Abstract
The epigenetic identity of oligodendrocytes is modulated by posttranslational modifications of histones. Acetylation of histone H3 results from the balance between the activity of histone acetyltransferases (HATs) and histone deacetylases and modulates transcriptional activation. We have previously shown that, in rodents, histone deacetylation favors oligodendrocyte differentiation, whereas acetylation is associated with increased levels of transcriptional inhibitors of oligodendrocyte differentiation. Here, we report, in humans brains, a shift toward histone acetylation in the white matter of the frontal lobes of aged subjects and in patients with chronic multiple sclerosis (MS). Increased immunoreactivity for acetylated histone H3 was observed in the nuclei of NogoA+ oligodendrocytes in a subset of MS samples. These changes were associated with high levels of transcriptional inhibitors of oligodendrocyte differentiation (i.e., TCF7L2, ID2, and SOX2) and higher HAT transcript levels (i.e., CBP, P300) in female MS patients compared with non-neurological controls and correlated with disease duration. Chromatin immunoprecipitation from samples of MS patients revealed enrichment of acetyl-histone H3 at the promoter of the increased target genes (i.e., TCF7L2). The data in chronic lesions contrasted with findings in early MS lesions, where a marked oligodendroglial histone deacetylation was observed. Together, these data suggest that histone deacetylation is a process that occurs at the early stages of the disease and whose efficiency decreases with disease duration.
Introduction
It is well established that the degree of compaction of chromatin and the transcriptional state of genes are associated with distinctive posttranslational modifications occurring on precise amino acid residues that occur mainly at the N terminus of the histones (Liu and Casaccia, 2010). These modifications include acetylation, methylation, sumoylation, and ubiquitination of lysine residues; methylation and citrullination of arginine residues; phosphorylation of serines and threonines; ADP-ribosylation and isomerization of prolines (Kouzarides, 2007). One of the best characterized histone modifications that modulate oligodendrocyte differentiation is the equilibrium between histone acetylation and deacetylation (Copray et al., 2009). In young animals, we and others have shown that pharmacological inhibitors of histone deacetylases (HDACs) increase histone acetylation and prevent differentiation of oligodendrocyte progenitors in vitro (Marin-Husstege et al., 2002; Hsieh et al., 2004) and in vivo (Shen et al., 2005; Liu et al., 2007). These data were supported by gene silencing experiments in primary cultures of oligodendrocyte progenitors (Shen et al., 2008b), the detection of defective oligodendrogliogenesis in hdac1 zebrafish mutants (Cunliffe and Casaccia-Bonnefil, 2006) and in mice with conditional ablation of Hdac1 and Hdac2 (Ye et al., 2009) or of the HDAC1 binding molecule Yy1 (He et al., 2007) in the oligodendrocyte lineage.
In aged rodent brains, the equilibrium was shifted toward histone acetylation (Shen et al., 2008a), and this correlated with heterotopic and heterochronic expression of genes characteristic of the myelinating phenotype (Shen et al., 2008a,b). Intriguingly, several of the genes downregulated by HDAC activity encoded for transcription factors downstream of the bone morphogenetic protein (BMP) (i.e., Id2 and Id4), Notch (i.e., Hes5), and Wnt (i.e., Tcf7l2/Tcf4) signaling pathways (Wang et al., 1998; Samanta and Kessler, 2004; He et al., 2007; Fancy et al., 2009; Li et al., 2009). Previous studies have shown that these pathways are downregulated during developmental myelination and reexpressed in progenitor cells after damage to myelin (John et al., 2002; Sim et al., 2006; Shen et al., 2008b; Fancy et al., 2009) and also in mature cells in the aged rodent brain (Shen et al., 2008a).
Increased histone acetylation has also been described in other organs and related to the presence of inflammatory conditions. In human lung, for instance, the pattern of histone acetylation detected in old subjects, resembled that observed in chronic obstructive pulmonary disease and led to the hypothesis that inflammation could mimic a condition of “premature” aging within different tissues (Ito and Barnes, 2009). Based on these premises, we aimed to test the hypothesis that changes of histone acetylation also occur in patients with multiple sclerosis (MS), an inflammatory disease of the CNS, and that these changes are associated with widespread aberrant modulation of gene expression.
Materials and Methods
Tissue samples.
We studied a total of 47 MS patients and 69 controls (Tables 1, 2). Chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR) were performed on frozen autoptic tissue specimens from the normal-appearing white matter (NAWM) samples of the frontal lobes of 23 MS patients with secondary progressive or chronic progressive type of the disease (12 females, 11 males; average age, 62.3 ± 7 years for males and 61.4 ± 9.2 years for females) and 19 non-neurological controls (7 females, 12 males; average age, 66.3 ± 10.3 years for males and 75.5 ± 3.8 years for females). These samples were obtained from the Human Brain and Spinal Fluid Resource Center (Veterans Affairs West Los Angeles Healthcare Center, Los Angeles, CA), which is sponsored by National Institute of Neurological Disorders and Stroke/National Institute of Mental Health, National Multiple Sclerosis Society, and Department of Veterans Affairs, and also from the German Brain-Net. Quantitative ChIP for acetyl-H3 was conducted in a subset of four MS patients (average age, 69.5 ± 7.8 years for males and 67.5 ± 9.2 years for females) and four controls (average age, 73 ± 2.8 years for males and 76.8 ± 4.2 years for females). Samples containing actively demyelinating or chronic lesions were not included in this study. RNA and chromatin quality was checked on agarose gel and only samples that did not show signs of degradation were pursued for additional analysis.
Table 1.
Summary of all the MS and non-neurological control samples used for qPCR and ChIP
| Patient ID | Age | Disease duration | Diagnosis | Sex | Postmortem interval (h) | Additional diagnosis | Experiment | |
|---|---|---|---|---|---|---|---|---|
| MS1 | 51 | 21 | MS | F | 6.5 | MS | qPCR | |
| MS2 | 51 | 32 | MS | F | 9 | CP | qPCR | |
| MS3 | 64 | 33 | MS | F | 17.3 | SP | qPCR | |
| MS4 | 61 | 39 | MS | F | 38 | SP, CxC | qPCR | |
| MS5 | 64 | 26 | MS | F | 9 | SP | qPCR | |
| MS6 | 50 | 24 | MS | F | 15 | SP | qPCR | |
| MS7 | 51 | 20 | MS | F | 19.5 | SP | qPCR | |
| MS8 | 61 | 17 | MS | F | 22.8 | PP | qPCR | ChIP |
| MS9 | 74 | 11 | MS | F | 9.5 | PP | qPCR | ChIP |
| MS10 | 72 | 17 | MS | F | 40.1 | SP, BnC | qPCR | |
| MS11 | 75 | 29 | MS | F | 13.8 | CP | qPCR | |
| MS12 | 63 | 18 | MS | F | 15 | MS | qPCR | |
| MS13 | 76 | 11 | MS | M | 25 | SP, AS | qPCR | |
| MS14 | 60 | 4 | MS | M | 17 | MS | qPCR | |
| MS15 | 59 | 14 | MS | M | 23 | MS | qPCR | |
| MS16 | 60 | 20 | MS | M | 9 | MS | qPCR | |
| MS17 | 59 | 12 | MS | M | 15 | PP | qPCR | |
| MS18 | 65 | 23 | MS | M | 29.4 | CP, Mel, CnC | qPCR | |
| MS19 | 54 | 25 | MS | M | 16.9 | CP | qPCR | |
| MS20 | 75 | 41 | MS | M | 12.9 | SP, PtC | qPCR | ChIP |
| MS21 | 71 | 39 | MS | M | 31.7 | SP | qPCR | |
| MS22 | 69 | ? | MS | M | 12.5 | CP, LgC | qPCR | |
| MS23 | 64 | 17 | MS | M | 12.5 | CP, COPD | qPCR | ChIP |
| N1 | 90 | N/A | N | F | 11.8 | CnC | qPCR | |
| N2 | 76 | N/A | N | F | 9 | CHD | qPCR | |
| N3 | 72 | 6 | N | F | 19.5 | BrC | qPCR | |
| N4 | 72 | 2 | N | F | 20.1 | BrC | qPCR | |
| N5 | 79 | 4 | N | F | 14 | CHD | qPCR | ChIP |
| N6 | 73 | 1 | N | F | 12 | COPD | qPCR | ChIP |
| N7 | 81 | N/A | N | F | 11.3 | AS, BrC | qPCR | |
| N8 | 80 | 9 | N | M | 11 | RnC | qPCR | |
| N9 | 68 | 1 | N | M | 10.5 | LgC, ischemia | qPCR | |
| N10 | 75 | 7 | N | M | 11.5 | PtC | qPCR | ChIP |
| N11 | 66 | 1 | N | M | 13.1 | LxC | qPCR | |
| N12 | 71 | 11 | N | M | 11.5 | COPD | qPCR | ChIP |
| N13 | 64 | 16 | N | M | 17.5 | Lymphoma | qPCR | |
| N14 | 54 | N/A | N | M | 19 | CHD, AS | qPCR | |
| N15 | 52 | N/A | N | M | 16 | LgC | qPCR | |
| N16 | 58 | 2 | N | M | 9 | CnC | qPCR | |
| N17 | 74 | N/A | N | M | 13.6 | LgC | qPCR | |
| N18 | 81 | N/A | N | M | 14 | Lymphoma | qPCR | |
| N19 | 53 | 3 | N | M | 15 | Mel | qPCR | |
| N20 | 90 | 4 | N | M | 17.8 | COPD | ||
Abbreviations: COPD, Chronic obstructive pulmonary disease; AS, atherosclerosis; CHD, coronary heart disease; Mel, melanoma; CnC, colon cancer; LgC, lung cancer; CvC, cervix cancer; PtC, prostate cancer; BnC, bone cancer; RnC, renal cancer; BrC, breast cancer; LxC, larynx cancer.
