Abstract
The fungal kingdom is vast, spanning ~1.5 to as many as 5 million species diverse as unicellular yeasts, filamentous fungi, mushrooms, lichens, and both plant and animal pathogens. The fungi are closely aligned with animals in one of the six to eight supergroups of eukaryotes, the opisthokonts. The animal and fungal kingdoms last shared a common ancestor ~1 billion years ago, more recently than other groups of eukaryotes. As a consequence of their close evolutionary history and shared cellular machinery with metazoans, fungi are exceptional models for mammalian biology, but prove more difficult to treat in infected animals. The last common ancestor to the fungal/metazoan lineages is thought to have been unicellular, aquatic, and motile with a posterior flagellum, and certain extant species closely resemble this hypothesized ancestor. Species within the fungal kingdom were traditionally assigned to four phyla, including the basal fungi (Chytridiomycota, Zygomycota) and the more recently derived monophyletic lineage, the dikarya (Ascomycota, Basidiomycota). The fungal tree of life project has revealed that the basal lineages are polyphyletic, and thus there are as many as eight to ten fungal phyla. Fungi that infect vertebrates are found in all of the major lineages, and virulence arose multiple times independently. A sobering recent development involves the species Batrachochytrium dendrobatidis from the basal fungal phylum, the Chytridiomycota, which has emerged to cause global amphibian declines and extinctions. Genomics is revolutionizing our view of the fungal kingdom, and genome sequences for zygomycete pathogens (Rhizopus, Mucor), skin-associated fungi (dermatophytes, Malassezia), and the Candida pathogenic species clade promise to provide insights into the origins of virulence. Here we survey the diversity of fungal pathogens and illustrate key principles revealed by genomics involving sexual reproduction and sex determination, loss of conserved pathways in derived fungal lineages that are retained in basal fungi, and shared and divergent virulence strategies of successful human pathogens, including dimorphic and trimorphic transitions in form. The overarching conclusion is that fungal pathogens of animals have arisen repeatedly and independently throughout the fungal tree of life, and while they share general properties, there are also unique features to the virulence strategies of each successful microbial pathogen.
Introduction
Infectious diseases remain one of the most significant threats to human health (Cohen, 2000; Jones et al., 2008; Morens et al., 2004). In contrast to chronic diseases such as heart disease and cancer, infectious diseases represent a threat capable of causing extinction and thus have the capacity to threaten the very survival of our species. In addition, infectious diseases are subject to rapid evolution and emergence, given the rapidity of their life cycles and large population sizes for disease-causing microbes. Among pathogenic microbes, the eukaryotic pathogens (fungi, parasites) are increasing in incidence, drug resistance is of mounting concern, and there are fewer drugs or vaccines available compared to bacteria and viruses.
The emergence of microbial pathogens involves several routes, including zoonotic transmission from animals to humans, changes in host or vector range, environmental change, and changes to the pathogen via genetic exchange. Genome reassortment of influenza leads to antigenic shifts, necessitating annual changes in vaccine strains and which can lead to pandemics (Lambert and Fauci, 2010). Bacteria predominantly exchange genetic material via horizontal gene transfer (transformation, conjugation, transduction). In both fungi and parasites, genetic exchange is mediated via sexual reproduction (Heitman, 2006, 2010). Sex generates genetic diversity, can promote and transmit drug resistance, and plays roles in pathogenesis and virulence cycles including infectious propagule production.
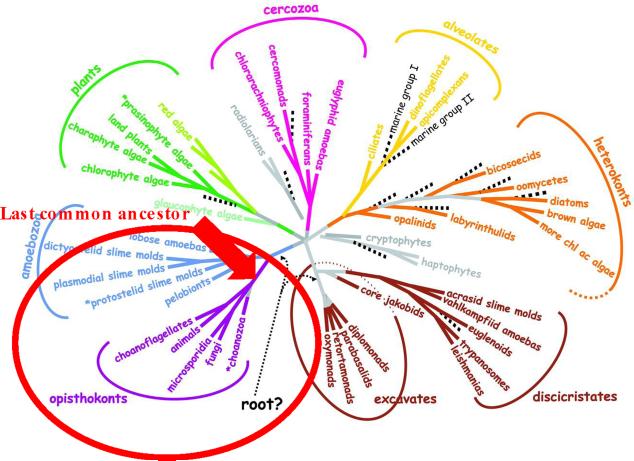
Our understanding of the origins of fungal microbial pathogens begins with the eukaryotic evolutionary tree of life, which has been redrawn over the past decade based on molecular phylogenetic studies (Baldouf and Palmer, 1993). A key insight was the realization that fungi and animals share a much more recent ancestor than had been appreciated and that the two are sister kingdoms within the opisthokont supergroup lineage of the eukaryotes, which shared a last common ancestor as recently as one billion years ago (Figure 1) (Baldouf and Palmer, 1993; Wainright et al., 1993). The last common ancestor from which all animals and fungi descend is thought to have been unicellular, aquatic, and motile, with a flagellum. There are extant species that closely resemble this hypothesized last common ancestor, and one is the unicellular choanoflagellate Monosiga brevicolis. Its genome is ~40 MB and encodes ~9,000 genes, which is about the size of a fungal genome but instead this organism is more closely related to the animal kingdom and serves as a pre-metazoan model for the evolution of metazoans (King et al., 2008). There are also extant fungi that look quite similar, and they are called the chytridiomycetes and are also aquatic with flagella. It is hypothesized that the flagellum was lost once in the fungal kingdom, as fungi exited the oceans and evolved to grow on land as mycorrhiza with the first land plants (Heckman et al., 2001; Liu et al., 2006; Simon et al., 1993). The ongoing UNICORN genome project seeks to understand how unicellular organisms evolved to be multicellular in both the animal and fungal kingdoms, and ten species are being targeted for sequencing (Ruiz-Trillo et al., 2007; Sebe-Pedros et al., 2010a; Sebe-Pedros et al., 2010b). Considering who the fungi are, how they evolved, and their relationships to the animal kingdom will be of general and broad significance.
Figure 1. The eukaryotic tree of life.
Recent molecular phylogenetic studies revealed the organization of the eukaryotic domains of life stemming from a last common ancestor. In particular, this analysis revealed a unique evolutionary relationship of the fungi and the animals as opisthokonts, sharing a more recent last common ancestor with each other to the exclusion of all other groups of eukaryotes. Modified from Baldauf et al, Science 2003.
