Abstract
Biosynthesis of the molybdenum cofactor in bacteria is described with a detailed analysis of each individual reaction leading to the formation of stable intermediates during the synthesis of molybdopterin from GTP. As a starting point, the discovery of molybdopterin and the elucidation of its structure through the study of stable degradation products are described. Subsequent to molybdopterin synthesis, the molybdenum atom is added to the molybdopterin dithiolene group to form the molybdenum cofactor. This cofactor is either inserted directly into specific molybdoenzymes or is further modified by the addition of nucleotides to the molybdopterin phosphate group or the replacement of ligands at the molybdenum center.
Keywords: molybdenum cofactor, molybdopterin, Precursor Z, molybdopterin guanine dinucleotide cofactor, urothione, dithiolene group
1. INTRODUCTION
The importance of molybdenum to living organisms was first reported in 1932 with the publication of studies suggesting that molybdenum was essential for plant growth and that the metal also plays a functional role in several bacterial, plant and animal enzymes [1]. The severity of the pathology associated with sulfite oxidase deficiency in humans was first described in 1967 [2], and subsequent characterization of sulfite oxidase as a molybdenum-containing protein in 1971 established the essentiality of molybdenum for normal human development [3]. These findings were followed by studies on the nature of the molybdenum cofactor (Moco) [4, 5], the metal-containing prosthetic group common to all molybdoenzymes with the exception of nitrogenase [6–8]. Structural studies of Moco associated with liver sulfite oxidase revealed the presence of a unique pterin component that was termed molybdopterin (MPT) [9, 10]. Later, several MPT-dinucleotide variants of Moco were identified in bacterial molybdoenzymes, including the molybdopterin guanine dinucleotide (MGD) form present in the majority of Escherichia coli molybdoenzymes [11].
A structure for MPT was first proposed in 1982 which revealed a reduced pterin with an unusual 6-alkyl side chain consisting of four carbons, a terminal phosphate ester and a unique dithiolene group critical for metal ligation [5]. The structural complexity of the MPT moiety coordinating the molybdenum atom suggested that Moco assembly was likely to be a complex process involving a number of enzymes catalyzing diverse chemical reactions [12]. Much of the early progress in understanding Moco biosynthesis was achieved through studies of E. coli strains containing lesions at various steps in the pathway [13, 14]. These studies were later amplified through purification and crystallization of its target enzymes and the proteins involved in the biosynthesis of Moco [6–8, 15]. This article summarizes the discovery of Moco and the elucidation of its structure as well as the dissection of the individual steps of Moco biosynthesis in E. coli.
2. THE DISCOVERY OF MOCO
The first evidence for the existence of a common Moco was obtained in the course of genetic studies characterizing gene mutations affecting nitrate reductase activity in Aspergillus nidulans [16]. Subsequent investigation of the expression of xanthine dehydrogenase (XDH) and nitrate reductase (NR) in A. nidulans led to the discovery of pleiotropic mutants which produced inactive forms of both enzymes [17]. These cnx strains (cofactor for nitrate reductase and xanthine dehydrogenase) were assumed to carry mutations in genes involved in the biosynthesis of a cofactor common to XDH and NR [17] which was distinct from the nitrogenase iron molybdenum cofactor [18]. Detailed studies were also performed with E. coli mutations leading to chlorate (chl) resistence, a property initially associated with the lack of nitrate reduction. Phenotypic characterization and genetic mapping of these chl mutants led to the identification of the genes for molybdenum cofactor biosynthesis and nitrate reductase [19]. The initial chl nomenclature was later changed to moa-mog for genes involved in molybdenum cofactor biosynthesis and nar for nitrate reductase genes [12, 19–22]. Subsequent to these germinal studies, similar pleiotropic mutant strains were also reported in Pseudomonas aeruginosa [23], Klebsiella aerogenes [24], Drosophila melanogaster [25, 26] and Nicotiana tabacum [27]. Additional studies by Nason and coworkers [28–31] identified a specific pleiotropic Neurospora crassa mutant termed nit-1, which was incapable of growth on nitrate or xanthine. In vitro studies using nit-1 extracts showed that the addition of denatured preparations of a variety of molybdoenzymes from different sources resulted in the reconstitution of nitrate reductase activity [29]. The totality of these studies lead to the understanding that with the exception of nitrogenase, Moco is the common element in all molybdoenzymes from different organisms.
2.1 Structural studies on Moco derivatives
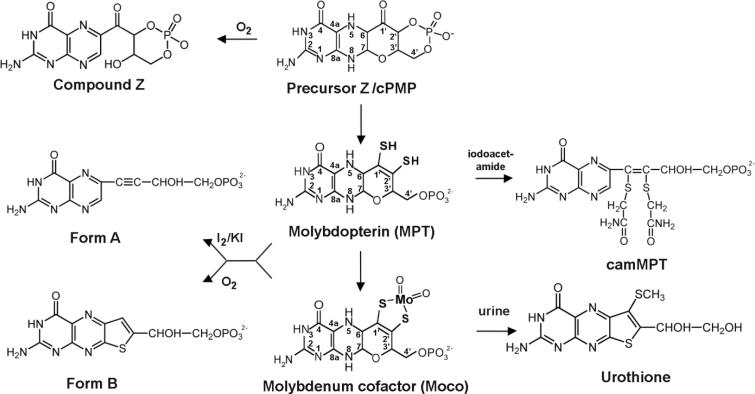
In addition to demonstrating the universality of Moco, the nit-1 extract assays also demonstrated that Moco is very labile with a lifetime of only a few minutes after release from molybdoenzymes [29, 32, 33], making chemical characterization of active Moco difficult [4]. Therefore, structural characterization of Moco was achieved through the analysis of stable degradation products [34]. For these studies, chicken liver sulfite oxidase was used as a Moco source, since this enzyme was easy to purify in relatively large quantities [35]. Using this source it was observed that oxidation of Moco under various acidic conditions generated two distinct fluorescent pterin derivatives (Form A and Form B) which could be used for structural determination studies of Moco [34, 36]. Form A is generated by iodine oxidization of Moco under acidic conditions, while Form B is generated by air-oxidization of Moco [32]. These two derivatives can be distinguished from each other by differences between the absorption and fluorescence spectra of their dephospho analogs [34]. For purification and characterization of the derivatives, HPLC in conjunction with sensitive detection methods provided separation for analytical quantities of samples. Additionally, since it was not possible to purify sufficient quantities of these Moco derivatives to permit elemental analysis, analytical methods such as energy dispersive analysis of X-rays (EDAX) were used for the quantification of sulfur and phosphorus in combination with NMR studies [9].
2.1.1 The structure of Form A
The initial clue to the chemical nature of Form A was provided by the finding that its absorption spectrum showed a red shift of 20 nm compared to the spectra of pterins with side chains containing saturated carbons [4, 36, 37]. This indicated that the pterin ring conjunction extended into the side chain, either via a carbonyl group on C1' or an unsaturated bond between C1' and C2'. Phosphate analysis of Form A revealed a single phosphate per pterin, and this phosphate could be removed with alkaline phosphatase. Periodate treatment of Form A produced pterin and formaldehyde, suggesting that the two terminal carbons of the side chain of Form A were present as a glycol function with one of the hydroxyls being esterified to a phosphate. In contrast to pterins with saturated side chains which yield pterin-6-carboxylic acid only after prolonged heating, Form A generated pterin-6-carboxylic acid even with cold alkaline permanganate [9]. Additionally, neither Form A nor dephospho Form A generated a hydazone when treated with 2,4-dinitrophenylhydrazine, indicating that the red shift observed in the absorption spectrum of Form A is not due to the presence of a carbonyl group on the C1' of the side chain but was caused by the presence of an unsaturated bond between C1' and C2' [9]. Mass spectral analysis of silylated dephospho Form A provided definitive evidence for the presence of four carbons in the 6-alkyl side chain [9], and the mass spectral data were in accordance with a basic molecular formula of C10H9O3N5 (MW = 247) for underivatized dephospho Form A [34]. From the NMR spectrum of Form A, it was determined that the molecule contained the oxidized form of the pterin ring. Based on these analyses, the structure shown in Fig. 1 was proposed for Form A [37]. While elucidation of the structure of Form A provided the basis for further Moco structural studies, it shed little light on those aspects of Moco which were important for its presumed interaction with molybdenum, since it seemed unlikely that either the oxidized pterin ring of Form A or its side chain would provide suitable ligands for molybdenum ligation. Thus, it was apparent that structural characterization of additional cofactor derivatives would be needed before a plausible structure for Moco itself could be inferred. The fact that Form B, the air oxidation derivative of Moco could be also obtained as a pure compound made it the material of choice for further studies [32].
Figure 1. Structures of Precursor Z, MPT, Moco and their derivatives.
Shown are the structures of Precursor Z, molybdopterin (MPT) and the molybdenum cofactor (Moco). Precursor Z is a cyclic pyranopterin monophosphate (cPMP) and Compound Z, its air-oxidized product, differs only in the reduction state of the pterin ring. MPT and Moco are both 5,6,7,8-tetrahydropyranopterins. A unique dithiolene group coordinates the molybdenum atom (in Moco), which is shown in its dioxo coordination. Form B is the air oxidized degradation product of MPT and Moco, while Form A is the iodine oxidized derivative. Form A and Form B are shown in their phosphorylated forms. Urothione is the degradation product of Moco in humans, which is excreted in the urine.
2.1.2 The structure of Form B
Investigations of Form B showed that many of its properties were similar to Form A [34]. Form B also contained an alkaline phosphatase-sensitive phosphate ester linkage and it was only sensitive to periodate oxidation after dephosphorylation. Surprisingly though, no pterin-6-carboxylic acid was formed after alkaline permanganate oxidation of Form B. Analysis of permanganate-treated Form B by thin-layer cellulose plate chromatography revealed two fluorescent products with the minor product possibly corresponding to a derivative of isoxanthopterin (7-oxopterin) with an anionic substituent [9]. Since previous EXAFS studies had provided evidence for ligation of molybdenum to at least two thiolate ligands in various molybdoenzymes, it was conceivable that the Moco itself might be the donor of one or more of these sulfur ligands [9]. The fact that the green-fluorescent acidic product of permanganate oxidation of Form B contained an anionic group with a pK lower than that of a carboxyl group suggested that it could be a sulfonate group arising from a sulfur atom of Form B [9].
In 1940, Koschara reported the discovery of a non-fluorescent sulfur-containing pterin named urothione in a study involving chromatographic analysis of components in human urine [38]. Thirty years later, a more detailed structural analysis by Goto et al. [39] determined that urothione had the structure shown in Fig. 1. To explore the possibility of structural similarities between Form B and urothione, urothione was oxidized with alkaline permanganate to yield the acidic, green-fluorescing compound pterin-6-carboxylic-7-sulfonate [5]. Heating this with 4 M HCl led to loss of the 6-carboxylic group and replacement of the 7-sulfonate function with an -OH yielding isoxanthopterin. Heating of the sulfonate compound in NaOH led to the formation of isoxanthopterin-6-carboxylate. Further analysis indicated that alkaline permanganate treatment of Form B yielded the same sulfonate generated by urothione and that this sulfonate had been partially degraded to yield isoxanthopterin-6-carboxylate [5].
The presence of sulfur in Form B was verified when EDAX analysis of the molecule yielded a P/S ratio of 1:1 [9]. NMR analysis found no evidence of the urothione −SCH3 substituent in Form B, and mass spectral analyses identified a difference of 46 in molecular weight between dephospho Form B and urothione, corresponding to the replacement of the −SCH3 group by a proton [5]. Thus it became apparent that Form B and urothione were structurally similar apart from the −SCH3 group and that the inherent high fluorescence of the thienopterin ring was totally quenched in urothione by the presence of the −SCH3 group [5]. In addition, urothione could be converted to dephospho Form B by desulfuration. The Form B structure derived from these results is shown in Fig. 1. The marked structural similarities between urothione and Form B implied a metabolic link between the two molecules, and conclusive evidence for this was obtained by the finding that the urine of patients with Moco deficiency did not contain urothione [5].
2.1.3 The proposed structure of MPT
In 1982, the structure for MPT shown in Fig. 1 was proposed using the information obtained from structural characterization of the three Moco derivatives (Form A, Form B and urothione) and the following rationalizations [5, 9]:
Form A and Form B both contain the same number of carbons.
The presence of two sulfurs in urothione must reflect the sulfur content of MPT. Form B and Form A are formed by the loss of one or both of these sulfurs, respectively.
The presence of sulfurs on adjacent carbons in urothione is indicative of the presence of vicinal thiols in MPT.
During the in vivo conversion of MPT to urothione, the phosphate group is removed and the methyl group of the −SCH3 is added to facilitate excretion.
The double bond in the thiophene ring of Form B and urothione indicates the presence of a dithiolene (enedithiol) grouping on C1' and C2' of the MPT side chain.
Although Form B and urothione are thienopterins, it is likely that their thiene rings are the result of cyclization of a linear MPT 6-alkyl pterin side chain.
2.1.4. The structure of alkylated MPT
The presence of a dithiolene group in MPT would be expected to contribute to the extreme lability of the molecule [40]. It therefore seemed probable that isolation of a stable MPT derivative retaining both sulfur atoms would require attenuation of the reactivity of the thiol groups [9]. This conclusion was corroborated by the successful isolation of a stable di-(carboxamidomethyl)-derivative of MPT (camMPT) from sulfite oxidase and XDH by a procedure involving treatment with iodoacetamide under mild conditions [41]. Determination of the structure of camMPT to be that shown in Fig. 1 confirmed the previously proposed structure of MPT as a 6-alkyl pterin with a four-carbon side chain containing an enedithiol at carbons-1' and -2', a hydroxyl at carbon-3', and a terminal phosphate group at carbon-4' [5]. Thirteen years later, the proposed MPT structure was confirmed by the publication of the first crystal structures of MPT-containing proteins, the tungsten-containing aldehyde-ferredoxin oxidoreductase from Pyrococcus furiosus by Chan et al. [42] and the aldehyde oxidoreductase from Desulfovibrio gigas by Ramão et al. [43]. The only difference between the proposed MPT structure and that observed in the crystal structures was that the pterin system was modified by an intramolecular cyclization resulting in a three-ringed pyranopterin structure in the crystals (Fig. 1).