Table 2.
Summary of all the MS and non-neurological control samples used for immunohistochemistry (IHC)
| Patient ID | Age | Disease duration | Diagnosis | Sex | Additional diagnosis | Experiment |
|---|---|---|---|---|---|---|
| MS24 | 57 | 15 years | MS | M | PP | IHC |
| MS25 | 59 | 41 years | MS | F | ? | IHC |
| MS26 | 66 | 34 years | MS | M | SP | IHC |
| MS27 | 59 | 9 years | MS | F | PP | IHC |
| MS28 | 57 | 12 years | MS | M | SP | IHC |
| MS29 | 59 | 16 years | MS | F | SP | IHC |
| MS30 | 22 | ? | N/A | F | IHC | |
| MS31 | 65 | 19 d | N/A | M | IHC | |
| MS32 | 32 | 58 d | N/A | M | IHC | |
| MS33 | 63 | 92 d | N/A | F | IHC | |
| MS34 | 64 | ? | N/A | M | IHC | |
| MS35 | 33 | ? | N/A | F | IHC | |
| MS36 | 43 | ? | N/A | F | IHC | |
| MS37 | 37 | ? | N/A | F | IHC | |
| MS38 | 28 | 31 d | N/A | M | IHC | |
| MS39 | 28 | ? | N/A | F | IHC | |
| MS40 | 45 | 45 d | N/A | F | IHC | |
| MS41 | 54 | ? | N/A | F | IHC | |
| MS42 | 54 | ? | N/A | F | IHC | |
| MS43 | 20 | ? | N/A | F | IHC | |
| MS44 | 46 | ? | N/A | F | IHC | |
| MS45 | 49 | ? | N/A | M | IHC | |
| MS46 | 42 | ? | N/A | F | IHC | |
| MS47 | 22 | ? | N/A | F | IHC | |
| N21 | 23 | N/A | N | F | IHC | |
| N22 | 24 | N/A | N | F | IHC | |
| N23 | 27 | N/A | N | F | IHC | |
| N24 | 28 | N/A | N | F | IHC | |
| N25 | 23 | N/A | N | F | IHC | |
| N26 | 73 | N/A | N | F | IHC | |
| N27 | 77 | N/A | N | F | IHC | |
| N28 | 78 | N/A | N | F | IHC | |
| N29 | 75 | N/A | N | F | IHC | |
| N30 | 30 | N/A | N | M | IHC | |
| N31 | 23 | N/A | N | M | IHC | |
| N32 | 27 | N/A | N | M | IHC | |
| N33 | 23 | N/A | N | M | IHC | |
| N34 | 78 | N/A | N | M | IHC | |
| N35 | 71 | N/A | N | M | IHC | |
| N36 | 71 | N/A | N | M | IHC | |
| N37 | 71–80 | N/A | N | F | IHC | |
| N38 | 71–80 | N/A | N | M | IHC | |
| N39 | 71–80 | N/A | N | M | IHC | |
| N40 | 71–80 | N/A | N | M | IHC | |
| N41 | 71–80 | N/A | N | M | IHC | |
| N42 | 71–80 | N/A | N | M | IHC | |
| N43 | 71–80 | N/A | N | F | IHC | |
| N44 | 71–80 | N/A | N | F | IHC | |
| N45 | 71–80 | N/A | N | F | IHC | |
| N46 | 71–80 | N/A | N | F | IHC | |
| N47 | 71–80 | N/A | N | F | IHC | |
| N48 | 71–80 | N/A | N | F | IHC | |
| N49 | 54 | N/A | N | M | IHC | |
| N50 | 59 | N/A | N | M | IHC | |
| N51 | 54 | N/A | N | M | IHC | |
| N52 | 55 | N/A | N | M | IHC | |
| N53 | 57 | N/A | N | M | IHC | |
| N54 | 54 | N/A | N | M | IHC | |
| N55 | 53 | N/A | N | M | IHC | |
| N56 | 62 | N/A | N | M | IHC | |
| N57 | 63 | N/A | N | M | IHC | |
| N58 | 61 | N/A | N | M | IHC | |
| N59 | 65 | N/A | N | M | IHC | |
| N60 | 57 | N/A | N | F | IHC | |
| N61 | 59 | N/A | N | F | IHC | |
| N62 | 57 | N/A | N | F | IHC | |
| N63 | 52 | N/A | N | F | IHC | |
| N64 | 56 | N/A | N | F | IHC | |
| N65 | 67 | N/A | N | F | IHC | |
| N66 | 68 | N/A | N | F | IHC | |
| N67 | 68 | N/A | N | F | IHC | |
| N68 | 68 | N/A | N | F | IHC | |
| N69 | 66 | N/A | N | F | IHC |
For immunohistochemistry, paraffin-embedded frontal tissue samples were obtained from 49 patients who died because of non-neurological diseases, aged 21–30 years (5 females, 4 males), 40–70 years (10 females, 11 males; mean age, 59.5 years), and 71–80 years (11 females, 8 males). Additionally, we included eight autopsy samples derived from frontal lobes of six patients with MS (3 females, 3 males; mean age, 59.5 years; disease duration between 9 and 41 years) and 19 biopsy samples (6 males, 13 females; the disease duration, only known in 5 patients, was between 19 and 92 d). Biopsies were performed for diagnostic reasons to exclude neoplastic or infectious disease. Demyelinating activity in biopsy tissue samples was determined as described previously (BRück et al., 1995). Actively demyelinating lesion areas (n = 6) were frequently located at the plaque border, partially demyelinated and infiltrated by T-cells and numerous macrophages containing myelin degradation products within the cytoplasm. In demyelinated lesion areas (n = 6), no myelin sheaths could be detected by either Luxol fast blue (LFB) staining or immunohistochemistry for different myelin proteins [e.g., myelin basic protein (MBP), proteolipid protein (PLP), 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP), myelin oligodendrocyte glycoprotein (MOG), myelin-associated glycoprotein (MAG)]. These lesion areas were infiltrated by numerous macrophages; however, they did not contain myelin degradation products. Remyelinating lesion areas (n = 12) were characterized by thin, irregularly formed myelin sheaths; the adjacent periplaque white matter (PPWM) (n = 10) showed no signs of demyelination; however, numerous reactive microglial cells and few T-cells were present. Paraffin-embedded MS autopsy tissues blocks were from the Institute of Neurology, McGill University (Montreal, Quebec, Canada), whereas MS biopsies and the nondiseased brain tissue samples were derived from the Department of Neuropathology, University Medical Center Göttingen (Göttingen, Germany). The study was approved by the Ethics Committee of the University of Göttingen.