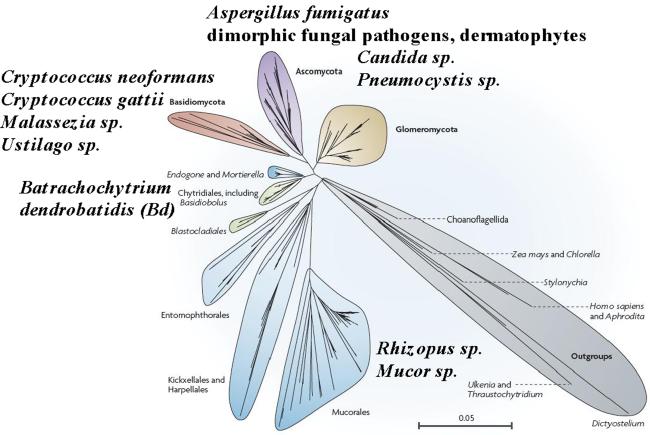
Traditionally the fungal kingdom has been divided into four phyla: two that share a monophyletic origin (the dikarya: Ascomycota, Basidiomycota) and two considered to be the basal fungi (Zygomycota, Chytridiomycota). However, the fungal tree of life project (AFTOL) revealed that the basal phyla are both polyphyletic, and thus as many as 8 to 10 phyla populate the fungal kingdom (James et al., 2006; Schüßler et al., 2001; Stajich et al., 2009). Microbial pathogens have evolved independently and repeatedly throughout the fungal phyla, and thus while some common shared virulence pathways have emerged such as that involving the protein phosphatase calcineurin (Blankenship et al., 2003; Chen et al., 2010b; Odom et al., 1997; Steinbach et al., 2007) many pathogens have unique virulence strategies, necessitating studies of each species in its own right.
Chytridiomycetes: Batrachochytrium dendrobatidis and global amphibian decline and extinction
One of the most sobering recent developments is that a fungus from the most obscure fungal phyla, the Chytridiomycota, has emerged and is globally causing devastating amphibian declines and numerous species extinctions (Figure 2) (Fisher et al., 2009). This species, Batrachochytrium dendrobatidis, or Bd for short, is aquatic with a flagellum. It infects the skin of frogs and other amphibians and perturbs their water balance and is often fatal. There is molecular phylogenetic evidence that this global outbreak is largely clonal (James et al., 2009; Morehouse et al., 2003), whereas studies of some unique populations reveal evidence for a higher level of diversity and the possibility of ongoing sexual reproduction (Morgan et al., 2007). Importantly, Koch's postulates have been satisfied from experimental infection studies establishing that Bd causes lethal infection of frogs (Lips et al., 2006; Nichols et al., 2001).
Figure 2. Microbial pathogens in the diverse phyla of the fungal kingdom.
Based on the AFTOL project, we now appreciate that the fungal kingdom spans as many as 10 phyla (the microsporidia are not depicted here) (James et al., 2006). Fungal pathogens have evolved repeatedly and independently throughout these phyla, and specific examples discussed in this review are depicted here. Adapted from Figure 1 from Schussler et al, Mycological Research 2001.
How Bd arose and swept the globe is an area of active investigation, but given the highly clonal nature of the outbreak isolates, the most parsimonious model is that the organism has been distributed from an ancestral source through some ubiquitous route rather than arising multiple times independently. A variety of models have been advanced, and one unifying hypothesis is that frogs and amphibians that can be colonized but do not succumb to disease serve as carriers. Xenopus laevis, commonly used for lab studies and also earlier for pregnancy testing, has been globally exported from Africa and has been proposed as the source and vector of transmission (Weldon et al., 2004). Further studies exploring other vectors, such as the American bull frog, and other possible ancestral geographic sources such as South America (Tim James and Russell Poulter, pers. comm..) are clearly warranted given the severity and implications of this pandemic. While it has been suggested that global warming may be contributing (Pounds et al., 2006)), other studies dissent from this opinion (Rohr et al., 2008), and it is also the case that higher temperature can promote clearing of the infection (Richards-Zawacki, 2010). Heroic efforts to treat infected animals with itraconazole or other antifungal drugs have thus far been successful in captivity (Forzan et al., 2008; Tamukai et al., 2010), but only partially successful in nature, as re-introduction to the wild frequently results in re-infection, or re-emergence of long term latent infection.
The role of Bd in ongoing devastating species collapse and extinction events (Crawford et al., 2010) may provide insights into historical events that profoundly altered the trajectory of the animal kingdom on Earth. Arturo Casadevall proposed a provocative model that fungi were involved in the extinction of the dinosaurs (Casadevall, 2005). We know that ~65 million years ago, a meteor struck the Yucatan Peninsula, and the Earth was enveloped by a resulting dust cloud that killed many of the animals and plants on the planet. As a consequence, a fungal bloom ensued that degraded the ubiquitous dead and decaying plant material. We know this because adjacent to the iridium peak in the fossil record we can see a layer of fungal spores at the KT boundary (Vajda and McLoughlin, 2004). This fungal bloom is thought to have given rise to a high density of aerosolized fungal spores, and conjectured to have infected the dinosaurs that were thought to be either poikilothermic (cold-blooded) or only partially homeothermic, and therefore unable to restrict the growth of fungi afforded by the higher core body temperature present in mammals (Bergman and Casadevall, 2010). As a consequence, the mammals flourished and emerged to become the dominant life form on the planet, replacing the dinosaurs and other reptiles. That Bd is causing contemporary extinction events lends support to the notion that infectious diseases caused by fungi may have dramatically shaped the evolutionary trajectory of life on our planet, including that of our own species.
Zygomycete pathogens--Rhizopus and Mucor
Our understanding of zygomycete fungi, including Phycomyces as a model of environmental sensing and sexual reproduction (Cerda-Olmedo and Lipson, 1987);(Idnurm et al., 2006; Idnurm et al., 2008; Sanz et al., 2009), and Rhizopus and Mucor as common and devastating pathogens of humans, is undergoing a renaissance with the impact of their completed genome sequences (Ma et al., 2009) (Figure 2). Rhizopus oryzae, Rhizopus delemar, and Mucor sp. cause devastating mucormycosis infections in humans that are difficult to treat because of resistance to many common antifungal drugs, and thus management can require surgical debridement and is in many cases fatal. Studies capitalizing on insights from the genome sequences have begun to explore virulence mechanisms involving iron acquisition or its therapeutic deprivation (Ibrahim et al., 2007; Ibrahim et al., 2010), the roles of mating type in sex determination and possibly in virulence (Gryganskyi et al., 2010; Lee et al., 2008)given precedents with other fungal pathogens such as Cryptococcus (Kwon-Chung et al., 1992; Nielsen et al., 2005b; Nielsen et al., 2005; Okagaki et al., 2010), and the roles of the dimorphic transition between hyphae and a multibudded yeast evoked by growth under anaerobic conditions and high CO2 (Bartnicki-Garcia and Nickerson, 1962; Pasteur, 1876). Finally, an interesting example of size dimorphism has emerged from studies of conidia, with links to virulence (Li et al., 2010a). Clearly much remains to be learned about the molecular and genetic basis of virulence evolution in the Mucorales phylum of the Zygomycota, but the future is a bright one with the advances afforded by the genome projects driven by the Broad Institute Fungal Genome Initiative and the DOE Fungal Kingdom genome project working in concert with community coordinators and collaborators.
