Abstract
Research in community genetics seeks to understand how the dynamic interplay between ecology and evolution shapes simple and complex communities and ecosystems. A community genetics perspective, however, may not be necessary or informative for all studies and systems. To better understand when and how intraspecific genetic variation and microevolution are important in community and ecosystem ecology, we suggest future research should focus on three areas: (i) determining the relative importance of intraspecific genetic variation compared with other ecological factors in mediating community and ecosystem properties; (ii) understanding the importance of microevolution in shaping ecological dynamics in multi-trophic communities; and (iii) deciphering the phenotypic and associated genetic mechanisms that drive community and ecosystem processes. Here, we identify key areas of research that will increase our understanding of the ecology and evolution of complex communities but that are currently missing in community genetics. We then suggest experiments designed to meet these current gaps.
Keywords: coevolution, community and ecosystem ecology, ecological genomics, extended phenotype, functional genomics, intraspecific genetic variation
1. Introduction
The fields of evolutionary ecology and ecological genetics have long sought to unify ecology and evolution into a single discipline to gain greater insights into the factors that influence the abundance and phenotypic diversity within and among species [1,2]. Historically, this effort focused on individual species and populations, but more recent studies in ‘community genetics’ and related disciplines recognize that it might be necessary to unite the diverse theories, concepts and techniques employed in community ecology, evolutionary biology, genetics and genomics to understand the ecology and evolution of interactions among species within communities [3–7]. Community genetics has lured many biologists with diverse interests with the hope that a successful unification of these seemingly disparate fields will bring novel insight and greater predictive power to address basic and applied problems in biology. Here, we consider the progress towards this goal and existing gaps in our knowledge. We then suggest several specific avenues for future research that we feel will be most important in advancing the field of community genetics.
Community genetics as a discipline was first articulated by Antonovics [3], who called for the formation of a new subdiscipline within evolutionary ecology that would ‘emphasize the analysis of evolutionary genetic processes that occur among interacting populations in communities’. Although similar ideas had been articulated [2,8,9], it was Antonovics' compelling arguments and examples that have lead to the explosion of interest into questions that bridge community ecology and evolutionary biology. Eighteen years after the introduction of the term, there is still confusion as to what community genetics entails. We operationally define community genetics as: the study of the dynamic interplay between ecology and evolution among multiple interacting populations. Thus, community genetics allows us to understand how intraspecific genetic variation, evolution and abiotic and biotic environmental factors influence natural selection, species interactions, community composition and ecosystem processes.
The first generation of studies under the moniker of community genetics largely focused on the role that genetic variation in basal plant populations has for associated arthropod and plant communities (see reviews [10–14]). These studies provide clear support for the hypothesis that intraspecific genetic variation and population-level genotypic diversity can have cascading effects on communities and ecosystems. For instance, genetic differences among individual plants (i.e. the effects of genotype identity) can alter the abundance, composition and diversity of herbivorous and predaceous arthropod species [15–21], the performance and coexistence of competing plant species [22–25] and the flow of energy and nutrients through ecosystems [26–30]. At the patch and population level, increasing genetic diversity (e.g. number of genotypes) increases diversity of associated arthropod communities, confers greater resilience to biotic and abiotic stressors, and affects key aspects of ecosystem function [21,27,28,31–35]. Recently, studies have begun to extend the scope of community genetics research to examine how genetic variation and population divergence within herbivorous insects, endosymbionts [36–38] and predaceous fishes [39–42] have cascading bottom-up and top-down effects within communities. And finally, a related area of research labelled ‘eco-evolutionary dynamics’ has shown that evolution within prey populations can influence predator–prey dynamics within microcosms [43–45].
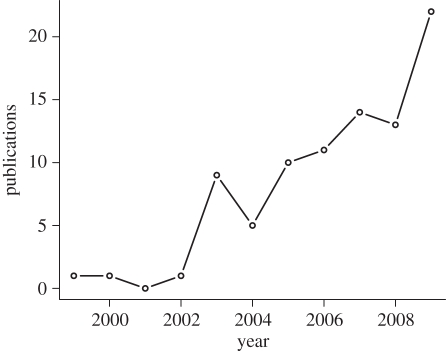
Given the growing interest and rapid advancements on this topic (figure 1), we think it is now important to ask: is a community genetics perspective needed in biology and does it bring novel insight and greater predictive power to answering basic and applied problems in biology? Community genetics research has shown that such an integrative approach could be important in understanding the ecology and evolution of species and communities in nature, but there still remain large gaps in our knowledge. To address these gaps and to assess whether and when a community genetics perspective is helpful, we suggest that future research should address three specific questions:
— What is the relative importance of intraspecific genetic variation compared with other ecological factors in affecting the structure and dynamics of communities?
— How do microevolutionary processes influence community ecology?
— What are the phenotypic and genetic mechanisms affecting community and ecosystem processes?
Figure 1.
The number of publications listed in web of science related to ecology that contain the phrase ‘community genetic’ or ‘eco-evolutionary dynamic’ in the title, abstract or keywords.
2. What is the relative importance of intraspecific genetic variation?
It is currently unclear whether a community genetics framework is needed to explain patterns and processes within community ecology [46,47]. This may seem like an odd statement given recent reviews showing that plant genotype identity and genotypic diversity can explain a large proportion of the variation in diverse aspects of community and ecosystem-level processes [12,14]. These studies provided an important and necessary first step to show that genetic variation can have extended effects beyond an individual's phenotype. However, the most commonly employed method used in these experiments has been to collect multiple genotypes from diverse and often distant environments, and to replicate these genotypes into single common environments where extraneous environmental variation is minimized. Thus, these studies provide little information about the importance of genetic variation and diversity compared with other factors that also influence multi-trophic communities and ecosystem processes. To address this gap, one must manipulate or measure other ecological factors, in addition to intraspecific genetic variation. Then, one could determine the relative importance of each factor and their interactions, in terms of the variance explained and/or the magnitude of effect of different variables for structuring communities and ecosystem processes.