Immunohistochemistry.
Tissue samples were cut in 4-μm-thick sections that were stained with hematoxylin and eosin (non-MS tissue samples) or LFB (MS tissue samples). Immunohistochemical staining was performed with an avidin–biotin technique. After deparaffinization, intrinsic peroxidase activity was blocked by incubation with 0.5% H2O2 in PBS for 20 min. Nonspecific antibody binding was inhibited with 10% FCS in PBS for 25 min. Microwave pretreatment for better antigen retrieval was performed before incubation with rabbit polyclonal anti-acetyl-H3 antibody (1:300) (catalog #06599; Abcam). As secondary antibody, anti-rabbit biotinylated Ig (1:400) (GE Healthcare) was used. For double staining, we combined anti-acetyl-H3 immunohistochemistry with mouse anti-NogoA immunohistochemistry (1:15,000) (11c7; generous gift from M. E. Schwab, Brain Research Institute, University of Zürich, and Department of Biology, Swiss Federal Institute of Technology Zürich, Zürich, Switzerland), mouse monoclonal anti-GFAP (1:50) (Dako) or mouse monoclonal anti-KiM1P (1: 5000) (a generous gift from H.-J. Radzun, Department of Pathology, University of Göttingen, Göttingen, Germany). Secondary antibodies were either biotinylated or conjugated to alkaline phosphatase. The number of acetyl-H3-positive cells was determined in at least 10 standardized microscopic fields of 10,000 μm2 each defined by an ocular morphometric grid and expressed as the percentage of total number of nuclei or as percentage of NogoA-positive mature oligodendrocytes. In the text and figures, the mean number of cells per square millimeter ± SEM is given.
RNA isolation and quantitative real-time PCR.
For RNA isolation, 50 mg of tissue was homogenized with 250 μl of Trizol using mortar and pestle and adjusting the volume to 1 ml. The mixture was transferred into Phase-Lock Gel Heavy 2 ml tubes (5 PRIME; 2302830) and centrifuged at 14,000 rpm for 15 min at 4°C. The aqueous phase containing the RNA was cleaned with 200 μl of chloroform, and the RNA was precipitated and washed with ethanol. The RNA samples were resuspended in water and further purified with RNeasy (QIAGEN; 74104) following the manufacturer's instructions. The quality of RNA was verified by agarose gel electrophoresis and measurement of the A260/A280 ratio and only those samples showing clear ribosomal RNA bands and A260/A280 ratio ∼2.0 were further processed for quantitative PCR analysis. For real-time PCR, 2 μg of total RNA were reverse transcribed using SuperScript First-Strand Synthesis System (Invitrogen; 11904018) and oligo-dT as primers for PCR. Quantitative real-time PCR was performed using Applied Biosystem SYBR Green master mix in a 7900HT Fast RT-PCR System (Applied Biosystems). The melting curve of each sample was measured to ensure that a single amplicon was obtained. The data were normalized using ribosomal protein 60S as internal control and analyzed by the Pfaffl ΔΔCt method (Pfaffl, 2001), using the equation ratio = E(target)ΔCt(target)(control-sample)/E(60S)ΔCt(60S)(control-sample), where the control is determined by the average of the normal samples from the same gender as the patients, and the efficiency was determined using a standard curve for each primer pair and the simplified Pfaffl equation for the efficiency = 10−1/slope. Primers used for qPCR are listed in Table 3.
Table 3.
Primers used for qPCR
| Gene | Approved symbol | Sequence sense | Sequence antisense | Gene accession no. |
|---|---|---|---|---|
| Myst3 | KAT6A | CAAGGCTGCCCAAATTGTAT | ATCTCATTGGCAGGAGGATG | NM_006766 |
| Myst4 | KAT6B | CCAGAAATCTCCACGGAAAA | GTTGTGGGATGGCTCTTCAT | NM_012330 |
| TIP60 | KAT5 | GGCTGAGGACAGCTCAAAAA | CCGGATCCCTTCTCACTGTA | NM_006388 |
| GCN5 | KAT2A | GTTATGGGACCCACCTGATG | CGGCGTAGGTGAGGAAGTAG | NM_021078 |
| PCAF | KAT2B | CCAGCAAAAGAAAGGCAAAC | ATCATTGGGAGATCGCAGTC | NM_003884 |
| CBP | KAT3A | GCCACGTCCCTTAGTAACCA | CCCCAAGTGTCCCTGATCTA | NM_004380 |
| ELP3 | KAT9 | GATTGGGGTGCAGAGTGTTT | ATCTTTGGCCAGGTGAAATG | NM_018091 |
| P300 | KAT3B | CGCTTTGTCTACACCTGCAA | TGCTGGTTGTTGCTCTCATC | NM_001429 |
| HDAC1 | HDAC1 | GATCTGCTCCTCTGACAAACGAA | CCCTCTCCCTCCTCTTCAGAA | NM_004964 |
| HDAC2 | HDAC2 | TGGAGGAGGTGGCTACACAAT | AATCTCACAATCAAGGGCAACTG | U31814 |
| HDAC3 | HDAC3 | TCTACCTCACTGACCGGGTCAT | ACCTGTGCCAGGGAAGAAGTAA | NM_003883 |
| HDAC8 | HDAC8 | GGATCCCATGTGCTCCTTTA | ATAGCCTCCTCCTCCCAAAA | NM_018486 |
| HDAC11 | HDAC11 | GAGCTGGCCCTTCCTCTACT | CTATGGGCTGGTGACTTCGT | NM_024827 |
| TCF7L2 | TCF7L2 | TGGTGCGGCCCTTGCAGATG | TCAAGCCCGAACAGTGCCCG | NM_030756 |
| TCF4 | TCF4 | CGAAATCTTCGGAGGACAAG | CTTCTCACGCTCTGCCTTCT | NM_003199 |
| JAG1 | JAG1 | AAGGGGTGCGGTATATTTCC | TCCCGTGAAGCCTTTGTTAC | NM_000214 |
| ID2 | ID2 | GACAGCAAAGCACTGTGTGG | TTCAACCATTTCACAAGGAGG | NM_002166 |
| SOX2 | SOX2 | AACCCCAAGATGCACAACTC | GCTTAGCCTCGTCGATGAAC | NM_003106 |
| MAL | MAL | GCCGTGGTGTTCTCCTACAT | ACACCATCTGGGTTTTCAGC | NM_002371 |
| MOG | MOG | TCACCTGCTTCTTCCGAGAT | GAGGAGAACCAGCACTCCAG | NM_001008228 |
| MBP | MBP | TCACAAGGGATTCAAGGGAG | TAGGTAACAGGGGCAAGTGG | NM_001025081 |
Chromatin immunoprecipitation.
