Abstract
Protein-design methodology can now generate models of protein structures and interfaces with computed energies in the range of those of naturally occurring structures. Comparison of the properties of native structures and complexes to isoenergetic design models can provide insight into the properties of the former that reflect selection pressure for factors beyond the energy of the native state. We report here that sidechains in native structures and interfaces are significantly more constrained than designed interfaces and structures with equal computed binding energy or stability, which may reflect selection against potentially deleterious non-native interactions.
Keywords: protein design, negative design, small-molecule binding, monomer design, Rossman fold design, aromatic residues, Boltzmann distribution, entropy of binding
Introduction
Protein-design methodology has been used to create new protein structures,1,2 redesign protein-protein interactions,3,4 and produce new enzymes.5,6 Although there is still more to improve in protein-design algorithms,7 the field has considerable promise for generating new molecules for use in a wide variety of areas of current interest. By providing a reference point for which all of the inputs are known, protein design can also help to cast into sharper focus the critical evolved properties of native biomolecules. For example, studies of the kinetics of folding of designed proteins have helped distinguish the features of folding that are simple consequences of having a unique folded state from those optimized by natural selection.8
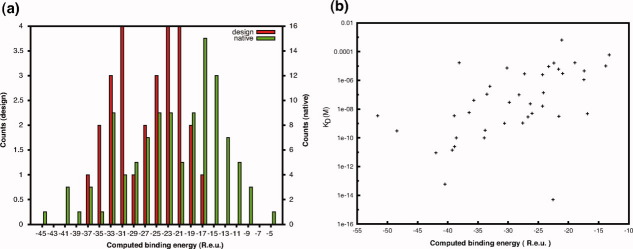
To gain insight into the properties of native structures and interfaces that result from more than simple selection for very low-energy native states, we used computational-design methodology to generate designed structures, protein-protein interfaces, and protein-ligand interfaces. As shown in Figure 1(a), designed protein-protein complexes have computed binding energies in the same range as those of native ones. Though the errors in computed energies are likely to be at least several kcal/mol, the computations do capture overall trends reasonably well; for example, the correlation between experimentally measured binding affinities and computed binding energies for protein-protein complexes [Fig. 1(b)]. Thus, although the errors in the computed energy of any one design may well be sufficiently large for the designed protein to not fold or the designed interface to not form, the distribution of interactions overall in the native and designed complexes are likely to be similar.
Figure 1.
(a) Computed binding energies for designed and native protein-protein interfaces. (b) Experimental versus computed binding energies in natural protein-protein complexes (Pearson correlation coefficient r = 0.53). Binding data from Ref.9.
With the initial aim of estimating the sidechain-entropy loss upon protein-complex formation, we developed a simple measure of the Boltzmann weight of sidechain conformations in native and designed interfaces. For protein interfaces, the method first separates the complex, and for each residue that makes an appreciable contribution to binding (see Methods section), iterates over all of its rotameric states as defined in the Dunbrack library of backbone-dependent rotamers,10 excluding rotamers that are predicted to clash with protein mainchain or Cβ atoms. For each rotamer placement, all residues within a 6 Å shell are repacked and minimized. The energy E of each such state is then evaluated using the Rosetta all-atom energy function,11 which is dominated by van der Waals, hydrogen bonding, and solvation terms. The probability of rotamer i, pi is then computed assuming a Boltzmann distribution:
| (1) |
where s is the rotameric state, kB is the Boltzmann constant, and T is the absolute temperature. E is the energy of the states.