2.1.5 Identification of dinucleotide analogs of MPT
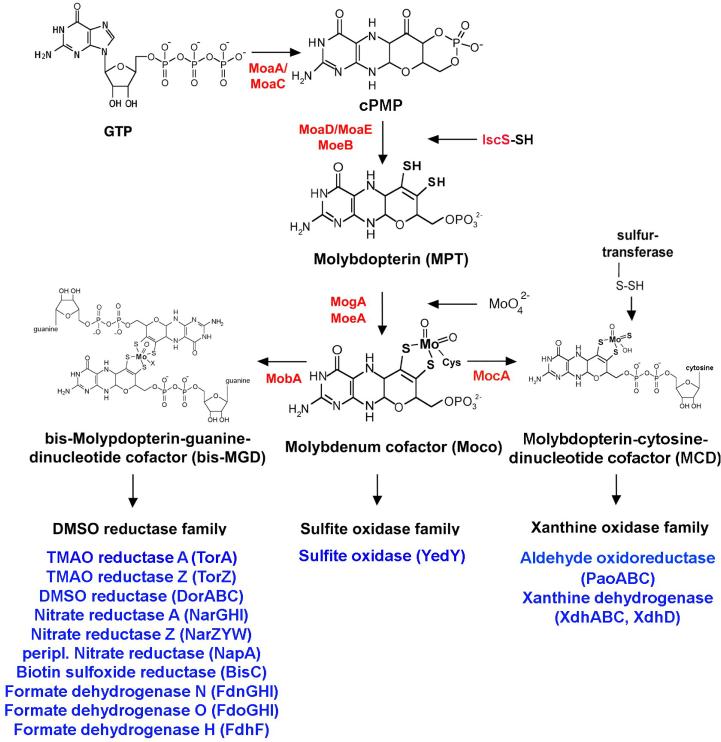
Early studies on Moco from various sources using the N. crassa nit-1 NR reconstitution assay let to the widespread belief of the universality of a single Moco structure [29]. However, as an increasingly large number of purified molybdoenzymes were studied, reports suggesting the possible existence of different forms of Moco in some of these enzymes became more common. For example, the cofactor from Methanobacterium formicium formate dehydrogenase was reported to be inactive in the nit-1 reconstitution assay, but was capable of producing Form A [44]. Additionally, studies by Meyer and coworkers [45, 46] showed that the CO dehydrogenase of Hydrogenophaga pseudoflava contained a more complex form of the pterin molecule. The study of alkylated pterin derivatives of molybdoenzymes from various sources, particularly the Moco in Rhodobacter sphaeroides DMSO reductase [47], eventually lead to the identification of a new form of MPT termed molybdopterin guanine dinucleotide (MGD) [9]. The structure of this phosphoric anhydride of MPT and 5'GMP shown in Fig. 2 was elucidated through detailed chemical investigations showing that the novel pterin contained two phosphates per pterin and that a 5'GMP moiety was liberated by acid hydrolysis or nucleotide pyrophosphatase treatment of the cofactor [11]. Further studies of the structure of the Moco present in molybdoenzymes from various sources determined that while enzymes from eukaryotic sources contain only MPT, eubacterial molybdoenzymes usually contain a dinucleotide derivative of the cofactor formed by the addition of GMP to yield MGD or by the addition of CMP to yield molybdopterin cytosine dinucleotide cofactor (MCD) as shown in Fig. 2 [11, 48, 49]. Since MGD has the same relationship to MPT as that of FAD to FMN, it was concluded that the purine nucleotide of MGD probably plays a passive role in Moco activity such as providing additional binding interactions with the protein [9].
Figure 2. The biosynthesis of Moco in E. coli.
Shown is a scheme of the biosynthetic pathway for Moco biosynthesis in E. coli and the proteins involved in this pathway. Moco is formed from 5'GTP with Precursor Z and MPT as intermediates. The molybdoenzymes are classified into three different enzyme families, the DMSO reductase family, the sulfite oxidase family and the xanthine oxidase family, a categorization based on the coordination of the molybdenum center. For enzymes of the DMSO reductase family, Moco is further modified by the attachment of GMP to form MGD, and two equivalents of MGD are bound to a single molybdenum to form the bis-MGD variant of the cofactor. Additional molybdenum ligands can be an oxo- or a sulfido group in addition to a serine, a cysteine, a selenocysteine or a hydroxide and/or water molecule. For enzymes of the sulfite oxidase family, Moco is directly inserted without further addition of a nucleotide and coordinated by an additional cysteine ligand in the enzyme. For enzymes in the E. coli xanthine oxidase family, Moco is further modified by the addition of a cytosine nucleotide to form the MPT-cytosine dinucleotide (MCD) form of the cofactor. Additionally, a terminal sulfur ligand is added to the molybdenum site, generating sulfurated molybdenum MCD. An additional ligand at the Mo-center usually is a hydroxo-group. The names of the proteins involved in the reactions are colored in red, and the identified and characterized molybdoenzymes of each E. coli molybdoenzyme family are shown in blue.
The crystal structure of a tungsten-containing aldehyde ferredoxin oxidoreductase from Pyrococcus reported by Chan et al. in 1995 [42] showed that the tungsten atom in this cofactor was coordinated by the four thiolate ligands from the dithiolenes of two MPT molecules, revealing yet another level of complexity in Moco structure. In 1996, the presence of a molybdenum atom coordinated by the four dithiolene ligands of two MGD molecules in the bis(MGD)-Mo form of Moco shown in Fig. 2 was identified in R. sphaeroides DMSO reductase [50]. Subsequently, crystallographic studies by Schindelin et al. [51] confirmed that bis-MGD is the form of Moco present in active DMSO reductase.
2.2 Determination of molecules involved in the early steps of Moco biosynthesis
2.2.1 Identification of Precursor Z
Investigations in the 1980s indicated that unlike wild type E. coli extracts, oxidation of E. coli extracts from cells containing mutations in either the moaD or moeB genes did not produce Form A, implying that no MPT was present in these extracts [14]. During additional analysis of these moaD− and moeB− strains, accumulation of a small Moco biosynthetic intermediate named Precursor Z that was converted to MPT when incubated with the protein fraction of extracts from moaA− cells was observed [14, 52]. Initial attempts to characterize Precursor Z were unsuccessful since it proved to be labile under aerobic conditions and was readily oxidized to a stable, inactive fluorescent molecule termed Compound Z. Emulating the method that had proven successful for structural determination of the unstable MPT molecule, structural characterization of Compound Z was undertaken to shed light on the structure of Precursor Z [53]. Through a combination of chemical and spectra analysis, it was determined that Compound Z is a 6-alkyl pterin with a 4-carbon side chain. The side chain had no sulfur, but did have a phosphate moiety in the form of a cyclic phosphate bound at C2' and C4' as seen in Fig. 1 [53].
Further studies demonstrated that Precursor Z oxidized directly to Compound Z without the accumulation of stable intermediates and that Precursor Z, like MPT, was a pyranopterin. EDAX analysis clearly showed that Precursor Z, like Compound Z, did not contain any sulfur atoms and mass spectral data indicated that Compound Z was just 2 Da smaller than Precursor Z [54]. The structure for Precursor Z shown in Fig. 1 was proposed in 1993 [54], and differs from Compound Z only in the reduction state of the pterin ring. An alternative structure for Precursor Z was more recently proposed by Santamaria-Araujo et al. [55] who found that Precursor Z analysis by 1H-NMR supported a structure with a geminal diol at the C1' position instead of a keto group. However, since it is known that geminal diols are readily formed by dehydration of a carbonyl, both models are chemically similar, and it has been proposed that Precursor Z can exist in either form depending on the local solvent and protein environment [56].
2.2.2 Determination of the initial molecule in Moco biosynthesis
It had previously been shown that GTP serves as the initial starting compound for the biosynthesis of the pterin/pteridine rings of folate, riboflavin and tetrahydrobiopterin in both plants and microorganisms [15, 57]. To determine if GTP served a similar function in Moco biosynthesis, studies involving in vivo labeling of Precursor Z were undertaken [58]. For these studies, uniformly 14C-labeled guanosine was added to cultures of an E. coli moeB− strain growing on minimal medium. The Precursor Z produced by these cells was then converted to Compound Z and purified [58]. In order to assess the distribution of label between the pterin ring and side chain carbons, Compound Z was subjected to alkaline potassium permanganate oxidation. This treatment resulted in loss of the phosphate and three terminal side chain carbons and yielded pterin-6-carboxylic acid. Extended UV illumination of this molecule released the remaining side chain carboxylate carbon in the form of CO2 leaving free pterin in solution. When Compound Z, pterin-6-carboxylic acid and pterin were analyzed for the presence of radioactivity, label was present in all three molecules, and the ratios of their specific radioactivities indicated that all of the side chain carbons had been labeled by the [U-14C] guanosine.
In the first step of the biosynthesis of biopterin, folate and riboflavin, the C8 carbon of GTP is released as formate in a reaction catalyzed by GTP cyclohydrolase I or II [59, 60]. Thus, no transfer of label from [8-14C] GTP to biopterin, folate or riboflavin would be observed. However, when moeB− cells were grown in medium containing [8-14C] guanosine and Compound Z was analyzed as described above, label was transferred to Compound Z, but solely at the C1'-position. Additionally, the label present in Compound Z purified from cultures grown on [8,5-3H] guanosine was lost by removal of the three terminal side chain carbons [58]. These results indicated that a guanosine derivative is indeed the initial substrate for the biosynthesis of Precursor Z, however, unlike the synthesis of all other previously studied pterins, during MPT biosynthesis, the C8-carbon of the initial guanosine precursor is retained and incorporated as the first carbon of the MPT side chain [58]. Consistent with this, the synthesis of Precursor Z was shown to be cyclohydrolase I independent [9]. In a similar set of experiments utilizing minimal media containing a variety of 13C and 15N multiply-labeled substrates and NMR analysis of the resulting labeled Compound Z, Rieder et al. subsequently verified both the fate of the GTP guanine C8 during Moco biosynthesis and the fact that all five of the guanosine ribose carbons were retained in MPT with two being incorporated into the pterin ring and the remaining three becoming C2', C3' and C4' of the Precursor Z side chain [61].
Following 1H-NMR results in 2004 by Santamaria-Araujo et al. [55] indicating that Precursor Z exists in the cyclo pyrano-form, the alternative name of cyclic pyranopterin monophosphate (cPMP) was proposed for Precursor Z. Both names have been used in more recent publications, however, since cPMP describes the nature of the molecule better than the original name Precursor Z, the term cPMP will be used for the remainder of this chapter.
3. BIOSYNTHESIS OF MPT AND MOCO
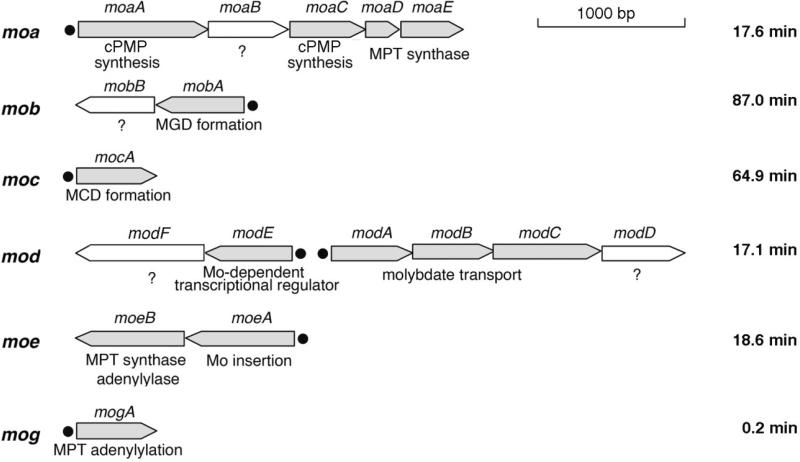
Using a combination of biochemical, genetic, and structural approaches, Moco biosynthesis in E. coli has been extensively studied for decades. These studies have identified at least 17 E. coli genes involved in Moco biosynthesis [6, 20]. As seen in Fig. 3, these genes are located in 6 different mo loci that are specific to Moco biosynthesis, and the corresponding proteins are highly conserved in other organisms [15, 20]. As seen in Fig. 2, E. coli Moco biosynthesis can be divided into four steps [6, 20]: (i) formation of cPMP, (ii) conversion of cPMP to MPT, (iii) insertion of molybdenum to form Moco, and (iv) additional modification of Moco via the attachment of different nucleotides such as CMP or GMP to form the MCD and MGD cofactors, respectively. The first three steps through the formation of the mononucleotide form of Moco are identical for all Moco-containing proteins in all microorganisms, while the final Moco modification steps vary between different Moco proteins and organisms. The proteins and biochemical reactions involved in each of these four steps are described in detail below.
Figure 3. Organization of the genes involved in the biosynthesis of Moco in E. coli.
In total, 17 genes are involved in the biosynthesis of Moco in E. coli. These are organized into 6 different gene loci termed moa, mob, moc, mod, moe and mog. Genes of known function are colored in grey. Black dots indicate promotor regions. The genes are drawn approximately to scale. Their locations on the E. coli chromosome are indicated in minutes at the far right. Additional operons involved in Moco biosynthesis, such as the isc operon, which is also involved in other biosynthetic pathways, are not shown.