(a). Manipulative factorial experiments
One way to assess the importance of multiple factors is to simultaneously manipulate genotypic identity or diversity and other ecological variables using multi-factorial experiments or multiple common gardens. The small number of studies that have taken this approach show that the relative importance of intraspecific genetic variation and genotype identity ranges along the continuum from being among the most important to being among the least important factors affecting communities [20,27,40,48–51]. Because the number of studies that have manipulated intraspecific genetic variation alongside other ecological factors are small, we cannot generalize about what the relative effects that genetic versus other ecological factors have on mediating community and ecosystem dynamics. It is also becoming clear that the relative importance of genetic variation and genotype identity can depend upon the spatial scale that is studied [20,51,52]. More manipulative experiments are needed to fill this important gap. In meeting this challenge, an added precedence should be placed on manipulating multiple factors and at multiple levels, rather than just their presence/absence, so that results can provide insight over the biologically relevant range of ecological factors and illuminate nonlinear and non-additive effects of multiple factors [53]. Ecological factors that could be manipulated in such experiments include: intensities of intra- and inter-specific interactions, disturbance, trophic complexity, species diversity and abiotic environmental conditions.
In addition to the factors listed above, future multi-factorial experiments should also consider the ecological effects of genetic variation in multiple species. Currently, we do not know how important genetic interactions between species are in structuring natural communities because most community genetics research has only manipulated genotype identity and genotypic diversity of one species. However, interactions between genotypes of multiple species at different trophic levels (i.e. genotype × genotype interactions; G × G) can also shape the ecology of communities [5] and ultimately determine the direction and rate of coevolutionary dynamics [54]. The nature of such G × G interactions can also be influenced by various factors of the environment, thus resulting in genotype × genotype × environment interactions (G × G × E), which has formed the foundation of Thompson's Geographic mosaic of coevolution [54]. Studies that simultaneously manipulate the genetic composition of multiple species and other environmental attributes will be a necessary first step towards understanding the importance of G × G × E interactions on demography and community structure (e.g. [55,56]).
(b). Measuring relative importance: combining approaches
After manipulating or measuring various factors using factorial, common garden or reciprocal transplant designs, the next step would be to tease apart the relative contribution that genetic versus additional ecological factors have on community assembly and ecosystem dynamics. Within a single study, a straightforward way to do this is to measure the relative amount of variance explained by each factor in terms of the coefficient of determination (r2). When using general linear models, r2 can be measured as the ratio of the treatment sums-of-squares (SS) divided by the total SS. A similar approach can be used to calculate r2 when using restricted maximum likelihood methods, which calculate the variance explained by each random effect included in the model. To determine the relative importance of each factor manipulated, it is first necessary to treat all factors (i.e. fixed and random alike) as ‘random’ and to use the variance component for each factor to estimate r2. For example, if aphid genotype and the presence of predators are manipulated to assess their relative effects on plant performance, all factors would be treated as random, and the variance explained by predators would be calculated as: (variance owing to predators)/(total variance), where total variance is the sum of all variance components. One could also measure effect sizes of specific factors using meta-analytical statistics (see [14,21,40]).
Aside from the tremendous amount of work that these types of studies entail, two obvious limitations to these approaches include: (i) deciding what ecological factors are relevant and thus should be manipulated; and (ii) determining how to measure relative effect sizes of complex ecological interactions (such as multiple species interactions). We suggest that combining the experimental approaches described above with structural equation modelling (SEM) could resolve these limitations. SEM is a statistical technique used to estimate relationships among variables and in many ways is similar to multiple regression, path, principal components and factor analyses [57]. However, SEM has some benefits over these other statistical approaches that are particularly relevant here. For instance, with SEM one can test the descriptive ability of different models to determine which factors are most important, and one can simultaneously construct and determine the effect sizes of unmeasured (a.k.a. latent) variables (e.g. species interactions). The use of SEM in ecological and evolutionary field studies is growing, and represents an exciting and important avenue of future community genetics research (i.e. [58–61]).
3. How does microevolution influence community ecology?
The diversification of species and their traits over macroevolutionary timescales influences what species can coexist in an area and the nature of their interactions [62–64]. Evolution over shorter timescales (i.e. microevolution) can also affect ecological processes and patterns within communities, but its role is not well understood [10,42,45,65]. Three general observations suggest that ongoing microevolution could play a prominent role in shaping complex communities: (i) heritable intraspecific variation in many different types of traits can have community and ecosystem-level consequences [12,20,66]; (ii) populations continually evolve and the rate of this evolution is often faster than originally appreciated [67–69]; and (iii) theoretical models and microcosm experiments of simple predator–prey communities show that evolution can affect ecological dynamics and species coexistence [44,70–74]. Despite these advances, it is not yet clear whether such phenomena occur in nature, where communities are infinitely more complex. To better understand the role that microevolution plays in shaping community ecology, we advocate two complementary experimental methods that can be implemented in both field and laboratory settings.
(a). Common environment approaches
A straightforward method to experimentally test the ecological consequences of microevolution involves testing the effect that recent evolutionary divergence within a species has on community or ecosystem properties. This method is most informative when there is a priori knowledge about how and why specific traits have diverged between two or more populations. Using this method, replicate individuals from diverged populations can be placed into a common environment and their impact on a natural or experimentally composed community can be quantified and compared. A weakness of this approach is that the ecological effects observed in a ‘common’ environment may not represent what has or will occur in the natural environment, in part, because coupled evolutionary change in other members of the community is not accounted for and the ‘common’ environment may not represent the natural environment of all diverged populations. Studies replicated in multiple environments, such as the respective environments of each divergent population, could help alleviate this weakness.
This approach has been employed to understand how recently diverged fish populations affect aquatic communities and ecosystems [39–41,75]. For example, Trinidadian guppies rapidly evolve morphological differences in response to variation in predation pressure [76], and mesocosm experiments and observations in native environments show that these diverged phenotypes affect aquatic invertebrates and algae biomass, and have cascading effects on ecosystem properties, such as nutrient flux and decomposition rate [40]. Similar approaches could be used to examine the ecological effects of divergence in plant and arthropod herbivores (e.g. [24]).
