Abstract
A ((1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl) succinamide derivative (here referred to as Compound 12) shows significant activity toward many matrix metalloproteinases (MMPs), including MMP-2, MMP-8, MMP-9, and MMP-13. Modeling studies had predicted that this compound would not bind to ADAMTS-5 (a disintegrin and metalloproteinase with thrombospondin motifs-5) due to its shallow S1′ pocket. However, inhibition analysis revealed it to be a nanomolar inhibitor of both ADAMTS-4 and −5. The observed inconsistency was explained by analysis of crystallographic structures, which showed that Compound 12 in complex with the catalytic domain of ADAMTS-5 (cataTS5) exhibits an unusual conformation in the S1′ pocket of the protein. This first demonstration that cataTS5 can undergo an induced conformational change in its active site pocket by a molecule like Compound 12 should enable the design of new aggrecanase inhibitors with better potency and selectivity profiles.
Keywords: aggrecanase, ADAMTS, metalloprotease, osteoarthritis
Introduction
A disintegrin and metalloproteinase with thrombospondin motifs-5 (ADAMTS-5), also known as aggrecanase-2, is currently, along with ADAMTS-4 (aggrecanase-1), the most attractive target for the development of Disease Modifying Osteoarthritis Drugs (DMOADs).1 Preclinical models of osteoarthritis (OA) in ADAMTS-5 null mice demonstrate that deletion of this metalloproteinase (MP) not only results in inhibition of aggrecan cleavage, a key characteristic in OA, but also blocks cleavage of Type II collagen, prevents the degeneration and fibrillation of articular cartilage, lessens subchondral bone sclerosis, stops osteophyte formation, and alleviates mechanical allodynia.2–5 Thus, the preclinical data indicate that selective inhibition of ADAMTS-5 results in total joint preservation giving rise to a new paradigm in OA, which purports that most of the phenotypes associated with this degenerative disease can be blocked by targeting a single gene or pathway. If these findings translate to the clinic, this paradigm has tremendous implications for drug discovery centered on developing aggrecanase inhibitors for clinical testing as DMOADs.
Due to the chronic nature of the disease, the MP selectivity profile is a key issue in developing aggrecanase inhibitors for OA. Achieving selectivity has been challenging because there are two related families of MPs sharing the zinc-binding consensus sequence, HEXXHXXXGXX, including 23 matrix metalloproteinases (MMPs), and 20 ADAMs.6,7 However, recent elucidation of crystal structures for ADAMTS-1,8 −4,9,10 and -59,11 can now enable the development of MP-sparing aggrecanase inhibitors by allowing direct comparison of the active site of ADAMTS-4/-5 with MMPs and ADAMs and applying the structure-based drug design approach.
Recently our group reported the mechanism of selectivity of a series of cis-1(S)2(R)-amino-2-indanol based compounds by generating the crystal structures of these molecules with the catalytic domain of ADAMTS-5.12 These structures reveal that subtle or secondary interactions, such as water bridging and polarity distribution, are responsible for selectivity, whereas the reaction pocket of cataTS5 in these structures is essentially unchanged.
The S1′ pockets in MPs have been shown to be flexible and readily changeable.13,14 However, more than 50 cataTS5 structures in complex with various inhibitors, including some with long P1′ moieties, have been solved and none of them have shown a significantly different S1′ pocket (unpublished results). One of the cis-1(S)2(R)-amino-2-indanol derivatives, Compound 12, ((1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl) succinamide derivative, shown in Figure 1, is quite nonselective among MMPs. Models predicted that this compound would not bind to cataTS5 due to its long and bulky P1′ moiety, but surprisingly, when analyzed, Compound 12 was found to be relatively potent in inhibiting ADAMTS-5 activity. To understand this discrepancy, we solved the co-crystal structure of cataTS5 with Compound 12, which was obtained by displacement of Compound 1, N-hydroxy-4-(4-(4-(trifluoromethyl)phenoxy)phenylsulfonyl)-tetrahydro-2H-pyran-4-carboxamide, from the cataTS5 crystal. It was observed that the S1′ loop of the active site undergoes significant conformational changes to accommodate the P1′ moiety of Compound 12. In this report, we describe the conformational change in detail and its potential implications for future inhibitor design.
Figure 1.
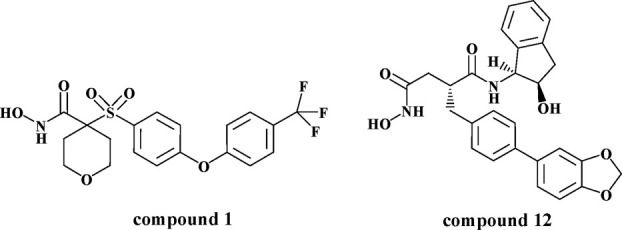
Inhibitors of aggrecanase. The molecular structures of Compounds 1 and 12 are illustrated.