For chromatin isolation, 400 mg of frozen brain samples were pulverized in dry ice and cross-linked with 1% formaldehyde for 7 min. After stopping the reaction by adding glycine to a final concentration of 125 mm, cells were lysed and the isolated nuclear fraction was lysed and sonicated using a Bioruptor sonicator (Bioruptor; Diagenode). Typically, 4–10 cycles of sonication of 5 min each (15 s ON/45 s OFF) at 4°C were used to reach chromatin fragments of ∼500 bp in size. Aliquots of isolated cross-linked chromatin (corresponding to ∼15 μg of DNA or 0.6 U/Ab 1 U = OD at 260 nm) were immunoprecipitated using 2 μg of antibody (anti-acetyl-H3; Millipore; 06-599) purified with protein A-Dynabeads (Invitrogen) and then reverse cross-linked by incubating at 65°C in a water bath in the presence of RNase A (final concentration, 40 μl/ml). The proteins of the immunocomplexes were digested with proteinase K (100 μg/ml final concentration); the recovered DNA was isolated by phenol-chloroform extraction followed by QIAquick PCR purification kit (QIAGEN) and analyzed by quantitative PCR amplification as previously described (Shen et al., 2008b). Primers used for ChIP were as follows: TCF7L2 Prom (−10/+125): sequence sense, TCGCCCTGTCAATAATCTCC; sequence antisense, CGCTCGGATTTGAGTGAGTT gene accession number, NM_030756; TUBA (−111/−15): sequence sense, GCACCAATCACCAGTCTCCT; sequence antisense, GATGCGGCTCCCTATATACAA gene accession number, NM_006009.
DNA recovered from chromatin that was not immunoprecipitated (input) and chromatin that underwent the same procedures in the absence of antibodies were both used as controls. The calculations were done using the standard curve method. The standard curve was performed using 0.5 U/μl input chromatin diluted fourfold with each dilution. The signal measured in the immunoprecipitated samples was referred to a standard curve of the signal from the input and thereby as percentage of input.
Slot blot.
To obtain the ratio of acetyl-H3/H3, nuclear fractions containing 5 μg of protein diluted to 100 ng/ml protein were loaded in triplicate onto wetted nitrocellulose membranes in a Biodot (slot format) ultrafiltration unit (Bio-Rad) under vacuum, as described by Mastronardi et al. (2006). The ratio of acetyl-H3/H3 was calculated from immunoslot blots after detection of acetyl-H3 and H3. Samples were loaded onto slots in triplicate.
Statistical analysis.
Results are expressed as mean ± SD. A t test with Welch's correction was performed to evaluate significance in the overall comparison of groups. Spearman's test was used to analyze the correlation of the transcript levels of different genes among each other and against disease duration. A linear regression was used to test whether slopes and intercepts included in the correlation were significantly different. Differences between individual groups were determined by the Scheffé test or the Mann–Whitney U test. In all cases, differences were considered to be significant when *p < 0.05; **p < 0.01.
Results
Acetyl-H3 in young and aged CNS
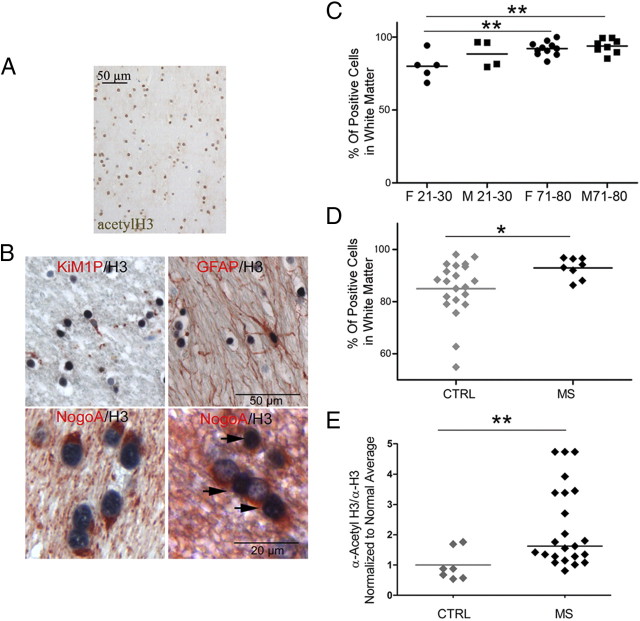
The premise of the current study was to characterize histone acetylation and deacetylation in the human aged brain, because our previous studies in old rodent brains had correlated these changes to myelin repair (Shen et al., 2008b). We focused our analysis on the percentage of acetyl-H3-positive nuclei over total cells in the frontal NAWM samples of 28 patients who died to non-neurological diseases aged 21–30 years (n = 9) or 71–80 years (n = 19) (Table 2). Analysis of the cellular composition of these samples revealed majority of oligodendroglial lineage cells. Astrocytes, identified by GFAP immunoreactivity, accounted for 1.2–5.3% of the cells, whereas microglial cells, identified using the marker KiM1P, ranged between 2.5 and 6.5% of the cells in white matter samples. On average, 90% of the astrocytes and 33.2% of the microglial cells expressed acetyl-H3 as demonstrated by double stainings combing acetyl-H3 with astrocytic or microglial markers. Since staining of the autopsy tissue samples with oligodendroglial markers, such as Olig2 or NogoA, resulted in insufficient staining quality (most likely because of the sensitivity of these markers to prolonged fixation times), we quantified the percentage of oligodendrocytes in white matter in biopsy samples from patients (n = 5) with pharmacoresistant seizures without any obvious pathology (except for mild gliosis). NogoA immunoreactivity accounted for 92.8% positive nuclei and confirmed that the vast majority of cells in the white matter were oligodendrocytes. A comparison of the cellular composition in young and old NAWM did not reveal any change in the percentage of astrocytes with age, although slightly higher numbers of microglial cells were detected in NAWM from young women and men (5.8 and 5.7% of white matter cells) compared with aged men (3.0%). Double immunoreactivity for NogoA and acetyl-H3 in tissue biopsies suggested that most of the acetyl-H3-positive cells in NAWM are oligodendrocytes (Fig. 1B). In control tissue samples, the lowest percentage of acetyl-H3-positive nuclei was observed in young females (79 ± 4.2%) and the highest percentage in older males (93 ± 1.7%) and females (92 ± 1.4%) (Fig. 1C).
Figure 1.
Histone H3 acetylation in the frontal cortex of MS and normal brains. A, Immunocytochemical analysis of histone H3 acetylation of white matter in the frontal lobes of control subjects, without neurological disease. B, Double stainings were performed with antibodies specific for acetyl-H3 (AcH3) (black) and either NogoA (red) to identify oligodendrocytes, KiM1P to identify microglia, and GFAP to label astrocytes. Examples of an AcH3-negative microglial cell, an AcH3-positive astrocyte, and AcH3-positive (arrows) and -negative oligodendrocytes are shown. C, Quantification of nuclei immunoreactive for acetylated histone H3 in the white matter of the frontal lobes of young (21–30 years) and old (71–80 years) male (squares) and female (circles) subjects. D, Quantification of total acetyl-H3+ nuclei in the frontal cortex of non-neurological (CTRL) and MS patients demonstrating a significantly increased percentage of acetyl-H3+ nuclei in chronic MS brains (Mann–Whitney test; *p < 0.05). E, Scatterplot of the amount of acetylated H3 protein levels isolated from CTRL and MS patients. Proteins were isolated from frozen frontal lobes perilesional and NAWM of control and MS patients and processed by slot blot using antibodies against acetyl-H3 and total H3 to measure global levels of acetylation on the tail of histone H3 (two-tailed t test; *p < 0.05; **p < 0.01).
Acetyl-H3 in the cerebral NAWM of patients with MS
A similar immunohistochemical analysis was then conducted with tissue samples from the NAWM of patients with chronic MS. We examined eight tissue blocks with NAWM of the frontal lobes from six MS patients (three females, three males; mean age, 59.5 years). As expected and described previously (Kutzelnigg et al., 2005), all tissue samples showed a mild-to-moderate activation of microglial cells, astrogliosis, and minimal acute axonal damage (Table 4). On average, 7.9% of the cells were KiM1P-positive microglial cells and 7.2% were GFAP-positive astrocytes in MS tissue, whereas in controls 4.7% of the cells were microglia and 3.2% astrocytes. Immunoreactivity for acetyl-H3 was detected in 92.9 ± 1.4% of the nuclei in the NAWM of patients with chronic MS, thereby suggesting that the vast majority of oligodendroglial lineage cells were immunoreactive for acetyl-H3. Tissue samples from non-neurological patients (mean age, 59.5 years; 11 males, 10 females) had a slight, but significantly lower percentage of acetyl-H3-positive nuclei (85.0 ± 2.4%) (Fig. 1D). To further quantify potential differences in the levels of histone acetylation between MS and control groups, we isolated proteins from frozen brain homogenates from 23 patients with chronic MS and 20 non-neurological controls. The relative levels of acetylation of histone H3 were quantified by slot blot analysis and normalized by dividing the signal intensity of acetyl-H3 by the total levels of histone H3. Similar to our immunohistochemical findings, the relative levels of histone H3 acetylation was significantly higher in MS patients compared with controls (Fig. 1E). Importantly, a subgroup of patients showed two to five times higher levels of acetyl-H3 than controls. This marked increase could not be explained by the relatively mild increase in the numbers of astrocytes and microglial cells in the NAWM of MS patients and therefore could be considered as an indirect measure of the global acetyl-H3 levels in cells.