Common garden experiments can also be used to examine the ecological effects of evolution within a single population by implementing a ‘resurrection protocol’ [77]. Resurrection protocols can be used for organisms that have dormant stages (i.e. plants and some invertebrate species), and in these experiments large numbers of seeds (or eggs) that represent the population genetic make-up are collected from the same population at multiple time points and then simultaneously ‘revived’ in one or more common environments. In this way, the community effects of evolution within a single population can be assessed.
(b). Experimental evolution and temporal changes in communities
The clearest evidence for the effects of evolution on temporal changes within communities and ecosystems will come from experiments that actually observe or manipulate evolution within populations and measure associated ecological changes. Observational studies that use morphological or molecular techniques to quantify evolution in natural populations and attempt to correlate evolution in one or more species to changes in community ecology are promising [78]; however, teasing apart the role of evolution versus concurrent ecological changes in the focal population (e.g. population size) as well as abiotic changes in the environment can be challenging [68]. Controlled evolution experiments are better able to account for environmental variation, changes in population size and evolution of other species in the community. Using selection experiments, one can manipulate evolution in species that have short generation times by either exposing populations to different abiotic or biotic environments for several generations and allowing evolution by natural selection to proceed, or by artificially selecting for divergent phenotypes [79]. As populations evolve, one can follow changes in associated communities and ecosystem properties, as well as the phenotypic traits and alleles that are correlated with any ecological changes. Experiments in the field can also directly simulate evolution by manually shifting the mean phenotypic trait value in populations over time; in doing so, the ecological effects of a dynamic evolutionary process can be observed. Although each of the approaches described above has various strengths and weaknesses, the implementation of the different approaches in a variety of systems will help to fill the existing gap in our current knowledge about whether evolution drives ecological changes in nature.
4. What are the phenotypic and genetic mechanisms affecting communities and ecosystems?
If genetic variation and evolution frequently affect community and ecosystem-level processes and patterns, then it will be important to identify the key genetic and phenotypic mechanisms responsible for these effects. With this information it will be possible to determine whether certain genes and phenotypic traits have similar consequences in diverse communities and ecosystems, and whether certain traits are more likely to evolve and affect ecological dynamics. In essence, a more mechanistic approach might enable us to make predictions about when a community genetics approach is warranted [5,11,61,80,81].
Progress towards a mechanistic understanding of community and ecosystem genetics can follow one of two paths. Either one can identify phenotypic traits that have significant affects on community and ecosystem processes, and then pinpoint the genetic mechanisms underlying such traits (‘top-down’ approach). Alternatively, one can study gene expression directly to identify candidate genes that show patterns of altered expression associated with ecological effects of interest (‘bottom-up’ approach). There has been considerable progress towards identifying ecologically relevant phenotypic traits, but unravelling genotype to phenotype links is still in its infancy.
(a). Top-down approach: identifying phenotypic traits of interest
A necessary first step towards unravelling genotype to phenotype links using the top-down approach is to identify phenotypic traits that influence community and ecosystem properties. Recent studies have identified many different types of traits that genetically vary and predict variation in community and ecosystem attributes, such as morphological, life history, chemical and physiological traits [18,20,24,25,28,48,82–84]. When phenotypic variation in specific traits is found to have important ecological effects, then one of several genetic approaches can be used to identify the gene(s) underlying these traits.
In the case of cottonwoods, Tom Whitham and colleagues have identified a quantitative trait locus (QTL) that is known to contain candidate genes that contribute to condensed tannin production—a class of secondary chemicals that has been shown to influence several cottonwood community and ecosystem properties [12,13,85]. To complete the genotype to phenotype to community/ecosystem link, the presumable next steps would be to fine map the QTL, identify and clone candidate genes for condensed tannin production, and to identify specific nucleotide polymorphisms within exons and promoter regions that covary with variation in tannin production and the associated community and ecosystem phenotypes. This would provide an elegant link between variation at specific regions of the genome, and community and ecosystem processes.
Using a functional genetics approach, Ian Baldwin and colleagues have also studied the genetic mechanisms that mediate multi-trophic species interactions. After identifying key biosynthetic pathways in plants that mediate interactions between plants and insects, these authors have created genetically modified transgenic lines in which single genes are silenced. These plants are then planted into the field, and their phenotypes and species interactions are compared with those of wild-type plants (i.e. [86–88]). This technique has successfully identified specific genes that mediate species interactions in the field.
These examples show that genomic approaches to community and/or ecosystem genetics are feasible in some systems. These top-down approaches, unfortunately, also have limitations that might limit their utility in non-model organisms. First, QTL analyses are time-consuming and expensive to implement because they require a mapping population with a dense linkage map based on many molecular markers. Even with these resources, it can be difficult to find a statistically significant correlation between the phenotype of interest and genetic variation in one or more narrow chromosomal regions (i.e. locus with a few genes) containing a candidate gene. As such, many QTLs with moderate and minor effects go undetected [89]. Second, all top-down approaches require a priori knowledge of the type of gene or biosynthetic pathway thought to mediate community or ecosystem pathways. Given the complexity of genetic regulation and pathways within organisms [90], many key genes and pathways are likely to be ignored if we rely solely on a priori predictions. Third, functional genetic approaches that target single genes will probably miss pleiotropic effects resulting from interactions with other genes that are also affected by selection and random mutational processes. Finally, all of these approaches rely heavily on genomic tools developed for model organisms; however, most ecologists do not study model organisms or their close relatives.
(b). Bottom-up approach: using next-generation sequencing technologies
Building a genotype to phenotype map has been notoriously difficult [89], but we believe that recent advances in molecular biology provide new opportunities for examining the genetic mechanisms underlying traits that affect biodiversity and ecosystem processes. By combining ultra-high throughput (a.k.a. ‘next generation’) sequencing technologies (454/Roche, Solexa/Illumina, SOLiD/ABI and Helicos/Biosciences) with recent advances in bioinformatics, these approaches are now available for studying sequence variation and transcriptome analysis (i.e. gene expression) in virtually any organism, and often at a much lower cost than traditional capillary-based or Sanger's sequencing and microarray methods [91–93]. Recently, these technologies have been successfully employed to identify genes within non-model organisms that are differentially expressed between species [94,95], between genotypes within species exposed to different herbicide treatments [91] and between individuals from the same genotype exposed to different pathogen environments [95].