3.1 Conversion of GTP to cPMP
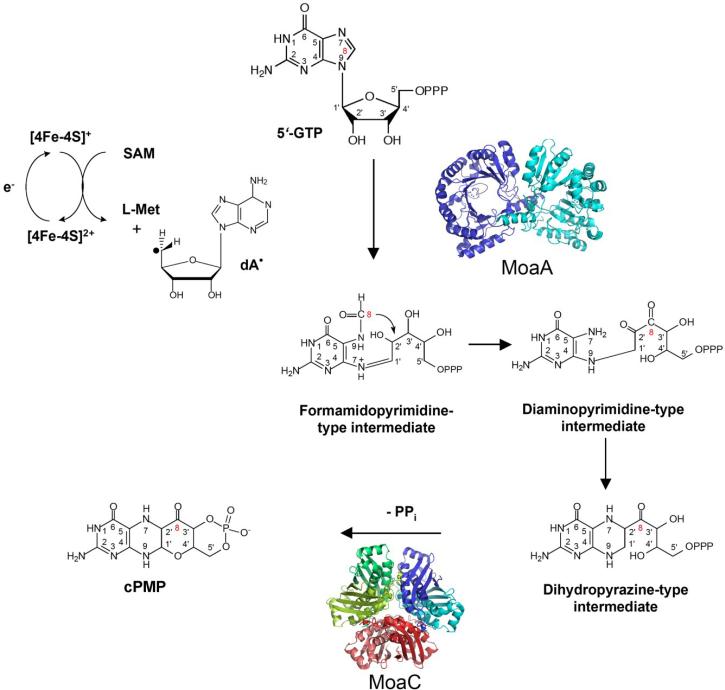
In E. coli, the moaA and moaC gene products are responsible for the complicated chemical reactions required to generate cPMP. As described earlier, labeling studies had determined that a guanosine derivative was the initial starting point for cPMP formation [58, 61]. More recently, Hänzelmann and Schindelin [62, 63] used an in vitro system for cPMP synthesis containing MoaA and MoaC proteins purified from Staphylococcus aureus to demonstrate that 5'-GTP is the specific initial substrate for Moco biosynthesis. As shown in Fig. 4, formation of cPMP from GTP involves several steps including opening of the guanine imidazole ring to generate a formyl-diaminopyrimidine nucleotide intermediate, insertion of the guanine C-8 formyl group between C-2' and C-3' of the ribose, closure of the new pterin ring, generation of a cyclic monophosphate with elimination of pyrophosphate, and formation of a pyran ring through attack by the 3' hydroxyl of the side chain on the pterin. The cooperative participation of the MoaA and MoaC proteins in this step of Moco biosynthesis bring about significant carbon rearrangements whose net result is that the two carbon atoms added into the newly formed cPMP pterin ring originated as C-1' and C-2' of the GTP ribose ring, and the 4-carbon side chain is formed from the GTP guanine C-8 and C-3', C-4' and C-5' of the GTP ribose in that order [58, 61].
Figure 4. The biosynthesis of Precursor Z from 5'GTP.
All the carbon and nitrogen atoms of the initial GTP molecule are retained within the Precursor Z during synthesis, and the numbering system matches that of the GTP carbons through all structures shown. The C8 atom of the guanine ring is transferred as a formyl group and is inserted between the C2' and C3' atoms of the ribose. Hypothetical formamidopyrimidine-type, diaminopyrimidine-type and dihydropyrazine-type intermediates that might be formed during the reaction are shown. This reaction is catalyzed by the MoaA protein, a S-adenosylmethionine (SAM)-dependent enzyme. MoaC is believed to cleave the pyrophosphate group of the dihydropyrazine intermediate. Precursor Z is shown in the dihydropyrano form. The structures of MoaA (S. aureus) and MoaC (E. coli) were derived using coordinates from the Protein Data Bank (accession numbers 1TV8 and 1EKR, respectively).
Based on sequence similarities to proteins such as biotin synthase, pyruvate-formate-lyase-activating enzyme and anaerobic ribonucleotide reductase-activating enzyme, MoaA has been classified as a member of the S-adenosylmethionine (SAM)-dependent radical enzyme superfamily [64]. In the reactions catalyzed by members of this family, SAM serves as the free radical initiator and undergoes cleavage to methionine and a 5'deoxyadenosyl radical that in turn initiates radical formation of substrate molecules or of glycyl residues within the target enzymes to activate them for radical-based chemistry. The source of the electron required for the cleavage of SAM is a reduced form of a conserved FeS cluster within the protein [64]. MoaA contains two oxygen-sensitive [4Fe-4S] clusters, one typical for SAM-dependent radical enzymes at the N-terminus and an additional C-terminal cluster unique to MoaA proteins [62].
Although MoaA is involved in the first step in the Moco biosynthesis pathway, it was actually the last Moco biosynthetic protein to be structurally characterized when in 2004, Hänzelmann and Schindelin reported the crystal structure of S. aureus MoaA [62]. The core of this protein was characterized by an incomplete (β)6 TIM barrel type fold formed by the N-terminal part of the protein, and this portion of the protein also contained the SAM characteristic [4Fe4S] cluster bound to three cysteine residues. SAM is the fourth ligand to the cluster and is bound to the Fe as an N/O chelate. The lateral opening of the incomplete TIM barrel is covered by the C-terminal portion of the protein containing the additional [4Fe4S] cluster, and the two clusters are separated by 17 Å [62]. The C-terminal [4Fe4S] cluster is also ligated by three cysteine ligands, with its non-cysteinyl-ligated Fe site possibly involved in the binding and activation of 5'-GTP [63]. The 5'desoxyadenosyl radical generated from SAM at the N-terminal cluster was proposed to be in close proximity to the 5'-GTP where it is probably involved in facilitating hydrogen abstraction at either the C8 of the guanine or the C2' or C3' atoms of the ribose [63, 65]. Insertion of the formyl group between the ribose C2' and C3' carbons might also require radical mediation. The proposed intermediates during the reaction catalyzed by MoaA are shown in Fig. 4.
The crystal structure of E. coli MoaC reported by Wuebbens, et al. in 2000 revealed that it is present in solution as a hexamer composed of a trimer of dimers with a putative active site located at each dimer interface [66]. It has been speculated that MoaC is involved in the cleavage of the dihydropyrazine-type intermediate pyrophosphate group and formation of the cPMP cyclic phosphate group [67]. Alternatively, data obtained from the structure of a co-crystal of Thermus thermophilus HB8 MoaC with bound 5'GTP has been interpreted to suggest that the formamidopyrimidine-type intermediate may be the MoaC substrate rather than 5'GTP [68]. Further studies are required to elucidate the exact nature of the involvement of both MoaA and MoaC in cPMP formation.
Recently, it has been reported that a human patient suffering from molybdenum cofactor deficiency was cured with cPMP (Precursor Z) purified from E. coli cells, confirming the universal structure of cPMP (Precursor Z) among the kingdoms of life [69], which additionally was shown by cPMP (Precursor Z) treatment of Moco deficient mice before [70].
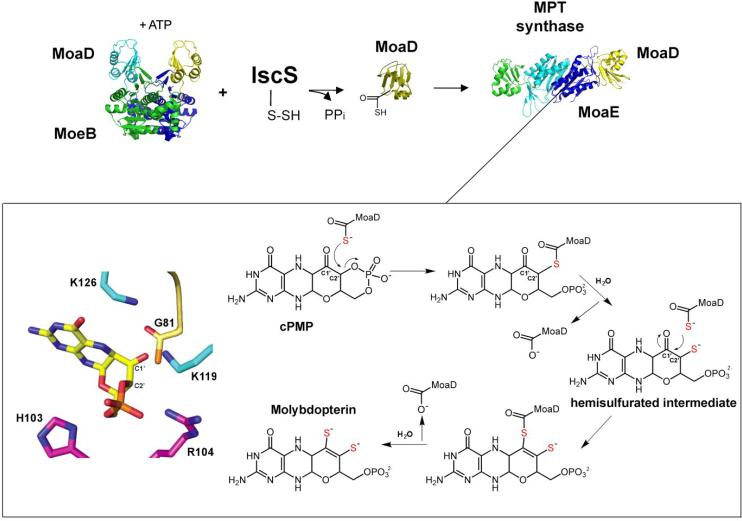
3.2 Insertion of sulfur into cPMP and formation of MPT
While cPMP is structurally quite similar to MPT, it lacks the dithiolene function essential for molybdenum ligation in Moco. Therefore, for the conversion of cPMP to MPT, two sulfur atoms must be incorporated at the C1' and C2' positions of cPMP (Fig. 2). In E. coli, this reaction is catalyzed by the MPT synthase protein (Fig. 5). MPT synthase activity was first identified in 1989, when it was reported that the N. crassa nit-1 strain accumulates cPMP that can be converted to Moco by an activity present in extracts from an E. coli moaA− strain [53]. Purification of this MPT synthase activity identified a heterotetrameric enzyme consisting of two small (~8,750 Da) and two larger (~16,850 Da) protein subunits, later shown to be encoded by the E. coli moaD and moaE loci, respectively [52]. Pitterle et al. were able to establish an in vitro system for MPT synthesis consisting solely of MPT synthase and cPMP [71]. They also demonstrated that the MoaD small subunit of MPT synthase carries the sulfur used for generation of Moco since pretreatment of MoaD with the sulfhydryl reagents N-ethylmaleimide, N-bromobimane, or iodoacetamide abolished activity in the in vitro assay, while similar treatment of MoaE had no effect on activity [72]. These data were confirmed by electrospray mass spectrometry experiments that found a mass increase of 16 Da when active MoaD was compared to inactive MoaD. This difference corresponds to the change in mass expected for the exchange of an oxygen atom for a sulfur atom at the C-terminal glycine carboxylate of MoaD [72]. These results were confirmed 8 years later when Gutzke et al. used an in vitro intein-based expression system to generate preparative amounts of thiocarboxylated MoaD and showed that this protein readily assembled with MoaE to form active MPT synthase which was able to convert cPMP to MPT [73].
Figure 5. The biosynthesis of Molybdopterin from Precursor Z.
In the MPT synthase mechanism, the initial attack and transfer of the first MoaD-SH sulfur atom occurs at the C2' position, coupled to the hydrolysis of the Precursor Z cyclic phosphate. An intermediate is formed in which the MoaD C-terminus is covalently linked to the substrate via a thioester linkage that is subsequently hydrolyzed by a water molecule and generates a hemisulfurated intermediate at C2'. Opening of the cyclic phosphate shifts the location of the intermediate within the complex to a position where C1' becomes more accessible. A new MoaD thiocarboxylate attacks the C1' resulting again in a second covalent intermediate which is converted to MPT via the elimination of a water molecule and hydrolysis of the thioester intermediate. On the left side, the structure of Precursor Z is shown in the middle with the C1' and C2' carbons labeled. Conserved E. coli MPT synthase residues in the active site are shown with MoaE residues in cyan and magenta and the C-terminus of MoaD in yellow with the thiocarboxylate sulfur in orange. The structures of MPT synthase and MoeB were derived using the coordinates from the Protein Data Bank (accession numbers 1NVI and 1JW9, respectively). The model was adapted after figures from Daniels et al. [56].
The high-resolution crystal structure of the MPT synthase complex published by Rudolph, et al. in 2001 revealed that within the MPT synthase heterotetramer, the two MoaE subunits form a central dimer, and one MoaD subunit is located at the opposite end of each dimer as seen in Fig. 5 [74]. The MoaD protein has a β-grasp fold characteristic of ubiquitin-like proteins, all of which contain a highly conserved double glycine motif at their C-termini [74]. The C-terminus of each MoaD monomer is inserted into a pocket at the edge of its adjacent MoaE partner. These pockets are lined with highly conserved MoaE amino acids, and serve as the cPMP binding sites [75].
Publication of the structure of MPT synthase prompted speculation as to whether the addition of both dithiolene sulfurs to a single cPMP molecule occurs independently at both active sites [76], or whether a hemisulfurated intermediate is transferred from one active site to the other within a single MPT synthase tetramer for addition of the second sulfur atom [73]. Based on purification of a hemisulfurated intermediate that was tightly associated with the MPT synthase complex, a model was proposed for the reaction. After transfer of its thiocarboxylate sulfur to cPMP, the MoaD subunit would dissociate from the MPT synthase complex [76]. This would be followed by subsequent binding of a second, thiocarboxylated MoaD (Moad-SH) to the same active site and transfer of the second sulfur to form MPT. In this model, each cPMP molecule remains bound at a single active site until conversion to MPT is completed by the exchange of carboxylated and thiocarboxylated MoaD molecules. In 2008, Daniels, et al. reported of the structure of a co-crystal of S. aureus MPT synthase with cPMP bound to the active site along with further chemical analysis of the reaction products [56]. In the co-crystal, cPMP was bound to the MoaE subunit of MPT synthase in its keto form rather than as a geminal diol, and a more detailed reaction mechanism was proposed in which initial attack and transfer of the first sulfur atom from a MoaD-SH occurs at the C2' position and is coupled to hydrolysis of the Precursor Z cyclic phosphate as seen in Fig. 5. During the course of this reaction, an intermediate is formed in which the MoaD C-terminus is covalently linked to the substrate via a thioester linkage which is subsequently hydrolyzed by a water molecule. The opening of the cyclic phosphate is proposed to shift the location of the intermediate within the protein so that the C1' position now becomes more accessible to attack by the second MoaD-SH resulting in a second covalent intermediate between the two that is converted to MPT via the elimination of a water molecule and hydrolysis of the thioester intermediate [56].
