Abstract
Within-species genetic variation is a potent factor influencing between-species interactions and community-level structure. Species of the hemi-parasitic plant genus Rhinanthus act as ecosystem engineers, significantly altering above- and below-ground community structure in grasslands. Here, we show the importance of genotypic variation within a single host species (barley—Hordeum vulgare), and population-level variation among two species of parasite (Rhinanthus minor and Rhinanthus angustifolius) on the outcome of parasite infection for both partners. We measured host fitness (number of seeds) and calculated parasite virulence as the difference in seed set between infected and uninfected hosts (the inverse of host tolerance). Virulence was determined by genetic variation within the host species and among the parasite species, but R. angustifolius was consistently more virulent than R. minor. The most tolerant host had the lowest inherent fitness and did not gain a fitness advantage over other infected hosts. We measured parasite size as a proxy for transmission ability (ability to infect further hosts) and host resistance. Parasite size depended on the specific combination of host genotype, parasite species and parasite population, and no species was consistently larger. We demonstrate that the outcome of infection by Rhinanthus depends not only on the host species, but also on the underlying genetics of both host and parasite. Thus, genetic variations within host and parasite are probably essential components of the ecosystem-altering effects of Rhinanthus.
Keywords: host–parasite interactions, community genetics, genetic variation, ecosystem engineer, Rhinanthus, barley
1. Introduction
For over 30 years, within-species genetic variation has been considered a key factor in influencing the structure of ecological communities [1]. Empirical evidence is now mounting that highlights the importance of genetic diversity, genotypic identity, genotype × genotype and genotype × environment interactions in organizing ecological communities.
Focusing on interactions among individuals, recent evidence from grassland studies demonstrates that higher within-species genetic diversity promotes the maintenance of species diversity over time, as well as the increased survival of individual species [2,3]. Reusch et al. [4] show that increased within-species genetic diversity in the eel grass, Zostera marina, enhances community recovery following a climatic perturbation event. Extensive studies of a North American cottonwood system indicate that genetically determined levels of condensed tannins within a hybridizing complex of two tree species (Populus fremontii and Populus angustifolia) are associated with different arthropod communities [5], and that tree chemistry and genetics have wide-ranging effects on whole community composition and dynamics [6–8]. Furthermore, arthropod communities on the evening primrose (Oenothera biennis) are found to assemble according to plant genotype as well as micro-environment [9], and host plant genetic diversity determines the abundance of an ecosystem engineer, the goldenrod bunch gall midge, on Solidago altissima [10]. In addition, work by Tétard-Jones et al. [11] shows that the performance of plants and aphids in an experimental system depends on the interaction between plant and aphid genotype and the community changes of rhizobacteria in the soil.
At the population scale, the geographical mosaic theory provides evidence of the importance of genetic variation in ecological communities, where variation among populations can determine the strength of selection and evolutionary trajectories between two coevolving species [12]. Alternatively, Palkovacs & Post [13] show that differential selection for foraging traits in landlocked and anadromous populations of alewives (the fish Alosa pseudoharengus) can cause cascading community-level effects, changing zooplankton community composition. Finally, Bangert et al. [14,15] show that the effects observed in North American cottonwood communities can be seen both within and across populations at the regional scale.
(a). Rhinanthus—a parasitic plant and ecosystem engineer
Plants in the genus Rhinanthus are facultative hemi-parasitic plants, distributed throughout grasslands in Europe and North America [16,17]. They are generalist parasites, and one species (Rhinanthus minor) has over 50 recorded host plants [18]. Rhinanthus are root parasites, attaching to their hosts via specialized structures called haustoria and extracting nutrients from the host xylem [16].
Parasitic plants, in general, and Rhinanthus in particular, can have an enormous impact on the structure and function of the natural communities in which they grow [19–21]. However, the effect of genetic variability in Rhinanthus systems on host response to infection and parasite attachment to hosts has rarely been considered (but see [22]). Rhinanthus plants act as ecosystem engineers, dramatically altering the diversity and productivity of the grasslands they inhabit [23,24]. Changes in community diversity are, at least in part, the result of differential resistance to infection between potential host plants [25,26], causing a shift in the competitive balance between species within the community [27].
The parasites generally act to suppress grasses, thereby facilitating forb proliferation and potentially enhancing biodiversity [19,27]. This has led to the parasite's use as a management tool for the restoration of degraded grasslands [28–31]. While the directionality of this trend (suppression of grasses and promotion of forbs) is conserved across a number of studies, the magnitude of the effect is highly variable, with grass biomass suppressed by 8–84% and forb abundance promoted by 5–57% [21]. Such unpredictability currently limits the use of Rhinanthus in restoration ecology.
In addition to the effects of Rhinanthus on associated plant communities, presence of the parasitic plant is associated with changes in soil microbial communities [32], long-term nutrient availability [33] and the outcome of host–mycorrhizal interactions [34,35]. Rhinanthus also influences host interactions with aphid herbivores [36] and above-ground arthropod community structure (S. Hartley 2010, personal communication).
(b). Host–parasite interactions
Interactions between hosts and parasites are often characterized by host resistance and tolerance, and parasite virulence and transmission, as these traits affect the fitness, and therefore evolutionary potential, of one or both partners. Host-resistance traits typically act to prevent either infection by the parasite (qualitative resistance) [37], as demonstrated for Rhinanthus–host interactions by Cameron et al. [25], or reduce the fitness of the parasite (quantitative resistance) [37]. Host-tolerance traits act to mitigate the negative impact of the parasite on host fitness and are measured as the fitness of an infected individual compared with an uninfected individual [38]. Tolerance traits may not affect parasite fitness directly, but can still have important evolutionary effects on parasites as well as hosts [39]. Parasite virulence is defined as (i) the rate at which a host becomes infected, and (ii) the damage inflicted to the host by infection with the parasite [40]. The latter definition is measured as the difference in host fitness between an infected and uninfected individual and is essentially the inverse of host tolerance, where the least-tolerant host supports the most virulent parasite. Virulence, and therefore tolerance, can change with the density of infection, and the relationship between density of infection and host fitness may not be linear. This means that depending on the density of infection, relative tolerance and virulence of infected hosts can change [39]. Finally, the transmission ability of the parasite defines how well a parasite can infect further host individuals [41], which in the case of Rhinanthus, is linked to the production of seed. All these traits are inextricably linked together, as host traits influence parasite traits and vice versa [39].