An interesting recent development has emerged from genomic comparisons of the basal and other fungi with the microsporidia, an unusual group of obligate intracellular eukaryotic microbial pathogens that are closely aligned with the fungi, either as true fungi or as a sister group just outside the gates of the fungal kingdom (Keeling, 2009). More than 1,200 microsporidian species are known, of which 13 infect humans and others are implicated as a causative agent for bee hive collapse (Bromenshenk et al., 2010). They harbor highly reduced genomes, as small as 2.9 MB and <2,000 genes for Encephalitozoon cuniculi (Katinka et al Science 2001). Moreover, they have lost multiple pathways that normally serve to generate metabolic energy and ATP including the TCA cycle, fatty acid beta oxidation, ATP synthase, and the respiratory electron transport chain. Instead they acquired via horizontal gene transfer bacterial/chlamydial ATP transporters that enable them to usurp ATP from the host cytoplasm (Tsaousis et al., 2008) and even import ATP into their remnant mitochondria, the mitosome (Williams et al., 2002). Recent whole genome comparisons based on gene synteny provide evidence for their shared ancestry and emergence from within the fungal kingdom, and analysis of gene fusions that are shared in metazoans, choanoflagellates, and filasterea, but not in the fungi or microsporidians, further support their alignment (Lee et al., 2008; Lee et al., 2010). As genomes from other fungal phyla are sampled for the first time, further whole genome comparisons of this evolutionary relationship will become possible to understand how such a unique and highly successful group of obligate eukaryotic intracellular pathogens emerged.
Basidiomycetes--Animal and plant fungal pathogens
Fungal pathogens of both animals and plants have evolved repeatedly and independently in the dikarya, both in the Basidiomycota and the Ascomycota. These two broadly successful phyla have a common shared origin estimated to be ~500 million years ago (Figure 2). Two species, Cryptococcus neoformans and Cryptococcus gattii, have risen to prominence as common pathogens of humans (Cryptococcus ASM Book, 2011). They are ubiquitous in the environment, associated with pigeon guano or trees as an arboreal niche. We have all been exposed by inhalation of desiccated yeast cells or spores, which are small enough to penetrate into the alveoli of the lung and cause an initial pulmonary infection that frequently disseminates to the CNS to cause meningoencephalitis. Recent studies from the US Centers for Disease Control reveal that Cryptococcus causes more than one million cases of infection annually, >620,000 attributable deaths, and ~one-third of all AIDS associated deaths, now surpassing tuberculosis as a cause of death in Africa (Park et al., 2009). The majority of these cases are attributable to C. neoformans causing infection in the context of the AIDS pandemic.
Also of considerable concern is an outbreak of C. gattii that began on Vancouver Island in 1999 in both humans and animals, and which has now expanded into the US in both Washington and Oregon (Bartlett et al., 2010; Datta et al., 2009). In this case, >50% of infected patients were otherwise healthy, and their clinical course can be complicated, resulting in up to 20–33% mortality (Bartlett et al., 2010; DeBess, 2010). We now know from the work of Karen Bartlett (an environmental microbiologist at the University of British Columbia, Vancouver) that the organism is endemic throughout the region, associated with soil and a variety of indigenous tree species, such as Douglas Fir, and that particles small enough to be spores are present in the air. Based on molecular phylogenetic analysis, three clonal isolates are responsible for the outbreak. Two are found on Vancouver Island and now also in the US (VGIIa/major, VGIIb/minor), and one is thus far unique to Oregon (VGIIc/novel) (Byrnes et al., 2009; Byrnes et al., 2010; Fraser et al., 2005a; Kidd et al., 2004). The outbreak isolates are sexually fertile, and population genetic studies provide evidence they are part of a recombining global population (Byrnes et al., 2010; Fraser et al., 2005a; Fraser et al., 2005b). That the outbreak isolates are unisexual and all of the α mating type, combined with the finding of a diploid intermediate and molecular analysis of the mating type locus, provides evidence for models in which an unusual form of sexual reproduction, unisexual or same-sex mating (Figure 3), has contributed to both the origins of hypervirulent clones and the ongoing production of infectious spores (Fraser et al., 2005a; Lin et al., 2005). Moreover, given that the VGIIb/minor lineage on Vancouver Island is indistinguishable from isolates from a fertile, recombining, unisexual population in Australia at 30 MLST loci provides evidence that this outbreak lineage may have originated in Australia (Campbell et al., 2005a; Campbell et al., 2005b; Fraser et al., 2005a), and may have been transported to Vancouver Island with imported eucalypts and then made the jump to indigenous host tree species with which it is now associated.
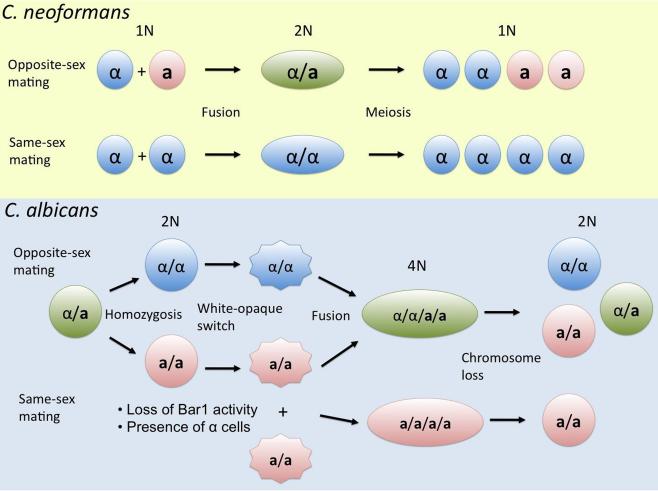
Figure 3. Opposite- and same-sex mating pathways in C. neoformans and C. albicans.
Recent studies reveal that these two common human fungal pathogens have retained extant sexual cycles involving cells of either opposite mating type or the same mating type. In C. neoformans, both sexual cycles are complete, including meiotic recombination, and generate infectious haploid spores. C. albicans is an obligate diploid, and mating first requires homozygosis of the MAT locus to produce or α/α or a/a cells, which undergo white to opaque switching and then fuse to produce a tetraploid zygote that undergoes concerted random chromosome loss to return to the diploid state by a currently recognized parasexual cycle. Same-sex mating can occur in strains lacking the Bar1 protease that destroys α factor, or in the presence of limiting α cells as a pheromone donor. The parasexual cycle of C. albicans involves Spo11-dependent recombination, and therefore may also involve cryptic versions of meiosis.
Another group of basidiomycete species, the Malassezia, has emerged as frequently associated with humans and linked to common skin disorders, including atopic dermatitis (eczema) and dandruff. The genomes for two species, M. restricta and M. globosa have recently been determined and reveal considerable insight (Xu et al., 2007). First, their genomes lack the enzyme fatty acid synthase, providing a neat explanation why these fungi are fastidious and must be cultured on media containing lipids. It also provides an interesting explanation for their ubiquitous association with human skin--it is there that they can scavenge lipids from our sebaceous secretions that they require for growth. As a consequence, they are uniquely specialized to survive on human skin and are readily transferred human to human, which impacts their evolutionary trajectory as highly successful commensals or pathogens of humans. Second, the genome and previous phylogenetic studies revealed a close relationship of the Malassezia sp. to Ustilago maydis, a highly successful basidiomycete pathogen of maize (Kamper et al., 2006). Remarkably, analysis of the secreted enzyme repertoire of Malassezia and Ustilago reveals marked differences, which likely occurred as one group evolved to colonize human skin (lipases, proteases) and the other plants (cutinases, glycosyl hydrolases). In fact, the secreted enzymes of Malassezia are more similar to those of the distant ascomycete human pathogen Candida albicans than they are to the more closely related plant pathogen. Third, and most provocatively, the Malassezia genomes reveal the potential for an extant sexual cycle in that a mating type locus and meiotic machinery are present. And while no sexual reproduction has yet been observed for this group of species, the fact that the mating type locus is organized similarly to the fused locus present in the bipolar species Ustilago hordei, a plant pathogen that infects barley and rye, suggests that there might be two mating types (Bakkeren et al., 2008; Hsueh and Heitman, 2008). A further speculation is that sex might occur on the skin of the infected human host, which is known to occur with C. albicans (Lachke et al., 2003), and this might lead to the production of novel antigens that stimulate immune responses in the skin linked to inflammatory diseases such as eczema. To quote the New Zealand Herald's popular press summary: “they found that not only does an icky fungus live on your head and cause dandruff, but it could be having sex. On your head. Right now.”