For MPT synthase to act catalytically, it is necessary to regenerate the transferable sulfurs at the C-terminus of the MoaD proteins after each sulfur transfer to cPMP [72]. In addition to strains bearing mutations in the moaD and moaE loci, the observation that an E. coli strain carrying a mutation in the moeB locus also accumulated cPMP suggested that the MoeB protein might be involved in the MPT synthase reaction [14]. This observation led to the eventual conclusion that MoeB serves as the MPT synthase sulfurase, the protein responsible for regenerating the thiocarboxylate group at the C-terminus of MoaD [77]. It was later demonstrated that MoeB-dependent activation of MoaD required ATP consumption [78], and sequence analysis indicated that MoeB shares significant sequence similarities to the ubiquitin-activating enzyme, E1 [77, 79]. The functional and sequence similarities of MoeB to E1-like enzymes combined with the structural similarity of MoaD to ubiquitin-like proteins led to the proposal that activation of MPT synthase by MoeB might resemble the process of ubiquitin-dependent protein degradation [78, 80]. The first step in this degradation process is activation of ubiquitin by formation of an acyl-adenylate intermediate at its C-terminal glycine. This is followed by formation of a thioester bond between the ubiquitin and a conserved cysteine residue on the E1-activating enzyme [80]. Further analysis determined that the activation of MoaD by MoeB did resemble the first step in ubiquitin activation [78]. These data were also confirmed by the MoaD-MoeB crystal structure which was solved in the apo-, ATP-bound and MoaD-adenylate forms [80]. The crystal structure of this complex revealed a (MoaD-MoeB)2 heterotetramer in which the MoeB subunits form a dimer. In the MoeB-MoaD-ATP ternary complex, ATP is bound in close proximity to the C-terminus of MoaD. The overall shape of the binding pocket distorts the ATP molecule and induces a kink at the α-phosphate. In the next step, the MoaD-acyladenylate intermediate is formed, consisting of the C-terminal MoaD-Gly81 covalently linked to the α-phosphate through a mixed anhydride and the release of PPi [78, 81].
In the ubiquitin-E1 system, it has been determined that the reaction proceeds via an absolutely ordered mechanism with ATP binding to E1 preceding ubiquitin binding which is followed by ubiquitin-adenylate formation [82–85]. Ubiquitin residue Arg72 and the terminal glycine (Gly76) are involved in interactions between ubiquitin-AMP and its E1, and Arg72 plays a particularly critical role in determining the ubiquitin specificity with respect to its E1 [86]. In contrast, the formation of the (MoaD-MoeB)2 heterotetramer clearly occurs in the absence of ATP [87]. A similar reaction also occurs between the ThiS and ThiF proteins during thiamine biosynthesis [88], showing a general mechanism of ubiquitin-like and E1-like proteins in bacteria. Thus, although members of the MoeB/E1 enzyme superfamily have diversifed to be used in seemingly unrelated pathways, their mechanism for acyl-adenylate formation has been evolutionarily conserved [81].
Although stable, thiocarboxylated MoaD (MoaD-SH) freely exists in solution, in the ubiquitin system, a free ubiquitin thiocarboxylate is never formed [81]. Rather, the ubiquitin acyladenylate reacts with a cysteine on E1 to form a thioester. This species then acts as the ubiquitin donor for proteins targeted for degradation [81]. In E. coli thiamine biosynthesis, a persulfurated ThiI protein displaces AMP from the ThiS acyl-adenylate to form a ThiS-ThiI acyl-disulfide [89, 90]. Through disulfide interchange, ThiF then plays a second role in the thiamine pathway by formation of a ThiS-ThiF acyldisulfide at ThiF Cys184 [89, 91]. This species serves as the thiazole donor for thiamine synthesis, and again, there is no evidence for a free ThiS thiocarboxylate. Conversely, in the MPT synthase system, no evidence has been found for the existence of a MoeB-bound sulfur or for a thioester or disufide linkage between MoeB and MoaD [78]. Additionally, individual substitution of each MoeB cysteine residue with alanine revealed that the only cysteine residues essential for MoeB activity were those involved in binding of the structurally stabilizing zinc atoms [78]. Given these data and the existence of free MoaD-SH in solution, it is clear that although their initial steps seem similar, the pathway for MoaD-SH formation differs greatly from both the ubiquitin and thiamine pathways after the first activation step [81].
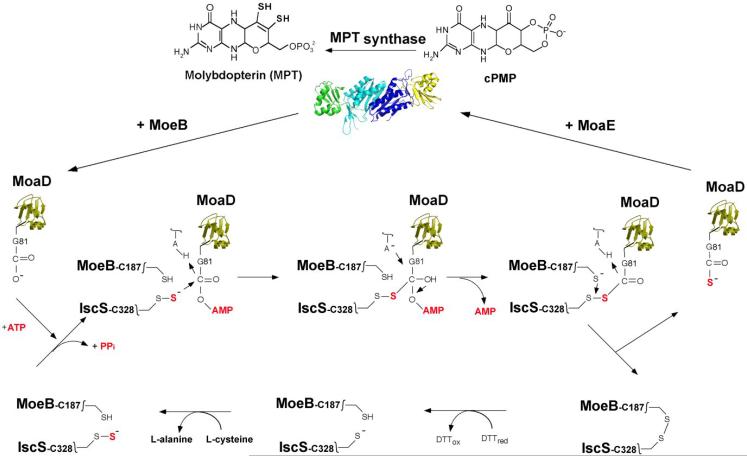
After the formation of the activated MoaD acyl-adenylate in the complex with MoeB, sulfur is inserted to form the terminal MoaD thiocarboxylate group on MoaD-Gly81 (Fig. 6). It was determined that in E. coli, L-cysteine serves as the origin of the MPT dithiolene sulfurs and that the cysteine sulfur is transferred to the activated MoaD acyl-adenylate by the action of a persulfide-containing protein [92]. Initial results showed that this protein could be any of the three pyridoxal-phosphate-dependent E. coli L-cysteine desulfurases IscS, CsdA or SufS, since all three were capable of transferring their protein-bound persulfide sulfur to MoaD in the MoaD-MoeB-complex in an in vitro reaction [92]. These results were in agreement with studies indicating that the persulfide sulfurs formed on an active site cysteine residue of pyridoxal 5-phosphate-dependent cysteine desulfurases are incorporated into the biosynthetic pathways of a variety of sulfur-containing cofactors and thionucleosides [93]. More detailed studies by Zhang et al. [94] of the L-cysteine desulfurase reaction in E. coli Moco biosynthesis indicated that of the three possible desulfurases, only IscS interacts specifically with MoeB and MoaD (Fig. 6). These results were confirmed by in vivo studies showing that IscS is the primary physiological sulfur-donating enzyme for the generation of the thiocarboxylate of MPT synthase in MPT biosynthesis [94].
Figure 6. Formation of the thiocarboxylate group on E. coli MoaD.
Precursor Z is converted to MPT by the transfer of two sulfur groups from the C-terminal thiocarboxylate of the MoaD subunit of MPT synthase. Regeneration of the MoaD-SH sulfur occurs in a MoaD/MoeB complex where adenylylated MoaD is formed by attachment of an AMP moiety at the MoaD C-terminus with loss of pyrophosphate. MoaD-AMP is then sulfurated by a protein-bound persulfide group on the IscS sulfurtransferase, likely by the formation of an IscS-MoaD disulfide intermediate. Reductive cleavage of the disulfide bond could then occur by attack of the thiol group of MoeB-C187, and in turn, MoaD-SH is generated and released from the ternary complex. The disulfide bond formed between MoeB and IscS is likely reduced by the thioredoxin system in vivo (in vitro reduction by DTT is shown). The sulfur donor for IscS is L-cysteine. After formation of the thiocarboxylate group, MoaD-SH dissociates from the MoeB dimer and reassociates with MoaE.
Summarizing these results, a sulfurtransfer mechanism shown in Fig. 6 for the formation of the MoaD thiocarboxylate group was proposed which is similar to the reaction postulated for the homologous human proteins [95]. In the (MoaD-MoeB)2 complex, MoeB activates MoaD by formation of an acyladenylate bond, and IscS in its persulfide-bound form is able to interact with this complex. A possible reaction could be that MoaD is transferred to the IscS-Cys328 persulfide group, forming a disulfide bond. Release of the MoaD thiocarboxylate requires a second thiol group, which is proposed to be MoeB-Cys187, but other reducing factors are also possible. In comparison, a disulfide intermediate for sulfur transfer exists between ThiS and ThiF and the rhodanese-like protein ThiI in thiamine and thiouridine biosynthesis [89, 90]. Whether in addition to IscS, a rhodanese like protein acts as a mediator for sulfurtransfer in Moco biosynthesis in E. coli remains to be determined.
After the sulfur transfer reaction, MoaD-SH dissociates from the complex and reassociates with MoaE to form active MPT synthase (Fig. 6). The binding constants within the different complexes of MoaD follow the order (MoaD-SH-MoaE)2 > (MoaD-MoeB)2 > (MoaD-MoaE)2 [87]. This order is mechanistically logical given that during the course of MPT biosynthesis, MoaD-SH first binds to MoaE to form the active MPT synthase complex where transfer of the MoaD-SH thiocarboxylate to cPMP yielding MPT and inactive MPT synthase occurs. MoaD must then dissociate from this inactive complex to form a new complex with MoeB, a prerequisite for the regeneration of MoaD-SH. In addition, the (MoaD-MoeB)2 complex is stabilized by ATP addition and the subsequent formation of the acyl-adenylate on MoaD. In this form, IscS-SH transfers the sulfur to MoaD, generating MoaD-SH. After the formation of the (MoaD-SH-MoaE)2 complex, introduction of the dithiolene moiety to MPT completes the formation of the chemical backbone necessary for binding and coordination of the molybdenum atom in Moco (Fig. 2).
3.3 Insertion of molybdenum into MPT
Molybdenum enters the cell as the soluble molybdate oxyanion and high affinity transporters for molybdate have been described in bacteria [96]. The first identified gene involved in E. coli molybdate transport, modC, was identified due to the fact that molybdoenzyme activity in modC− strains could be restored by addition of molybdenum at a 1000-fold higher concentration (>0.1 mM) than that sufficient for wild-type cells [21]. The modC− phenotype is pleiotropic for all molybdoenzymes. It has since been determined that E. coli ModC is the ATP-binding component of a high-affinity molybdenum uptake system. This system belongs to the ATP-binding cassette (ABC) superfamily of proteins responsible for the transport of a wide variety of molecules in prokaryotes and eukaryotes [97, 98]. In their most common bacterial form, ABC transporters consist of a dimer of integral membrane proteins (ModB for molybdate transport in E. coli), and each ModB subunit binds one hydrophilic peripheral membrane protein (the ATP-binding protein ModC for molybdate transport in E. coli). In Gram-negative bacteria, a periplasmic binding protein, in this case ModA, captures the substrate (molybdate) and delivers it to the transporter complex in the inner membrane. Both molybdate and tungstate bind to ModA in the periplasm in a tetrahedral complex stabilized by seven hydrogen bonds formed between the oxygen of the bound anion and the protein groups from two different ModA domains [99].
Expression from the modABC operon is subject to molybdate-dependent regulation controlled by the ModE protein since when bound to molybdate, ModE binds to the operator region of modABC [100]. ModE/molybdate also enhances the transcription of molybdenum-dependent enzymes such as DMSO reductase [101] and nitrate reductase A [102] as well as the molybdenum cofactor biosynthesis operon moaABCDE [103]. Schüttelkopf et al. [104] determined that ModE is a homodimer and each monomer can be subdivided into four structural domains: the N-terminal DNA-binding domain, a linker domain and two molybdate-binding (Mop) domains. ModE binds two molybdate molecules per dimer and molybdate binding results in extensive conformational changes enabling DNA binding.
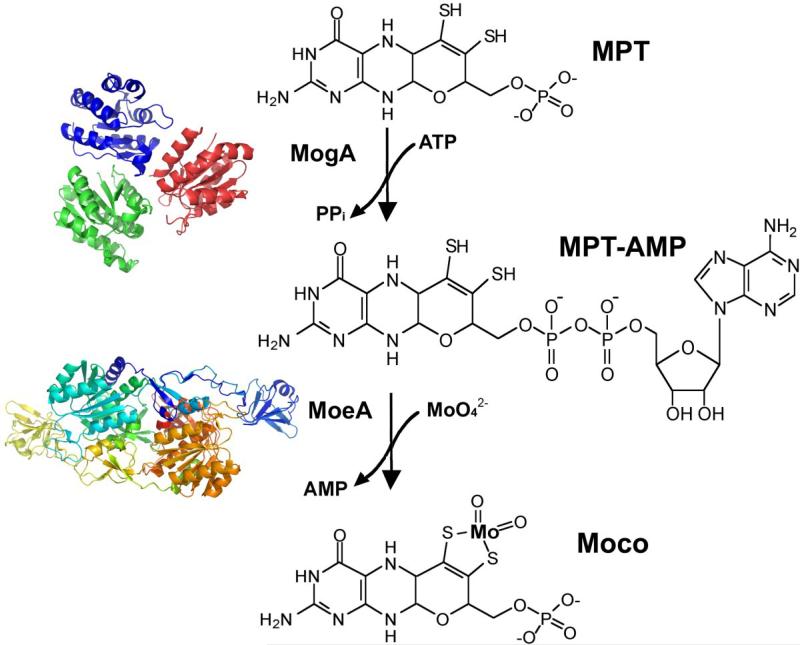
In E. coli, insertion of the metal into the MPT is accomplished by the moeA and mogA gene products (Fig. 2 and 7) [14, 105]. The structure of E. coli MogA shown in Fig. 7 was published by Liu et al. in 2000 [106] and was the first Moco biosynthesis protein crystal structure to be reported. MogA is a trimer in solution with each monomer folded into a single compact domain, and MogA binds MPT with high affinity [106]. Two crystal structures for E. coli MoeA published in 2001 by Xiang et al. (94) and Schrag et al. [107, 108] both reported a dimeric structure with an elongated monomer consisting of four distinct domains, one of which was structurally related to MogA (Fig. 7). Nichols and Rajagopalan [109] reported that the proteins performed different functions in the chelation reaction. MoeA appeared to mediate molybdenum ligation to newly synthesized MPT in vitro at low concentrations of MoO42−. This reaction was strongly inhibited by MogA in the absence of ATP, but in the presence of ATP, MogA doubled the rate of molybdenum ligation [109]. Llamas et al. [110–112] were the first to verify the catalytic formation of an MPT-AMP intermediate during the reaction, which was identified in the crystal structure of a Cnx1 variant to the Arabidopsis thaliana Cnx1-S583A protein [112]. The accumulation of a comparable MPT-AMP intermediate in E. coli moeA− extracts has also been verified (Leimkühler, unpublished data). Thus, it appears that in E. coli, MPT is only activated by MogA-mediated adenylation prior to molybdenum insertion by MoeA under physiological molybdenum concentrations (Fig. 7), a reaction which is not required under high molybdenum concentrations [113].