Table 4.
Histological changes in normal appearing white matter from MS patients
| Autopsy | KiM1P (cells/mm2) | GFAP (cells/mm2) | Fiber gliosis | APP (spheroids/mm2) |
|---|---|---|---|---|
| MS1 | 33 | 75.7 | + | 2 |
| 34.7 | 87.5 | + | 0 | |
| MS2 | 29 | 72.5 | + | 0 |
| MS3 | 69.3 | 115.4 | + | ND |
| MS5 | 41.1 | 41.1 | − | 2.56 |
| 144 | 61 | + | 0 | |
| MS6 | 59.6 | 82.5 | + | 1.28 |
| MS7 | 83.2 | 96 | + | 2.24 |
APP, Amyloid precursor protein.
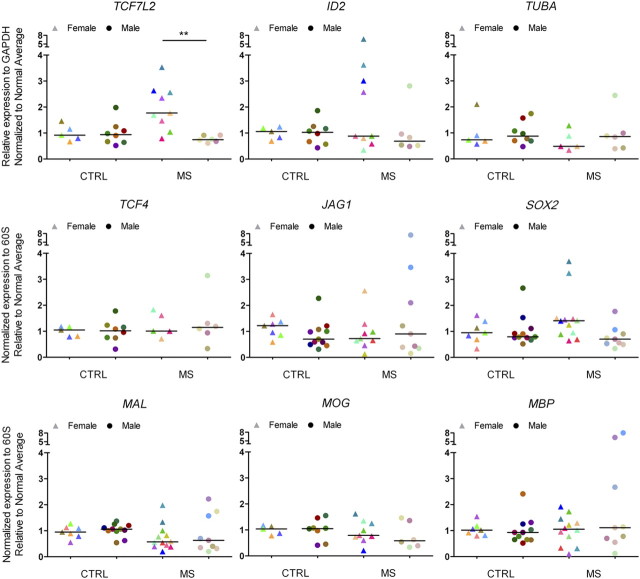
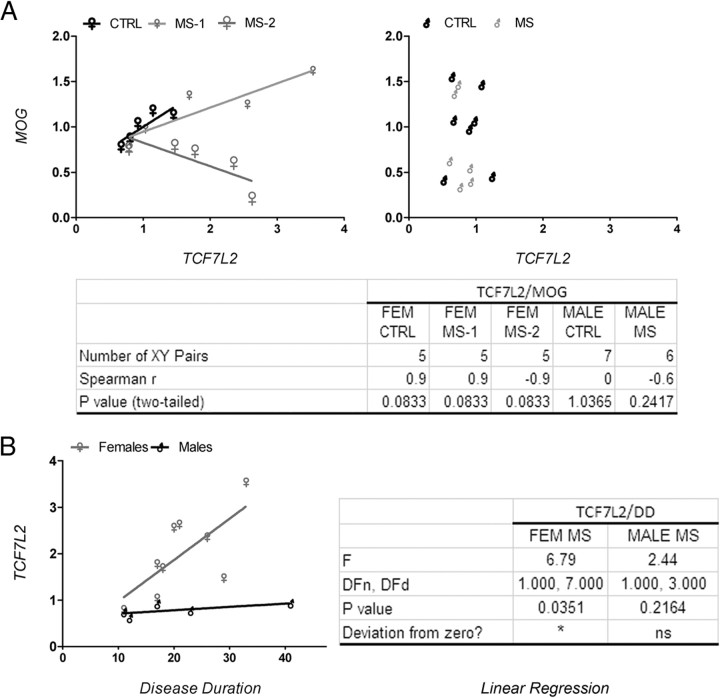
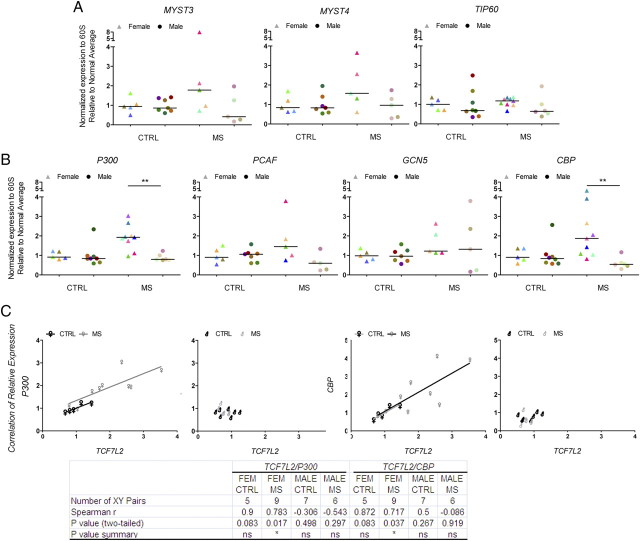
Previous studies have reported the importance of histone acetylation in modulating the levels of downstream effectors of the Notch, BMP4, and Wnt signaling pathway and negatively impact oligodendrocyte differentiation (Li et al., 2009). For this reason, we obtained RNA from the NAWM of the frontal lobes of 21 chronic MS patients and 18 age- and sex-matched non-neurological controls (Table 1) and analyzed the transcript levels of downstream signaling components of the BMP4 (i.e., ID2) and Wnt (i.e., TCF7L2) pathway. Additional transcript levels were assessed for myelin genes (i.e., MAL, MOG) and other transcription regulators (i.e., SOX2). Tubulin-α (i.e., TUBA) was used as a control (Fig. 2). Despite the interindividual variability characteristic of postmortem human material, we noticed that 8 of 10 female and 0 of 6 male patients consistently showed statistically significant higher levels of TCF7L2 transcripts compared with sex-matched controls (Fig. 2). The subgroup of female MS patients with increased levels of TCF7L2 also showed higher levels of SOX2 and ID2 transcripts compared with controls. The age of these female patients ranged between 50 and 60 years, and the disease duration was longer than 20 years. To begin understanding the potential implications of increased levels of TCF7L2 in female patients, we assessed the existence of a correlation between the levels of TCF7L2 and those of the myelin gene MOG (Fig. 3A). Also in this case, two subgroups of patients could be identified within the female MS population: one with a positive, although statistically not significant correlation between TCF7L2 and MOG, the other one with a negative correlation (Fig. 3A). None of these correlations were detected in male patients (Fig. 3A). The subgroup of female patients with higher transcript levels for TCF7L2 was also the one showing a statistically significant positive correlation with disease duration (Fig. 3B). Together, these data suggest the existence of gender-related mechanisms that impact the functional output of increased histone acetylation and modulate the transcriptional network, so that only female MS patients with a long history of the disease show increased levels of downstream signaling effectors such as Wnt and BMP4 pathways.
Figure 2.
Dysregulated expression of downstream effectors of developmental myelination pathway in the NAWM of a subset of patients with chronic MS. Quantitative real-time PCR analysis of transcripts involved in the Wnt (i.e., TCF7L2), BMP4 (i.e., ID2), and Notch (i.e., JAG1) pathway. The levels of the neural stem cell marker SOX2, myelin genes (i.e., MAL, MOG, and MBP), and tubulin α (TUBA) were also determined. The RNA was isolated from NAWM samples from the frontal lobes of patients with chronic MS and non-neurological controls. The values of the qRT-PCR of the target genes were referred to those of transcripts for the ribosomal protein 60S subunit. Note that the same patients with high levels of TCF7L2 were also the ones with high levels of ID2 and SOX2. Each patient is identified by the same color in all diagrams (two-tailed t test with Welch's correction; *p < 0.05; **p < 0.01).