Results and Discussion
Inhibition profile of Compound 12
Compound 12 was tested for potency against ADAMTS-4 and −5 and for activity against a panel of MPs (Table I). Compound 12 was potent in blocking aggrecanase-1 and -2 with an IC50 of 10 and 129 nM, respectively, and was as active against every MMP tested except for MMP-1 and -7. The compound was next tested for efficacy in blocking aggrecan catabolism in cytokine-stimulated bovine cartilage, which is mediated by the aggrecanases.15 As expected, Compound 12 effectively blocked cartilage aggrecan breakdown with an IC50 value of 500 nM, through inhibition of the aggrecanases as previously shown.15 In vitro kinetic analysis using a peptide substrate revealed that Compound 12 is a competitive, reversible inhibitor of both ADAMTS-4 and -5 (data not shown).
Table I.
Profile of Compound 12 Against a Panel of Metalloproteases
| Metalloproteinase | Compound 12 |
|---|---|
| ADAMTS-4 (IC50) | 10 ± 3 |
| ADAMTS-5 (IC50) | 129 ± 11 |
| MMP-1 | 7560 |
| MMP-2 | 3.4 |
| MMP-3 | 35 |
| MMP-7 | 622 |
| MMP-8 | 2.2 |
| MMP-9 | 6.3 |
| MMP-13 | 7.3 |
| MT1-MMP | 37 |
| BCA (EC50) | 500 ± 115 |
Calculated Ki values (nM) ± Standard Error for three determinations. When the standard error is not reported, the values represent single determinations.
Ki values are reported for each MP with the exception of ADAMTS-4 and −5 where IC50 values are reported and BCA where EC50 values are reported.
Structure and the catalytic site
High resolution cataTS5 structures with Compound 12 (1.6 Å) and Compound 1 (1.4 Å) were obtained and are presented in Figure 2(a) along with a stereo “omit” difference electron density map for Compound 12 in the cataTS5 active site, Figure (2b). Both crystals have similar overall structures, as shown in Figure (2a). However, careful examination of the structures reveals that they have different S1′ pockets at the catalytic site. The structure of cataTS5 with Compound 12 represents a unique and new category, very different from other known cataTS5 structures generated to date. The S1′ loops in all other cataTS5 structures are similar to each other and to that of the cataTS5/Compound 1 structure, assumed to be its native conformation.11 Hence, cataTS5 coupled to Compound 12 is a new discovery and a novel display of the S1′ conformation.
Figure 2.
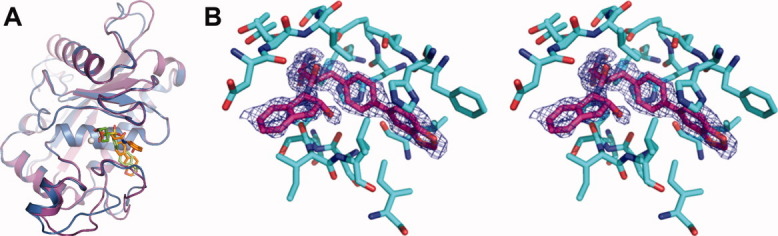
Structures of cataTS5. (a) The crystal structure of cataTS5 with Compound 1 is shown in purple and with Compound 12 displayed in pale blue. (b) An “omit” difference electron density map for Compound 12 in the cataTS5 active site is given in stereo. It is a 2|Fo|-|Fc| “omit” map (with calculated phase angles) around the ligand binding site. The map was calculated using 1.6 Å resolution data and contoured at 1.0 σ. Protein atoms within 4.0 Å of the ligand are shown.
The S1′ pocket of cataTS5 is defined by the area delineated by the immediate loop entering helix C (residues L402 and H403), first half of helix C (residues A404 to H410), and the S1′ loop (residues R437 to I446), as shown in Figure 3. Helix C (404-411) is quite conserved in all MPs, though the loop, that is, residue 402 and 403 entering to the helix can be quite different. The S1′ loops are believed to be flexible. However, this flexibility had not been shown in any ADAMTS member until the structure of cataTS5 with Compound 12 was determined. Many compounds with long P1′ moieties, 4-(3-(2-(p-tolyloxy)ethyl)phenyl)pyridine, 5-(6-(piperidin-4-yl)pyridin-3-yl)pent-4-ynyl acetate), N-(3-(p-tolyloxy)propyl)quinoline-2-carboxamide), 4-(2-(2-fluoro-4-methylphenoxy)ethoxy)benzamide), 4-(3-(3-(p-tolyloxy)propyl)phenyl)pyridine), shown in Figure 4, have been used to probe the S1′ pockets of MPs. These P1′ moieties, when co-crystallized with cataTS5 did not produce any detectable S1′ pocket changes in the protein (unpublished results).
Figure 3.
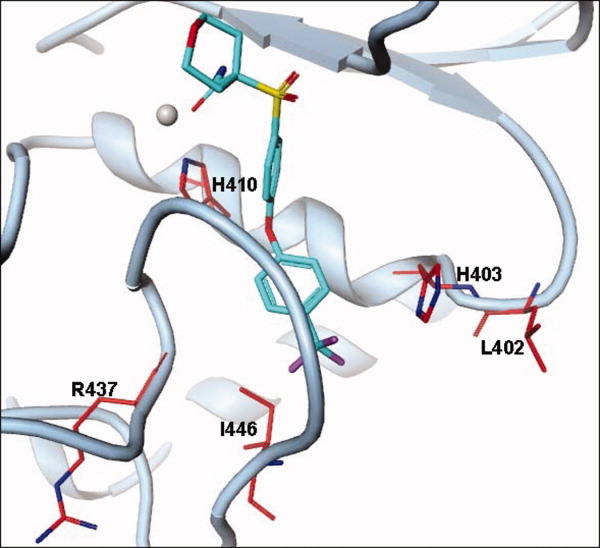
Residues defining the S1′ pocket in cataTS5. The S1′ pocket of cataTS5 is defined by a loop entering helix C (L402 and H403), first half of helix C (A404 to H410), and the S1′ loop (R437 to I446).