Studies that employ these next-generation sequencing technologies can be used to transform our understanding of how genetic variation mediates community and ecosystem processes (‘bottom-up approach’). For example, to understand why certain genotypes with distinct phenotypes influence community and ecosystem processes more than others, these distinct genotypes can be exposed to different ecological environments. Once such experimental designs have been established, RNA can be extracted from genotypes of interest that were exposed to different treatments, and ultra-high sequencing technologies and bioinformatics can be used to identify genes that are differentially expressed between genotypes and between treatments. The function of these genes can then be determined using existing databases (e.g. National Center for Biotechnology Information, The Arabidopsis Information Resource, Gene Ontology) and recently developed software specifically designed for transcriptome data and cross platform analysis [91,96]. Finally, one can simultaneously measure ecologically relevant phenotypic traits and then correlate variation in these ‘traits’ with gene expression profiles, similar to what has been done with QTL and microarray analyses [97]. In this way it is possible to identify candidate genes that are most ecologically significant. Once these candidate genes are identified, ideally one would explicitly test the phenotypic and ecological effects of these genes using functional genomic approaches.
What is particularly appealing about this sequencing and trait-based bottom-up approach is that we do not need a priori knowledge about the types of genes involved in the expression of phenotypes of interest, and it will allow us to identify multiple non-exclusive candidate genes that simultaneously affect species interactions, biodiversity and contribute to the effects of biodiversity on ecosystem functioning. Therefore, these bottom-up approaches might be better suited to embracing the quantitative variation that contributes to most observed heritable variation in nature. Most importantly, this approach can be used in model and non-model organisms alike.
5. Concluding remarks
The first generation of community genetics research has shown that intraspecific genetic variation can be an important factor determining community and ecosystem processes. Genetics and microevolution of populations should also be considered as factors affecting community ecology, and by considering these factors in experimental designs, we will be able to assess their relative importance in structuring communities and ecosystems. A second generation of research is now needed that includes a mechanistic genes-to-ecosystem understanding of natural systems, where the biological importance of the ecological and ecosystem-level consequences of genetic variation and evolution are rigorously assessed in a wide array of ecosystems, including complex tropical systems (e.g. [30]). Broad-scale empirical and theoretical (e.g. [74]) efforts that address the relative importance of inter- and intra-specific effects and variation on community processes should also try to complement these fine-scale genetic mechanistic approaches [61]. As such, this second generation of research will require an interdisciplinary toolkit in model and non-model organisms and will help to build a cohesive and predictive framework between community ecology, evolutionary biology, genetics and genomics. In other words, it will move us towards a modern synthesis of evolutionary ecology.
Acknowledgements
We thank the editors for extending the invitation to submit this article and the input from two anonymous reviewers. This work was supported by grants from the National Science Foundation DEB-0919869 and DEB-0950486 to M.T.J.J.
Footnotes
One contribution of 13 to a Theme Issue ‘Community genetics: at the crossroads of ecology and evolutionary genetics’.
References
- 1.Ford E. B. 1964. Ecological genetics. London, UK: Broadwater Press [Google Scholar]
- 2.Lack D. 1965. Evolutionary ecology. J. Ecol. 53, 237–245 10.2307/2257975 (doi:10.2307/2257975) [DOI] [Google Scholar]
- 3.Antonovics J. 1992. Toward community genetics. In Plant resistance to herbivores and pathogens: ecology, evolution, and genetics (eds Fritz R. S., Simms E. L.), pp. 426–449 Chicago, IL: University of Chicago Press [Google Scholar]
- 4.Agrawal A. A. 2003. Community genetics: new insights into community ecology by integrating population genetics. Ecology 84, 543–544 10.1890/0012-9658(2003)084[0543:CGNIIC]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[0543:CGNIIC]2.0.CO;2) [DOI] [Google Scholar]
- 5.Whitham T. G., Difazio S. P., Schweitzer J. A., Shuster S. M., Allan G. J., Bailey J. K., Woolbright S. A. 2008. Extending genomics to natural communities and ecosystems. Science 320, 492–495 10.1126/science.1153918 (doi:10.1126/science.1153918) [DOI] [PubMed] [Google Scholar]
- 6.Rowntree J. K., Shuker D. M., Preziosi R. F. 2011. Forward from the crossroads of ecology and evolution. Phil. Trans. R. Soc. B 366, 1322–1328 10.1098/rstb.2010.0357 (doi:10.1098/rstb.2010.0357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade M. J. 2007. The co-evolutionary genetics of ecological communities. Nat. Rev. Genet. 8, 185–195 10.1038/nrg2031 (doi:10.1038/nrg2031) [DOI] [PubMed] [Google Scholar]
- 8.Pimentel D. 1961. Animal population regulation by the genetic feed-back mechanism. Am. Nat. 95, 65–79 10.1086/282160 (doi:10.1086/282160) [DOI] [Google Scholar]