Recent efforts have advanced our understanding of local adaptation and coevolution between hosts and parasites, and much host–parasite work is set within this context (e.g. [42–47]). However, local adaptation and coevolutionary dynamics are difficult to detect [22] and understand for generalist parasites with multiple [48], and in the case of Rhinanthus, simultaneous hosts, as there are complex interactions among hosts as well as between the parasite and the hosts. Community genetics offers an alternative structure in which to consider complex host–parasite interactions, where simple coevolution between two species does not exist. In his seminal paper, Antonovics [49] introduced community genetics as a means to free us ‘from the overly restrictive frame of reference, the reciprocality, that coevolutionists would choose for their own discipline…’ thus enabling us to ‘…generalize community processes in terms of interactions that occur among genotypes as individuals…’ From this point of view, in combination with the fact that Rhinanthus can have far-reaching effects on ecological communities, the interactions among Rhinanthus and its hosts are an excellent model with which to investigate community genetic effects.
(c). Aims
Here, we investigate the role that genetic variation within host and parasitic plants might play in determining the outcome of the interaction between the two and determine the value of such a system in answering community genetics questions. We use an established barley (Hordeum vulgare)–Rhinanthus model system [50–52] to investigate the effects of within and between species diversity using four genotypes of the barley host, and individuals from two populations of two Rhinanthus species (R. minor L. and Rhinanthus angustifolius C.C.Gmel.). The system we used was not natural, but was chosen as a functional Rhinanthus–host system that had genetic tools available for the hosts (all host genotypes are doubled haploid parental genotypes for quantitative trait loci (QTL) mapping lines [53,54]). While not directly applicable for this study, these tools facilitate future mechanistic work on resistance, tolerance and virulence of host–parasitic plant systems. It is also worth noting that many of the recent theoretical advances in evolutionary biology have been based on non-natural laboratory-based systems (e.g. [55,56]), which have enabled the development of hypotheses that can then be tested in more complex field environments.
We used a common garden experimental design, where differences in response among populations of the same species indicate an underlying genetic difference. We determined the effect of parasite infection on host plants by counting the number of seed (a measure of fitness) produced by infected and uninfected barley. We calculated parasite virulence as the difference in seed set between infected and uninfected hosts (sensu [41]) in order to investigate whether host genotype, parasite species and parasite population affected host response to infection. We also measured the height of Rhinanthus plants as a proxy for parasite size and fecundity (as size and fecundity are closely positively related factors; [57]) to determine if host genotype, parasite species and parasite population affected parasite response to host attachment.
2. Material and methods
We obtained seeds of four barley (H. vulgare L.) doubled haploid genotypes (Morex, Steptoe, Oregon Wolfe Dominant and Oregon Wolfe Recessive) from P. Hayes (Oregon State University, OR, USA). Genotypes Morex and Steptoe are barley cultivars and the Oregon Wolfe barleys originate from multiple marker stock [53,54]. We obtained seeds of R. angustifolius C.C.Gmel from two Dutch populations (Doode Bemde and Wageningen) from R. Wesselingh and V. Ducarme (Université Catholique de Louvain, Belgium) and seeds of R. minor L. from two UK populations (Wiltshire and Somerset) from Emorsgate Seeds (Kings Lynn, Norfolk, UK).
We surface-sterilized the Rhinanthus seeds (3% v/v sodium hypochlorite solution, 2–3 min) and germinated them in the dark at 4°C over a three- to four-month period in sealed Petri dishes (9 cm diameter) containing moist, sterile filter paper and capillary matting. We planted and germinated barley seeds in soil (John Innes no. 1) in the dark at 20°C one week before they were required and then moved the seedlings into the light (20°C, 16 L : 8 D) 2 days before transplanting them into the experimental pots.
We transplanted single barley seedlings at the one or two fully expanded leaf-stage, into the centre of large plastic pots (15 cm diameter) filled with horticultural sand. Then, we planted Rhinanthus seedlings (2–4 per pot) with approximately 1 cm radicles at a distance of 2–3 cm from the barley near the surface of the sand. We lightly covered Rhinanthus seedlings with sand and sprayed with water. We planted more than one Rhinanthus seedling per pot, to ensure attachment of at least one parasite to the host plant, as not all seedlings successfully attach to the host plant. We also prepared uninfected controls of each barley genotype. We placed pots on upturned saucers in a greenhouse with supplementary lighting to provide a 16 L : 8 D photoperiod and watered them every day with 100 ml of 1/4 strength Hoaglands solution [58] for the duration of the experiment.
Two weeks after planting, we scored the Rhinanthus plants for morphological characteristics associated with attachment and continued monitoring levels of attachment for two weeks more. Attached Rhinanthus plants show inflated leaves and rapid growth when compared with unattached plants. Leaves also change colour upon attachment from dark green to yellowish-green [59]. We reduced Rhinanthus density to a single plant per pot at four weeks post-planting or when 80 per cent of the pots in a treatment group contained a minimum of one attached plant, whichever occurred sooner. Additional Rhinanthus plants were removed as soon as practicable at the start of the experiment to reduce the impact of multiple attachments to a single host plant. The experimental pots contained either a single barley plant and a single Rhinanthus plant (treatments) or single barley plants without Rhinanthus (negative controls). After four months, when the barley plants had set seed, we collected plants from all pots (shoots and roots) and left them to air-dry in paper bags. Once dry, we separated the Rhinanthus and barley plants from each other. We determined the total above-ground dry weight (shoot and fruit) for the barley, counted the number of barley seeds and recorded the height of the Rhinanthus. As density of the parasite infection was held constant at a single plant per host, we calculated parasite virulence as the difference in individual host plant fitness from the mean of the appropriate control group using the following equation:
 |
where 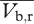 is the mean virulence of Rhinanthus from population (r) growing on barley genotype (b),
is the mean virulence of Rhinanthus from population (r) growing on barley genotype (b),  is the seed set of an individual of barley genotype (b) infected by a plant from Rhinanthus population (r),
is the seed set of an individual of barley genotype (b) infected by a plant from Rhinanthus population (r),  the average seed set of the uninfected control of barley genotype (b) and nb,r the number of replicates per treatment.
the average seed set of the uninfected control of barley genotype (b) and nb,r the number of replicates per treatment.