Figure 7. Insertion of molybdate into MPT.
The E. coli MoeA and MogA proteins catalyze the specific incorporation of molybdenum into MPT in a multistep reaction with an adenylated MPT intermediate (MPT-AMP), which was first identified in the A. thaliana Cnx1-G protein [112]. While MogA forms the MPT-adenylate intermediate, MoeA mediates molybdenum ligation to MPT at low concentrations of MoO42−. The structures of MogA and MoeA were derived using the coordinates from the Protein Data Bank (accession numbers 1DI6 and 1G8L, respectively)
Interestingly, when Kuper et al. [112] reported the crystal structure of the A. thaliana Cnx1-S583A protein in complex with MPT-AMP, they found copper bound to the MPT dithiolene sulfurs. They proposed that the function of this copper was to protect the dithiolene group prior to molybdenum insertion. In subsequent studies, a comparable copper-bound MPT-AMP intermediate could not be detected in E. coli [114]. However, it was shown that bivalent copper and cadmium ions as well as trivalent arsenite ions could all insert nonspecifically into MPT without the presence of either MoeA or MogA and that copper had a higher affinity for the dithiolene group of MPT than molybdate. It was additionally found that under physiological molybdate concentrations (1–10 μM), MogA is required in E. coli to form an MPT-AMP intermediate that facilitates molybdate insertion on the dithiolene sulfurs. Under high molybdate concentrations (> 1 mM), MPT-AMP formation by MogA is not required and molybdate can be directly inserted into MPT with the aid of the MoeA protein [114]. Thus, ATP-dependent activation of MPT and molybdenum is only required under physiological molybdenum concentrations. Copper is not needed to protect the dithiolene group of MPT, and it actually decreases molybdoenzyme activity when present at high concentrations due to nonspecific insertion into MPT [113]. These results were confirmed by in vivo studies by Morrison et al. [115] who investigated the effect of copper-limiting media on the synthesis of molybdoenzymes in E. coli and R. sphaeroides. They demonstrated that the activities of DMSO reductase and nitrate reductase were not repressed under copper starvation, verifying that copper is not required for the biosynthesis of Moco in bacteria [115].
3.4 ADDITIONAL MODIFICATION OF MOCO
3.4.1 Attachment of GMP and CMP
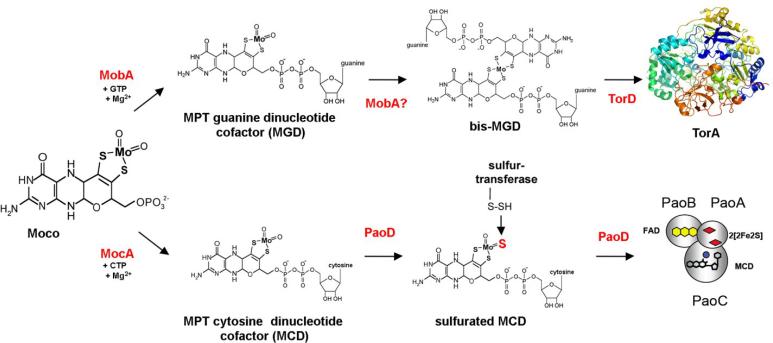
To date, the YedY protein, an E. coli sulfite oxidase homologue with unknown function, is the only E. coli molybdoenzyme known to bind the simple Mo-MPT form of Moco [116, 117]. Indeed, prior to the identification of YedY in 2004 [104], it was commonly believed that all E. coli molybdoenzymes contained the bis-MGD form of Moco [6, 20]. In E. coli, MGD is formed by covalent addition of the GMP moiety from GTP to the C4' phosphate of MPT via a pyrophosphate bond leading to release of the β- and γ-phosphates of GTP as pyrophosphate as shown in Fig. 8 [11, 118]. This reaction is catalyzed by MobA, one of two proteins encoded by the mob locus (Fig. 3) [119]. While MobA is essential for this reaction [120], the role of MobB, the second protein encoded by the mob locus, remains uncertain. Based on its crystal structure, McLuskey et al. [121] postulated that MobB could be an adapter protein, acting in concert with MobA to achieve the efficient biosynthesis and utilization of MGD. In vitro, however, MobA, GTP, MgCl2 and Mo-MPT are sufficient for the formation and insertion of bis-MGD into R. sphaeroides DMSO reductase without MobB [118, 122]. Significantly, it was shown that addition of the nucleotide to the cofactor occurs only after insertion of molybdenum into MPT [123]. The crystal structure of monomeric MobA showed a nucleotide-binding Rossman fold formed by the N-terminal half of the protein that also contains the GTP binding site. A possible MPT binding site was localized to the C-terminal half of the protein [118, 124].
Figure 8. Molybdopterin dinucleotide biosynthesis.
Synthesized Moco can be further modified in E. coli by the addition of GMP or CMP to the C4' phosphate of MPT via a pyrophosphate bond. MGD is formed by the MobA protein, which specifically binds GTP, while MCD is formed by the MocA protein, which acts specifically on CTP. In both cases, it has been shown that addition of the dinucleotide to the cofactor occurs after insertion of molybdenum into MPT. In all E. coli MGD-containing molybdoenzymes, the molybdenum atom is coordinated by the dithiolene groups of two MGD molecules, forming the bis-MGD cofactor. The protein involved in this reaction has not yet been identified. This step may be catalyzed by MobA, an MGD binding chaperone such as TorD or directly on the target enzyme. The structure of Shewanella massilia TorA is shown (accession number 1TMO). Once formed, MCD is further modified in E. coli by exchange of the equatorial oxygen to a sulfido ligand, forming sulfurated MCD. This step is carried out by the PaoD protein in conjunction with an as yet unidentified sulfurtransferase. After sulfuration, MCD is inserted into the specific target enzyme, here PaoABC.
As mentioned earlier, in all E. coli MGD-containing molybdoenzymes, a single molybdenum atom is coordinated by the dithiolene groups of two MGD molecules, forming the bis-MGD cofactor [50]. The structure of bis-MGD Moco was verified when the crystal structure of DMSO reductase from R. sphaeroides was reported by Schindelin et al. in 1996 [51]. The formation of bis-MGD is one of the most enigmatic steps in E. coli Moco biosynthesis. It is still not known whether the two MGD molecules assemble on MobA or at the cofactor binding sites of the molybdoenzymes themselves. Crystal structures for both monomeric and octameric forms of MobA have been reported by Lake et al. [118]. While a binding groove that could accomodate MGD was identified in the monomer, there was no comparable position in the monomer large enough for bis-MGD binding. However, the central cavity of the octameric form of MobA is large enough to support bis-MGD formation, and hydrophilic residues in the octamer channel could facilitate bis-MGD binding and storage prior to insertion into molybdoenzymes [118]. Alternatively, MGD binding to proteins involved in the maturation of several molybdoenzymes such as TorD protein, the system-specific chaperone for E. coli TMAO reductase has been observed, so it is also possible that the bis form of the MGD cofactor is formed in conjunction with these types of proteins [125, 126].
In prokaryotes other than E. coli, variants of the cofactor containing CMP, AMP, or IMP dinucleotide forms of MPT have been identified [11, 127]. For many years, it was believed that E. coli was incapable of synthesizing dinucleotide forms of the cofactor other than MGD, an idea supported by the fact that heterologous expression of MCD-containing enzymes in E. coli was unsuccessful [128]. Recently, however, three E. coli enzymes in the xanthine oxidase family named XdhABC, XdhD, and PaoABC contain the MCD form of the cofactor [129, 130], and the existence of MCD in E. coli molybdoenzymes was clearly demonstrated by the thorough characterization of the periplasmic aldehyde oxidoreductase PaoABC [131]. Additionally, using amino acid sequence alignments based on the E. coli MobA sequence, the specific protein involved in E. coli MCD biosynthesis was identified [132]. This protein, named MocA (for molybdopterin cytosine dinucleotide synthesis) (Fig. 2), exhibits 22% amino acid sequence identity to E. coli MobA [132]. The catalytic reaction of MocA is similar to the reaction of MobA, in that it acts as a CTP:MPT cytidylyl transferase and covalently links MPT and CMP with the concomitant release of the β- and γ-phosphates of CTP as pyrophosphate (Fig. 8) [132].
Comparison of the two transferases revealed that MobA is highly specific for binding of the purine nucleotide GTP, while MocA is specific for binding of the pyrimidine nucleotide CTP. The most significant sequence differences between the two proteins were observed in two conserved motifs at their N-terminal domains. The crystal structure of MobA with bound GTP showed that the guanine moiety is mainly bound by the 12LAGG15 and 78GPLAG82 amino acid sequence segments [118, 133]. In MocA, these sequences are altered to 12TAAG15 and 78GLLTS82. Site directed mutagenesis studies revealed that the introduction of only 5 amino acid exchanges in the two N-terminal regions of either MobA or MocA was sufficient to cause loss of specificity for the pyrimidine or purine nucleotides so that both proteins were able to bind either CTP or GTP to almost the same extent. In addition, the C-terminal domains of MocA and MobA play an important role in determining the specificity of their interaction with the target molybdoenzymes [134]. This domain had previously been implicated in MPT binding [133].
3.4.2 Formation of sulfurated Moco
Apart from the attachment of additional nucleotides in prokaryotes, Moco can be further modified by an exchange of the equatorial molybdenum oxygen ligand with a sulfur atom. The result is the sulfurated or mono-oxo form of Moco shown in Figs. 2 and 8. Sulfurated Moco is the form of the cofactor present in all members of the XO family [7]. Cofactor sulfuration is mediated by the help of members of the XdhC family of molecular chaperones. These molecular chaperones are required for the maturation of molybdoenzymes of the xanthine oxidase family but are not part of the active holo-molybdoenzymes themselves [135]. Investigation of Rhodobacter capsulatus XdhC showed that it binds the Moco produced by MoeA/MogA and protects it from oxidation until the terminal molybdenum sulfur ligand is inserted [123, 136]. XdhC also interacts with the R. capsulatus L-cysteine desulfurase, NifS4, the protein that actually replaces the cofactor equatorial oxygen ligand with a sulfido ligand [137]. The sulfur for this reaction originates from L-cysteine, and a NifS4 persulfide group is formed during the course of the reaction. After the sulfuration reaction, it is believed that XdhC with its bound sulfurated Moco dissociates from NifS4 and forms a new interaction with the XdhB subunits of the R. capsulatus (αβ)2 XDH heterotetramer [123, 138]. Dimerization via the XdhB β-subunits of this protein is required to stabilize a structure that is suitable for Moco insertion [139]. Since XdhC is a dimer itself, it has been suggested that the XdhC dimer interacts with the XdhB dimer to simultaneously insert mature, sulfurated Moco at both XdhB active sites. Following cofactor insertion, XDH folds into its correct conformation and XdhC dissociates from the complex. Since XdhC is extremely labile in the absence of Moco, it is believed to degrade in the cell at this point. Although the mode of control has not yet been identified, insertion of Moco into XDH must be strictly regulated in R. capsulatus, since only sulfurated Moco is inserted into XDH [138].
From the R. capsulatus studies it appears that XdhC-like proteins perform a number of functions in the late steps of Moco synthesis including stabilization of the newly formed Moco, interaction with an L-cysteine desulfurase to ensure that Moco sulfuration occurs [137] as well as interaction with their specific target proteins for insertion of the sulfurated Moco. Because Moco is deeply buried in the protein, it is also believed that the XdhC proteins may act as chaperones to facilitate the proper folding of the target proteins after Moco insertion [135]. This model implies that molybdoenzymes requiring the sulfurated form of Moco exist in a Moco competent ìopenì apo-molybdoenzyme conformation until the insertion of sulfurated Moco. After insertion, the protein adapts the final active îclosedì conformation that can no longer accept Moco [135].
The E. coli aldehyde-oxidoreductase PaoABC protein complex contains the sulfurated MCD cofactor [131], and the PaoD protein is essential for the insertion of sulfurated MCD into PaoABC (Fig. 8). Since PaoD belongs to the XdhC family, it would be expected to play a role similar to that of XdhC with the only difference being that PaoD facilitates sulfuration and insertion of an MCD cofactor rather than an MPT cofactor (Fig. 8). The specific L-cysteine desulfurase involved in E. coli sulfuration of PaoD-bound MCD has not yet been identified.