Figure 4.
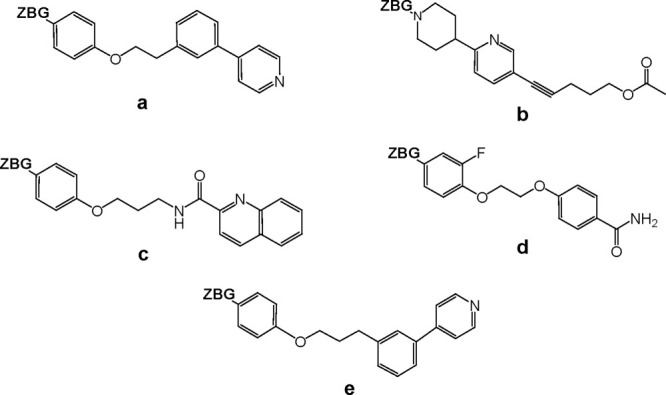
Molecular Structures of various long P1′ moieties. Structures of several P1′ moieties are shown including: (a) 4-(3-(2-(p-tolyloxy) ethyl)phenyl)pyridine, (b) 5-(6-(piperidin-4-yl)pyridin-3-yl)pent-4-ynyl acetate), (c) N-(3-(p-tolyloxy)propyl)quinoline-2-carboxamide), (d) 4-(2-(2-fluoro-4-methylphenoxy)ethoxy)benzamide), and (e) 4-(3-(3-(p tolyloxy)propyl)phenyl)pyridine). The label “ZBG” stands for “Zinc Binding Group.”
In Figure 5(a), the S1′ pocket in cataTS5 with Compound 1 is shown. This represents the normal state of the pocket, which is quite hydrophobic as illustrated by color coding (red for acidity, blue for basicity, and white for neutrality). There are three residues, F406, L438, and I446, at the bottom of the pocket. These three residues not only define the pocket but are also responsible for its shape. One significant role of these three residues is to force the pocket to turn sideways. Therefore, a straight and bulky group like the benzodioxole-tolyl group (the P1′ moiety of Compound 12) is predicted not to fit into the pocket of cataTS5, as shown in Figure 5(b). This prediction was based on modeling using the coordinates of Compound 12 obtained from a co-crystal structure with MMP-13. However, counter to the model's prediction, enzyme inhibition data clearly show that Compound 12 inhibits both MMP-13 and ADAMTS-5 activity (Table I). Thus, the model incorrectly predicted that Compound 12 would not bind to cataTS5. The reason for this discrepancy is made clear by the crystal structure of cataTS5 with Compound 12, which shows that the compound induces significant changes in the S1′ loop of cataTS5, creating a proper pocket to accommodate binding of the molecule. This observation is a novel finding that has not been observed before in any ADAMTS protein. However, the dynamic nature of the active site of ADAMTS enzymes has been suggested by Mosyak et al.,9 who point out that conformational mobility of the active site is a feature of mature aggrecanases that may be required for high affinity association with its substrates.
Figure 5.
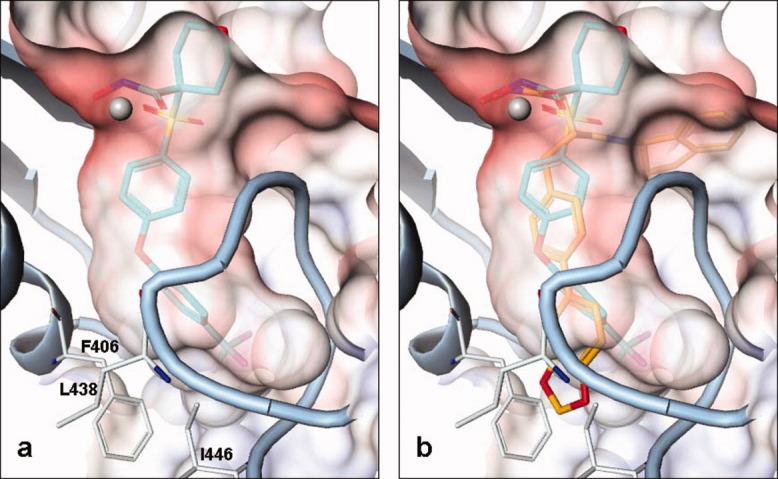
Delineation of the S1′ pocket of cataTS5. (a) The crystal structure of the S1′ pocket bound to Compound 1 and (b) the S1′ pocket with Compound 12 docked is based on orientation and conformation from a co-crystal structure of MMP-13/Compound 12. The benzodioxole-tolyl group of Compound 12 is clearly seen stretching outside the pocket.