We used a fully factorial experimental design with four barley genotypes, five Rhinanthus treatments (two populations per species and an uninfected control) and eight replicates, giving an initial total of 160 experimental pots (128 host–parasite pairs and 32 controls). Pots were fully randomized on a single bench within the greenhouse. Rhinanthus plants from all populations failed to grow in 15 pots across all host genotypes and we removed these from the analyses, giving a final total of n = 145 (113 host–parasite pairs and 32 controls).
In order to determine the effect of infection by Rhinanthus on the barley genotypes, we analysed barley fitness (number of seeds) using a partially nested three-way analysis of variance (ANOVA) with barley genotype, Rhinanthus treatment (both species and uninfected controls) and Rhinanthus population nested within Rhinanthus treatment as fixed effects (n = 145). We used Bonferroni-corrected multiple contrast tests to compare the effect of Rhinanthus infection with the uninfected controls.
In order to determine if there was an interaction between Rhinanthus population and barley genotype on either the host or parasite, we analysed parasite virulence and Rhinanthus height using two separate three-way partially nested ANCOVAs with barley genotype, Rhinanthus species and Rhinanthus population nested within-species as fixed factors (n = 113). We included Rhinanthus height as a covariate in the analysis of parasite virulence and the total above-ground barley dry weight as a covariate in the analysis of Rhinanthus height. The covariates were included as an indicator of either parasite or host plant size to determine if larger parasites were more virulent and larger hosts supported larger parasites. We used post hoc Tukey–Kramer tests to analyse main effects further. We performed all statistical analyses in JMP v. 8. The following statistical models were used:
— Barley fitness = barley genotype + Rhinanthus treatment + Rhinanthus population (Rhinanthus treatment) + barley genotype × Rhinanthus treatment + barley genotype × Rhinanthus population (Rhinanthus treatment).
— Parasite virulence = Rhinanthus height + barley genotype + Rhinanthus species + Rhinanthus population (Rhinanthus species) + barley genotype × Rhinanthus species + barley genotype × Rhinanthus population (Rhinanthus species).
— Rhinanthus height = barley total above-ground dry weight + barley genotype + Rhinanthus species + Rhinanthus population (Rhinanthus species) + barley genotype × Rhinanthus species + barley genotype × Rhinanthus population (Rhinanthus species).
3. Results
Four weeks after planting, attachment levels for all R. angustifolius treatments were over 80 per cent. Most R. minor treatments were also over 80 per cent attachment, except for two R. minor treatments from the Wiltshire population (Oregon Wolfe Recessive (75%), Morex (63%)) and two from the Somerset population (Steptoe (75%), Oregon Wolfe Dominant (75%)).
(a). The effect of Rhinanthus infection on barley fitness
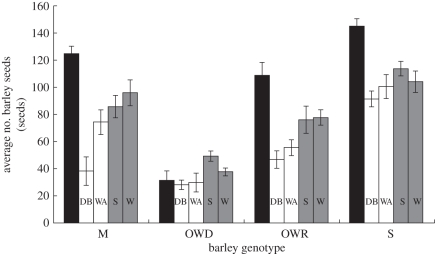
Barley fitness was significantly affected by barley genotype (three-way partially nested ANOVA: F3,125 = 95.27, p < 0.0001), Rhinanthus treatment (F2,125 = 54.06, p < 0.0001) and an interaction between barley genotype and Rhinanthus treatment (F6,125 = 6.15, p < 0.0001). There was neither a significant effect of Rhinanthus population nested within treatment (F2,125 = 1.76, p = 0.18), nor an interaction between population nested within treatment and barley genotype (F6,125 = 1.35, p = 0.24). Barley fitness is therefore influenced by a combination of barley genotype and Rhinanthus treatment. Of the uninfected controls, genotype Steptoe was the most fecund (145 seeds ± 16; mean ± s.d.) and genotype Oregon Wolfe Dominant the least fecund (31 seeds ± 20; mean ± s.d.). When compared with uninfected control plants, both species of Rhinanthus reduced host fitness for barley genotypes Steptoe, Morex and Oregon Wolfe Recessive (p < 0.001). However, infection with either species of Rhinanthus did not have an impact on the fitness of barley genotype Oregon Wolfe Dominant (p = 0.63). Despite this, fitness of infected barley genotype Oregon Wolfe Dominant remained low in comparison with the majority (but not all) of the other infected barley genotypes (figure 1).
Figure 1.
Average number of barley seeds for four genotypes of barley (Morex (M), Oregon Wolfe Dominant (OWD), Oregon Wolfe Recessive (OWR) and Steptoe (S) infected with Rhinanthus angustifolius (white bars) from two Dutch populations (Doode Bemde (DB) and Wageningen (WA)) and Rhinanthus minor (grey bars) from two UK populations (Somerset (S) and Wiltshire (W)). Black bars are uninfected barley controls. Error bars are ±1 s.e.
(b). Effect of barley genotype, Rhinanthus species and population on parasite virulence
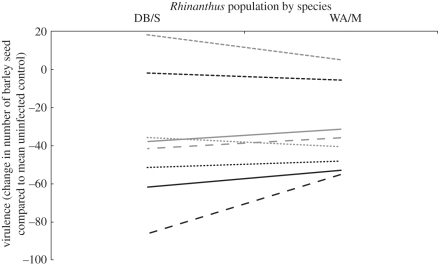
Parasite virulence (measured as the difference in individual host plant fitness from the mean of the appropriate control group) was significantly affected by barley genotype (three-way partially nested ANCOVA: F3,96 = 46.38, p < 0.0001) and Rhinanthus species (F1,96 = 20.30, p < 0.0001). There was no significant effect of Rhinanthus population nested within-species (F2,96 = 1.18, p = 0.31) and there were no significant interactions between barley genotype and either Rhinanthus species (F3,96 = 1.24, p = 0.30) or population nested within-species (F6,96 = 1.47, p = 0.20). Rhinanthus height was not a significant covariate in the analysis (F1,96 = 0.94, p = 0.33), indicating that parasite size does not influence the virulence of the respective parasites. Instead, virulence is influenced by barley genotype and Rhinanthus species. Of the two parasite species, R. angustifolius was significantly more virulent than R. minor (Tukey–Kramer test: p < 0.0001) and all parasites infecting Oregon Wolfe Dominant barley were significantly less virulent than the parasites infecting the other three barley genotypes (Tukey–Kramer test: all p < 0.05; figure 2). As parasite virulence can be seen as the inverse of host tolerance, under the circumstances herein, barley genotype Oregon Wolfe Dominant can also be described as the most tolerant host.