Figure 3.
Correlation between TCF7L2 and MOG transcripts levels. A, Linear regression curve addressing the relations between TCF7L2 and MOG transcript levels in control and MS patients. Note the presence of two subgroups of female patterns, one displaying a linear positive correlation between the two variables and one with a negative correlation. No correlation was detected in males. B, Correlation plot between TCF7L2 transcripts levels and disease duration in MS patients. Linear regression of the female but not male transcript levels is significantly different from zero; p < 0.05.
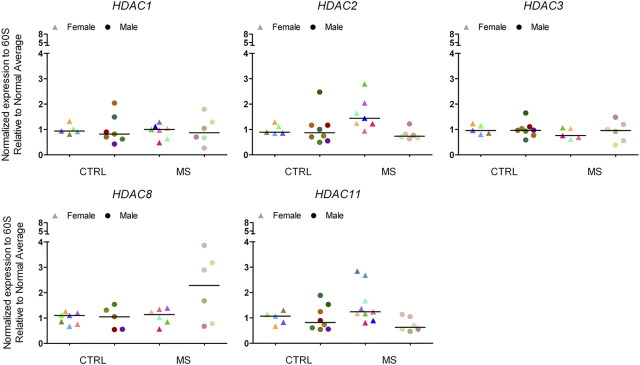
We and others have previously shown that the expression of Tcf7l2, Sox2, and Ids in rodent oligodendrocyte progenitors is modulated by histone acetylation, which depends on the equilibrium between HDACs and histone acetyltransferases (HATs) (Shen et al., 2008b; Ye et al., 2009). Therefore, we asked whether the transcript levels for these two major classes of enzymes were altered in the NAWM of chronic MS patients (Fig. 4). Of the several HDAC family members, we opted to focus on members of class I because of their predominant nuclear localization in oligodendrocyte lineage cells (Shen et al., 2005), and on Hdac11 because of its enrichment in oligodendrocytes (Liu et al., 2009). Despite the increased levels of histone acetyl-H3 in the oligodendrocytes of the NAWM of MS patients, we did not detect a significant decrease in the transcript levels of HDAC1 and HDAC2, two isoforms previously linked to successful oligodendrogliogenesis (Shen et al., 2008b; Ye et al., 2009). However, we observed a tendency toward increased HDAC8 (in males) and HDAC11 (in females) transcript levels in the NAWM of the frontal lobes of MS patients compared with controls (Fig. 4).
Figure 4.
Selective changes of histone deacetylase transcript levels in the frontal lobes of chronic MS brains. Quantitative RT-PCR of RNA isolated from white matter samples in the frontal cortex of non-neurological controls and MS patients to detect transcript levels of histone deacetylases. The relative levels of expression were first normalized to the transcript levels of the ribosomal protein of the 60S subunit and then expressed as fold changes over the average values calculated in male or female non-neurological controls. The individual values are shown as triangles for females and circles for males. The median value of the ranking distribution is shown by a line.
Because acetylation results from the equilibrium between HATs and HDACs, we also measured the transcript levels of several HAT isoforms in the NAWM of female and male MS patients. We detected statistically significant higher levels of CBP and P300, in white matter samples from the same subgroup of female MS patients that also showed increased TCF7L2 compared with controls (Fig. 5A,B). Interestingly, the levels of these HATs showed a positive correlation with TCF7L2 levels only in female, but not in male MS patients (Fig. 5C). Thus, gender plays an important role in modulating the transcriptional output of increased histone acetylation.
Figure 5.
Increased levels of histone acetyltransferase transcripts in the frontal lobes of a subset of patients with chronic MS. Quantitative RT-PCR of RNA isolated from white matter samples in the frontal cortex of non-neurological controls and MS patients to detect transcript levels of distinct classes of histone acetyltransferases, including MYST3, MYST4, and TIP60 (A), and P300, PCAF, GCN5, and CBP (B). The relative levels of expression were normalized to the levels of 60S RNA protein transcripts and then expressed as fold changes over the average values calculated in male or female non-neurological controls. The individual values are shown as triangles for females and circles for males. The median value of the ranking distribution is shown by a line. Note the increased levels of P300 and CBP in female MS patients compared with controls (two-tailed t test with Welch's correction; *p < 0.05; **p < 0.01). C, Correlation between P300 or CBP levels and TCF7L2. Linear regression of female but not male MS patients is significantly different from zero; p < 0.05.
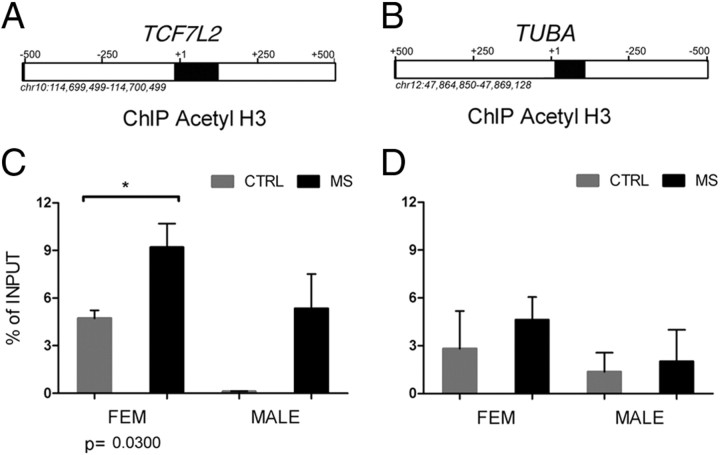
To prove further that acetylation of histones was directly related to the increased transcript levels of transcriptional inhibitors, we performed ChIP. Briefly, chromatin samples were isolated from pulverized NAWM of the frontal lobes of frozen samples from non-neurological and chronic MS patient brains. Samples were cross-linked and then immunoprecipitated with antibodies that bind to acetyl-H3. After digestion of the chromatin-associated proteins, the DNA was used as a template for quantitative PCR with primers specific for the region surrounding the start sites of the genes encoding TCF7L2. TUBA, a constitutively expressed cytoskeletal component, was used as control (Fig. 6). The amount of DNA immunoprecipitated with acetyl-H3 antibody was expressed as percentage of input control. The levels of acetylation of histone H3 in regions corresponding to the TCF7L2 gene were statistically significantly (p = 0.0034) higher in MS patient (x = 7.9089 ± 3.964945; n = 9) compared with the levels detected in non-neurological controls (x = 2.080 ± 2.522142; n = 7). No significant changes were detected in constitutively regulated genes, such as TUBA.
Figure 6.
Increased histone H3 acetylation in the region corresponding to TCF7L2, and TUBA, in chromatin samples from MS patients. A, B, TCF7L2 and TUBA gene loci from UCSC genome browser. The regions amplified in the ChIP experiment are shown as black boxes. C, D, After isolation of chromatin from the NAWM of the frontal lobes of patients with chronic MS and non-neurological controls, samples were immunoprecipitated with antibodies against acetyl-H3 and then assayed using quantitative PCR. The values are referred as percentage of enrichment of the signal in the immunoprecipitated samples relative to nonprecipitated chromatin (i.e., input control). Note the more prominent acetylation in female MS patients compared with controls detected at the TCF7L2 locus (C), but not at the constitutively expressed TUBA locus (D) patients (two-tailed t test with Welch's correction; *p < 0.05; **p < 0.01). Error bars indicate SEM.
The higher levels of histone acetylation at the TCF7L2 locus were more prominent in samples from female MS patients than corresponding samples from male patients (Fig. 6B). Thus, additional factors in male MS patients (i.e., hormone receptors or hormone receptor effect on chromatin or microRNAs, as discussed) might dampen the effect of histone acetylation on the activation of the TCF7L2 promoter.
