease (COPD) pathogenesis. [DOI] [PubMed] [Google Scholar]
- 12.Hodge S, Hodge G, Holmes M, Reynolds PN. Increased airway epithelial and T-cell apoptosis in COPD remains despite smoking cessation. Eur Respir J. 2005;25:447–454. doi: 10.1183/09031936.05.00077604. [DOI] [PubMed] [Google Scholar]
- 13.Segura-Valdez L, Pardo A, Gaxiola M, Uhal BD, Becerril C, Selman M. Upregulation of gelatinases A and B, collagenases 1 and 2, increased parenchymal cell death in COPD. Chest. 2000;117:684–694. doi: 10.1378/chest.117.3.684. [DOI] [PubMed] [Google Scholar]
- 14.Yokohori N, Aoshiba K, Nagai A. Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest. 2004;125:626–632. doi: 10.1378/chest.125.2.626. [DOI] [PubMed] [Google Scholar]
- 15.Imai K, Mercer BA, Schulman LL, Sonett JR, D’Armiento JM. Correlation of lung surface area to apoptosis and proliferation in human emphysema. Eur Respir J. 2005;25:250–258. doi: 10.1183/09031936.05.00023704. [DOI] [PubMed] [Google Scholar]
- 16.Slebos DJ, Ryter SW, van der Toorn M, et al. Mitochondrial localization and function of heme oxygenase-1 in cigarette smoke-induced cell death. Am J Respir Cell Mol Biol. 2007;36:409–417. doi: 10.1165/rcmb.2006-0214OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Park JW, Kim HP, Lee S-J, et al. Protein kinase Cα and ζ differentially regulate death-inducing signaling complex formation in cigarette smoke extract-induced apoptosis. J Immunol. 2008;180:4668–4678. doi: 10.4049/jimmunol.180.7.4668. [DOI] [PubMed] [Google Scholar]
- 18.Aoshiba K, Tamaoki J, Nagai A. Acute cigarette smoke exposure induces apoptosis of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1392–L1401. doi: 10.1152/ajplung.2001.281.6.L1392. [DOI] [PubMed] [Google Scholar]
- 19•.Kim HP, Wang X, Chen Z-H, et al. Autophagic proteins regulate cigarette smoke induced apoptosis: protective role of heme oxygenase-1. Autophagy. 2008;4(7):887–895. doi: 10.4161/auto.6767. Examines the relationship between stress protein responses and autophagy in epithelial cells exposed to cigarette smoke. [DOI] [PubMed] [Google Scholar]
- 20••.Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. Documents observation of increased autophagic markers in human clinical samples from chronic obstructive pulmonary disease patients. A comparative analysis of autophagic activation and the underlying molecular mechanism is provided in models of cigarette smoke-induced emphysema and lung cell injury. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. Comprehensive review on the current state of mammalian autophagy research. State-of-the-art methodology is reviewed and discussed with critical guidelines for interpretative analysis and common pitfalls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. Updated review on the functional significance of autophagy. [DOI] [PubMed] [Google Scholar]
- 23•.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. Updated comprehensive review on the molecular machinery of autophagy in mammalian systems and yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 25.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 28.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1–PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 35.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008;15:171–182. doi: 10.1038/sj.cdd.4402233. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gison SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 38.Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 39.Ito K, Ito M, Elliott WM, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 40.Chen ZH, Kim H, Lam H, Choi AM. Caveolin-1 protects against cigarette smoking induced autophagic cell death and emphysema in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179:A1001. [Google Scholar]
- 41.Schlegel A, Lisanti MP. The caveolin triad: caveolae biogenesis, cholesterol trafficking, and signal transduction. Cytokine Growth Factor Rev. 2001;12:41–51. doi: 10.1016/s1359-6101(00)00022-8. [DOI] [PubMed] [Google Scholar]
- 42.Ryter SW, Chen ZH, Kim HP, Choi AM. Autophagy in chronic obstructive pulmonary disease: homeostatic or pathogenic mechanism? Autophagy. 2009;5:235–237. doi: 10.4161/auto.5.2.7495. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 46.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 47.Lee JA. Autophagy in neurodegeneration: two sides of the same coin. BMB Rep. 2009;42:324–330. doi: 10.5483/bmbrep.2009.42.6.324. [DOI] [PubMed] [Google Scholar]
- 48.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 49.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinet W, Agostinis P, Vanhoecke B, Dewaele M, De Meyer GR. Autophagy in disease: a double-edged sword with therapeutic potential. Clin Sci (Lond) 2009;116:697–712. doi: 10.1042/CS20080508. [DOI] [PubMed] [Google Scholar]
- 51.Martinet W, De Meyer GR. Autophagy in atherosclerosis: a cell survival and death phenomenon with therapeutic potential. Circ Res. 2009;104:304–317. doi: 10.1161/CIRCRESAHA.108.188318. [DOI] [PubMed] [Google Scholar]