- 9.Price P. 1983. Hypotheses on organization and evolution in herbivorous insect communities. In Variable plants and herbivores in natural and managed systems (eds Denno R. F., McClure M. S.), pp. 559–595 New York, NY: Academic Press [Google Scholar]
- 10.Johnson M. T. J., Stinchcombe J. R. 2007. An emerging synthesis between community ecology and evolutionary biology. Trends Ecol. Evol. 22, 250–257 10.1016/j.tree.2007.01.014 (doi:10.1016/j.tree.2007.01.014) [DOI] [PubMed] [Google Scholar]
- 11.Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623 10.1111/j.1461-0248.2008.01179.x (doi:10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 12.Whitham T. G., et al. 2006. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523 10.1038/nrg1877 (doi:10.1038/nrg1877) [DOI] [PubMed] [Google Scholar]
- 13.Whitham T. G., et al. 2003. Community and ecosystem genetics: a consequence of the extended phenotype. Ecology 84, 559–573 10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2) [DOI] [Google Scholar]
- 14.Bailey J. K., et al. 2009. From genes to ecosystems: synthesizing the effects of plant genetic factors across systems. Phil. Trans. R. Soc. B 364, 1607–1616 10.1098/rstb.2008.0336 (doi:10.1098/rstb.2008.0336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stiling P., Rossi A. M. 1996. Complex effects of genotype and environment on insect herbivores and their enemies. Ecology 77, 2212–2218 10.2307/2265714 (doi:10.2307/2265714) [DOI] [Google Scholar]
- 16.Maddox G. D., Root R. B. 1987. Resistance to 16 diverse species of herbivorous insects within a population of goldenrod, Solidago altissima: genetic variation and heritability. Oecologia 72, 8–14 10.1007/BF00385037 (doi:10.1007/BF00385037) [DOI] [PubMed] [Google Scholar]
- 17.Fritz R. S., Price P. W. 1988. Genetic variation among plants and insect community structure: willows and sawflies. Ecology 69, 845–856 10.2307/1941034 (doi:10.2307/1941034) [DOI] [Google Scholar]
- 18.Dungey H. S., Potts B. M., Whitham T. G., Li H. F. 2000. Plant genetics affects arthropod community richness and composition: evidence from a synthetic eucalypt hybrid population. Evolution 54, 1938–1946 [DOI] [PubMed] [Google Scholar]
- 19.Wimp G. M., Young W. P., Woolbright S. A., Martinsen G. D., Keim P., Whitham T. G. 2004. Conserving plant genetic diversity for dependent animal communities. Ecol. Lett. 7, 776–780 10.1111/j.1461-0248.2004.00635.x (doi:10.1111/j.1461-0248.2004.00635.x) [DOI] [Google Scholar]
- 20.Johnson M. T. J., Agrawal A. A. 2005. Plant genotype and the environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86, 874–885 10.1890/04-1068 (doi:10.1890/04-1068) [DOI] [Google Scholar]
- 21.Crutsinger G., Collins M., Fordyce J., Gompert Z., Nice C., Sanders N. 2006. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313, 966–968 10.1126/science.1128326 (doi:10.1126/science.1128326) [DOI] [PubMed] [Google Scholar]
- 22.Proffitt C. E., Chiasson R. L., Owens A. B., Edwards K. R., Travis S. E. 2005. Spartina alterniflora genotype influences facilitation and suppression of high marsh species colonizing an early successional salt marsh. J. Ecol. 93, 404–416 10.1111/j.0022-0477.2005.00983.x (doi:10.1111/j.0022-0477.2005.00983.x) [DOI] [Google Scholar]
- 23.Iason G. R., Lennon J. J., Pakeman R. J., Thoss V., Beaton J. K., Sim D. A., Elston D. A. 2005. Does chemical composition of individual Scots pine tree determine the biodiversity of their associated ground vegetation? Ecol. Lett. 8, 364–369 10.1111/j.1461-0248.2005.00732.x (doi:10.1111/j.1461-0248.2005.00732.x) [DOI] [Google Scholar]
- 24.Crutsinger G., Strauss S. Y., Rudgers J. A. 2010. Genetic variation within a dominant shrub species determines plant species colonization in a coastal dune ecosytem. Ecology 91, 1237–1243 10.1890/09-0613.1 (doi:10.1890/09-0613.1) [DOI] [PubMed] [Google Scholar]
- 25.Lankau R. A., Strauss S. Y. 2007. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science 307, 1561–1563 10.1126/science.1147455 (doi:10.1126/science.1147455) [DOI] [PubMed] [Google Scholar]
- 26.Madritch M. D., Greene S. L., Lindroth R. L. 2009. Genetic mosaics of ecosystem functioning across aspen-dominated landscapes. Oecologia 160, 119–127 10.1007/s00442-009-1283-3 (doi:10.1007/s00442-009-1283-3) [DOI] [PubMed] [Google Scholar]
- 27.Madritch M., Donaldson J. R., Lindroth R. L. 2006. Genetic identity of Populus tremuloides litter influences decomposition and nutrient release in a mixed forest stand. Ecosystems 9, 528–537 10.1007/s10021-006-0008-2 (doi:10.1007/s10021-006-0008-2) [DOI] [Google Scholar]
- 28.Schweitzer J. A., Bailey J. K., Hart S. C., Whitham T. G. 2005. Nonadditive effects of mixing cottonwood genotypes on litter decomposition and nutrient dynamics. Ecology 86, 2834–2840 10.1890/04-1955 (doi:10.1890/04-1955) [DOI] [Google Scholar]
- 29.Schweitzer J. A., Bailey J. K., Fischer D. G., Leroy C. J., Lonsdorf E. V., Whitham T. G., Hart S. C. 2008. Soil microorganism–plant interactions: a heritable relationship between plant genotype and associated soil microorganisms. Ecology 89, 773–781 10.1890/07-0337.1 (doi:10.1890/07-0337.1) [DOI] [PubMed] [Google Scholar]