Acetyl-H3 in demyelinating and remyelinating MS lesions
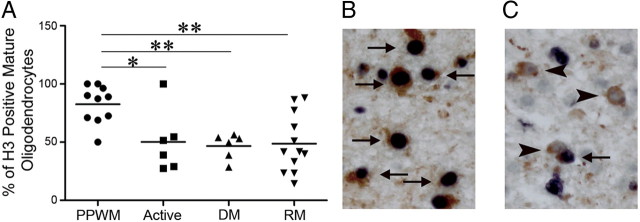
Molecular studies in rodent models had suggested that histone acetylation is associated with the maintenance of an undifferentiated state of oligodendrocyte progenitor cells and that differentiation is associated with decreased levels of histone acetylation. Our studies in chronic MS samples suggested that deacetylation is impaired with disease duration, but did not reveal any information regarding the initial stages of the disease. To further analyze the acetylation/deacetylation of histone H3 in oligodendrocytes in early MS lesions, we determined the percentage of acetyl-H3-positive oligodendrocytes in newly developed MS lesions with ongoing demyelination and/or remyelination. Whereas in the periplaque white matter the vast majority of NogoA-positive oligodendrocytes expressed acetyl-H3 (83 ± 5%), this percentage was significantly decreased in actively demyelinating, demyelinated, and remyelinating lesion areas (between 47 ± 4 and 50 ± 11%) (Fig. 7A–C). In seven biopsies, remyelinating lesion areas and adjacent periplaque white matter were available for quantitative analysis; whereas four biopsies showed a markedly reduced percentage of acetyl-H3- and NogoA-positive oligodendrocytes in remyelinating areas, the other three biopsies revealed a similar fraction of acetyl-H3-expressing mature oligodendrocytes compared with the periplaque white matter. These data suggest that deacetylation of histone H3 is a process that can be detected in early MS lesions and that it is less efficient in patients with long disease duration.
Figure 7.
Reduced histone H3 acetylation in oligodendrocyte lineage cells in early MS lesions. A, The percentage of acetyl-H3+ oligodendrocyte lineage cells is significantly reduced in actively demyelinating, demyelinated, and remyelinating lesion areas compared with the periplaque white matter (ANOVA with Bonferroni's correction; *p < 0.05; **p < 0.01). B, C, Double staining for acetyl-H3 (black) and NogoA (red) in PPWM (B) and a remyelinating lesion area (C) are shown; both pictures are taken from the same patient. In the PPWM, numerous acetyl-H3+ NogoA+ oligodendrocytes are found (arrows), whereas in remyelinating areas numerous acetyl-H3− NogoA+ oligodendrocytes (arrowheads) are observed. PPWM, Periplaque white matter; active, actively demyelinating; DM, demyelinated; RM, remyelinating.
Discussion
In this study, we analyzed the equilibrium between histone acetylation, mediated by HATs, and histone deacetylation, mediated by HDACs, in the NAWM of chronic MS brains. Similar to what was reported for the old rodent brain, also in the NAWM of human brains from aged individuals and chronic MS patients we detected a shift toward acetylation. This shift toward acetylation detected in a subset of female patients correlated with the consistent and reproducible increase of several histone acetyltransferase family members, including CBP, P300, MYST3, and MYST4. It is worth noting that, although we also detected increased levels of HDAC11 in this subpopulation, the increased of the acetyltransferases was much greater and likely determined the shift in favor of increased acetylation. These differences were most prominent in a subset of female MS patients and were associated with high levels of developmentally regulated genes (i.e., TCF7L2, SOX2, ID2) compared with controls. Several other genes (i.e., TUBA, HDAC1, HDAC3) were not changed and served as internal controls. In contrast to our findings in NAWM, in early MS lesions with ongoing demyelinating and remyelinating activity the percentage of acetyl-H3-positive mature oligodendrocytes was significantly reduced compared with adjacent periplaque white matter, thereby suggesting that the equilibrium between histone acetylation and deacetylation is affected by multiple parameters, including disease duration.
Oligodendrocyte lineage cells within NAWM in the brain of patients with chronic MS were characterized by high levels of histone acetylation, whereas cells within early MS lesions showed variable patterns of deacetylation or acetylation of histone H3. The global increase of histone acetylation in the NAWM of MS subjects was consistent with the detection of genes in the BMP4 (Sim et al., 2002), Notch (John et al., 2002), and Wnt signaling (Fancy et al., 2009) pathways and with other elegant studies reporting dysregulated gene expression in white matter that lacks clear signs of demyelination (Graumann et al., 2003; Zeis et al., 2009). Together, the global changes of histone acetylation and the effects on gene expression can be interpreted in terms of widespread modifications of the chromatin landscape in demyelinating disorders (Huynh and Casaccia, 2010) affecting all the cells present in the white matter regions, even in the absence of active signs of demyelination.
Although the levels of histone acetylation were consistently higher in the brain of MS patients compared with controls, it is worth mentioning that the higher transcript levels for some histone acetyltransferases (i.e., P300 and CBP) or downstream Wnt effectors (i.e., TCF7L2) were only detected in a subset of female patients and correlated with long disease duration. One possible explanation for these gender-based differences was that histone acetylation regulates the promoter for these genes only in female patients and that in male patients the increased levels of HDAC8 might be compensatory. However, the results of the ChIP experiments suggested that the chromatin at the TCF7L2 locus in both male and female MS subjects is more acetylated than in controls. An alternative explanation is a gender-based differential regulation of microRNAs and long noncoding RNA, driven by sex steroids. It has been suggested that estrogen binding to estrogen receptors downregulates the expression of a subset of microRNA in Drosophila and in mammals (Yamagata et al., 2009). Conversely, androgens have been reported to upregulate micro RNAs in prostate and muscle cells (Narayanan et al., 2010) and regulate transcription of long noncoding RNA (Louro et al., 2007). Acetylation modulates the transcriptional competence of chromatin. Our data suggest that MS disease duration modulates the overall chromatin landscape, rendering it more accessible to other modulating factors, such as androgen or estrogen receptors bound to their respective ligands. The overall effect on transcription, however, is determined by the type of complexes that are recruited to specific genes. The differential regulation of microRNAs by estrogens and androgens indicates that estrogens might recruit activating complexes and androgen repressive complexes, thereby accounting for the observed gender-based differences of TCF7L2 transcript levels.
The NAWM in chronic MS is characterized by a diffuse inflammatory environment (e.g., mild microglial activation and astrogliosis). In other organs, such as the lung, the pattern of increased histone acetylation detected in chronic obstructive pulmonary disease follows the one detected in the lungs of older subjects and led to the concept of inflammation-associated premature aging (Ito and Barnes, 2009). Similar to the lung, the inflammatory environment in the NAWM might induce a progressive increase of global acetylation of histone H3 in patients with chronic MS, which resembles the changes observed in the aged CNS and positively correlates with disease duration. The overall change in the total number of acetyl-H3+ nuclei in the normal appearing white matter is relatively modest. However, it is important to consider that immunohistochemistry provides a global and semiquantitative assessment of acetylation. Additional quantification of the protein levels by slot blot or chromatin immunoprecipitation has clearly shown that the amount of acetylation is increased severalfold and that these changes are highly localized at the level of specific gene loci. This would explain why the apparently modest change in the number of immunoreactive acetyl-H3+ nuclei results in important changes in gene expression.