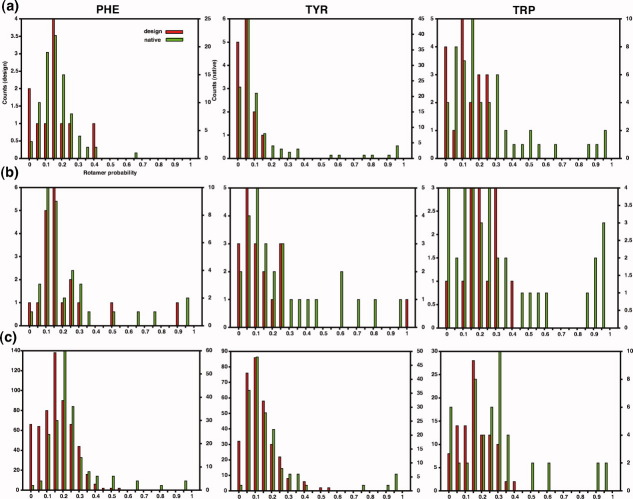
We estimated the probabilities in the unbound state of aromatic sidechain conformations observed in bound complexes and designed binders according to Eq. (1) [Fig. 2(a,b)]. Both designs and native complexes have low-probability sidechains at the interface that contribute significantly to the binding energy. However, very high-probability conformations (p > 50%) are almost exclusively found in natives, whereas designs are over-represented in the very low-probability bin (p < 5%). The trend extends to monomeric structures: we observe higher probabilities for aromatic residues in native conformations than in designs [Fig. 2(c)]. Thus, although the overall computed energies of the designed structures and complexes are similar to those of native structures, aromatic sidechains in the former are considerably less restricted in conformation than those in the latter. We found no correlation between the computed stability of individual residues in the cores of designed and native monomers and their conformational probability (not shown). The lack of correlation demonstrates that the conformational restrictions we observe in native proteins compared to designs are not a simple consequence of natural proteins having more stable cores than designs.
Figure 2.
The probabilities in the unbound state of residue conformations observed in the bound state in (a) protein-protein interactions; and (b) protein-ligand interaction. (c) The probabilities of sidechain conformations within the cores monomers. Left, center, and right panels correspond to Phe, Tyr, and Trp residues, respectively. The probabilities in the bound state (not shown) of complexes are highly correlated with those in the unbound state, implying that the monomers contribute most to conformational restriction, even in the bound state.
What is the origin of these differences in sidechain-conformational restriction? The answer may lie in the fact that most current design methods, including those used to generate the structures and complexes in this study, focus on optimizing the energy of the desired target structure or complex. Such “positive design” strategies suffer from the flaw that even though the energy of the target structure is optimized to be extremely low, the target conformation or complex might be high in energy relative to alternative conformations, and hence could be poorly populated.12 Negative-design methods have been developed that contain explicit bias for sequence substitutions that favor a target structure versus a set of alternative structures,13,14 but the latter have to be explicitly modeled and thus for computational tractability the set must not be too large.
A likely explanation for the differences in sidechain flexibility between the designed and native structures and complexes is that evolutionary pressures on native structures involve aspects of negative design. Functioning cell circuitry, for example, requires not only that binding proteins bind their targets with high affinity, but just as importantly, that they not bind other cellular proteins, which will almost always be in vast excess over the former.15 Likewise, enzymes and ligand-binding proteins must bind the small molecules relevant to their function but not other small molecules present in the cell. From a free-energy standpoint, the preordering of sidechains in unbound complexes is beneficial as it reduces the loss of entropic degrees of freedom upon binding. The interactions in monomeric proteins must stabilize the folded state, but not the much larger number of non-native states.
How can evolutionary pressure accomplish this high selectivity for native structures and interactions? To gain insight into this issue, we investigated the structural contexts of the low-probability sidechains in designed structures and complexes and the high-probability sidechains in native conformations (Supporting Information Figures). The computed energies of these low- and high-probability sidechains are all very favorable. We found that designed low-probability aromatic sidechains tend to be exposed in the unbound state and make few if any interactions. In contrast, native high-probability sidechains interact with several residues within the host monomer and are often conformationally restricted by contacts with backbone or Cβ atoms. Native proteins appear to have dense interaction networks between nearby sidechain and backbone atoms that favor native conformations over non-native conformations and are not found in isoenergetic designed conformations. Evolution appears to have used negative design to ensure that bulky aromatic residues are constrained to very specific conformations within the cores of monomers and at interfaces.
Our results highlight an important aspect of evolved native structures—the conformational restriction of large sidechains to disfavor non-native interactions—and have immediate relevance to protein design. The reference set of designed structures is necessary in this case to show that conformational restriction is not a simple consequence of selection for a very low-energy structure, which would otherwise have been a quite reasonable assumption. Further comparison of the properties of native and designed complexes will undoubtedly continue to reveal the complex and intricate features engrained in native structures and complexes by evolution. On the design side, the sidechain conformational-probability metric provides a computationally tractable alternative to explicit negative design, which as noted above, becomes prohibitive when the number of alternative structures is large. The sidechains in designs can be assessed using the metric, and overly flexible aromatic sidechains can be restricted by incorporating rigidifying interactions. The metric can also be used to guide the development of design strategies which produce more conformationally restricted sidechains by construction.