- 30.Zytynska S. E., Fay M. F., Penney D., Preziosi R. F. 2011. Genetic variation in a tropical tree species influences the associated epiphytic plant and invertebrate communities in a complex forest ecosystem. Phil. Trans. R. Soc. B 366, 1329–1336 10.1098/rstb.2010.0183 (doi:10.1098/rstb.2010.0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson M. T. J., Lajeunesse M. J., Agrawal A. A. 2006. Additive and interactive effects of plant genotypic diversity on arthropod communities and plant fitness. Ecol. Lett. 9, 24–34 [DOI] [PubMed] [Google Scholar]
- 32.Crutsinger G., Souza L., Sanders N. 2008. Intraspecific diversity and dominant genotypes resist plant invasions. Ecol. Lett. 11, 16–23 10.1111/j.1461-0248.2007.01118.x (doi:10.1111/j.1461-0248.2007.01118.x) [DOI] [PubMed] [Google Scholar]
- 33.Reusch T. B. H., Ehlers A., Hammerli A., Worm B. 2005. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl Acad. Sci. USA 102, 2826–2831 10.1073/pnas.0500008102 (doi:10.1073/pnas.0500008102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes A. R., Stachowicz J. J. 2004. Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl Acad. Sci. USA 101, 8998–9002 10.1073/pnas.0402642101 (doi:10.1073/pnas.0402642101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford K. M., Whitney K. D. 2010. Population genetic diversity influences colonization success. Mol. Ecol. 19, 1253–1263 10.1111/j.1365-294X.2010.04550.x (doi:10.1111/j.1365-294X.2010.04550.x) [DOI] [PubMed] [Google Scholar]
- 36.Ferrari J. C., Muller B., Kraaijeveld A. R., Godfray H. C. J. 2001. Clonal variation and covariation in aphid resistance to parasitoids and a pathogen. Evolution 55, 1805–1814 [DOI] [PubMed] [Google Scholar]
- 37.Hazell S. P., Fellowes M. D. E. 2009. Intra-specific variation affects the structure of the natural enemy assemblage attacking pea aphid colonies. Ecol. Entomol. 34, 34–42 10.1111/j.1365-2311.2008.01051.x (doi:10.1111/j.1365-2311.2008.01051.x) [DOI] [Google Scholar]
- 38.Ferrari J., Vavre F. 2011. Bacterial symbionts in insects or the story of communities affecting communities. Phil. Trans. R. Soc. B 366, 1389–1400 10.1098/rstb.2010.0226 (doi:10.1098/rstb.2010.0226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harmon L. J., Matthews B., Des Roches S., Chase J. M., Shurin J. B., Schluter D. 2009. Evolutionary diversification in stickleback affects ecosystem functioning. Nature 458, 1167–1170 10.1038/nature07974 (doi:10.1038/nature07974) [DOI] [PubMed] [Google Scholar]
- 40.Bassar R. D., et al. 2010. Local adaptation in Trinidadian guppies alters ecosystem processes. Proc. Natl Acad. Sci. USA 107, 3616–3621 10.1073/pnas.0908023107 (doi:10.1073/pnas.0908023107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palkovacs E. P., Marshall M. C., Lamphere B. A., Lynch B. R., Weese D. J., Fraser D. F., Reznick D. N., Pringle C. M., Kinnison M. T. 2009. Experimental evaluation of evolution and coevolution as agents of ecosystem change in Trinidadian streams. Phil. Trans. R. Soc. B 364, 1617–1628 10.1098/rstb.2009.0016 (doi:10.1098/rstb.2009.0016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Post D. M., Palkovacs E. P. 2009. Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Phil. Trans. R. Soc. B 364, 1629–1640 10.1098/rstb.2009.0012 (doi:10.1098/rstb.2009.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida T., Ellner S. P., Jones L. E., Bohannan B. J. M., Lenski R. E., Hairston N. G. 2007. Cryptic population dynamics: rapid evolution masks trophic interactions. PLoS Biol. 5, 1868–1879 10.1371/journal.pbio.0050235 (doi:10.1371/journal.pbio.0050235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fussmann G. F., Loreau M., Abrams P. A. 2007. Eco-evolutionary dynamics of communities and ecosystems. Funct. Ecol. 21, 465–477 10.1111/j.1365-2435.2007.01275.x (doi:10.1111/j.1365-2435.2007.01275.x) [DOI] [Google Scholar]
- 45.Pelletier F., Garant D., Hendry A. P. 2009. Eco-evolutionary dynamics. Phil. Trans. R. Soc. B 364, 1483–1489 10.1098/rstb.2009.0027 (doi:10.1098/rstb.2009.0027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morin P. J. 2003. Community ecology and the genetics of interacting species. Ecology 84, 577–580 10.1890/0012-9658(2003)084[0577:CEATGO]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[0577:CEATGO]2.0.CO;2) [DOI] [Google Scholar]
- 47.Ricklefs R. C. 2003. Genetics, evolution, and ecological communities. Ecology 84, 588–591 10.1890/0012-9658(2003)084[0588:GEAEC]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[0588:GEAEC]2.0.CO;2) [DOI] [Google Scholar]
- 48.Johnson M. T. J. 2008. Bottom-up effects of plant genotype on aphids, ants and predators. Ecology 89, 145–154 10.1890/07-0395.1 (doi:10.1890/07-0395.1) [DOI] [PubMed] [Google Scholar]
- 49.Stiling P., Rossi A. M. 1995. Coastal insect herbivore communities are affected more by local environmental conditions than by plant genotype. Ecol. Entomol. 20, 184–190 10.1111/j.1365-2311.1995.tb00444.x (doi:10.1111/j.1365-2311.1995.tb00444.x) [DOI] [Google Scholar]
- 50.Johnson M. T. J., Dinnage R., Zhou A. Y., Hunter M. D. 2008. Environmental variation has stronger effects than plant genotype on competition among plant species. J. Ecol. 96, 947–955 10.1111/j.1365-2745.2008.01410.x (doi:10.1111/j.1365-2745.2008.01410.x) [DOI] [Google Scholar]
- 51.Tack A. J. M., Ovaskainen O., Pulkkinen P., Roslin T. 2010. Spatial location dominates over host plant genotype in structuring an herbivore community. Ecology 91, 2660–2672 10.1890/09-1027.1 (doi:10.1890/09-1027.1) [DOI] [PubMed] [Google Scholar]