Ascomycetes--- Roles of sex, dimorphism, RNAi, and light sensing in virulence
We will close with the most successful, and most populated, phyla of the fungal kingdom, the Ascomycota, in which there are myriad pathogens of both animals and plants (Figure 2). Given the success of this group both as fungi and as microbial pathogens, we could devote an entire lecture or review to just this group. Instead we will consider a brief survey and the general themes that emerge. First, it is clear that microbial pathogens have emerged not only independently in the different phyla of the kingdom, but also multiple times independently even within phyla. In the Ascomycota this spans organisms as diverse as: 1) the archiascomycete Pneumocystis sp., which are unculturable, infect the lungs to cause pneumonia, are transmitted animal to animal, and have speciated in concert with their hosts such that there are unique pathogens restricted to humans, or mice, or rats, and so on, 2) the hemiascomycetes including Candida sp. that are part of our normal GI and skin microbiota and which cause systemic, mucocutaneous, and cutaneous infection, and 3) the euascomycetes including Aspergillus sp., the dimorphic fungal pathogens Histoplasma capsulatum and Coccidioides immitis and others, and the dermatophytes such as Trichophyton rubrum, the causative agent of athlete's foot.
Studies on the fungal pathogens in the Ascomycota are legion, and critical given the ubiquity and frequency with which they cause human disease. Recent highlights from an evolutionary perspective include the Candida pathogenic species comparative genome project enabling cross-species studies of biology and virulence strategies (Butler et al., 2009), and the discovery that C. albicans has at least an extant parasexual cycle, including both heterothallic mating and the recent discovery of homothallism involving same-sex mating (Figure 3) (Alby et al., 2009; Heitman, 2009). Given the previous discovery of same-sex mating in Cryptococcus (Lin et al., 2005; Lin et al., 2010; Lin et al., 2007; Lin et al., 2009), the finding that two of the three most successful human pathogens have both retained sexual cycles including both opposite-sex mating heterothallic out-crossing and homothallic same-sex mating that can promote inbreeding suggests a benefit of both forms of sexual reproduction. This may involve the amount of genetic admixture that occurs, giving rise to diversity vs. clonal population structures that may be uniquely adapted to changing vs. more static niches, such as the host. Same-sex mating arose independently in the two species given the ~500 million year evolutionary divide between them, and thus it seems quite likely that there will be other examples of same-sex mating that remain to be discovered in the fungal kingdom, and possibly among those that infect animals given the precedent set by the first two paradigms (Figure 3).
Possible virulence roles for mating pathways beyond sex have also begun to emerge. For example, pheromone signaling in C. albicans can promote biofilm formation which may, in some cases, serve as a prelude to mating, but in other settings may serve to facilitate formation of adherent, drug-resistant biofilms (Daniels et al., 2006). And recent studies have revealed a remarkable example of size dimorphism in which the basidiomycete Cryptococcus forms giant cells as large as 50 microns in the lungs of infected animals. Giant cell formation is enhanced during co-infection with cells of opposite mating type, and genetic analysis provides evidence for a paracrine signaling pathway evoked by mating pheromone acting via the Ste3a receptor on a-cells, analogous to quorum sensing in bacteria (Okagaki et al., 2010; Zaragoza et al., 2010). Hence, cell signaling circuits may dually govern both mating and virulence, and therefore be subject to distinct evolutionary pressures to serve two functions.
The ongoing dermatophyte genome projects at the Broad Institute and the University of Jena promise to revolutionize our understanding of this ubiquitous group of highly successful species that have a monophyletic origin. Genetic and genomic tools are advancing (White et al., 2008), and the identification of the mating type locus and recapitulation of sexual cycles advances both classic genetic approaches and enables tests of when and where sex might occur to impact the organism (Li et al., 2010b). As just one example, the T. rubrum population is known to be highly clonal and unisexual. Studies of T. rubrum sexual capacity are underway (Anzawa et al., 2010)and promise to be of considerable interest, possibly as a third example of unisexual same-sex mating either on its own or with assistance from closely aligned species in which both mating types remain extant via cross-species pheromone signals. In fact, just this type of ménage a trios mating in which one partner stimulates fusion of two cells of the same mating type has been well documented in both Cryptococcus (Hull et al., 2002; Hull and Heitman, 2002; Lin et al., 2005) and C. albicans (Alby et al., 2009). We would be remiss to not herald the recent discovery of an extant sexual cycle for Aspergillus fumigatus (O'Gorman et al., 2009), which involves culture on oatmeal agar in the dark for incubation periods as long as six months! With this discovery, the triumvirate of the three most successful systemic human fungal pathogens (Cryptococcus, Candida, Aspergillus) have all been revealed to have extant sexual cycles, and unusual ones involving unisexual, parasexual, or delayed sexual reproduction.
The general theme that has emerged from fungal genomics is that there appear to be few, if any, truly asexual fungi. Instead, each genome has revealed, even for anamorphic fungi with no known sexual cycle, that the machinery for both mating and meiosis, including the mating type locus, are conserved. We are drawn to the conclusion that the vast majority of fungi, perhaps even all, have a sexual nature that in many cases remains to be discovered under laboratory conditions. For the pathogenic fungi, these sexual cycles are often rare or cryptic, leading to clonal populations punctuated by limited recombination, with broad implications for the evolution of eukaryotic microbial pathogens including fungi, parasites, and oomycetes (Heitman, 2006, 2010).
Transitions in form are common throughout the fungal kingdom (Bastidas and Heitman, 2009; Odds, 1988). For example, Mucor is dimorphic growing commonly as a multinucleated hyphal form, yet switches to a multi-budded yeast under anaerobic conditions and elevated CO2 (Bartnicki-Garcia and Nickerson, 1962). Saccharomyces cerevisiae undergoes a dimorphic transition from yeast to pseudohyphae in response to nitrogen limitation (Gimeno et al., 1992). Yet other fungi such as A. fumigatus grow strictly as a filamentous fungus, a growth mode shared with ~80% of fungal species. These transitions in form are not only fascinating, but intimately linked to virulence. For example, the hyphae of A. fumigatus are critical for tissue invasion, and in C. albicans the yeast cell is essential for dissemination, whereas the hyphal form allows the organism to battle its way out of a macrophage following phagocytosis (Lorenz and Fink, 2002; Rocha et al., 2001; Uppuluri et al., 2010). There is no simple rule, for example that hyphae are pathogenic and yeasts are not. C. neoformans infections are caused by the yeast mode, whereas the hyphal mode occurs during sexual reproduction in nature. For C. albicans, both the yeast and the hyphae are essential for infection, as mutants locked in either growth mode are avirulent. Thus, dimorphic transitions in both directions can be critical, enabling a yeast to switch to hyphae and invade tissue, and a hyphal biofilm to switch to yeast that are released to seed distant tissues.