Changes in the S1′ loop and pocket
Compound 12 induces conformational changes in the S1′ loop in the active site of cataTS5, and the changes are not due to the packing environment. Even though the space group did show a change from P21 for Compound 1 to P212121 for Compound 12, there is no intermolecular contact observed around this region in either structure. Furthermore, many other structures, including those coupled to molecules containing long P1′ moieties do not show any observable changes. The alterations seen with Compound 12 are specific to this inhibitor. We believe the reasons for the compound's ability to change the cataTS5 S1′ loop are due to its strong anchoring groups located on the indanolsuccinamide moiety that interacts with cataTS5, as described by Tortorella et al.12 as well as to the rigidity and hydrophobicity of the P1′ phenylbenzodioxole moiety.
The changes can be described as a two-step movement, as shown in Figure 6. In Step 1, L438, one of the three (L438, F406, and I446) crucial S1′ lower-pocket-defining residues was unchanged while the ring of F406 rotated ∼45° and I446 was totally moved. This may have been the initialization point responsible for propagating the overall conformational shift in the pocket. A torsion comparison (Table II) clearly indicates that the change starts from the χ angle of L443 and ends with the ϕ angle of S445. The conformation for I446 is unchanged. This step results in the segment containing I442-K450 to undergo significant movements. The new segment will have a head-on collision with K436, therefore the next action in the conformational change, referred to as Step 2, is to move away the loop comprising F430-R437 (as shown in Figure 6). The torsion comparison (Table II) shows a series of peptide conformational angle changes from χ angle of S432 to χ angle of R437 with several angle changes near 180°.
Figure 6.
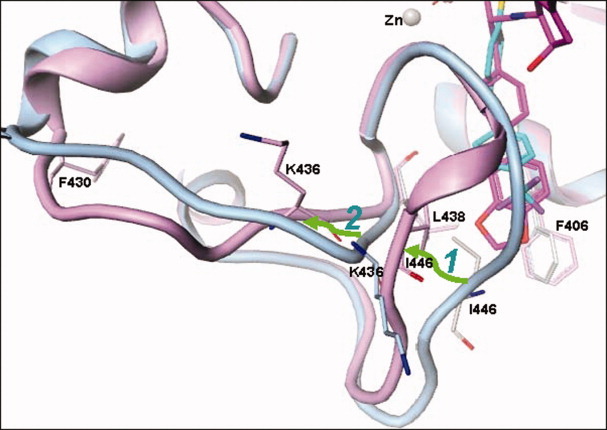
Conformational transition of cataTS5. cataTS5 in complex with Compound 1 is used as a reference structure and is highlighted in blue and cataTS5 coupled with Compound 12 is shown in purple. The transition is indicated by two green arrows with the labels “1” and “2” for Step 1 and 2, respectively. Compound 12 induces the right portion of the loop to shift towards left (Step 1). The shifted loop fragment pushes the opposite stretch of the loop to also change position (Step 2).
Table II.
Torsion Angles
| F430 | G431 | S432 | T433 | E434 | D435 | K436 | R437 | L438 | M439 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Torsion angle | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X |
| 1a | −114 | −19 | 89 | 0 | −161 | 168 | −128 | 153 | −108 | 132 | −93 | 165 | 67 | 31 | −118 | 167 | −67 | −29 | −90 | 15 |
| 12 | −109 | −12 | 57 | 17 | −95 | 12 | 61 | −48 | −131 | 57 | −122 | 61 | −102 | 101 | −119 | −144 | −59 | −31 | −85 | 9 |
| S440 | S441 | I442 | L443 | T444 | S445 | I446 | D447 | A448 | S449 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Torsion angle | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X | ϕ | X |
| 1a | −63 | 143 | −87 | −8 | −112 | 153 | −97 | 102 | −123 | −92 | −158 | 175 | −118 | 131 | −75 | 108 | −64 | −25 | −80 | −14 |
| 12 | −63 | 146 | −75 | −23 | −134 | 131 | −69 | −20 | −68 | −27 | −83 | −7 | −75 | 133 | −101 | 120 | −82 | 7 | −70 | −19 |
Comparison between the two cataTS5 structures in complex with Compound 1 and 12, separately. The angles are highlighted in bold when the differences are greater than 40°.