Methods
Computational design of proteins that bind native proteins
Twenty-six proteins were designed to bind to surfaces on native proteins that have been cocrystallized with other proteins. The design method followed the general procedure described by Jha et al.16 Briefly, the coordinates of natural scaffold proteins were obtained from the Protein Data Bank and docked against the target surface using the feature-matching algorithm PatchDock.17 Subsequent iterations of RosettaDock18 and RosettaDesign2 were invoked along with backbone and sidechain minimization to enhance the stability of the complex. Candidate complexes were then selected based on computed binding energy19 and surface shape complementarity20 and were manually refined to increase predicted binding energy. Evaluation of sidechain-conformational probabilities was restricted to residues on the designed proteins and did not include the target proteins.
Computational design of small-molecule binding proteins
Thirty proteins were computationally redesigned to bind to the small-molecule ligands biotin and dopamine using the Rosetta enzyme-design methodology.6 Briefly, a set of ∼250 ligand-binding scaffolds was searched for appropriate placements of amino acid binding groups for the ligand using RosettaMatch.21 Initial matches were refined using RosettaDesign,2 and minimization of the rest of the binding pocket. Candidate designs were chosen and characterized as described earlier.
Computational models and native structures of monomeric proteins
Sixty-one proteins of Rossmann-like folds, which were computationally de novo designed (unpublished), were used. The structures were built up completely from scratch, based on previously published methods.2 39 native monomeric proteins including all-β, all α, α/β, and α+β were used.
Parameters used in computing sidechain probabilities
All calculations assumed kBT = 0.8. The rotamer set used in all calculations was expanded to include rotamers one standard deviation away from the base rotamers in the Dunbrack library.10
Energy function
The energy function used in all evaluations reported here was the default all-atom Rosetta energy2 known as score 12.
Source code
All code was written in C++ within the Rosetta macromolecular-modeling software suite and is freely available to academic users under the Rosetta Commons agreement (http://www.rosettacommons.org). RosettaScripts and commandlines for running these evaluations are available as Supporting Information. Scripts for generating the histograms using gnuplot are provided as Supporting Information as well.
Acknowledgments
The authors thank Jacob E. Corn and Erik Procko for providing some of the computational designs of protein-protein complexes used in this analysis. The authors thank Paul Bates, Alexandre Bonvin, Howook Hwang, Joel Janin, Iain Moal, and Zhiping Weng for providing experimental-binding data for this analysis before publication. S.J.F. was supported by a long-term fellowship from the Human Frontier Science Program and N.K. by a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad.
Supplementary material
References
- 1.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 2.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 3.Shifman JM, Mayo SL. Exploring the origins of binding specificity through the computational redesign of calmodulin. Proc Natl Acad Sci USA. 2003;100:13274–13279. doi: 10.1073/pnas.2234277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 5.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watters AL, Deka P, Corrent C, Callender D, Varani G, Sosnick T, Baker D. The highly cooperative folding of small naturally occurring proteins is likely the result of natural selection. Cell. 2007;128:613–624. doi: 10.1016/j.cell.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Kastritis PL, Bonvin AM. Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J Proteome Res. 2010;9:2216–2225. doi: 10.1021/pr9009854. [DOI] [PubMed] [Google Scholar]
- 10.Dunbrack RL, Jr, Karplus M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat Struct Biol. 1994;1:334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 11.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 12.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 13.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 14.Richardson JS, Richardson DC, Tweedy NB, Gernert KM, Quinn TP, Hecht MH, Erickson BW, Yan Y, McClain RD, Donlan ME, Surles MC. Looking at proteins: representations, folding, packing, and design. Biophysical Society National Lecture, 1992. Biophys J. 1992;63:1185–1209. [PMC free article] [PubMed] [Google Scholar]
- 15.Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426:676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 16.Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. Computational design of a PAK1 binding protein. J Mol Biol. 2010;400:257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 19.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE2004. 2004 doi: 10.1126/stke.2192004pl2. pl2. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 21.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.