Figure 2.
Reaction norms of parasite virulence (number of barley seeds normalized by controls) for four genotypes of barley (Morex, Oregon Wolfe Dominant (OWD), Oregon Wolfe Recessive (OWR) and Steptoe) infected with Rhinanthus angustifolius (RA, black lines) and Rhinanthus minor (RM, grey lines). Each line represents the mean reaction norm for a single genotype infected by two populations of a single species of Rhinanthus. Rhinanthus angustifolius originates from two Dutch populations (Doode Bemde (DB) and Wageningen (WA)) and R. minor from two UK populations (Somerset (S) and Wiltshire (W)). OWR, solid lines; Morex, long dashed lines; OWD, short dashed lines; Steptoe, dotted lines.
(c). Effect of barley genotype, Rhinanthus species and population on parasite size
Parasite size (measured as height) was marginally significantly affected by barley genotype (three-way partially nested ANCOVA: F3,96 = 2.77, p = 0.046), significantly affected by Rhinanthus species (F1,96 = 10.28, p = 0.0018), Rhinanthus population nested within-species (F2,96 = 4.36, p = 0.016) and an interaction between barley genotype and Rhinanthus population nested within-species (F6,96 = 2.89, p = 0.012). Total above-ground barley dry weight was not a significant covariate in the analysis (F1,96 = 3.38, p = 0.069), indicating that the size of barley plants does not influence the size of the parasite. Rather, parasite size is determined by the specific combination of host genotype, Rhinanthus species and population (figure 3). As parasite size and fecundity are closely related, the larger parasites should also be the most fecund. The largest parasite was R. angustifolius from population Doode Bemde growing on Oregon Wolfe Recessive (height = 54.4 cm ± 8.6; mean ± s.d.) and the smallest was R. minor from Somerset growing on Oregon Wolfe Dominant (height = 31.6 cm ± 10.5; mean ± s.d.), suggesting that these two parasites should also have the highest and lowest seed set and therefore transmission ability, respectively, of the combinations tested. As quantitative resistance can also be defined as a reduction in host fitness, Oregon Wolfe Dominant has the highest resistance to R. minor from Somerset, and Oregon Wolfe Recessive has the lowest resistance to R. angustifolius from Doode Bemde of all the combinations tested.
Figure 3.
Reaction norms of Rhinanthus height (cm) for four genotypes of barley (Morex, Oregon Wolfe Dominant (OWD), Oregon Wolfe Recessive (OWR) and Steptoe) infected with Rhinanthus angustifolius (RA, black lines) and Rhinanthus minor (RM, grey lines). Each line represents the mean reaction norm between two populations of the same species of Rhinanthus growing on a single genotype of barley. Rhinanthus angustifolius originates from two Dutch populations (Doode Bemde (DB) and Wageningen (WA)) and R. minor from two UK populations (Somerset (S) and Wiltshire (W)). OWR, solid lines; Morex, long dashed lines; OWD, short dashed lines; Steptoe, dotted lines.
4. Discussion
Here we demonstrate, for the first time in a Rhinanthus–barley system, that the outcome of infection for both host and parasite, in terms of host fitness, host tolerance, parasite virulence and transmission ability, depends on genetic variation within both partners.
(a). Ecosystem effects
We know from previous studies that Rhinanthus can have cascading effects on grassland communities that reach far beyond its immediate impact on an individual host plant. Presence of Rhinanthus can influence plant [60,61], soil [32] and arthropod [36] community structure as well as the cycling of nutrients within the system [33]. From other research we know that genetic variation within a focal plant can change the outcome of interactions with arthropods, soil microbes [6,11] and the effects of an associated ecosystem engineer [10]. Finally, we know genetic variation to be an important factor driving competition among, and long-term survival of, grassland plant species [2,3,62]. A proportion of the effects of Rhinanthus on plant community species composition is attributed to the differences in resistance to, and tolerance of, parasite infection by potential host species [25]. Here, we show that genetic variation in the host can change its tolerance to infection and the virulence of the parasite. We also show that an interaction between genetic variation in the host and parasite changes the size, potential seed set and hence transmission ability of Rhinanthus and resistance of the host. Therefore, it follows that the effects of Rhinanthus on community composition probably depend on genetic variation within host and parasitic plants. Some variation in the plant community diversity response to Rhinanthus has been noted [21] and we propose that this is due, at least in part, to the genetic variation within host and parasite species.
We used a non-natural host for this study, so caution needs to be exercised when extrapolating these results to natural host–Rhinanthus communities. However, in an earlier study on local adaptation in R. angustifolius (syn. serotinus), Mutikainen et al. [22] showed similar patterns of variation in the response of different populations of the natural grass host Agrostis capillaris to infection as we found among the genotypes of barley. Similarly, in an alternative natural host–parasitic plant system (Urtica dioica–Cuscuta europaea), Koskela et al. [37] demonstrated genetic variation in host resistance and tolerance traits. Thus, it seems likely that the genetic variation we observed in response to infection between Rhinanthus and barley should translate to a more natural community.
We examined variation among two hybridizing species of Rhinanthus and two populations of each species growing on distinct barley genotypes. Ideally, we would have used distinct genotypes of Rhinanthus as well, but these are not available currently for either parasite species. Hybrid complexes have been used previously in similar studies to good effect where the two parental species represent morphological and genetic extremes of types [6]. The pure species of Rhinanthus we used fall into two distinct morphological groups based on their floral characteristics. However, when hybrid and backcross individuals are included in the analysis, phenotypic variation across species and hybrids is continuous [63,64]. The two Rhinanthus species are genetically distinct and genetic differences between the parent and the hybrid groups can also be detected [64]. Analysis using recently developed microsatellite markers for R. minor indicates that there is genetic variation among populations [65]. Therefore, it is reasonable to assume that phenotypic differences among populations are due, in part, to genetic differences.
Common garden experiments are an established technique for separating the effect of within-species genetic variation from environmental-based variation [66]. However, the phenotypic variation observed among populations in a common garden also includes maternal effects, for which we did not control. While maternal effects are an important factor controlling the traits of seeds and young plants, these effects generally decrease with age [67]. Maternal effects are, therefore, not likely to be a major cause of variation in final adult height of Rhinanthus (as measured here). However, we cannot completely rule out the possibility that variation among populations of Rhinanthus in our experiment is caused by a mixture of genetic variation and maternal effects.