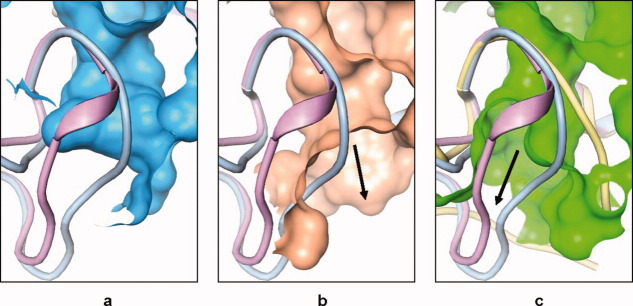
In the normal cataTS5 structures, the S1′ pocket is limited and has a side-pocket [Figures 5 and 7(a)]. The new pocket, induced by Compound 12, opens up and connects to the solvent region [Figure 7(b)]. This new S1′ pocket bears similarities to the MMP-13 pocket as evidenced by the comparison of Figures 7(b,c). However, the direction of the tunnel toward solvent shows angle differences between the two structures. Hence, the trajectory of Compound 12 in these two structures is reflected in this change (Figure 8). In the structural alignment, the starting divergence points of the S1′ loop in cataTS5 and MMP-13 are I442 and I222, respectively, and the ending divergence points are roughly P451 and P234. In this sense, MMP-13 has a longer S1′ loop than cataTS5 and in fact, Compound 12 penetrates the S1′ loop in MMP-13 while it bypasses the loop in cataTS5. These variations and changes might contain key elements by which inhibitors can be designed to distinguish between these two proteins and other MPs. Further comparison of the two cataTS5 structures reveals some other changes that occurred in other parts of the structure away from the catalytic site. They are in the region containing amino acids L321 to S327 and N344 to E357. These alterations are related to the surface flexibility and crystalline packing and bear no importance to the catalytic pocket important for ligand and substrate binding.
Figure 7.
Pocket comparison. Crystal structures of the active site pockets of (a) cataTS5 with Compound 1 featuring the loop colored pale blue and the pocket colored blue, which looks quite enclosed; (b) showing cataTS5with Compound 12 with the loop colored pink, and the pocket colored brown which opens up on the left; and (c) showing MMP-13 with Compound 12 with the loop colored yellow, and the pocket in green which opens up on the right.
Figure 8.
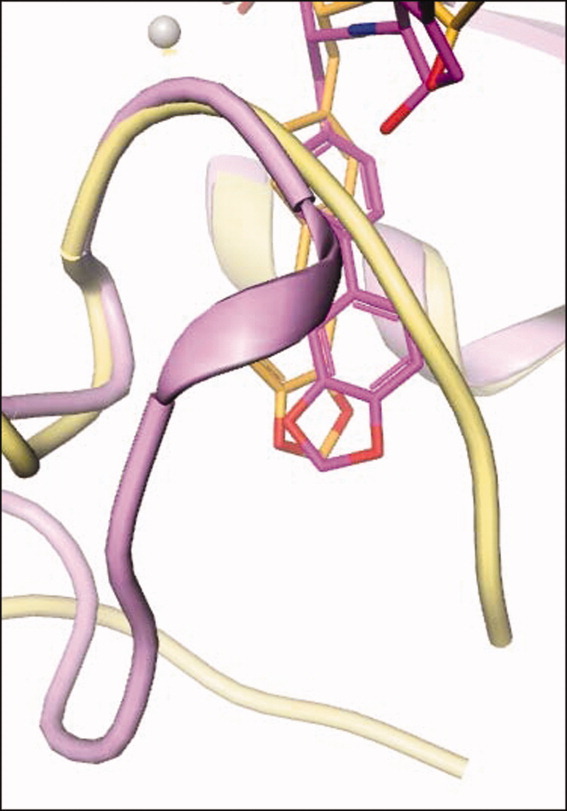
Trajectory change. The figure highlights the trajectory alteration for bound Compound 12 seen in both cataTS5 and MMP-13 structures.
The question arises whether the significant conformational changes at the bottom of the S1′-pocket observed in the present work with the cataTS5 would in fact occur in the cataTS5-disintegrin-like domain construct as the bottom of the S1′-pocket packs closely against the disintegrin-like domain.9 To answer this question, we have overlaid the structure of the cataTS5 complexed to Compound 12 obtained in the present studies to that of the cataTS5-disintegrin-like domain complexed to batimastat of Mosyak et al.,9 Figure 9. From the overlaid structures, we observed that the shortest distance between the S1′ loop of ADAMTS-5 and the disintegrin-like domain is 6.4 Å, which impedes clash between these two regions. We conclude that the newly discovered S1′ loop conformation may occur also in the extended structure. The structure of the cataTS5-Compound 12 complex was solved with crystals obtained by soaking methods to exchange Compound 1 with 12 rather than by a direct co-crystallization of cataTS5 complexed to Compound 12. This approach was dictated by our inability to produce reasonably good crystals of either apo-cataTS5 or cataTS5 complexed to inhibitor, in spite of the fact that numerous inhibitors were tested and extensive crystallization conditions explored.11 As pointed out in our earlier work,11 observations made by us and others suggested that a number of proteins are more prone to co-crystallize with inhibitors that produce a significant increase in protein unfolding temperature (Tm). CataTS5 complexed to a variety of inhibitors gave the most significant change with Compound 1, specifically, an increase of 27°C (Tm = 43° →Tm = 70°C) (11). This inhibitor enabled crystals of the complex to be grown and allowed a successful crystal soaking-displacement approach with many inhibitors, including Compound 12. There might be some concerns with this method as the Ki values of Compound 1 and 12 are similar for cataTS5: 290 nM and 129 nM, respectively (Table I). These concerns are removed by the high resolution structure of cataTS5 complexes with Compound 12 that unambiguously shows that Compound 1 has been completely replaced by 12. Moreover, the substantial conformational change that occurred as a consequence of swapping molecules did not damage the crystals as evidenced by the fact that, under the microscope, they were intact and yielded high resolution X-ray diffractions.