4. THE REGULATION OF MOCO BIOSYNTHESIS
Genetic analysis of the regulation of Moco biosynthesis determined that the moaABCDE genes are organized in one operon structure [140] and that this is the main target for transcriptional and translational regulation of Moco biosynthesis [103]. Expression of this operon is enhanced under anaerobic conditions and in the presence of molybdate. The dimeric ModE protein binds two molybdate ions, and this complex acts as a positive regulator that binds to the moa promotor region and enhances transcription of the operon as seen in Fig. 9. Dimeric ModE binds two molybdate ions and has a positive effect on molybdoenzyme transcription, e.g. on the operons encoding DMSO reductase and nitrate reductase A in E. coli [141]. It can also act as a transcriptional repressor of the modABCD molybdate transport operon when molybdate is already present in sufficient quantities in the cell [102, 104]. In addition to the ModE/molybdate complex, transcription of the moaABCDE operon in E. coli is also upregulated by the FNR protein (fumarate and nitrate reduction regulatory protein) as seen in Fig. 9 [103]. FNR is a transcriptional regulator that is essential for expressing the proteins involved in anaerobic respiratory processes [142]. The fnr gene was initially identified by Spiro and Guest [143] through the isolation of a number of pleiotropic mutants which lacked the ability to use fumarate and nitrate as reducible substrates for growth under anaerobic conditions. FNR directly senses the ambient oxygen concentration via the disassembly and reassembly of its [4Fe-4S] clusters [144]. The combined regulation of moaABCDE by both ModE and FNR ensures that transcription of the Moco biosynthesis proteins only occurs under conditions where they are both needed (anaerobic growth conditions as sensed by FNR) and capable of producing active Moco (sufficient molybdate levels as sensed by ModE) [103]. Regulation of the moeAB operon is more complicated, since it is regulated by FNR, ArcA (the DNA-binding response regulator in a two-component regulatory system in which ArcB represses transcription of aerobic genes and activates some anaerobic genes) and NarL (part of a two-component regulator protein system for nitrate/nitrite response that acts with the sensor protein NarX) [145]. Surprisingly, FNR represses the nitrate-dependent transcription of the moe operon, but it is assumed that under anaerobic conditions, an intermediate level of transcription from the moeAB operon is ensured by the antagonistic effects of FNR and ArcA-P. In addition, NarL regulates a nitrate-dependent increase in moeAB expression [145].
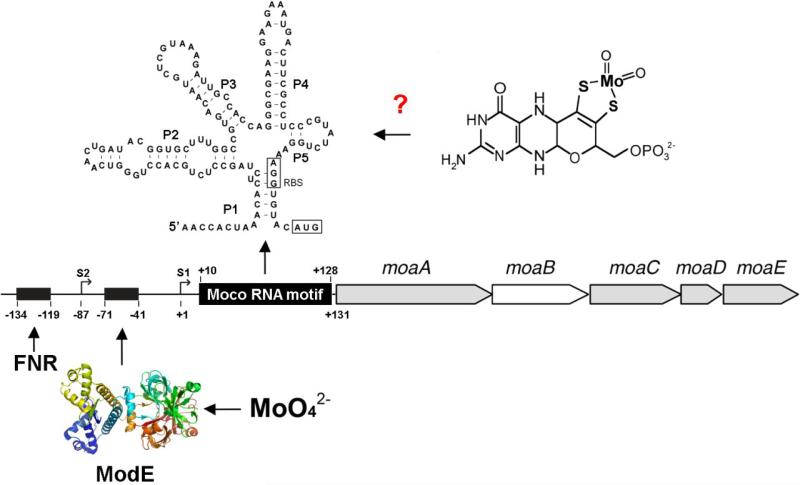
Figure 9. Schematic representation of the moaABCDE gene region in E. coli.
The moaABCDE operon and its upstream regulatory region are shown. S1 defines the first transcriptional start site at +1 and S2 defines the second transcriptional start site at −87. The predicted locations of the FNR and ModE binding sites are indicated by filled boxes. The structure of ModE was derived using the coordinates from the Protein Data Bank (accession number 1B9M). The region designated as the Moco RNA motif is shown in addition to it's proposed secondary structure model. The structure of the Moco RNA motif was adapted from figures shown in Regulski et al. [146]. Numberings and locations of various features are as reported previously [103].
The observation that transcription of the moaABCDE operon is decreased when Moco is present in excess implied there was an additional level of regulation in the system that was not mediated by FNR or ModE [103]. The possible existence of a regulatory region located upstream of the moaA start codon that was responsive to the availability of Moco was first described by Anderson et al. in 2000 [103]. Its existence was confirmed in 2008 when Regulski et al. [146] identified a highly conserved RNA motif upstream of the moa operon in E. coli which was also present upstream of genes encoding molybdoenzymes in a number of different organisms. They determined that the RNA molecule encoded by this sequence controlled gene expression in response to Moco production and suggested that this family of RNA sequences represented a novel class of riboswitches that sense Moco [146]. Riboswitches are structured RNA domains that selectively bind metabolites or metal ions and function as gene control elements [147, 148]. Prior to the identification of the Moco RNA family, four previously identified classes of riboswitch aptamers were known to exist in E. coli. These respond to coenzyme B12, thiamine pyrophosphate, flavin mononucleotide and lysine [147].
Regulski et al. [146] determined that the architectural features of the Moco RNA, which spans 138 nucleotides and forms at least five conserved stem-loop elements as seen in Fig. 9, are similar to other classes of riboswitches. They found that translation of the moa operon is prevented when the Moco RNA structure and Moco are present, although a direct binding of Moco to this RNA motif has not been demonstrated to date due to the lability of isolated Moco [146]. They also suggested that only the molybdenum derivative of Moco binds to the RNA region and not the tungsten derivative which would have its own class of riboswitches This would imply that Moco RNAs are able to distinguish between these two nearly identical cofactors [146]. A Moco sensing riboswitch would ensure that translation of the proteins encoded by the moaABCDE operon is down regulated when Moco is already present in sufficient amounts. Thus riboswitch regulation, in combination with ModE and FNR regulation, would allow for a rapid and efficient response to the changing demand for Moco within the cell.
5. OUTLOOK
Since conserved genes for Moco biosynthesis are found in bacteria, archaea, fungi, plants and animals, it has long been proposed that, with the exception of some organisms like yeast that apparently possesses no molybdoenzymes [149], the biosynthesis of Moco is conserved in all organisms. Thus it seems that determination of the Moco biosynthetic pathway would have been a relative straightforward endeavor. However, more than fifteen years after the report of the first crystal structure of a Moco-containing enzyme, and almost 30 years after publication of the first proposed structure for Moco, a number of questions remain about the pathway of Moco biosynthesis as discussed in this chapter. In addition, elucidation of the details of the pathway for Moco biosynthesis in a variety of organisms has revealed that significant differences in the biosynthetic pathways do exist [15, 20]. For example, in higher eukaryotes, many individual Moco biosynthesis proteins appear to be involved in several different biosynthetic pathways and perform several roles as a consequence of gene sharing [15, 20]. Although a wide range of variations on the basic Moco structure exists in bacteria, no dinucleotide forms of Moco have been identified in eukaryotes to date. Additionally, bacteria contain a large variety of molybdoenzymes that catalyze specific, usually non-essential, redox-reactions. In humans however, only four molybdoenzymes have been identified, and a defect in Moco biosynthesis is lethal due to the loss of sulfite oxidase activity.
Acknowledgements
The authors thank all current and former members of their research groups in addition to collaboration partners who were involved in this work over the past years and decades. The authors were supported by continuous grants from the Deutsche Forschungsgemeinschaft (to S.L.) and the NIH (to K.V.R. and M.M.W.). The Deutsche Forschungsgemeinschaft Cluster of Excellence “Unifying Concepts in Catalysis” coordinated by the Technische Universit‰t Berlin (funding for S.L.) is also acknowledged.
Abbreviations
- cPMP
cyclic pyranopterin monophosphate
- EDAX
energy dispersive analysis of X-rays
- EXAFS
extended X-ray absorption fine structure
- MCD
molybdopterin cytosine dinucleotide cofactor
- MGD
molybdopterin guanine dinucleotide cofactor
- Moco
molybdenum cofactor
- MPT
molybdopterin
- NR
nitrate reductase
- XDH
xanthine dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ter Meulen H. Distribution of Molybdenum. Nature. 1932;130:966. [Google Scholar]
- [2].Mudd SH, Irreverre F, Laster L. Sulfite oxidase deficiency in man: demonstration of the enzymatic defect. Science. 1967;156:1599–1602. doi: 10.1126/science.156.3782.1599. [DOI] [PubMed] [Google Scholar]
- [3].Cohen HJ, Fridovich I, Rajagopalan KV. Hepatic sulfite oxidase. A functional role for molybdenum. J. Biol. Chem. 1971;246:374–382. [PubMed] [Google Scholar]
- [4].Johnson JL, Hainline BE, Rajagopalan KV. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine oxidase and nitrate reductase. Identification of a pteridine as a structural component. J. Biol. Chem. 1980;255:1783–1786. [PubMed] [Google Scholar]
- [5].Johnson JL, Rajagopalan KV. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc. Natl. Acad. Sci. U. S. A. 1982;79:6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rajagopalan KV. Biosynthesis of the molybdenum cofactor, in Escherichia coli and Salmonella. In: Neidhardt FC, editor. Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 674–679. [Google Scholar]
- [7].Hille R. The mononuclear molybdenum enzymes. Chemical Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- [8].Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- [9].Rajagopalan KV. Novel aspects of the biochemistry of the molybdenum cofactor. In: Meister A, editor. Advances in Enzymology and Related Areas of Molecular Biology. John Wiley and Sons; New York: 1991. pp. 215–290. [DOI] [PubMed] [Google Scholar]
- [10].Rajagopalan KV, Johnson JL. The pterin molybdenum cofactors. J. Biol. Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- [11].Johnson JL, Bastian NR, Rajagopalan KV. Molybdopterin guanine dinucleotide: a modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3190–3194. doi: 10.1073/pnas.87.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shanmugam KT, Stewart V, Gunsalus RP, Boxer DH, Cole JA, Chippaux M, DeMoss JA, Giordano G, Lin ECC, Rajagopalan KV. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol. Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- [13].Stewart V, MacGregor CH. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J. Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnson ME, Rajagopalan KV. Involvement of chlA, E, M, and N loci in Escherichia coli molybdopterin biosynthesis. J. Bacteriol. 1987;169:117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fischer M, Thöny B, Leimkühler S. The Biosynthesis of Folate and Pterin and their Enzymology. Comprehensive natural products chemistry II Volume. 2010;7:599–648. [Google Scholar]
- [16].Cove DJ, Pateman JA. Independently segregating genetic loci concerned with nitrate reductase activity in Aspergillus nidulans. Nature. 1963;198:262–263. doi: 10.1038/198262a0. [DOI] [PubMed] [Google Scholar]
- [17].Pateman JA, Cove DJ, Rever BM, Roberts DB. A common cofactor for nitrate reductase and xanthine dehydrogenase which also regulates the synthesis of nitrate reductase. Nature. 1964;201:58–60. doi: 10.1038/201058a0. [DOI] [PubMed] [Google Scholar]
- [18].Shah VK, Brill WJ. Isolation of an iron-molybdenum cofactor from nitrogenase. Proc Natl Acad Sci U S A. 1977;74:3249–3253. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Glaser JH, DeMoss JA. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116:1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- [20].Schwarz G. Molybdenum cofactor biosynthesis and deficiency. Cell Mol Life Sci. 2005;62:2792–2810. doi: 10.1007/s00018-005-5269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miller JB, Amy NK. Molybdenum cofactor in chlorate-resistant and nitrate reductase-deficient insertion mutants of Escherichia coli. J. Bacteriol. 1983;155:793–801. doi: 10.1128/jb.155.2.793-801.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Amy NK, Rajagopalan KV. Characterization of molybdenum cofactor from Escherichia coli. J. Bacteriol. 1979;140:114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jeter RM, Sias SR, Ingraham JL. Choromosomal location and function affecting Pseudomonas aeruginosa nitrate assimilation. J Bacteriol. 1984;157:673–677. doi: 10.1128/jb.157.2.673-677.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stouthamer AH. Genetics and biochemistry of reductase formation in enteribacteriaceae. Antonie van Leeuwenhoek. 1970;36:181. [Google Scholar]
- [25].Scazzocchio C. The genetics of the molybdenum-containing enzymes. In: Coughlan MP, editor. Molybdenum and Molybdenum-Containing Enzymes. Pergamon Press; Oxford: 1980. pp. 487–515. [Google Scholar]
- [26].Warner CK, Finnerty V. Molybdenum hydroxylases in Drosophila II. Molybdenum cofactor in xanthine dehydrogenase, aldehyde oxidase and pyridoxal oxidase. Mol. Gen. Genet. 1981;184:92–96. doi: 10.1007/BF00271201. [DOI] [PubMed] [Google Scholar]