(b). Host–parasite interactions
Previous studies concentrate on the reduction of host biomass following infection by Rhinanthus species (see [19] for review). Here, we show that infection also reduced host fitness in terms of seed production to three of the four host genotypes tested. Unsurprisingly, different host genotypes vary in their fecundity, with Oregon Wolfe Dominant producing substantially fewer seeds overall than Oregon Wolfe Recessive, Morex or Steptoe. Seed production was not reduced in infected Oregon Wolfe Dominant hosts when compared with uninfected controls, which showed that the least virulent Rhinanthus infections occurred on this genotype. This suggests that Oregon Wolfe Dominant barley is highly tolerant to infection by Rhinanthus, while the other genotypes are less so. All barley genotypes are doubled haploid lines. The two Oregon Wolfe barleys were constructed to form one strain with multiple dominant traits (Oregon Wolfe Dominant) and another strain with multiple recessive traits (Oregon Wolfe Recessive) from an original population of barley [68]. Some of the dominant traits can be attributed to wild barley (H. vulgare L. subsp. spontaneum (K. Koch) Thell.), while most of the recessive traits originate from agricultural barley strains [68]. Both the genotypes Steptoe and Morex are strains of barley that were developed to have a number of good agronomic traits [53]. The high tolerance of infection but low fitness of Oregon Wolfe Dominant suggests that there is a trade-off between fitness and tolerance of parasite attack, and that the agricultural strains, bred to maximize fitness, may have lost tolerance to parasite infection.
In contrast to resistant host genotypes, tolerant hosts may not reduce the fitness of an infecting parasite [39]. This defence strategy allows the parasites to persist and spread in a population. For tolerance to be an evolutionarily stable strategy, there must be some fitness advantage to tolerant hosts, i.e. an infected tolerant host should have a higher fitness than an infected non-tolerant host. The most tolerant host in our study, Oregon Wolfe Dominant, also had the lowest overall fitness. This was the case both before and after infection, although its relative fitness when compared with the other barley genotypes changed with infection status. We kept infection density constant at a single parasite per host, but other studies have shown that host tolerance can change with parasite density, with more tolerant hosts at one parasite density becoming relatively less tolerant at another [39]. The effect on host plant tolerance of multiple infections with Rhinanthus is unknown. However, in natural populations, multiple Rhinanthus infections of a single host are possible and differential tolerance gradients with changing infection density would probably also influence host community dynamics.
In terms of the parasites, R. angustifolius has a larger impact on host fitness than R. minor for all barley genotypes and is thus the more virulent parasite species under our experimental conditions. We hypothesized that this might be the result of R. angustifolius being a more robust plant than R. minor [69], better able to abstract host resources. However, we found that although R. angustifolius is generally larger than R. minor, this is not always the case. For example, R. minor from the Somerset population, growing on barley genotype Steptoe, is a similar size to many R. angustifolius–host genotype combinations, while R. angustifolius from the Wageningen population growing on barley genotypes Oregon Wolfe Dominant and Steptoe is similar size to many of the R. minor–host genotype combinations. In addition, when included as a covariate in the analysis, parasite size had no significant effect on virulence, suggesting the virulence was independent of parasite size.
Parasite size gives an indication of potential seed set [25] or transmission ability of the parasites [41] and also host resistance to infection [37]. In our study, parasite size depended on the specific combination of parasite population and host genotype, suggesting that genetic variation within both host and parasite affects parasite fitness. Tolerance and resistance have been proposed as complementary plant-defence strategies, where a fully tolerant genotype has no need of resistance and vice versa. While in extreme cases this may be so, evidence for a clear negative relationship between tolerance and resistance traits in plants is conflicting [38,70]. In our study, the largest parasite was R. angustifolius from Doode Bemde growing on Oregon Wolfe Recessive barley and the smallest R. minor from Somerset growing on Oregon Wolfe Dominant. These represent the least and most resistant host–parasite combinations, respectively. Virulence of R. angustifolius from Doode Bemde growing on Oregon Wolfe Recessive was relatively high, suggesting that tolerance of this combination was low. Virulence of R. minor from Somerset growing on Oregon Wolfe Dominant was low, suggesting that tolerance of this combination was high. These results indicate a positive, rather than negative relationship between tolerance and resistance in barley. This corroborates evidence from a meta-analysis by Leimu & Koricheva [70], who found positive correlations between tolerance and resistance traits in crop plants but negative correlations in wild plants.
Acknowledgements
We thank Pat Hayes, Renate Wesselingh and Véronique Ducarme for barley and Rhinanthus seeds; Patrick Lunt, Charlotte McHugh, Nicola Brennan and Abigail Rumsey for helping to collect data and water plants. We also thank two anonymous reviewers for comments on an earlier draft of the manuscript. Experimental work was undertaken at the Firs Experimental Botanical Grounds, University of Manchester. D.D.C. is supported by a Natural Environment Research Council independent fellowship (Award number: NE/E014 070/1).
Footnotes
One contribution of 13 to a Theme Issue ‘Community genetics: at the crossroads of ecology and evolutionary genetics’.