Figure 9.
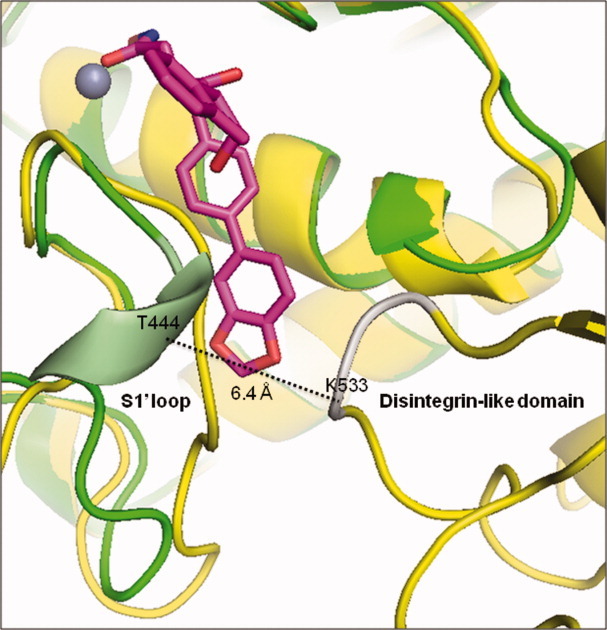
Overlay of the 3-D structure of cataTS5 (green) complexed to Compound 12 (magenta) to that of cataTS5 plus its disintegrin-like domain (yellow) complexed to batimastat (not shown). The shortest distance, 6.4 Å, between the cataTS5 S1′ loop (pale green) formed by Compound 12 binding and the disintegrin-like domain segment (silver) is indicated by the dotted line connecting the α-carbon atoms of T444 and K533.
Implication for drug design
The primary structures of ADAMTS-5 and MMPs are overall different with the exception of the zinc-binding region. Their S1′ loops are flexible and highly specific and can assume several different conformational states. This new finding of various conformational states in cataTS5, an ADAMTS family member, in our opinion is crucial for inhibitor design. We believe that the conformational flexibility must be addressed during the designing process to achieve the desired and needed selectivity profile required for the development of aggrecanase inhibitors.
The potency and specificity of cis-1(S)2R-amino-2-indanol-based compounds have been discussed in recent studies.12 This new discovery reveals that the P1′ moieties of these inhibitors bear important factors that recognize or exclude binding to MPs. The uniqueness of this newly discovered S1′ pocket in cataTS5 generated by Compound 12 and its likely formation in the two-domain structure, cataTS5-disintegrin-like domain, upon binding Compound 12, is important for drug design. Specifically, the easily obtainable single domain cataTS5 protein should enable the design of new aggrecanase inhibitors with better potency and selectivity profiles against the full-length protein present in vivo by exploiting the unusual conformation in the S1′ pocket.
Future studies will focus on designing inhibitors of ADAMTS-5 that will benefit from the flexibility of the S1′ pocket.
Conclusions
This is the first report of a small molecule inhibitor capable of shifting the conformational structure of the active site of ADAMTS-5. Understanding the altered shape and size of this newly discovered S1′ pocket may pave the way for designing new and better DMOADs aimed at treating OA by selectively targeting aggrecanase activity.
Experimental Procedures
Aggrecanase inhibition assay
Full-length recombinant human ADAMTS-4 and −5 were expressed in SF9 cells and isolated as previously described.16,17 Inhibition of ADAMTS-4 or ADAMTS-5 mediated cleavage of bovine aggrecan at the Glu373/Ala374 site was determined as reported in previous work.12
Metalloproteinase inhibition assays
The expression and purification of full-length MMP-1, −2, −3, −9, −13, and the catalytic domain of MMP-7, -8, and MT1-MMP (MMP-14) was carried out as described earlier.18,19 Inhibition assays using specific fluorogenic peptides as substrates, determinations of Ki(app) and calculation of Ki values taking into account substrate concentration and Km, were performed as previously reported.12
Bovine cartilage assay (BCA)
Cartilage was dissected from the metatarsophalangeal joints of young cows obtained from the slaughterhouse.12 Subsequently, cartilage was cut into explants, weighing ∼15 mg each, and incubated in 96-well plates for 48 h with either serum-free Dulbecco's Modified Eagle Medium as a control, medium containing interleukin (IL)-1β (produced at Pfizer as described20) (100 ng/mL) or medium containing IL-1β (100 ng/mL) plus Compound 12 at concentrations ranging from 30 to 10,000 nM. Glycosaminoglycan content of the supernatants was determined using the dimethylmethylene blue assay, as described by Farndale et al.21
Expression and purification of cataTS5
The catalytic domain of ADAMTS-5 comprising amino acids S263 to I480 preceded by a Met-Ala sequence and incorporating the single point mutation L282K was expressed in Escherichia coli inclusion bodies, refolded, and purified to homogeneity by size exclusion chromatography as described earlier.11 The criteria used in our previous work11 were applied here to assess enzyme purity, activity, and authenticity. Specifically, SDS-PAGE established that the protein was >98% pure and mass spectrometry revealed the presence of the two species including AS262ISRARQ…I480 and I263SRARQ…I480 of 24,261 and 24,103 daltons, respectively. The peptide substrate, K-[6FAM]-DVQE↓FRGVTAVIRC-[Qsy9]-KGK, (↓ indicates the cleavage site) verified that the enzyme was active. Moreover, enzymatic titration using tissue inhibitor of matrix metalloproteinase-3 determined that ∼80% of the cataTS5 was catalytically competent.11
Preparation of inhibitors
The synthesis of Compound 1 is described in patents WO 2000050396, WO 2000038719, WO 2000038718, WO 2000038717, WO 2000037107, and WO 9925687; while the synthesis of hydroxyindanylbutanediamides and related compounds has been described in WO 9909000 19990225 CAN 130:209514.