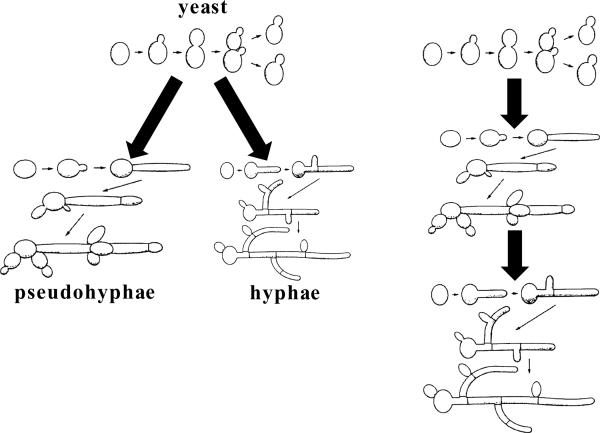
There is considerable debate on whether the ancestral fungus was a yeast or a hyphae, but given that the last common ancestor was a unicellular aquatic creature with a flagella, that would seem to favor origin as a yeast rather than a hyphae. If so, that suggests that the evolution of hyphal growth was a highly successful one given that only ~20% of species are known to grow as yeasts, many of which are dimorphic and produce pseudohyphae, hyphae, or both. C. albicans is one such trimorphic fungus, able to grow as a yeast, pseudohyphae, or hyphae. Whether pseudohyphae or hyphae were distinct developmental fates or part of a continuum (Figure 4) was unclear until recent studies examining the regulated expression of Ume6 provided evidence that yeast can form filaments that contain both pseudohyphae and hyphae (Carlisle et al., 2009). Low levels of Ume6 evoke yeast growth, intermediate levels produce pseudohyphae, and high levels drive hyphae. Moreover, increasing Ume6 from intermediate to higher levels converted pseudohyphae to hyphae, even producing hyphal/pseudohyphal composite filaments. When Ume6 was repressed in hyphal cells, they first produce pseudohyphae and then yeast. Thus, both hyphal growth modes are reversibly orchestrated by the levels of a single key regulatory factor, and this supports models in which pseudohyphae are a way station between yeast and hyphae and not a distinct fate (Figure 4). Recent studies also provide insight into how hyphae can produce yeast cells (Shen et al., 2008). Kohler and colleagues discovered that the pescadillo protein is required for C. albicans hyphae to produce yeast cells, and strains lacking pescadillo are locked in the hyphal growth mode, providing insight into how strictly filamentous fungi may have evolved from ancestral yeasts.
Figure 4. Trimorphic transitions in C. albicans.
The left panel depicts the transition of C. albicans yeast cells to pseudohyphae (left) and hyphae (right) as two distinct developmental fates. The right panel depicts this trimorphic transition as a continuum from yeast to pseudohyphae to hyphae. Recent studies support the continuum model of development. Modified from Figures 5.2, 5.3, and 5.4 from Odds 1988 with permission.
Fungal genomics reveals poignant examples of gene loss and retention linked to virulence, and just two are considered here: RNAi and light sensing. The RNAi pathway is central to the ability of organisms to mount defenses to RNA viruses, and also silence transposons. Remarkably, the RNAi pathway has been lost in U. maydis, a highly successful pathogen of maize, but retained in the closely related species U. hordei (Laurie et al., 2008). Similarly, the RNAi pathway has been lost in S. cerevisiae, but retained in the closely related species S. castelli and in the pathogen C. albicans (albeit with a novel form of Dicer) (Drinnenberg et al., 2009). Many other human pathogens have retained the RNAi machinery, including Mucor and Cryptococcus, and novel and key roles in silencing both exogenous and endogenous DNA elements have emerged (de Haro et al., 2009; Janbon et al., 2010; Nicolas et al., 2007; Nicolas et al., 2010; Nicolas et al., 2003; Wang et al., 2010). Similar examples of RNAi loss and retention have recently been discovered in Leishmania parasite species (Lye et al., 2010), illustrating the generality of this theme.
Light is one of the most pervasive environmental signals that impinges on life, and the ability to sense light has been central to the evolutionary trajectory of animals (vision), plants (photosynthesis), and also the fungi. A panoply of fungal light sensors have been discovered, including phytochromes (red light), opsins (green light), and the white collar protein complex (Wc1–Wc2) (blue light) (Corrochano, 2007; Corrochano and Garre, 2010; Idnurm et al., 2005; Idnurm et al., 2010; Rodriguez-Romero et al., 2010)). These photoperception systems are tuned to photons of different wavelengths across the visible light spectrum. Why might fungi sense not only photons but also their color? One thought is that sensing both red and blue light enables fungi to coordinate their behavior with the circadian rhythm of the earth, sensing red light at dawn, the blue sky of day, and then red at sunset. There is even recent evidence that the proteins for blue and red light sensing form a protein complex, serving as something of a primitive eye for the fungi (Blumenstein et al., 2005; Purschwitz et al., 2008). The white collar complex first came to light from studies on photosensory properties of Neurospora crassa (Ballario et al., 1996; Chen et al., 2010a; Linden and Macino, 1997). From studies on Cryptococcus (Idnurm and Heitman, 2005) and the zygomycetes Phycomyces and Mucor (Idnurm et al., 2006; Sanz et al., 2009; Silva et al., 2008; Silva et al., 2006), we now appreciate that the white collar complex is an ancient one, and extant in at least the Ascomycota, Basidiomycota, and Zygomycota phyla. It is quite remarkable that while many fungi have found it of evolutionary benefit to retain light sensing, others have lost it entirely, including S. cerevisiae and C. albicans, apparently as they became specialized to thrive in niches that are predominantly dark (the GI tract) or in which light sensing is not essential, analogous to the blind cave fish that have evolved repeatedly and independently around the globe. It is further quite striking that the model yeast S. cerevisiae has lost both the RNAi pathway and light sensing, and fortuitous then that we have other fungi to study for these critical biological processes. This is a recurrent theme throughout mycology, that it is essential to choose a system in which one can explore the biology of interest and to not be limited to model systems or species in which a process or pathway may have been either entirely lost or in which it may be impractical to study.