References
- 1.Antonovics J. 1976. The input from population genetics: ‘the new ecological genetics’. Syst. Bot. 1, 233–245 10.2307/2418718 (doi:10.2307/2418718) [DOI] [Google Scholar]
- 2.Booth R. E., Grime J. P. 2003. Effects of genetic impoverishment on plant community diversity. J. Ecol. 91, 721–730 10.1046/j.1365-2745.2003.00804.x (doi:10.1046/j.1365-2745.2003.00804.x) [DOI] [Google Scholar]
- 3.Whitlock R., Grime J. P., Booth R., Burke T. 2007. The role of genotypic diversity in determining grassland community structure under constant environmental conditions. J. Ecol. 95, 895–907 10.1111/j.1365-2745.2007.01275.x (doi:10.1111/j.1365-2745.2007.01275.x) [DOI] [Google Scholar]
- 4.Reusch T. B. H., Ehlers A., Hammerli A., Worm B. 2005. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl Acad. Sci. USA 102, 2826–2831 10.1073/pnas.0500008102 (doi:10.1073/pnas.0500008102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimp G. M., Martinsen G. D., Floate K. D., Bangert R. K., Whitham T. G. 2005. Plant genetic determinants of arthropod community structure and diversity. Evolution 59, 61–69 [PubMed] [Google Scholar]
- 6.Whitham T. G., et al. 2006. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523 10.1038/nrg1877 (doi:10.1038/nrg1877) [DOI] [PubMed] [Google Scholar]
- 7.Schweitzer J. A., Bailey J. K., Rehill B. J., Martinsen G. D., Hart S. C., Lindroth R. L., Keim P., Whitham T. G. 2004. Genetically based trait in a dominant tree affects ecosystem processes. Ecol. Lett. 7, 127–134 10.1111/j.1461-0248.2003.00562.x (doi:10.1111/j.1461-0248.2003.00562.x) [DOI] [Google Scholar]
- 8.Schweitzer J. A., Bailey J. K., Fischer D. G., Leroy C. J., Lonsdorf E. V., Whitham T. G., Hart S. C. 2008. Plant–soil–microorganism interactions: heritable relationship between plant genotype and associated soil microorganisms. Ecology 89, 773–781 10.1890/07-0337.1 (doi:10.1890/07-0337.1) [DOI] [PubMed] [Google Scholar]
- 9.Johnson M. T. J., Agrawal A. A. 2005. Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86, 874–885 10.1890/04-1068 (doi:10.1890/04-1068) [DOI] [Google Scholar]
- 10.Crawford K. M., Crutsinger G. M., Sanders N. J. 2007. Host-plant genotypic diversity mediates the distribution of an ecosystem engineer. Ecology 88, 2114–2120 10.1890/06-1441.1 (doi:10.1890/06-1441.1) [DOI] [PubMed] [Google Scholar]
- 11.Tétard-Jones C., Kertesz M. A., Gallois P., Preziosi R. F. 2007. Genotype-by-genotype interactions modified by a third species in a plant–insect system. Am. Nat. 170, 492–499 10.1086/520115 (doi:10.1086/520115) [DOI] [PubMed] [Google Scholar]
- 12.Thompson J. N. 2005. Coevolution: the geographic mosaic of coevolutionary arms races. Curr. Biol. 15, R992–R994 10.1016/j.cub.2005.11.046 (doi:10.1016/j.cub.2005.11.046) [DOI] [PubMed] [Google Scholar]
- 13.Palkovacs E. P., Post D. M. 2009. Experimental evidence that phenotypic divergence in predators drives community divergence in prey. Ecology 90, 300–305 10.1890/08-1673.1 (doi:10.1890/08-1673.1) [DOI] [PubMed] [Google Scholar]
- 14.Bangert R. K., Allan G. J., Turek R. J., Wimp G. M., Meneses N., Martinsen G. D., Keim P., Whitham T. G. 2006. From genes to geography: a genetic similarity rule for arthropod community structure at multiple geographic scales. Mol. Ecol. 15, 4215–4228 10.1111/j.1365-294X.2006.03092.x (doi:10.1111/j.1365-294X.2006.03092.x) [DOI] [PubMed] [Google Scholar]
- 15.Bangert R. K., et al. 2006. A genetic similarity rule determines arthropod community structure. Mol. Ecol. 15, 1379–1391 10.1111/j.1365-294X.2005.02749.x (doi:10.1111/j.1365-294X.2005.02749.x) [DOI] [PubMed] [Google Scholar]
- 16.Westbury D. B. 2004. Rhinanthus minor L. J. Ecol. 92, 906–927 10.1111/j.0022-0477.2004.00929.x (doi:10.1111/j.0022-0477.2004.00929.x) [DOI] [Google Scholar]
- 17.Preston C. D., Pearman D. A., Dines T. D. 2002. New atlas of the British and Irish flora. Oxford, UK: Oxford University Press [Google Scholar]
- 18.Gibson C. C., Watkinson A. R. 1989. The host range and selectivity of a parasitic plant—Rhinanthus minor L. Oecologia 78, 401–406 10.1007/BF00379116 (doi:10.1007/BF00379116) [DOI] [PubMed] [Google Scholar]
- 19.Ameloot E., Verheyen K., Hermy M. 2005. Meta-analysis of standing crop reduction by Rhinanthus spp. and its effect on vegetation structure. Folia Geobot. 40, 289–310 10.1007/BF02803241 (doi:10.1007/BF02803241) [DOI] [Google Scholar]
- 20.Press M. C., Phoenix G. K. 2005. Impacts of parasitic plants on natural communities. New Phytol. 166, 737–751 10.1111/j.1469-8137.2005.01358.x (doi:10.1111/j.1469-8137.2005.01358.x) [DOI] [PubMed] [Google Scholar]
- 21.Cameron D. D., Hwangbo J. K., Keith A. M., Geniez J. M., Kraushaar D., Rowntree J., Seel W. E. 2005. Interactions between the hemiparasitic angiosperm Rhinanthus minor and its hosts: from the cell to the ecosystem. Folia Geobot. 40, 217–229 10.1007/BF02803236 (doi:10.1007/BF02803236) [DOI] [Google Scholar]
- 22.Mutikainen P., Salonen V., Puustinen S., Koskela T. 2000. Local adaptation, resistance, and virulence in a hemiparasitic plant–host plant interaction. Evolution 54, 433–440 [DOI] [PubMed] [Google Scholar]
- 23.Davies D. M., Graves J. D., Elias C. O., Williams P. J. 1997. The impact of Rhinanthus spp. on sward productivity and composition: implications for the restoration of species-rich grasslands. Biol. Conserv. 82, 87–93 10.1016/S0006-3207(97)00010-4 (doi:10.1016/S0006-3207(97)00010-4) [DOI] [Google Scholar]