Crystallization, data collection and structure determination
A 10 mg/mL solution of the cataTS5 in 25 mM Hepes, containing 5 mM CaCl2 and 6 mM Compound 1 (as surrogate) was used in the crystallization experiments. Crystals were obtained by vapor diffusion with a well solution consisting of 30% PEG 3350 and 0.1M Tris pH 8.5 at room temperature. The crystal growth was completed within one week. Then, the crystals were transferred to a solution containing 2 mM Compound 12 in 33% PEG 3350 and 0.1M Tris pH 8.5 for overnight soaking. Compound 12 displaced Compound 1 from the cataTS5 crystal. Prior to data collection, the crystal was transferred to a solution consisting of three to four parts of the stabilizing solution and one part of ethylene glycol before flash-cooling in liquid nitrogen. X-ray diffraction data were collected at the LS-CAT beam line 21-IDG of Advanced Photon Source, Argonne National Laboratory, at a wavelength of 1.0 Å, with a MAR300 detector. The diffraction data were integrated and scaled by HKL2000. The structures were determined, fitted and refined by AFITT, REFMAC, and COOT. A summary of the diffraction data and refinement statistics is presented in Table III.
Table III.
Data Collection and Refinement Statistics
| Data collection | cataTS5/Compound 12 | MMP13/Compound 12 |
|---|---|---|
| Space group | P212121 | P212121 |
| Cell dimensions | 51.40, 44.52, 76.25 (Å) | 73.60, 94.93, 120.40 (Å) |
| Z | 4 | 16 |
| Resolution (Å) | 50.0–1.60 (1.66–1.60)a | 20.0–2.00 (2.07–2.00)a |
| Total unique reflections | 20,954 (1542) | 53,516 (4064) |
| Rmerge | 0.073 (0.356) | 0.090 (0.251) |
| I/σI | 17.6 (2.2) | 13.4 (5.9) |
| Completeness (%) | 88.0 (65.8) | 93.1 (71/4) |
| Redundancy | 5.2 (2.8) | 4.2 (1.9) |
| Refinement | ||
| Resolution (Å) | 38.12–1.60 | 20.00–2.00 |
| No. reflections | 19,840 | 49,487 |
| Rwork/Rfree | 0.159/0.238 | 0.240/0.267 |
| No. nonhydrogen atoms (molecule-A/B/C/D) | ||
| Protein | 1709 | 1286/1290/1270/1300 |
| Ligand | 35 | 35/35/35/35 |
| Ion | 4 | 4/4/4/4 |
| EGL | 4 | |
| HEPES | 30 | |
| Sulfate | 45 | |
| Water | 293 | 351 |
| B-factors (average) (molecule-A/B/C/D) | ||
| Protein (backbone atoms only) | 11.38 | 19.40/16.75/21.06/17.71 |
| Ligand | 19.02 | 25.75/15.87/22.03/19.84 |
| Ion | 15.94 | 35.17/27.70/38.17/31.76 |
| EGL | 10.79 | |
| HEPES | 46.34 | |
| Sulfate | 47.92 | |
| Water | 28.45 | 28.68 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.011 | 0.010 |
| Bond angles (°) | 1.4 | 1.4 |
Values in parentheses are for highest resolution shell. The data were obtained using one crystal.
Accession code
The structure of Compound 12 complexed with cataTS5 has been deposited to RCSB with pdb code 3ljt.
Acknowledgments
We thank Dr. Ravi Kurumbail (Pfizer Inc., Groton, CT) and Tom Emmons (Pfizer, Inc., Chesterfield, MO) for discussions and help with graphic work.