- [27].Buchanan RJ, Wray JL. Isolation of molybdenum cofactor defective cell lines of Nicotiana tabacum. Mol Gen Genet. 1982;188:228–234. [Google Scholar]
- [28].Nason A, Antoine AD, Ketchum PA, Frazier WA, III, Lee DK. Formation of assimilatory nitrate reductase by in vitro inter-cistronic complementation in Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 1970;65:137–144. doi: 10.1073/pnas.65.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nason A, Lee K-Y, Pan S-S, Erickson RH. Evidence for a molybdenum cofactor common to all molybdenum enzymes based on the in vitro assembly of assimilatory NADPH-nitrate reductase using the Neurospora mutant nit-1. J. Less Common Metals. 1974;36:449–459. [Google Scholar]
- [30].Nason A, Lee K-Y, Pan S-S, Ketchum PA, Lamberti A, DeVries J. In vitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: nitrate reductase from a Neurospora mutant and a component of molybdenum-enzymes. Proc. Natl. Acad. Sci. U. S. A. 1971;68:3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ketchum PA, Cambier HY, Frazier WA, III, Madansky CH, Nason A. In vitro assembly of Neurospora assimilatory nitrate reductase from protein subunits of a Neurospora mutant and the xanthine oxidizing or aldehyde oxidase systems of higher animals. Proc. Natl. Acad. Sci. U. S. A. 1970;66:1016–1023. doi: 10.1073/pnas.66.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hagemann RV, Rajagopalan KV. Assay and detection of the molybdenum cofactor. Methods Enzymol. 1986;122:399–412. doi: 10.1016/0076-6879(86)22200-4. [DOI] [PubMed] [Google Scholar]
- [33].Kramer S, Hageman RV, Rajagopalan KV. In vitro reconstitution of nitrate reductase activity of the Neurospora crassa mutant nit-1: specific incorporation of molybdopterin. Arch Biochem Biophys. 1984;233:821–829. doi: 10.1016/0003-9861(84)90511-3. [DOI] [PubMed] [Google Scholar]
- [34].Johnson JL, Hainline BE, Rajagopalan KV, Arison BH. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J. Biol. Chem. 1984;259:5414–5422. [PubMed] [Google Scholar]
- [35].Johnson JL, Jones HP, Rajagopalan KV. In vitro reconstitution of demolybdosulfite oxidase by a molybdenum cofactor from rat liver and other sources. J. Biol. Chem. 1977;252:4994–5003. [PubMed] [Google Scholar]
- [36].Rajagopalan KV, Johnson JL, Hainline BE. The pterin of the molybdenum cofactor. Fed. Proc. 1982;41:2608–2612. [PubMed] [Google Scholar]
- [37].Taylor EC, Ray PS, Darwish IS, Johnson JL, Rajagopalan KV. Studies on the molybdenum cofactor. Determination of the structure and absolute configuration of Form A. J. Am. Chem. Soc. 1989;111:7664–7665. [Google Scholar]
- [38].Koschara W. Urothion, ein gelber, schwefelreicher Farbstoff aus Menschenharn. Hoppe-Seyler's Z. Physiol. Chem. 1940;263:78–79. [Google Scholar]
- [39].Goto M, Sakurai A, Ohta K, Yamakami H. Die struktur des urothions. J. Biochem. 1969;65:611–620. doi: 10.1093/oxfordjournals.jbchem.a129054. [DOI] [PubMed] [Google Scholar]
- [40].Mendel RR, Alikulov ZA. Reversible mobilization of molybdenum cofactor on a gel matrix via sulphydryl groups. J. Chromatogr. 1983;267:409–413. [Google Scholar]
- [41].Kramer SP, Johnson JL, Ribeiro AA, Millington DS, Rajagopalan KV. The structure of the molybdenum cofactor. Characterization of di-(carboxamidomethyl)molybdopterin from sulfite oxidase and xanthine oxidase. J. Biol. Chem. 1987;262:16357–16363. [PubMed] [Google Scholar]
- [42].Chan MK, Mukund S, Kletzin A, Adams MWW, Rees DC. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science. 1995;267:1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- [43].Ramão MJ, Archer M, Moura I, Moura JJG, LeGall J, Engh R, Schneider M, Hof P, Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxidoreductase from D. gigas. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- [44].May HD, Schauer NL, Ferry JG. Molybdopterin cofactor from Methanobacterium formicium formate dehydrogenase. J Bacteriol. 1986;166:500–504. doi: 10.1128/jb.166.2.500-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Krüger B, Meyer O. Structural elements of bactopterin from Pseudomonas carboxydoflava carbon monoxide dehydrogenase. Biochim. Biophys. Acta. 1987;912:357–364. doi: 10.1016/0167-4838(87)90040-9. [DOI] [PubMed] [Google Scholar]
- [46].Krüger B, Meyer O, Nagel M, Andreesen JR, Meincke M, Bock E, Blümle S, Zumft WG. Evidence for the presence of bactopterin in the eubacterial molybdoenzymes nicotinic acid dehydrogenase, nitrite oxidoreductase, and respiratory nitrate reductase. FEMS Microbiol. Lett. 1987;48:225–227. [Google Scholar]
- [47].Satoh T, Kurihara FN. Purification and properties of dimethylsulfoxide reductase containing a molybdenum cofactor from a photonitrifier, Rhodopseudomonas sphaeroides f.s. denitrificans. J. Biochem. 1987;102:191–197. doi: 10.1093/oxfordjournals.jbchem.a122032. [DOI] [PubMed] [Google Scholar]
- [48].Meyer O, Rajagopalan KV. Molybdopterin in carbon monoxide oxidase from carboxydotrophic bacteria. J. Bacteriol. 1984;157:643–648. doi: 10.1128/jb.157.2.643-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Johnson JL, Rajagopalan KV, Meyer O. Isolation and characterization of a second molybdopterin dinucleotide: molybdopterin cytosine dinucleotide. Arch. Biochem. Biophys. 1990;283:542–545. doi: 10.1016/0003-9861(90)90681-n. [DOI] [PubMed] [Google Scholar]
- [50].Hilton JC, Rajagopalan KV. Identification of the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides f. sp. denitrificans as bis(molybdopterin guanine dinucleotide)molybdenum. Arch. Biochem. Biophys. 1996;325:139–143. doi: 10.1006/abbi.1996.0017. [DOI] [PubMed] [Google Scholar]
- [51].Schindelin H, Kisker C, Hilton J, Rajagopalan KV, Rees DC. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- [52].Pitterle DM, Rajagopalan KV. Two proteins encoded at the chlA locus constitute the converting factor of Escherichia coli chlA1. J. Bacteriol. 1989;171:3373–3378. doi: 10.1128/jb.171.6.3373-3378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Johnson JL, Wuebbens MM, Rajagopalan KV. The structure of a molybdopterin precursor. Characterization of a stable, oxidized derivative. J. Biol. Chem. 1989;264:13440–13447. [PubMed] [Google Scholar]
- [54].Wuebbens MM, Rajagopalan KV. Structural characterization of a molybdopterin precursor. J. Biol. Chem. 1993;268:13493–13498. [PubMed] [Google Scholar]
- [55].Santamaria-Araujo JA, Fischer B, Otte T, Nimtz M, Mendel RR, Wray V, Schwarz G. The tetrahydropyranopterin structure of the sulfur- and metal-free molybdenum cofactor precursor. J Biol Chem. 2004;279:15994–15999. doi: 10.1074/jbc.M311815200. [DOI] [PubMed] [Google Scholar]
- [56].Daniels JN, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal Structure of a Molybdopterin Synthase-Precursor Z Complex: Insight into Its Sulfur Transfer Mechanism and Its Role in Molybdenum Cofactor Deficiency. Biochemistry. 2008;47:615–626. doi: 10.1021/bi701734g. [DOI] [PubMed] [Google Scholar]
- [57].Bacher A, Eberhardt S, Eisenreich W, Fischer M, Herz S, Illarionov B, Kis K, Richter G. Biosynthesis of riboflavin. Vitam Horm. 2001;61:1–49. doi: 10.1016/s0083-6729(01)61001-x. [DOI] [PubMed] [Google Scholar]
- [58].Wuebbens MM, Rajagopalan KV. Investigation of the early steps of molybdopterin biosynthesis in Escherichia coli through the use of in vivo labeling studies. J. Biol. Chem. 1995;270:1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- [59].Pfleiderer W. Chemistry of naturally occurring pterins. In: Blakley RL, Benkovic SJ, editors. Folates and Pterins. John Wiley and Sons; New York: 1985. pp. 43–114. [Google Scholar]
- [60].Blau N, Niederwieser A. GTP-cyclohydrolases: a review. J Clin Chem Clin Biochem. 1895;23:169–176. [PubMed] [Google Scholar]
- [61].Rieder C, Eisenreich W, O'Brien J, Richter G, Götze E, Boyle P, Blanchard S, Bacher A, Simon H. Rearrangement reactions in the biosynthesis of molybdopterin. An NMR study with multiply 13C/15N labelled precursors. Eur. J. Biochem. 1998;255:24–36. doi: 10.1046/j.1432-1327.1998.2550024.x. [DOI] [PubMed] [Google Scholar]
- [62].Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hanzelmann P, Schindelin H. Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci U S A. 2006;103:6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lees NS, Hänzelmann P, Hernandez HL, Subramanian S, Schindelin H, Johnson MK, Hoffman BM. ENDOR spectroscopy shows that guanine N1 binds to [4Fe-4S] cluster II of the S-adenosylmethionine-dependent enzyme MoaA: mechanistic implications. J Am Chem Soc. 2009;131:9184–9185. doi: 10.1021/ja903978u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wuebbens MM, Liu MT, Rajagopalan K, Schindelin H. Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC. Structure Fold Des. 2000;8:709–718. doi: 10.1016/s0969-2126(00)00157-x. [DOI] [PubMed] [Google Scholar]
- [67].Hanzelmann P, Hernandez HL, Menzel C, Garcia-Serres R, Huynh BH, Johnson MK, Mendel RR, Schindelin H. Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis. J Biol Chem. 2004;279:34721–34732. doi: 10.1074/jbc.M313398200. [DOI] [PubMed] [Google Scholar]
- [68].Kanaujia SP, Jeyakanthan J, Nagawaka N, Balasubramaniam S, Shinkai A, Kuramitsu S, Yokoyama S, Sekar K. Sructure of apo and GTP-bound molydenum cofactor biosnthesis protein MoaC from Thermus thermophilus HB8. Acta Crystallograph. 2010;D66:821–833. doi: 10.1107/S0907444910019074. [DOI] [PubMed] [Google Scholar]
- [69].Veldman A, Santamaria-Araujo JA, Sollazzo S, Gianello PJ,R, Yaplito-Lee J, Wong F, Ramsden CA, Reiss J, Cook I, Fairweather J, Schwarz G. Successful treatment of molybdenum cofactor deficiency type A with cPMP. Pedriatics. 2010;125:e1249–1254. doi: 10.1542/peds.2009-2192. [DOI] [PubMed] [Google Scholar]
- [70].Schwarz G, Santamaria-Araujo JA, Wolf S, Lee HJ, Adham IM, Grone HJ, Schwegler H, Sass JO, Otte T, Hanzelmann P, Mendel RR, Engel W, Reiss J. Rescue of lethal molybdenum cofactor deficiency by a biosynthetic precursor from Escherichia coli. Hum Mol Genet. 2004;13:1249–1255. doi: 10.1093/hmg/ddh136. [DOI] [PubMed] [Google Scholar]
- [71].Pitterle DM, Johnson JL, Rajagopalan KV. In vitro synthesis of molybdopterin from precursor Z using purified converting factor. Role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 1993;268:13506–13509. [PubMed] [Google Scholar]
- [72].Pitterle DM, Rajagopalan KV. The biosynthesis of molybdopterin in Escherichia coli. Purification and characterization of the converting factor. J. Biol. Chem. 1993;268:13499–13505. [PubMed] [Google Scholar]
- [73].Gutzke G, Fischer B, Mendel RR, Schwarz G. Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins. J. Biol. Chem. 2001;276:36268–36274. doi: 10.1074/jbc.M105321200. [DOI] [PubMed] [Google Scholar]
- [74].Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- [75].Rudolph MJ, Wuebbens MM, Turque O, Rajagopalan KV, Schindelin H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. J. Biol. Chem. 2003;278:14514–14522. doi: 10.1074/jbc.M300449200. [DOI] [PubMed] [Google Scholar]
- [76].Wuebbens MM, Rajagopalan KV. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J Biol Chem. 2003;278:14523–14532. doi: 10.1074/jbc.M300453200. [DOI] [PubMed] [Google Scholar]
- [77].Rajagopalan KV. Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans. 1997;25:757–761. doi: 10.1042/bst0250757. [DOI] [PubMed] [Google Scholar]
- [78].Leimkühler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of MPT synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 2001;276:34695–34701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- [79].McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- [81].Schindelin H. Evolutionary Origin of the Activation Step During Ubiquitin-dependent Protein Degradation. In: Mayer JR, Ciechanover A, Rechsteiner M, editors. Protein Degradation 1. Ubiquitin and the Chemistry of Life (Vol. 1) Wiley-VCH Verlag; Weinheim: 2005. pp. 21–43. [Google Scholar]
- [82].Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- [83].Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- [84].Haas AL, Warms JV, Rose IA. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry. 1983;22:4388–4394. doi: 10.1021/bi00288a007. [DOI] [PubMed] [Google Scholar]
- [85].Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- [86].Burch TJ, Haas AL. Site-directed mutagenesis of ubiquitin. Differential roles for arginine in the interaction with ubiquitin-activating enzyme. Biochemistry. 1994;33:7300–7308. doi: 10.1021/bi00189a035. [DOI] [PubMed] [Google Scholar]
- [87].Schmitz J, Wuebbens MM, Rajagopalan KV, Leimkühler S. Role of the C-Terminal Gly-Gly Motif of Escherichia Coli MoaD, a Molybdenum Cofactor Biosynthesis Protein with a Ubiquitin Fold. Biochemistry. 2007;46:909–916. doi: 10.1021/bi062011w. [DOI] [PubMed] [Google Scholar]