- 24.Joshi J., Matthies D., Schmid B. 2000. Root hemiparasites and plant diversity in experimental grassland communities. J. Ecol. 88, 634–644 10.1046/j.1365-2745.2000.00487.x (doi:10.1046/j.1365-2745.2000.00487.x) [DOI] [Google Scholar]
- 25.Cameron D. D., Coats A. M., Seel W. E. 2006. Differential resistance among host and non-host species underlies the variable success of the hemi-parasitic plant Rhinanthus minor. Ann. Bot. Lond. 98, 1289–1299 10.1093/aob/mcl218 (doi:10.1093/aob/mcl218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron D. D., Seel W. E. 2007. Functional anatomy of haustoria formed by Rhinanthus minor: linking evidence from histology and isotope tracing. New Phytol. 174, 412–419 10.1111/j.1469-8137.2007.02013.x (doi:10.1111/j.1469-8137.2007.02013.x) [DOI] [PubMed] [Google Scholar]
- 27.Gibson C. C., Watkinson A. R. 1991. Host selectivity and the mediation of competition by the root hemiparasite Rhinanthus minor. Oecologia 86, 81–87 10.1007/BF00317393 (doi:10.1007/BF00317393) [DOI] [PubMed] [Google Scholar]
- 28.Westbury D. B., Davies A., Woodcock B. A., Dunnett N. P. 2006. Seeds of change: the value of using Rhinanthus minor in grassland restoration. J. Veg. Sci. 17, 435–446 [Google Scholar]
- 29.Bullock J. M., Pywell R. F. 2005. Rhinanthus: a tool for restoring diverse grassland? Folia Geobot. 40, 273–288 10.1007/BF02803240 (doi:10.1007/BF02803240) [DOI] [Google Scholar]
- 30.Pywell R. F., Bullock J. M., Walker K. J., Coulson S. J., Gregory S. J., Stevenson M. J. 2004. Facilitating grassland diversification using the hemiparasitic plant Rhinanthus minor. J. Appl. Ecol. 41, 880–887 10.1111/j.0021-8901.2004.00940.x (doi:10.1111/j.0021-8901.2004.00940.x) [DOI] [Google Scholar]
- 31.Ameloot E., Hermy M., Verheyen K. 2006. Rhinanthus: an effective tool in reducing biomass of road verges? An experiment along two motorways. Belg. J. Bot. 139, 173–187 [Google Scholar]
- 32.Bardgett R. D., Smith R. S., Shiel R. S., Peacock S., Simkin J. M., Quirk H., Hobbs P. J. 2006. Parasitic plants indirectly regulate below-ground properties in grassland ecosystems. Nature 439, 969–972 10.1038/nature04197 (doi:10.1038/nature04197) [DOI] [PubMed] [Google Scholar]
- 33.Ameloot E., Verlinden G., Boeckx P., Verheyen K., Hermy M. 2008. Impact of hemiparasitic Rhinanthus angustifolius and R. minor on nitrogen availability in grasslands. Plant Soil 311, 255–268 10.1007/s11104-008-9640-2 (doi:10.1007/s11104-008-9640-2) [DOI] [Google Scholar]
- 34.Stein C., Rissmann C., Hempel S., Renker C., Buscot F., Prati D., Auge H. 2009. Interactive effects of mycorrhizae and a root hemiparasite on plant community productivity and diversity. Oecologia 159, 191–205 10.1007/s00442-008-1192-x (doi:10.1007/s00442-008-1192-x) [DOI] [PubMed] [Google Scholar]
- 35.Davies D. M., Graves J. D. 1998. Interactions between arbuscular mycorrhizal fungi and the hemiparasitic angiosperm Rhinanthus minor during co-infection of a host. New Phytol. 139, 555–563 10.1046/j.1469-8137.1998.00211.x (doi:10.1046/j.1469-8137.1998.00211.x) [DOI] [Google Scholar]
- 36.Hartley S. E., Bass K. A., Johnson S. N. 2007. Going with the flow: plant vascular systems mediate indirect interactions between plants, insect herbivores, and parasitic plants. In Ecological communities—plant mediation in indirect interaction webs (eds Ohgushi T., Craig T. P., Price P. W.), pp. 51–74 Cambridge, UK: Cambridge University Press [Google Scholar]
- 37.Koskela T., Puustinen S., Salonen V., Mutikainen P. 2002. Resistance and tolerance in a host plant–holoparasitic plant interaction: genetic variation and costs. Evolution 56, 899–908 [DOI] [PubMed] [Google Scholar]
- 38.Strauss S. Y., Agrawal A. A. 1999. The ecology and evolution of plant tolerance to herbivory. Trends Ecol. Evol. 14, 179–185 10.1016/S0169-5347(98)01576-6 (doi:10.1016/S0169-5347(98)01576-6) [DOI] [PubMed] [Google Scholar]
- 39.Little T. J., Shuker D. M., Colegrave N., Day T., Graham A. L. 2010. The coevolution of virulence: tolerance in perspective. PLoS Pathog 6, e1001006. 10.1371/journal.ppat.1001006 (doi:10.1371/journal.ppat.1001006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandon S., Van Baalen M., Jansen V. A. A. 2002. The evolution of parasite virulence, superinfection, and host resistance. Am. Nat. 159, 658–669 10.1086/339993 (doi:10.1086/339993) [DOI] [PubMed] [Google Scholar]
- 41.Salvaudon L., Heraudet V., Shykoff J. A. 2007. Genotype-specific interactions and the trade-off between host and parasite fitness. BMC Evol. Biol. 7, 189. 10.1186/1471-2148-7-189 (doi:10.1186/1471-2148-7-189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandon S. 2002. Local adaptation and the geometry of host–parasite coevolution. Ecol. Lett. 5, 246–256 10.1046/j.1461-0248.2002.00305.x (doi:10.1046/j.1461-0248.2002.00305.x) [DOI] [Google Scholar]
- 43.Gandon S., Agnew P., Michalakis Y. 2002. Coevolution between parasite virulence and host life-history traits. Am. Nat. 160, 374–388 10.1086/341525 (doi:10.1086/341525) [DOI] [PubMed] [Google Scholar]
- 44.Best A., White A., Boots M. 2009. The implications of coevolutionary dynamics to host–parasite interactions. Am. Nat. 173, 779–791 10.1086/598494 (doi:10.1086/598494) [DOI] [PubMed] [Google Scholar]
- 45.Boots M., Best A., Miller M. R., White A. 2009. The role of ecological feedbacks in the evolution of host defence: what does theory tell us? Phil. Trans. R. Soc. B 364, 27–36 10.1098/rstb.2008.0160 (doi:10.1098/rstb.2008.0160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mutikainen P., Koskela T. 2002. Population structure of a parasitic plant and its perennial host. Heredity 89, 318–324 10.1038/sj.hdy.6800142 (doi:10.1038/sj.hdy.6800142) [DOI] [PubMed] [Google Scholar]