Glossary
Abbrevations
- ADAM
a disintegrin and metalloproteinase
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- BCA
Bovine Cartilage Assay
- cataTS5
catalytic domain of ADAMTS-5
- DMOADs
Disease Modifying Osteoarthritis Drugs
- MMP
matrix metalloproteinases
- OA
osteoarthritis
References
- 1.Fosang AJ, Little CB. Drug insight: aggrecanases as therapeutic targets for osteoarthritis. Nat Clin Pract Rheumatol. 2008;4:420–427. doi: 10.1038/ncprheum0841. [DOI] [PubMed] [Google Scholar]
- 2.Glasson S, Askew R, Sheppard B, Carito B, Blanchet T, Ma H, Flannery CR, Peluso D, Kanki K, Yang Z, Majumdar MK, Morris EA. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 3.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, Fourie AM, Fosang AJ. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 4.Botter SM, Glasson SS, Hopkins B, Clockaerts S, van Leeuwen JP, Weinans H, van Leeuwen JP, van Osch GJ. ADAMTS5-/- mice have less subchondral bone changes after induction of osteoarthritis through surgical instability: implications for a link between cartilage and subchondral bone changes. Osteoarthr Cartil. 2009;17:636–645. doi: 10.1016/j.joca.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Malfait AM, Ritchie J, Gil AS, Austin JS, Hartke J, Qin W, Tortorella MD, Mogil JS. ADAMTS-5 deficient mice do not develop mechanical allodynia associated with osteoarthritis following medial meniscal destabilization. Osteoarthr Cartil. 2010;18:572–80. doi: 10.1016/j.joca.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Hooper NM, Uwe L. Vol. 4. Birkhaűser: Verlag; 2005. The ADAM Family of Proteases. [Google Scholar]
- 7.Lagente V, Boichot E. Matrix metalloproteinases in tissue remodelling and inflammation, XI. Birkhaűser: Verlag; 2008. [Google Scholar]
- 8.Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, Hargreaves D, Ting A, Pauptit RA, Parker AE, Abbott WM. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, Kriz R, Reifenberg EL, Collins-Racie LA, Corcoran C, Freeman B, Zollner R, Marvell T, Vera M, Sum PE, Lavallie ER, Stahl M, Somers W. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopper DW, Vera MD, How D, Sabatini J, Xiang JS, Ipek M, Thomason J, Hu Y, Feyfant E, Wang Q, Georgiadis KE, Reifenberg E, Sheldon RT, Keohan CC, Majumdar MK, Morris EA, Skotnicki J, Sum PE. Synthesis and biological evaluation of ((4-keto)-phenoxy)methyl biphenyl-4-sulfonamides: a class of potent aggrecanase-1 inhibitors. Bioorg Med Chem Lett. 2009;19:2487–2491. doi: 10.1016/j.bmcl.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 11.Shieh H, Mathis K, Williams J, Hills R, Wiese J, Benson T, Kiefer JR, Marino MH, Carroll JN, Leone JW, Malfait AM, Arner EC, Tortorella MD, Tomasselli A. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- 12.Tortorella MD, Tomasselli AG, Mathis KJ, Schnute ME, Woodard SS, Munie G, Williams JM, Caspers N, Wittwer AJ, Malfait AM, Shieh HS. Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors. J Biol Chem. 2009;284:24185–24191. doi: 10.1074/jbc.M109.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovejoy B, Welch AR, Carr S, Luong C, Broka C, Hendricks RT, Campbell JA, Walker KA, Martin R, Van Wart H, Browner MF. Crystal structures of MMP-1 and −13 reveal the structural basis for selectivity of collagenase inhibitors. Nat Struct Biol. 1999;6:217–221. doi: 10.1038/6657. [DOI] [PubMed] [Google Scholar]
- 14.Pochetti G, Montanari R, Gege C, Chevrier C, Taveras A, Mazza F. Extra binding region induced by non-zinc chelating inhibitors into the S1′ subsite of matrix metalloproteinase 8 (MMP-8) J Med Chem. 2009;52:1040–1049. doi: 10.1021/jm801166j. [DOI] [PubMed] [Google Scholar]
- 15.Tortorella MD, Malfait AM, Decicco C, Arner E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthr Cartil. 2001;9:539–552. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- 16.Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Duke JL, George HJ, Hillman MC, Jr, Murphy K, Wiswall BH, Copeland RA, Decicco CP, Bruckner R, Nagase H, Itoh Y, Newton RC, Magolda RL, Trzaskos JM, Hollis GF, Arner EC, Burn TC. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 17.Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, Rosenfeld SA, Copeland RA, Decicco CP, Wynn R, Rockwell A, Yang F, Duke JL, Solomon K, George H, Bruckner R, Nagase H, Itoh Y, Ellis DM, Ross H, Wiswall BH, Murphy K, Hillman MC, Jr, Hollis GF, Newton RC, Magolda RL, Trzaskos JM, Arner EC. Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 18.Tortorella MD, Arner EC, Hills R, Gormley J, Fok K, Pegg L, Munie G, Malfait AM. ADAMTS-4 (aggrecanase-1): N-terminal activation mechanisms. Arch Biochem Biophys. 2005;444:34–44. doi: 10.1016/j.abb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld SA, Ross OH, Corman JI, Pratta MA, Blessington DL, Feeser WS, Freimark BD. Production of human matrix metalloproteinase 3 (stromelysin) in Escherichia coli. Gene. 1994;139:281–286. doi: 10.1016/0378-1119(94)90770-6. [DOI] [PubMed] [Google Scholar]
- 20.Yem AW, Curry KA, Tomich CS, Deibel MR., Jr A two step purification of recombinant human interleukin-1 beta expressed in E. coli. Immunol Invest. 1988;17:551–559. doi: 10.3109/08820138809030588. [DOI] [PubMed] [Google Scholar]
- 21.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]