- 52.Bangert R. K., Lonsdorf E. V., Wimp G. M., Shuster S. M., Fischer D. G., Schweitzer J. A., Allan G. J., Bailey J. K., Whitham T. G. 2008. Genetic structure of a foundation species: scaling community phenotypes from the individual to the region. Heredity 100, 121–131 10.1038/sj.hdy.6800914 (doi:10.1038/sj.hdy.6800914) [DOI] [PubMed] [Google Scholar]
- 53.Abrams P. A. 2001. Describing and quantifying interspecific interactions: a commentary on recent approaches. Oikos 94, 201–218 10.1034/j.1600-0706.2001.940201.x (doi:10.1034/j.1600-0706.2001.940201.x) [DOI] [Google Scholar]
- 54.Thompson J. T. 2005. The geographic mosaic of coevolution. Chicago, IL: University of Chicago Press [Google Scholar]
- 55.Tétard-Jones C., Kertesz M. A., Gallois P., Preziosi R. F. 2007. Genotype-by-genotype interactions modified by a third species in a plant–insect system. Am. Nat. 170, 492–499 10.1086/520115 (doi:10.1086/520115) [DOI] [PubMed] [Google Scholar]
- 56.Zytynska S. E., Fleming S., Tétard-Jones C., Kertesz M. A., Preziosi R. F. 2010. Community genetic interactions mediate indirect ecological effects between a parasitoid wasp and rhizobacteria. Ecology 91, 1563–1568 10.1890/09-2070.1 (doi:10.1890/09-2070.1) [DOI] [PubMed] [Google Scholar]
- 57.Grace J. B. 2006. Structural equation modelling and natural systems. Cambridge, UK: Cambridge University Press [Google Scholar]
- 58.Mitchell R. J. 1992. Testing evolutionary and ecological hypotheses using path analysis and structural equation modelling. Funct. Ecol. 6, 123–129 10.2307/2389745 (doi:10.2307/2389745) [DOI] [Google Scholar]
- 59.Cronin J. P., Tonsor S. J., Carson W. P. 2010. A simultaneous test of trophic interaction models: which vegetation characteristic explains herbivore control over plant community mass? Ecol. Lett. 13, 202–212 10.1111/j.1461-0248.2009.01420.x (doi:10.1111/j.1461-0248.2009.01420.x) [DOI] [PubMed] [Google Scholar]
- 60.Sutton-Griera A. E., Kenney M. A., Richardson C. J. 2010. Examining the relationship between ecosystem structure and function using structural equation modelling: a case study examining denitrification potential in restored wetland soils. Ecol. Model. 221, 761–768 10.1016/j.ecolmodel.2009.11.015 (doi:10.1016/j.ecolmodel.2009.11.015) [DOI] [Google Scholar]
- 61.Wolf J. B., Mutic J. J., Kover P. X. 2011. Functional genetics of intraspecific ecological interactions in Arabidopsis thaliana. Phil. Trans. R. Soc. B 366, 1358–1367 10.1098/rstb.2010.0239 (doi:10.1098/rstb.2010.0239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vellend M. 2010. Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206 10.1086/652373 (doi:10.1086/652373) [DOI] [PubMed] [Google Scholar]
- 63.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 10.1111/j.1461-0248.2009.01314.x (doi:10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 64.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 10.1146/annurev.ecolsys.33.010802.150448 (doi:10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 65.Thompson J. N. 2009. Which ecologically important traits are most likely to evolve rapidly? Oikos 118, 1281–1283 10.1111/j.1600-0706.2009.17835.x (doi:10.1111/j.1600-0706.2009.17835.x) [DOI] [Google Scholar]
- 66.Shuster S. M., Lonsdorf E. V., Wimp G. M., Bailey J. K., Whitham T. G. 2006. Community heritability measures the evolutionary consequences of indirect genetic effects on community structure. Evolution 60, 991–1003 [PubMed] [Google Scholar]
- 67.Thompson J. N. 1998. Rapid evolution as an ecological process. Trends Ecol. Evol. 13, 329–332 10.1016/S0169-5347(98)01378-0 (doi:10.1016/S0169-5347(98)01378-0) [DOI] [PubMed] [Google Scholar]
- 68.Hairston N. G., Ellner S. P., Geber M. A., Yoshida T., Fox J. A. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127 10.1111/j.1461-0248.2005.00812.x (doi:10.1111/j.1461-0248.2005.00812.x) [DOI] [Google Scholar]
- 69.Kinnison M. T., Hendry A. P. 2001. The pace of modern life. II. From rates of contemporary microevolution to pattern and process. Genetica 112, 145–164 10.1023/A:1013375419520 (doi:10.1023/A:1013375419520) [DOI] [PubMed] [Google Scholar]
- 70.Abrams P. A., Matsuda H. 1997. Prey adaptation as a cause of predator–prey cycles. Evolution 51, 1742–1750 10.2307/2410997 (doi:10.2307/2410997) [DOI] [PubMed] [Google Scholar]
- 71.Bohannan B. J. M., Lenski R. E. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377 10.1046/j.1461-0248.2000.00161.x (doi:10.1046/j.1461-0248.2000.00161.x) [DOI] [Google Scholar]
- 72.Yoshida T., Jones L. E., Ellner S. P., Fussmann G. F., Hairston N. G. 2003. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306 10.1038/nature01767 (doi:10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 73.Meyer J. R., Kassen R. 2007. The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435 10.1038/nature05599 (doi:10.1038/nature05599) [DOI] [PubMed] [Google Scholar]
- 74.Moya-Laraño J. 2011. Genetic variation, predator–prey interactions and food web structure. Phil. Trans. R. Soc. B 366, 1425–1437 10.1098/rstb.2010.0241 (doi:10.1098/rstb.2010.0241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Post D. M., Palkovacs E. P., Schielke E. G., Dodson S. I. 2008. Intraspecific variation in a predator affects community structure and cascading trophic interactions. Ecology 89, 2019–2032 10.1890/07-1216.1 (doi:10.1890/07-1216.1) [DOI] [PubMed] [Google Scholar]
- 76.Reznick D. N., Shaw F. H., Rodd F. H., Shaw R. G. 1997. Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata). Science 275, 1934–1937 10.1126/science.275.5308.1934 (doi:10.1126/science.275.5308.1934) [DOI] [PubMed] [Google Scholar]