In developing MS lesions, in contrast, we detected a marked downregulation of acetylated histone H3 in oligodendrocytes compared with the periplaque white matter. This downregulation was obvious in all lesion areas, suggesting that the presence of inflammatory cells in acute MS lesions modulates histone acetylation in oligodendrocyte lineage cells. We have previously shown that developmental myelination in the rodent brain requires a shift toward deacetylation of nucleosomal histones. In remyelinating lesion areas, we could detect oligodendrocyte lineage cells with two distinct patterns of histone acetylation. High levels of acetylation of histone H3 were found in oligodendrocytes within the PPWM, whereas reduced histone H3 acetylation was mainly found in remyelinating areas. One possibility is that these two patterns reflect two distinct stages of remyelination. Alternatively, they could represent intrinsic differences in the capability of oligodendrocytes to remyelinate. However, remyelination is a complex process that, in addition to a potential modification by histone acetylation, is also dependent on the extent of axonal damage (Frischer et al., 2009), the presence of oligodendroglial progenitor cells (Wolswijk, 1998; Chang et al., 2002; Kuhlmann et al., 2008), the composition of the inflammatory infiltrates (Arnett et al., 2003; Bieber et al., 2003; Kotter et al., 2005), the anatomical localization of the lesions (Patrikios et al., 2006; Goldschmidt et al., 2009), as well as the presence or absence of cell signals which inhibit or promote remyelination (Charles et al., 2000; Mi et al., 2005, 2007; Bachelin et al., 2010).
Future studies are required to dissect the functional relevance of histone modifications for myelin gene expression in healthy controls as well as in patients with MS and to analyze the functional significance of “reactivating” developmental pathways in the chronic MS brain.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant 2R01NS042925-08, NIH–American Recovery and Reinvestment Act Grant 3R01NS042925-07S1, and National Multiple Sclerosis Society Grant RG-4134 (P.C.); Ministerio de Ciencia y Tecnología Grants CSD2006-00049 and PR2008-0243 (G.L.-R.); and Deutsche Forschungsgemeinschaft Grant KU 1477/5-1 (T.K.). We thank Dr. Jack Antel (Neuroimmunology Unit, Montreal Neurological Institute, McGill University, Montreal, Quebec, Canada), the Brain and Spinal Fluid Resource Center, Veterans Affairs West Los Angeles Healthcare Center (Los Angeles, CA), and the Brain-Net Germany supported by Federal Ministry of Health Grant 01GI0505 for providing multiple sclerosis tissue and control samples.
Inception Research and Therapeutics has no commerical interest in the work that was conducted by F.M. at The Hospital for Sick Children in Toronto, Canada.
References
- Arnett HA, Wang Y, Matsushima GK, Suzuki K, Ting JP. Functional genomic analysis of remyelination reveals importance of inflammation in oligodendrocyte regeneration 2. J Neurosci. 2003;23:9824–9832. doi: 10.1523/JNEUROSCI.23-30-09824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelin C, Zujovic V, Buchet D, Mallet J, Baron-Van Evercooren A. Ectopic expression of polysialylated neural cell adhesion molecule in adult macaque Schwann cells promotes their migration and remyelination potential in the central nervous system. Brain. 2010;133:406–420. doi: 10.1093/brain/awp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber AJ, Kerr S, Rodriguez M. Efficient central nervous system remyelination requires T cells. Ann Neurol. 2003;53:680–684. doi: 10.1002/ana.10578. [DOI] [PubMed] [Google Scholar]
- Brück W, Porada P, Poser S, Rieckmann P, Hanefeld F, Kretzschmar HA, Lassmann H. Monocyte/macrophage differentiation in early multiple sclerosis lesions. Ann Neurol. 1995;38:788–796. doi: 10.1002/ana.410380514. [DOI] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Charles P, Hernandez MP, Stankoff B, Aigrot MS, Colin C, Rougon G, Zalc B, Lubetzki C. Negative regulation of central nervous system myelination by polysialylated-neural cell adhesion molecule. Proc Natl Acad Sci U S A. 2000;97:7585–7590. doi: 10.1073/pnas.100076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia. 2009;57:1579–1587. doi: 10.1002/glia.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischer JM, Bramow S, Dal-Bianco A, Lucchinetti CF, Rauschka H, Schmidbauer M, Laursen H, Sorensen PS, Lassmann H. The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain. 2009;132:1175–1189. doi: 10.1093/brain/awp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt T, Antel J, König FB, Brück W, Kuhlmann T. Remyelination capacity of the MS brain decreases with disease chronicity. Neurology. 2009;72:1914–1921. doi: 10.1212/WNL.0b013e3181a8260a. [DOI] [PubMed] [Google Scholar]
- Graumann U, Reynolds R, Steck AJ, Schaeren-Wiemers N. Molecular changes in NAWM in multiple sclerosis are characteristic of neuroprotective mechanisms against hypoxic insult. Brain Pathol. 2003;13:554–573. doi: 10.1111/j.1750-3639.2003.tb00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JL, Casaccia P. Defining the chromatin landscape in demyelinating disorders. Neurobiol Dis. 2010;39:47–52. doi: 10.1016/j.nbd.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Barnes PJ. COPD as a disease of accelerated lung aging. Chest. 2009;135:173–180. doi: 10.1378/chest.08-1419. [DOI] [PubMed] [Google Scholar]
- John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, Brosnan CF. Multiple sclerosis: re-expression of a developmental pathway that restricts oligodendrocyte maturation. Nat Med. 2002;8:1115–1121. doi: 10.1038/nm781. [DOI] [PubMed] [Google Scholar]
- Kotter MR, Zhao C, van Rooijen N, Franklin RJ. Macrophage-depletion induced impairment of experimental CNS remyelination is associated with a reduced oligodendrocyte progenitor cell response and altered growth factor expression. Neurobiol Dis. 2005;18:166–175. doi: 10.1016/j.nbd.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cuo Q, Wegner C, Antel J, Brück W. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- Kutzelnigg A, Lucchinetti CF, Stadelmann C, Brück W, Rauschka H, Bergmann M, Schmidbauer M, Parisi JE, Lassmann H. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128:2705–2712. doi: 10.1093/brain/awh641. [DOI] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, Casaccia-Bonnefil P. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. J Neurosci. 2007;27:7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hu Q, D'ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R, Nakaya HI, Amaral PP, Festa F, Sogayar MC, da Silva AM, Verjovski-Almeida S, Reis EM. Androgen responsive intronic non-coding RNAs. BMC Biol. 2007;5:4. doi: 10.1186/1741-7007-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Mi S, Hu B, Hahm K, Luo Y, Kam Hui ES, Yuan Q, Wong WM, Wang L, Su H, Chu TH, Guo J, Zhang W, So KF, Pepinsky B, Shao Z, Graff C, Garber E, Jung V, Wu EX, Wu W. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med. 2007;13:1228–1233. doi: 10.1038/nm1664. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Jiang J, Gusev Y, Jones A, Kearbey JD, Miller DD, Schmittgen TD, Dalton JT. MicroRNAs are mediators of androgen action in prostate and muscle. PLoS One. 2010;5:e13637. doi: 10.1371/journal.pone.0013637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrikios P, Stadelmann C, Kutzelnigg A, Rauschka H, Schmidbauer M, Laursen H, Sorensen PS, Brück W, Lucchinetti C, Lassmann H. Remyelination is extensive in a subset of multiple sclerosis patients. Brain. 2006;129:3165–3172. doi: 10.1093/brain/awl217. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, Casaccia-Bonnefil P. Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiol Aging. 2008a;29:452–463. doi: 10.1016/j.neurobiolaging.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008b;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Zhao C, Penderis J, Franklin RJ. The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J Neurosci. 2002;22:2451–2459. doi: 10.1523/JNEUROSCI.22-07-02451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Lang JK, Waldau B, Roy NS, Schwartz TE, Pilcher WH, Chandross KJ, Natesan S, Merrill JE, Goldman SA. Complementary patterns of gene expression by human oligodendrocyte progenitors and their environment predict determinants of progenitor maintenance and differentiation. Ann Neurol. 2006;59:763–779. doi: 10.1002/ana.20812. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci. 1998;18:601–609. doi: 10.1523/JNEUROSCI.18-02-00601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [Erratum (2010)37:588] [DOI] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeis T, Probst A, Steck AJ, Stadelmann C, Brück W, Schaeren-Wiemers N. Molecular changes in white matter adjacent to an active demyelinating lesion in early multiple sclerosis. Brain Pathol. 2009;19:459–466. doi: 10.1111/j.1750-3639.2008.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]