Fungal light sensing has recently been linked to virulence. First, the white collar orthologs Bwc1 and Bwc2 were found to contribute to virulence of C. neoformans in mice, and mutants lacking either protein are attenuated (Idnurm and Heitman, 2005). Thus, fungi may sense light in the environment, and then the relative darkness that surrounds them when they in a host. Analogously, the Wc1 ortholog was found to contribute to the infection of mice by Fusarium oxysporum but not for infection of tomato plants (Ruiz-Roldan et al., 2008). Strikingly, recent studies of bacterial pathogens have similarly highlighted a role for photoperception in virulence (Idnurm and Crosson, 2009; Swartz et al., 2007). Recent comparative studies fueled by genome advances reveal that the while the fungal light sensors have been retained in some pathogenic species (Cryptococcus, Mucor and Rhizopus, Magnaporthe oryzae, H. capsulatum), strikingly, they have been lost in many others, including the dimorphic fungi C. immitis and B. dermatitidis, the dermatophytes T. rubrum and Microsporum gypseum, C. albicans and other Candida sp., Malassezia globosa, E. cuniculi, and B. dendrobatidis (in this last case, either lost or never present) (Idnurm et al., 2010). While it is understandable why C. albicans, which has adapted to survive in the dark environment of the mammalian GI tract, might have eschewed light sensing, it is quite remarkable that fungi that are well adapted to survive on human skin (dermatophytes, Malassezia) have apparently lost the capacity to sense light. This is all the more remarkable given that blue light often serves as a proxy for the concomitant presence of DNA damaging UV light, and bwc1 and bwc2 mutants of C. neoformans are UV sensitive (Idnurm and Heitman, 2005). One possible theory is that these skin adapted fungi lost visible light/UV sensing as a virulence strategy, and now visible light/UV serve to restrain their growth on skin, both promoting commensalism by keeping them at bay and also causing them to survive better in skin microenvironments that are less sun exposed, such as the interdigital toe webs on our feet where they will be shielded from harmful UV rays.
Clearly, much remains to be learned, both about fungal light sensing in general and with respect to virulence strategies of fungal pathogens, but the future promises to be a bright one.
Coda on the routes to evolution of human fungal pathogens
It is clear that fungal pathogens of animals evolved repeatedly and independently throughout the fungal kingdom, but three general themes emerge. First, for some organisms, their evolutionary fate was from commensal to pathogen. These examples include the pathogenic Candida sp., which are part of the normal human microbiota resident on our skin and mucosal surfaces (oropharynx, GI tract, vagina). These organisms run the gauntlet of exposure to immune cells and have evolved and adapted to survive in this milieu, enabling them to survive to varying extents and cause disease in humans and to be transmitted from human to human. C. albicans is readily transmitted from mother to fetus during birth, or mother to infant during nursing, and between sex partners. C. parapsilosis is frequently transferred from the hands of health care workers to patients. Second, for a series of other fungal pathogens it is debatable whether they are considered part of the normal microbiota, but they commonly colonize or infect humans and are also transmitted human to human. These include the dermatophytes and Malassezia sp. resident on our skin that are transferred person to person or via fomites, and Pneumocystis sp. in the lung that are transferred by aerosol and inhalation.
Finally, there are many fungal pathogens that are environmental, and each human encounter is unique without evidence for human to human transmission. This includes A. fumigatus, C. neoformans, C. gattii, and the dimorphic human fungal pathogens, among others. This example is the most perplexing. How is it that these organisms can apparently be so well adapted to a human host, despite the seeming lack of human to human transmission? It is frequently posited that these are “accidental” pathogens (Casadevall and Pirofski, 2007). While that is possible, it also seems unsatisfying given the vast numbers of fungi and the vanishly small number that are pathogenic to animals, unless of course these are the few that have just by chance happened upon a successful virulence strategy. However, there are other considerations. First, Arturo Casadevall and others have championed the idea that human fungal pathogens evolved in an evolutionary crucible of heterologous hosts: amoeba, nematodes, insects, and even plants that formed a staging ground for the evolution of virulence (Steenbergen and Casadevall, 2003; Steenbergen et al., 2003, 2004; Steenbergen et al., 2001). One might consider this the Muhammad Ali model of fungal pathogenesis--most fights involve a sparring partner that prepares one for the rare prize fight for the title. Second, there may be more complex animal-environment-animal cycles, or even cryptic animal-animal cycles, that promote virulence evolution. For example, in nature when infected animals are consumed by predation, or die and their carcass decays or is consumed by scavengers, this may return the pathogen to the environment or lead to transmission to a new host. Other modes of animal behavior may also contribute. For example, C. gattii frequently colonizes the nares of both Koala and cats, and in this setting may be more readily transmitted animal to animal than currently appreciated during grooming or other activities (Connolly et al., 1999; Krockenberger et al., 2002). If so, these routes may lead to adaptation for survival on and in animals, and thereby select for virulence attributes. Perhaps this begins to provide an explanation as to why Cryptococcus is a well adapted facultative intracellular pathogen that survives in macrophages, and elaborates a complex polysaccharide capsule virulence factor in response to host signals, including high CO2 (Granger et al., 1985). In the specific case of the dimorphic human fungal pathogen C. immitis, it is found sporadically in the desert soil, most commonly in association with a decaying carcass. Analysis of its genome reveals a vast expansion of protease genes, and those encoding other enzymes that would be expected to mediate decay of animal tissues, and a loss of enzymes associated with plant degradation (Sharpton et al., 2009). Thus, C. immitis appears to have evolved from a plant associated ancestor to adapt to growth on dead or living animal tissue, and this may have promoted the evolution of virulence strategies that render this an evolved rather than an accidental pathogen. Hence, other environmental fungi may have similarly evolved the capacity to infect animals, despite not appearing to be frequently contagious.
In closing, these musings about the evolution of human fungal pathogens throughout the fungal kingdom necessarily restrict our thoughts to just a handful of interesting species, but what this perspective reveals is just how powerful a cross species comparative approach is to provide profound insight. Thus, no matter the origin of your particular fascination with the fungal kingdom (pathogens, symbiosis, genetic models, lichens, mushrooms), our potential as the mycology community is in broadly considering what we have to learn not only from those fungi that are the objects of our detailed studies, but what we have to learn from all of the fungi currently under investigation, the legions that remain to be discovered and characterized, and from each other. The future for mycology, and for mycologists, is indeed a bright one.