- 77.Franks S. J., Avise J. C., Bradshaw W. E., Conner J. K., Etterson J. R., Mazer S. J., Shaw R. G., Weis A. E. 2008. The resurrection initiative: storing ancestral genotypes to capture evolution in action. Bioscience 58, 870–873 10.1641/B580913 (doi:10.1641/B580913) [DOI] [Google Scholar]
- 78.Grant P. R., Grant B. R. 2006. Evolution of character displacement in Darwin's finches. Science 313, 224–226 10.1126/science.1128374 (doi:10.1126/science.1128374) [DOI] [PubMed] [Google Scholar]
- 79.Conner J. K. 2003. Artificial selection: a powerful tool for ecologists. Ecology 84, 1650–1660 10.1890/0012-9658(2003)084[1650:ASAPTF]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[1650:ASAPTF]2.0.CO;2) [DOI] [Google Scholar]
- 80.McGill B., Enquist B., Weiher E., Westoby M. 2006. Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185 10.1016/j.tree.2006.02.002 (doi:10.1016/j.tree.2006.02.002) [DOI] [PubMed] [Google Scholar]
- 81.Dalziel A. C., Rogers S. M., Schulte P. M. 2009. Linking genotypes to phenotypes to fitness: how mechanistic biology can inform molecular ecology. Mol. Ecol. 18, 4997–5017 10.1111/j.1365-294X.2009.04427.x (doi:10.1111/j.1365-294X.2009.04427.x) [DOI] [PubMed] [Google Scholar]
- 82.Agrawal A. A. 2005. Natural selection on common milkweed (Asclepias syriaca) by a community of specialized insect herbivores. Evol. Ecol. Res. 7, 651–667 [Google Scholar]
- 83.Wimp G. M., Wooley S., Bangert R. K., Young W. P., Martinsen G. D., Keim P., Rehill B., Lindroth R. L., Whitham T. G. 2007. Plant genetics predicts intra-annual variation in phytochemistry and arthropod community structure. Mol. Ecol. 16, 5057–5069 10.1111/j.1365-294X.2007.03544.x (doi:10.1111/j.1365-294X.2007.03544.x) [DOI] [PubMed] [Google Scholar]
- 84.Barbour R. C., Forster L. G., Baker S. C., Steane D. A., Potts B. M. 2009. Biodiversity consequences of genetic variation in bark characteristics within a foundation tree species. Conser. Biol. 23, 1146–1155 10.1111/j.1523-1739.2009.01247.x (doi:10.1111/j.1523-1739.2009.01247.x) [DOI] [PubMed] [Google Scholar]
- 85.Woolbright S. A., Difazio S. P., Yin T., Martinsen G. D., Zhang X., Allan G. J., Whitham T. G., Keim P. 2008. A dense linkage map of hybrid cottonwood (Populus fremontii × P. angustifolia) contributes to long-term ecological research and comparison mapping in a model forest tree. Heredity 100, 59–70 10.1038/sj.hdy.6801063 (doi:10.1038/sj.hdy.6801063) [DOI] [PubMed] [Google Scholar]
- 86.Kessler D., Gase K., Baldwin I. T. 2008. Field experiments with transformed plants reveal the sense of floral scents. Science 321, 1200–1202 10.1126/science.1160072 (doi:10.1126/science.1160072) [DOI] [PubMed] [Google Scholar]
- 87.Kessler A., Halitschke R., Baldwin I. T. 2004. Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305, 665–668 10.1126/science.1096931 (doi:10.1126/science.1096931) [DOI] [PubMed] [Google Scholar]
- 88.Meldau S., Wu J., Baldwin I. T. 2009. Silencing two herbivory-activated MAP kinases, SIPK and WIPK, does not increase Nicotiana attenuata's susceptibility to herbivores in the glasshouse and in nature. New Phytol. 181, 161–173 10.1111/j.1469-8137.2008.02645.x (doi:10.1111/j.1469-8137.2008.02645.x) [DOI] [PubMed] [Google Scholar]
- 89.Mackay T., Stone E., Ayroles J. 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10, 565–577 10.1038/nrg2612 (doi:10.1038/nrg2612) [DOI] [PubMed] [Google Scholar]
- 90.Ecker J. R., Cook D. 2004. Genome studies and molecular genetics: unwrapping new layers of complexity in plant genomes. Curr. Opin. Plant Biol. 7, 99–101 10.1016/j.pbi.2004.01.017 (doi:10.1016/j.pbi.2004.01.017) [DOI] [Google Scholar]
- 91.Zhou X., Su Z., Sammons R., Peng Y., Tranel P. J., Stewart C. N., Yuan J. S. 2009. Novel software package for cross-platform transcriptome analysis (CPTRA). BMC Bioinform. 10(Suppl. 11), S16. 10.1186/1471-2105-10-S11-S16 (doi:10.1186/1471-2105-10-S11-S16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morozova O., Marra M. 2008. Applications of next-generation sequencing technologies in functional genomics. Genomics 92, 255–264 10.1016/j.ygeno.2008.07.001 (doi:10.1016/j.ygeno.2008.07.001) [DOI] [PubMed] [Google Scholar]
- 93.Cannon C., Kua C.-S., Zhang D., Harting J. 2010. Assembly free comparative genomics of short-read sequence data discovers the needles in the haystack. Mol. Ecol. 19(Suppl. 1), 147–161 10.1111/j.1365-294X.2009.04484.x (doi:10.1111/j.1365-294X.2009.04484.x) [DOI] [PubMed] [Google Scholar]
- 94.Toth A. L., et al. 2007. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 318, 441–444 10.1126/science.1146647 (doi:10.1126/science.1146647) [DOI] [PubMed] [Google Scholar]
- 95.Barakat A., Diloreto D. S., Zhang Y., Smith C., Baier K., Powell W. A., Wheeler N., Sederoff R., Carlson J. E. 2009. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 9, 1–11 10.1186/1471-2229-9-1 (doi:10.1186/1471-2229-9-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barker M. S., Dlugosch K. M., Reddy A. C. C., Amyotte S. N., Rieseberg L. H. 2009. SCARF: maximizing next-generation EST assemblies for evolutionary and population genomic analyses. Bioinformatics 25, 535–536 10.1093/bioinformatics/btp011 (doi:10.1093/bioinformatics/btp011) [DOI] [PubMed] [Google Scholar]
- 97.Ayroles J. F., et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41, 299–307 10.1038/ng.332 (doi:10.1038/ng.332) [DOI] [PMC free article] [PubMed] [Google Scholar]