- [88].Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Taylor SV, Kelleher NL, Kinsland C, Chiu H-J, Costello CA, Backstrom AD, McLafferty FW, Begley TP. Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- [90].Kambampati R, Lauhon CT. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J Biol Chem. 2000;275:10727–10730. doi: 10.1074/jbc.275.15.10727. [DOI] [PubMed] [Google Scholar]
- [91].Xi J, Ge Y, Kinsland C, McLafferty F, Begley TP. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: Identification of an acyldisulfide-linked protein-protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc Natl Acad Sci U S A. 2001;98:8513–8518. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Leimkühler S, Rajagopalan KV. An Escherichia coli NifS-like sulfurtransferase is required for the transfer of cysteine sulfur in the in vitro synthesis of molybdopterin from precursor Z. J. Biol. Chem. 2001;276:22024–22031. doi: 10.1074/jbc.M102072200. [DOI] [PubMed] [Google Scholar]
- [93].Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- [94].Zhang W, Urban A, Mihara H, Leimkühler S, Kurihara T, Esaki N. IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J Biol Chem. 2010;285:2302–2308. doi: 10.1074/jbc.M109.082172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Matthies A, Rajagopalan KV, Mendel RR, Leimkühler S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc Natl Acad Sci U S A. 2004;101:5946–5951. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- [97].Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- [98].Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- [99].Hu Y, Rech S, Gunsalus RP, Rees DC. Crystal structure of the molybdate binding protein ModA. Nat Struct Biol. 1997;4:703–707. doi: 10.1038/nsb0997-703. [DOI] [PubMed] [Google Scholar]
- [100].McNicholas PM, Rech SA, Gunsalus RP. Characterization of the ModE DNA-binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol Microbiol. 1997;23:515–524. doi: 10.1046/j.1365-2958.1997.d01-1864.x. [DOI] [PubMed] [Google Scholar]
- [101].McNicholas PM, Mazzotta MM, Rech SA, Gunsalus RP. Functional dissection of the molybdate-responsive transcription regulator, ModE, from Escherichia coli. J Bacteriol. 1998;180:4638–4643. doi: 10.1128/jb.180.17.4638-4643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Self WT, Grunden AM, Hasona A, Shanmugam KT. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology. 1999;145(Pt 1):41–55. doi: 10.1099/13500872-145-1-41. [DOI] [PubMed] [Google Scholar]
- [103].Anderson LA, McNairn E, Lubke T, Pau RN, Boxer DH. ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J Bacteriol. 2000;182:7035–7043. doi: 10.1128/jb.182.24.7035-7043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Schuttelkopf AW, Boxer DH, Hunter WN. Crystal structure of activated ModE reveals conformational changes involving both oxyanion and DNA-binding domains. J Mol Biol. 2003;326:761–767. doi: 10.1016/s0022-2836(02)01358-x. [DOI] [PubMed] [Google Scholar]
- [105].Joshi MS, Johnson JL, Rajagopalan KV. Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J. Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Liu MT, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli. J. Biol. Chem. 2000;275:1814–1822. doi: 10.1074/jbc.275.3.1814. [DOI] [PubMed] [Google Scholar]
- [107].Xiang S, Nichols J, Rajagopalan KV, Schindelin H. The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin. Structure. 2001;9:299–310. doi: 10.1016/s0969-2126(01)00588-3. [DOI] [PubMed] [Google Scholar]
- [108].Schrag JD, Huang W, Sivaraman J, Smith C, Plamondon J, Larocque R, Matte A, Cygler M. The crystal structureof Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway. J Mol Biol. 2001;310:419–431. doi: 10.1006/jmbi.2001.4771. [DOI] [PubMed] [Google Scholar]
- [109].Nichols JD, Rajagopalan KV. In vitro molybdenum ligation to molybdopterin using purified components. J Biol Chem. 2005;280:7817–7822. doi: 10.1074/jbc.M413783200. [DOI] [PubMed] [Google Scholar]
- [110].Llamas A, Mendel RR, Schwarz G. Synthesis of adenylated molybdopterin: an essential step for molybdenum insertion. J Biol Chem. 2004;279:55241–55246. doi: 10.1074/jbc.M409862200. [DOI] [PubMed] [Google Scholar]
- [111].Llamas A, Otte T, Multhaup G, Mendel RR, Schwarz G. The Mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J Biol Chem. 2006;281:18343–18350. doi: 10.1074/jbc.M601415200. [DOI] [PubMed] [Google Scholar]
- [112].Kuper J, Llamas A, Hecht HJ, Mendel RR, Schwarz G. Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature. 2004;430:803–806. doi: 10.1038/nature02681. [DOI] [PubMed] [Google Scholar]
- [113].Leimkühler S, Rajagopalan KV. In vitro incorporation of nascent molybdenum cofactor into human sulfite oxidase. J Biol Chem. 2001;276:1837–1844. doi: 10.1074/jbc.M007304200. [DOI] [PubMed] [Google Scholar]
- [114].Neumann M, Leimkuhler S. Heavy metal ions inhibit molybdoenzyme activity by binding to the dithiolene moiety of molybdopterin in Escherichia coli. Febs J. 2008;275:5678–5689. doi: 10.1111/j.1742-4658.2008.06694.x. [DOI] [PubMed] [Google Scholar]
- [115].Morrison MS, Cobine PA, Hegg EL. Probing the role of copper in the biosynthesis of the molybdenum cofactor in Escherichia coli and Rhodobacter sphaeroides. J Biol Inorg Chem. 2007 doi: 10.1007/s00775-007-0279-x. [DOI] [PubMed] [Google Scholar]
- [116].Loschi L, Brokx SJ, Hills TL, Zhang G, Bertero MG, Lovering AL, Weiner JH, Strynadka NC. Structural and biochemical identification of a novel bacterial oxidoreductase. J Biol Chem. 2004;279:50391–50400. doi: 10.1074/jbc.M408876200. [DOI] [PubMed] [Google Scholar]
- [117].Brokx SJ, Rothery RA, Zhang G, Ng DP, Weiner JH. Characterization of an Escherichia coli Sulfite Oxidase Homologue Reveals the Role of a Conserved Active Site Cysteine in Assembly and Function. Biochemistry. 2005;44:10339–10348. doi: 10.1021/bi050621a. [DOI] [PubMed] [Google Scholar]
- [118].Lake MW, Temple CA, Rajagopalan KV, Schindelin H. The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis. J Biol Chem. 2000;275:40211–40217. doi: 10.1074/jbc.M007406200. [DOI] [PubMed] [Google Scholar]
- [119].Palmer T, Goodfellow IP, Sockett RE, McEwan AG, Boxer DH. Characterisation of thee mob locus from Rhodobacter sphaeroides required for molybdenum cofactor biosynthesis. Biochem Biophys Acta. 1998;1395:135–140. doi: 10.1016/s0167-4781(97)00145-0. [DOI] [PubMed] [Google Scholar]
- [120].Palmer T, Santini C-L, Iobbi-Nivol C, Eaves DJ, Boxer DH, Giordano G. Involvement of the narJ and mob gene products in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- [121].McLuskey K, Harrison JA, Schuttelkopf AW, Boxer DH, Hunter WN. Insight into the role of Escherichia coli MobB in molybdenum cofactor biosynthesis based on the high resolution crystal structure. J Biol Chem. 2003;278:23706–23713. doi: 10.1074/jbc.M301485200. [DOI] [PubMed] [Google Scholar]
- [122].Temple CA, Rajagopalan KV. Mechanism of assembly of the Bis(Molybdopterin guanine Dinucleotide)Molybdenum cofactor in Rhodobacter sphaeroides dimethyl sulfoxide reductase. J Biol Chem. 2000;275:40202–40210. doi: 10.1074/jbc.M007407200. [DOI] [PubMed] [Google Scholar]
- [123].Neumann M, Stöcklein W, Leimkühler S. Transfer of the Molybdenum Cofactor Synthesized by Rhodobacter capsulatus MoeA to XdhC and MobA. J Biol Chem. 2007;282:28493–28500. doi: 10.1074/jbc.M704020200. [DOI] [PubMed] [Google Scholar]
- [124].Stevenson CE, Sargent F, Buchanan G, Palmer T, Lawson DM. Crystal structure of the molybdenum cofactor biosynthesis protein MobA from Escherichia coli at near-atomic resolution. Structure Fold Des. 2000;8:1115–1125. doi: 10.1016/s0969-2126(00)00518-9. [DOI] [PubMed] [Google Scholar]
- [125].Genest O, Neumann M, Seduk F, Stocklein W, Mejean V, Leimkühler S, Iobbi-Nivol C. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J Biol Chem. 2008;283:21433–21440. doi: 10.1074/jbc.M802954200. [DOI] [PubMed] [Google Scholar]
- [126].Genest O, Mejan V, Iobbi-Nivol C. Multiple roles of TorD-like chaperones in the biogenesis of molybdoenzymes. FEMS Microbiol Lett. 2009;297:1–9. doi: 10.1111/j.1574-6968.2009.01660.x. [DOI] [PubMed] [Google Scholar]
- [127].Börner G, Karrasch M, Thauer RK. Molybdopterin adenine dinucleotide and molybdopterin hypoxanthine dinucleotide in formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum (Marburg) FEBS Lett. 1991;290:31–34. doi: 10.1016/0014-5793(91)81218-w. [DOI] [PubMed] [Google Scholar]
- [128].Schübel U, Kraut M, Mörsdorf G, Meyer O. Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans. J. Bacteriol. 1995;177:2197–2203. doi: 10.1128/jb.177.8.2197-2203.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Xi H, Schneider BL, Reitzer L. Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J Bacteriol. 2000;182:5332–5341. doi: 10.1128/jb.182.19.5332-5341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Kozmin SG, Schaaper RM. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutat Res. 2007;619:9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Neumann M, Mittelstädt G, Iobbi-Nivol C, Miguel Saggu M, Lendzian F, Hildebrandt P, Leimkühler S. A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 2009;276:2762–2774. doi: 10.1111/j.1742-4658.2009.07000.x. [DOI] [PubMed] [Google Scholar]
- [132].Neumann M, Mittelstädt G, Seduk F, Iobbi-Nivol C, Leimkühler S. MocA is a specific cytidylyl transferase involved in molybdopterin cytosine dinucleotide biosynthesis in Escherichia coli. J Biol Chem. 2009;284:21891–21898. doi: 10.1074/jbc.M109.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Guse A, Stevenson CE, Kuper J, Buchanan G, Schwarz G, Giordano G, Magalon A, Mendel RR, Lawson DM, Palmer T. Biochemical and structural analysis of the molybdenum cofactor biosynthesis protein MobA. J Biol Chem. 2003;278:25302–25307. doi: 10.1074/jbc.M302639200. [DOI] [PubMed] [Google Scholar]
- [134].Neumann M, Seduk F, Iobbi-Nivol C, Leimkuhler S. Molybdopterin dinucleotide biosynthesis in Escherichia coli: Identification of amino acid residues of molybdopterin dinucleotide transferases that determine specificity for binding of guanine or cytosine nucleotides. J Biol Chem. 2010 doi: 10.1074/jbc.M110.155671. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Leimkühler S, Klipp W. Role of XDHC in Molybdenum cofactor insertion into xanthine dehydrogenase of Rhodobacter capsulatus. J Bacteriol. 1999;181:2745–2751. doi: 10.1128/jb.181.9.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Leimkühler S, Angermüller S, Schwarz G, Mendel RR, Klipp W. Activity of the molybdopterin-containing xanthine dehydrogenase of Rhodobacter capsulatus can be restored by high molybdenum concentrations in a moeA mutant defective in molybdenum cofactor biosynthesis. J Bacteriol. 1999;181:5930–5939. doi: 10.1128/jb.181.19.5930-5939.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Neumann M, Stöcklein W, Walburger A, Magalon A, Leimkühler S. Identification of a Rhodobacter capsulatus L-cysteine desulfurase that sulfurates the molybdenum cofactor when bound to XdhC and before its insertion into xanthine dehydrogenase. Biochemistry. 2007;46:9586–9595. doi: 10.1021/bi700630p. [DOI] [PubMed] [Google Scholar]
- [138].Neumann M, Schulte M, Jünemann N, Stöcklein W, Leimkühler S. Rhodobacter capsulatus XdhC is involved in molybdenum cofactor binding and insertion into xanthine dehydrogenase. J Biol Chem. 2006;281:15701–15708. doi: 10.1074/jbc.M601617200. [DOI] [PubMed] [Google Scholar]
- [139].Schumann S, Saggu M, Möller N, Anker SD, Lendzian F, Hildebrandt P, Leimkühler S. The mechanism of assembly and cofactor insertion into Rhodobacter capsulatus xanthine dehydrogenase. J Biol Chem. 2008;283:16602–16611. doi: 10.1074/jbc.M709894200. [DOI] [PubMed] [Google Scholar]
- [140].Rivers SL, McNairn E, Blasco F, Giordano G, Boxer DH. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol. Microbiol. 1993;8:1071–1081. doi: 10.1111/j.1365-2958.1993.tb01652.x. [DOI] [PubMed] [Google Scholar]
- [141].McNicholas PM, Chiang RC, Gunsalus RP. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol Microbiol. 1998;27:197–208. doi: 10.1046/j.1365-2958.1998.00675.x. [DOI] [PubMed] [Google Scholar]
- [142].Unden G, Achebach S, Holighaus G, Tran HG, Wackwitz B, Zeuner Y. Control of FNR function of Escherichia coli by O2 and reducing conditions. J Mol Microbiol Biotechnol. 2002;4:263–268. [PubMed] [Google Scholar]
- [143].Spiro S, Guest JR. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;6:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- [144].Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- [145].Hasona A, Self WT, Sahanmugam KT. Transcriptional regulation of moe (molybdate metabolism) operon of Escherichia coli. Arch Microbiol. 2001;175:178–188. doi: 10.1007/s002030100252. [DOI] [PubMed] [Google Scholar]
- [146].Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68:918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- [148].Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang Y, Glagyshev VN. Molybdoproteomes and evolution of molybdenum utilization. J Mol Biol. 2008;379:881–899. doi: 10.1016/j.jmb.2008.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]