- 47.Koskela T., Salonen V., Mutikainen P. 2000. Local adaptation of a holoparasitic plant, Cuscuta europaea: variation among populations. J. Evol. Biol. 13, 749–755 10.1046/j.1420-9101.2000.00226.x (doi:10.1046/j.1420-9101.2000.00226.x) [DOI] [Google Scholar]
- 48.Gandon S. 2004. Evolution of multihost parasites. Evolution 58, 455–469 [PubMed] [Google Scholar]
- 49.Antonovics J. 1992. Toward community genetics. In Plant resistance to herbivores and pathogens—ecology, evolution and genetics (eds Fritz R. S., Simms E. L.), pp. 426–449 Chicago, IL: University of Chicago Press [Google Scholar]
- 50.Jiang F., Jeschke W. D., Hartung W. 2003. Water flows in the parasitic association Rhinanthus minor–Hordeum vulgare. J. Exp. Bot. 54, 1985–1993 10.1093/jxb/erg212 (doi:10.1093/jxb/erg212) [DOI] [PubMed] [Google Scholar]
- 51.Seel W. E., Jeschke W. D. 1999. Simultaneous collection of xylem sap from Rhinanthus minor and the hosts Hordeum and Trifolium: hydraulic properties, xylem sap composition and effects of attachment. New Phytol. 143, 281–298 10.1046/j.1469-8137.1999.00461.x (doi:10.1046/j.1469-8137.1999.00461.x) [DOI] [Google Scholar]
- 52.Jiang F., Jeschke W. D., Hartung W., Cameron D. D. 2008. Mobility of boron–polyol complexes in the hemiparasitic association between Rhinanthus minor and Hordeum vulgare: the effects of nitrogen nutrition. Physiol. Plant. 134, 13–21 10.1111/j.1399-3054.2008.01116.x (doi:10.1111/j.1399-3054.2008.01116.x) [DOI] [PubMed] [Google Scholar]
- 53.Kleinhofs A., et al. 1993. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86, 705–712 10.1007/BF00222660 (doi:10.1007/BF00222660) [DOI] [PubMed] [Google Scholar]
- 54.Costa J. M., et al. 2001. Molecular mapping of the Oregon Wolfe barleys: a phenotypically polymorphic doubled-haploid population. Theor. Appl. Genet. 103, 415–424 10.1007/s001220100622 (doi:10.1007/s001220100622) [DOI] [Google Scholar]
- 55.Houle D., Rowe L. 2003. Natural selection in a bottle. Am. Nat. 161, 50–67 10.1086/345480 (doi:10.1086/345480) [DOI] [PubMed] [Google Scholar]
- 56.Mousseau T. A., Roff D. A. 1987. Natural-selection and the heritability of fitness components. Heredity 59, 181–197 10.1038/hdy.1987.113 (doi:10.1038/hdy.1987.113) [DOI] [PubMed] [Google Scholar]
- 57.Cameron D. D., White A., Antonovics J. 2009. Parasite–grass–forb interactions and rock-paper-scissor dynamics: predicting the effects of the parasitic plant Rhinanthus minor on host plant communities. J. Ecol. 97, 1311–1319 10.1111/j.1365-2745.2009.01568.x (doi:10.1111/j.1365-2745.2009.01568.x) [DOI] [Google Scholar]
- 58.Hoagland D. R., Arnon D. I. 1950. The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn Circ. 347, 1–32 [Google Scholar]
- 59.Klaren C. H., Janssen G. 1978. Physiological changes in hemiparasite Rhinanthus serotinus before and after attachment. Physiol. Plant. 42, 151–155 10.1111/j.1399-3054.1978.tb01556.x (doi:10.1111/j.1399-3054.1978.tb01556.x) [DOI] [Google Scholar]
- 60.Gibson C. C., Watkinson A. R. 1992. The role of the hemiparasitic annual Rhinanthus minor in determining grassland community structure. Oecologia 89, 62–68 10.1007/BF00319016 (doi:10.1007/BF00319016) [DOI] [PubMed] [Google Scholar]
- 61.Ameloot E., Verheyen K., Bakker J. P., De Vries Y., Hermy M. 2006. Long-term dynamics of the hemiparasite Rhinanthus angustifolius and its relationship with vegetation structure. J. Veg. Sci. 17, 637–646 [Google Scholar]
- 62.Fridley J. D., Grime J. P., Bilton M. 2007. Genetic identity of interspecific neighbours mediates plant responses to competition and environmental variation in a species-rich grassland. J. Ecol. 95, 908–915 10.1111/j.1365-2745.2007.01256.x (doi:10.1111/j.1365-2745.2007.01256.x) [DOI] [Google Scholar]
- 63.Kwak M. M. 1980. Artificial and natural hybridization and introgression in Rhinanthus (Scrophulariaceae) in relation to bumblebee pollination. Taxon 29, 613–628 10.2307/1220333 (doi:10.2307/1220333) [DOI] [Google Scholar]
- 64.Ducarme V., Wesselingh R. A. 2005. Detecting hybridization in mixed populations of Rhinanthus minor and Rhinanthus angustifolius. Folia Geobot. 40, 151–161 10.1007/BF02803231 (doi:10.1007/BF02803231) [DOI] [Google Scholar]
- 65.Houston K., Wolff K. 2009. Eight polymorphic microsatellite markers for Rhinanthus minor. Mol. Ecol. Resour. 9, 174–176 10.1111/j.1755-0998.2008.02415.x (doi:10.1111/j.1755-0998.2008.02415.x) [DOI] [PubMed] [Google Scholar]
- 66.Krebs C. J. 2001. Ecology: the experimental analysis of distribution and abundance, 5th edn San Francisco, CA: Benjamin Cummings [Google Scholar]
- 67.Roach D. A., Wulff R. D. 1987. Maternal effects in plants. Annu. Rev. Ecol. Syst. 18, 209–235 10.1146/annurev.es.18.110187.001233 (doi:10.1146/annurev.es.18.110187.001233) [DOI] [Google Scholar]
- 68.Wolfe R. I., Franckowiak J. D. 1991. Multiple dominant and recessive genetic marker stocks in spring barley. Barley Genet. Newsl. 20, 117–121 [Google Scholar]
- 69.Rose F. 1991. The wild flower key—British Isles–N.W. Europe. London, UK: Frederick Warne [Google Scholar]
- 70.Leimu R., Koricheva J. 2006. A meta-analysis of tradeoffs between plant tolerance and resistance to herbivores: combining the evidence from ecological and agricultural studies. Oikos 112, 1–9 10.1111/j.0030-1299.2006.41023.x (doi:10.1111/j.0030-1299.2006.41023.x) [DOI] [Google Scholar]