- 52.De Meyer GR, Martinet W. Autophagy in the cardiovascular system. Biochim Biophys Acta. 2009;1793:1485–1495. doi: 10.1016/j.bbamcr.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. [DOI] [PubMed] [Google Scholar]
- 54.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 55.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 57.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 58.Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 59.Verheye S, Martinet W, Kockx MM, et al. Selective clearance of macrophages in atherosclerotic plaques by autophagy. J Am Coll Cardiol. 2007;49:706–715. doi: 10.1016/j.jacc.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 60.Lee SJ, Kim HP, Choi AMK. Autophagy represents an adaptive stress response to offset the development of pulmonary arterial hypertension. Am J Respir Crit Care Med. 2009;179:A1813. [Google Scholar]
- 61.Kasahara Y, Tuder RM, Cool CD, Lynch DA, Flores SC, Voelkel NF. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am J Respir Crit Care Med. 2001;163:737–744. doi: 10.1164/ajrccm.163.3.2002117. [DOI] [PubMed] [Google Scholar]
- 62.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galluzzi L, Vicencio JM, Kepp O, Tasdemir E, Maiuri MC, Kroemer G. To die or not to die: that is the autophagic question. Curr Mol Med. 2008;8:78–91. doi: 10.2174/156652408783769616. [DOI] [PubMed] [Google Scholar]
- 64.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 65.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu L, Wan F, Dutta S, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem. 2006;281:19179–19187. doi: 10.1074/jbc.M513377200. [DOI] [PubMed] [Google Scholar]
- 70.Madden DT, Egger L, Bredesen DE. A calpain-like protease inhibits autophagic cell death. Autophagy. 2007;3:519–522. doi: 10.4161/auto.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 72.Wu YT, Tan HL, Huang Q, et al. Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy. 2008;4:457–466. doi: 10.4161/auto.5662. [DOI] [PubMed] [Google Scholar]
- 73.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 74.Ding WX, Ni HM, Gao W, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 75.Ullman E, Fan Y, Stawowczyk M, et al. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 2008;15:422–425. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Singh R, Massey AC, et al. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem. 2008;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 79.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 80.Crighton D, Wilkinson S, O’Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 81.Shimizu S, Konishi A, Nishida Y, et al. Involvement of JNK in the regulation of autophagic cell death. Oncogene. 2010;29:2070–2082. doi: 10.1038/onc.2009.487. [DOI] [PubMed] [Google Scholar]
- 82.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu Y, Jagannath C, Liu XD, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Delgado MA, Elmaoued RA, Davis AS, et al. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signaling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 86.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Augustin S, Berard M, Kellaf S, et al. Matrix metalloproteinases are involved in both type I (apoptosis) and type II (autophagy) cell death induced by sodium phenylacetate in MDA-MB-231 breast tumour cells. Anticancer Res. 2009;29:1335–1343. [PubMed] [Google Scholar]
- 89.Tyagi N, Vacek JC, Givvimani S, et al. Cardiac specific deletion of N-methyl-D-aspartate receptor 1 ameliorates mtMMP-9 mediated autophagy/mitophagy in hyperhomocysteinemia. J Recept Signal Transduct Res. 2010;30:78–87. doi: 10.3109/10799891003614808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou B, Boudreau N, Coulber C, et al. Microtubule-associated protein 1 light chain 3 is a fibronectin mRNA-binding protein linked to mRNA translation in lamb vascular smooth muscle cells. J Clin Invest. 1997;100:3070–3082. doi: 10.1172/JCI119862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou B, Rabinovitch M. Microtubule involvement in translational regulation of fibronectin expression by light chain 3 of microtubule-associated protein 1 in vascular smooth muscle cells. Circ Res. 1998;83:481–489. doi: 10.1161/01.res.83.5.481. [DOI] [PubMed] [Google Scholar]
- 92.Ying L, Lau A, Alvira CM, et al. LC3-mediated fibronectin mRNA translation induces fibrosarcoma growth by increasing connective tissue growth factor. J Cell Sci. 2009;122:1441–1451. doi: 10.1242/jcs.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- 94.Seidenbecher CI, Landwehr M, Smalla KH, et al. Caldendrin but not calmodulin binds to light chain 3 of MAP1A/B: an association with the microtubule cytoskeleton highlighting exclusive binding partners for neuronal Ca2+-sensor proteins. J Mol Biol. 2004;336:957–970. doi: 10.1016/j.jmb.2003.12.054. [DOI] [PubMed] [Google Scholar]