Acknowledgements
I thank Nick Read for the invitation to present a plenary lecture at the International Congress on Mycology in Edinburgh August 2009 (which formed the basis for this review) and for the invitation to contribute this review, and Cecelia Shertz, Min Ni, and Eddie Byrnes for prescient comments on the manuscript. Our research is supported by grants from the NIH/NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzawa K, Kawasaki M, Mochizuki T, Ishizaki H. Successful mating of Trichophyton rubrum with Arthroderma simii. Med Mycol. 2010;48:629–634. doi: 10.3109/13693780903437884. [DOI] [PubMed] [Google Scholar]
- Bakkeren G, Kamper J, Schirawski J. Sex in smut fungi: Structure, function and evolution of mating-type complexes. Fungal Genet Biol. 2008;45(Suppl 1):S15–21. doi: 10.1016/j.fgb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Baldouf SL, Palmer JD. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Bartlett K, Byrnes EJ, III, Duncan C, Fyfe M, Galanis E, Heitman J, Hoang L, Kidd S, MacDougall L, Mak S, Marr K, Morshed M, West S, Kronstad J. The emergence of Cryptococcus gattii infections on Vancouver Island and expansion in the Pacific Northwest. In: Heitman J, Kozel TR, Kwon-Chung JK, Perfect JR, Casadevall A, editors. Cryptococcus from Human Pathogen to Model Yeast. ASM Press; Washington, D.C.: 2010. pp. 313–325. [Google Scholar]
- Bartnicki-Garcia S, Nickerson WJ. Induction of yeast-like development in Mucor by carbon dioxide. J Bacteriol. 1962;84:829–840. doi: 10.1128/jb.84.4.829-840.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Heitman J. Trimorphic stepping stones pave the way to fungal virulence. Proc Natl Acad Sci U S A. 2009;106:351–352. doi: 10.1073/pnas.0811994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Casadevall A. Mammalian endothermy optimally restricts fungi and metabolic costs. MBio. 2010;1:e00212–e00210. doi: 10.1128/mBio.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryotic Cell. 2003;2:422–430. doi: 10.1128/EC.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, Fischer R. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Bromenshenk JJ, Henderson CB, Wick CH, Stanford MF, Zulich AW, Jabbour RE, Deshpande SV, McCubbin PE, Seccomb RA, Welch PM, Williams T, Firth DR, Skowronski E, Lehmann MM, Bilimoria SL, Gress J, Wanner KW, Cramer RA., Jr. Iridovirus and microsporidian linked to honey bee colony decline. PLoS ONE. 2010;5:e13181. doi: 10.1371/journal.pone.0013181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ, 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ, 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryotic Cell. 2005a;4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, Heitman J. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryotic Cell. 2005b;4:1410–1419. doi: 10.1128/EC.4.8.1410-1419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A. Fungal virulence, vertebrate endothermy, and dinosaur extinction: is there a connection? Fungal Genet Biol. 2005;42:98–106. doi: 10.1016/j.fgb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Olmedo E, Lipson ED. Phycomyces. Cold Spring Harbor Laboratory; New York: 1987. [Google Scholar]
- Chen CH, Dunlap JC, Loros JJ. Neurospora illuminates fungal photoreception. Fungal Genet Biol. 2010a;47:922–929. doi: 10.1016/j.fgb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-L, Kozubowski L, Cardenas ME, Heitman J. On the roles of calcineurin in fungal growth and pathogenesis. Curr Fungal Infect Rep. 2010b;4:244–255. [Google Scholar]
- Cohen ML. Changing patterns of infectious disease. Nature. 2000;406:762–767. doi: 10.1038/35021206. [DOI] [PubMed] [Google Scholar]
- Connolly JH, Krockenberger MB, Malik R, Canfield PJ, Wigney DI, Muir DB. Asymptomatic carriage of Cryptococcus neoformans in the nasal cavity of the koala (Phascolarctos cinereus) Med Mycol. 1999;37:331–338. doi: 10.1046/j.1365-280x.1999.00236.x. [DOI] [PubMed] [Google Scholar]
- Corrochano LM. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- Corrochano LM, Garre V. Photobiology in the Zygomycota: multiple photoreceptor genes for complex responses to light. Fungal Genet Biol. 2010;47:893–899. doi: 10.1016/j.fgb.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci U S A. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haro JP, Calo S, Cervantes M, Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot Cell. 2009;8:1486–1497. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBess E.e.a. Emergence of Cryptococcus gattii-- Pacific Northwest, 2004–2010. MMWR Morb Mortal Wkly Rep. 2010;59:865–868. [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Garner TW, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- Forzan MJ, Gunn H, Scott P. Chytridiomycosis in an aquarium collection of frogs: diagnosis, treatment, and control. J Zoo Wildl Med. 2008;39:406–411. doi: 10.1638/2007-0091.1. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005a;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Huang JC, Pukkila-Worley R, Alspaugh JA, Mitchell TG, Heitman J. Chromosomal translocation and segmental duplication in Cryptococcus neoformans. Eukaryotic Cell. 2005b;4:401–406. doi: 10.1128/EC.4.2.401-406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth - regulation by starvation and Ras. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito GM, Porter TM, Anishchenko IM, Heitman J, Vilgalys R. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS ONE. 2010;5:1–12. doi: 10.1371/journal.pone.0015273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- Heitman J. Microbial genetics: Love the one you're with. Nature. 2009;460:807–808. doi: 10.1038/460807a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe. 2010;8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y-P, Heitman J. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr Opin Microbiol. 2008;11:517–524. doi: 10.1016/j.mib.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, Heitman J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1alpha. Genes Dev. 2002;16:3046–3060. doi: 10.1101/gad.1041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu Rev Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- Ibrahim AS, Gebermariam T, Fu Y, Lin L, Husseiny MI, French SW, Schwartz J, Skory CD, Edwards JE, Jr., Spellberg BJ. The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J Clin Invest. 2007;117:2649–2657. doi: 10.1172/JCI32338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AS, Gebremariam T, Lin L, Luo G, Husseiny MI, Skory CD, Fu Y, French SW, Edwards JE, Jr., Spellberg B. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol Microbiol. 2010;77:587–604. doi: 10.1111/j.1365-2958.2010.07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Crosson S. The photobiology of microbial pathogenesis. PLoS Pathog. 2009;5:e1000470. doi: 10.1371/journal.ppat.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Rodriguez-Romero J, Corrochano LM, Sanz C, Iturriaga EA, Eslava AP, Heitman J. The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc Natl Acad Sci U S A. 2006;103:4546–4551. doi: 10.1073/pnas.0600633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Verma S, Corrochano LM. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet Biol. 2010;47:881–892. doi: 10.1016/j.fgb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Walton FJ, Floyd A, Heitman J. Identification of the sex genes in an early diverged fungus. Nature. 2008;451:193–196. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O'Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O'Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- James TY, Litvintseva AP, Vilgalys R, Morgan JA, Taylor JW, Fisher MC, Berger L, Weldon C, du Preez L, Longcore JE. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009;5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Maeng S, Yang DH, Ko YJ, Jung KW, Moyrand F, Floyd A, Heitman J, Bahn YS. Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet Biol. 2010;47:1070–1080. doi: 10.1016/j.fgb.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HA, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho EC, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Keeling P. Five questions about microsporidia. PLoS Pathog. 2009;5:e1000489. doi: 10.1371/journal.ppat.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci U S A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krockenberger MB, Canfield PJ, Malik R. Cryptococcus neoformans in the koala (Phascolarctos cinereus): colonization by C n. var. gattii and investigation of environmental sources. Med Mycol. 2002;40:263–272. doi: 10.1080/mmy.40.3.263.272. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med. 2010;363:2036–2044. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- Laurie JD, Linning R, Bakkeren G. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr Genet. 2008;53:49–58. doi: 10.1007/s00294-007-0165-7. [DOI] [PubMed] [Google Scholar]
- Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Corradi N, Doan S, Dietrich FS, Keeling PJ, Heitman J. Evolution of the sex-related locus and genomic features shared in microsporidia and fungi. PLoS ONE. 2010;5:e10539. doi: 10.1371/journal.pone.0010539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Cervantes M, Springer DJ, Ruiz-Vazquez RM, Torres-Martinez S, Heitman J, Lee SC. Analysis of the mating type locus and spore size dimorphism linked to virulence in the pathogenic zygomycete, Mucor circinelloides species subcomplex. PLoS Pathog. 2010a doi: 10.1371/journal.ppat.1002086. submitted Sept. 17, 2010, in revision as of Nov. 5, 2010, in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Metin B, White TC, Heitman J. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryotic cell. 2010b;9:46–58. doi: 10.1128/EC.00259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP. Emerging infectious disease and the loss of biodiversity in a neotropical amphibian community. Proc Natl Acad Sci U S A. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Hodson MC, Hall BD. Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom fungi inferred from RNA polymerase II subunit genes. BMC Evol Biol. 2006;6:74. doi: 10.1186/1471-2148-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryotic cell. 2002;1:657–662. doi: 10.1128/EC.1.5.657-662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, Abe A, Calvo SE, Corrochano LM, Engels R, Fu J, Hansberg W, Kim JM, Kodira CD, Koehrsen MJ, Liu B, Miranda-Saavedra D, O'Leary S, Ortiz-Castellanos L, Poulter R, Rodriguez-Romero J, Ruiz-Herrera J, Shen YQ, Zeng Q, Galagan J, Birren BW, Cuomo CA, Wickes BL. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5:e1000549. doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse EA, James TY, Ganley AR, Vilgalys R, Berger L, Murphy PJ, Longcore JE. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol Ecol. 2003;12:395–403. doi: 10.1046/j.1365-294x.2003.01732.x. [DOI] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Vredenburg VT, Rachowicz LJ, Knapp RA, Stice MJ, Tunstall T, Bingham RE, Parker JM, Longcore JE, Moritz C, Briggs CJ, Taylor JW. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A. 2007;104:13845–13850. doi: 10.1073/pnas.0701838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DK, Lamirande EW, Pessier AP, Longcore JE. Experimental transmission of cutaneous chytridiomycosis in dendrobatid frogs. J Wildlife Dis. 2001;37:1–11. doi: 10.7589/0090-3558-37.1.1. [DOI] [PubMed] [Google Scholar]
- Nicolas FE, de Haro JP, Torres-Martinez S, Ruiz-Vazquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, Torres-Martinez S, Moulton V, Ruiz-Vazquez RM, Dalmay T. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010;38:5535–5541. doi: 10.1093/nar/gkq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK, Jr., Giles SS, Mitchell TG, Casadevall A, Perfect JR, Heitman J. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 2005b;73:4922–4933. doi: 10.1128/IAI.73.8.4922-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, Heitman J. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis: a review and bibliography. 2nd edi. ed. Bailliere Tindall; London: 1988. [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Pasteur L. Ettudes sur la biere. Gauthier-Villars; Paris: 1876. [Google Scholar]
- Pounds JA, Bustamante MR, Coloma LA, Consuegra JA, Fogden MP, Foster PN, La Marca E, Masters KL, Merino-Viteri A, Puschendorf R, Ron SR, Sanchez-Azofeifa GA, Still CJ, Young BE. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature. 2006;439:161–167. doi: 10.1038/nature04246. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Muller S, Kastner C, Schoser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Richards-Zawacki CL. Thermoregulatory behaviour affects prevalence of chytrid fungal infection in a wild population of Panamanian golden frogs. Proc Biol Sci. 2010;277:519–528. doi: 10.1098/rspb.2009.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Romero J, Hedtke M, Kastner C, Muller S, Fischer R. Fungi, hidden in soil or up in the air: light makes a difference. Annu Rev Microbiol. 2010;64:585–610. doi: 10.1146/annurev.micro.112408.134000. [DOI] [PubMed] [Google Scholar]
- Rohr JR, Raffel TR, Romansic JM, McCallum H, Hudson PJ. Evaluating the links between climate, disease spread, and amphibian declines. Proc Natl Acad Sci U S A. 2008;105:17436–17441. doi: 10.1073/pnas.0806368105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Roldan MC, Garre V, Guarro J, Marine M, Roncero MI. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryotic Cell. 2008;7:1227–1230. doi: 10.1128/EC.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Sanz C, Rodriguez-Romero J, Idnurm A, Christie JM, Heitman J, Corrochano LM, Eslava AP. Phycomyces MADB interacts with MADA to form the primary photoreceptor complex for fungal phototropism. Proc Natl Acad Sci U S A. 2009;106:7095–7100. doi: 10.1073/pnas.0900879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüßler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: evolution and phylogeny. Mycological Research. 2001;105:1413–1421. [Google Scholar]
- Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2010a Nov. 17 doi: 10.1093/molbev/msq309. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010b;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, Hung CY, McMahan C, Muszewska A, Grynberg M, Mandel MA, Kellner EM, Barker BM, Galgiani JN, Orbach MJ, Kirkland TN, Cole GT, Henn MR, Birren BW, Taylor JW. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009;19:1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR. The C. albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc. Natl. Acad. Sci. USA. 2008;105:20918–20923. doi: 10.1073/pnas.0809147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva F, Navarro E, Penaranda A, Murcia-Flores L, Torres-Martinez S, Garre V. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a white collar-1-like activator. Mol Microbiol. 2008;70:1026–1036. doi: 10.1111/j.1365-2958.2008.06470.x. [DOI] [PubMed] [Google Scholar]
- Silva F, Torres-Martinez S, Garre V. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol Microbiol. 2006;61:1023–1037. doi: 10.1111/j.1365-2958.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Simon L, Bousquet J, Levesque RC, Lalonde M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature. 1993;363:67–69. [Google Scholar]
- Stajich JE, Berbee ML, Blackwell M, Hibbett DS, James TY, Spatafora JW, Taylor JW. The fungi. Curr Biol. 2009;19:R840–845. doi: 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Casadevall A. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 2003;5:667–675. doi: 10.1016/s1286-4579(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun. 2003;71:4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Interaction of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infect Immun. 2004;72:3478–3488. doi: 10.1128/IAI.72.6.3478-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A. 2001;98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Reedy JL, Cramer RA, Jr., Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- Tamukai K, Une Y, Tominaga A, Suzuki K, Goka K. Treatment of spontaneous chytridiomycosis in captive amphibians using itraconazole. J Vet Med Sci. 2010 Sept. 14 doi: 10.1292/jvms.10-0261. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda V, McLoughlin S. Fungal proliferation at the Cretaceous-Tertiary boundary. Science. 2004;303:1489. doi: 10.1126/science.1093807. [DOI] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: An evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- Wang X, Hsueh YP, Li W, Floyd A, Skalsky R, Heitman J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010;24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon C, du Preez LH, Hyatt AD, Muller R, Spears R. Origin of the amphibian chytrid fungus. Emerg Infect Dis. 2004;10:2100–2105. doi: 10.3201/eid1012.030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TC, Oliver BG, Graser Y, Henn MR. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot Cell. 2008;7:1238–1245. doi: 10.1128/EC.00100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, Deangelis YM, Reeder NL, Johnstone KR, Leland M, Fieno AM, Begley WM, Sun Y, Lacey MP, Chaudhary T, Keough T, Chu L, Sears R, Yuan B, Dawson TL., Jr. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci U S A. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010;6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]