Abstract
TRIM5 is a RING domain-E3 ubiquitin ligase that restricts infection by HIV-1 and other retroviruses immediately following virus invasion of the target cell cytoplasm1,2. Antiviral potency correlates with TRIM5 avidity for the retrovirion capsid lattice3,4 and several reports indicate that TRIM5 plays a role in signal transduction5–7, but the precise mechanism of restriction is unknown8. Here we demonstrate that TRIM5 promotes innate immune signaling and that this activity is amplified by retroviral infection and interaction with the capsid lattice. Acting with the heterodimeric, ubiquitin-conjugating enzyme UBC13/UEV1A, TRIM5 catalyzes the synthesis of unattached K63-linked ubiquitin chains that activate the TAK1 (MAP3K7) kinase complex and stimulate AP-1 and NFκB signaling. Interaction with the HIV-1 capsid lattice greatly enhances the UBC13/UEV1A-dependent E3 activity of TRIM5 and challenge with retroviruses induces the transcription of AP-1 and NFκB-dependent factors with a magnitude that tracks with TRIM5 avidity for the invading capsid. Finally, TAK1 and UBC13/UEV1A contribute to capsid-specific restriction by TRIM5. Thus, the retroviral restriction factor TRIM5 has two additional activities that are linked to restriction: it constitutively promotes innate immune signaling and it acts as a pattern recognition receptor specific for the retrovirus capsid lattice.
To determine if TRIM5 contributes to signal transduction, the effect of ectopic human TRIM5α expression on transcriptional reporters in HEK-293 cells was examined. TRIM5 stimulated either of two luciferase reporters for AP-1 with a magnitude comparable to that of MAVS or the AP-1 transcription factor c-Jun (Fig. 1a and Supplementary Fig. 1a). TRIM5 also stimulated NF-κB (Fig. 1b) but minimally activated IFN-β-, or IRF3-dependent, luciferase reporters (Fig. 1c and Supplementary Fig. 1b and c). The TRIM5-cyclophilin A fusion protein from owl monkey1 activated AP-1 and NF-κB to similar levels as human TRIM5α (Supplementary Fig. 1d and e). Though TRIM5 was not sufficient to activate IFN-β, induction of IFN-β by IRF3 was greatly enhanced by TRIM5 (Fig. 1c), consistent with the fact that IFN-β transcription requires NF-κB and AP-1, as well as IRF3 (Supplementary Fig. 1f)9.
Figure 1. TRIM5 promotes innate immune signaling.
a –c, HEK-293 cells transfected with the indicated pcDNA-based expression plasmids and luciferase reporters for AP-1 (a), NF-κB (b) or IFNB1 (c). Bars show mean luciferase activity +/− s.d. (n = 6). d, Global expression profile comparing TRIM5 KD to control KD THP-1 macrophages. Triangles indicate inflammatory genes significantly downregulated in TRIM5 KD. e, qRT-PCR for the indicated mRNAs harvested 2 to 8 hrs after LPS-treatment, depending on the peak values for that gene. Shown are the means +/− s.e.m (n = 3) relative to untreated cells. f, Concentration of the indicated proteins in the culture supernatant, 24 hrs after LPS-treatment (mean +/− s.d., n = 3). RNA and protein data are representative of at least 3 separate donors. g–j, THP-1 macrophages transduced with miR30-based lentivirus KD vectors targeting either TRIM5 (g and i), IRF3 (h), or STAT2 (i), were treated 24 hrs with the indicated compounds and challenged with VSV-G pseudotyped HIV-1 luciferase reporter virus (g–i) or with the indicated GFP reporter viruses (j). Data are expressed as fold-change compared to control KD cells, with s.e.m (n = 4). All data are representative of at least 3 independent experiments.
To determine if endogenous TRIM5 regulates AP-1 and NF-κB signaling pathways, the effect of TRIM5 knockdown (KD) was assessed in myeloid cells. THP-1 cells were transduced with lentiviral vectors engineered to confer puromycin-resistance and to express Pol II-driven, microRNA-based shRNAs targeting either TRIM5 or control RNAs (Supplementary Fig. 2a–c). Pools of puromycin-resistant cells were generated with each KD vector and global expression profiles were assessed. The effect of TRIM5 KD was extraordinarily specific in that, of 25,000 genes probed, only 33 were significantly decreased (Fig. 1d). The majority of these were NF-κB and AP-1-responsive inflammatory mediators, 70% being inflammatory chemokines and cytokines (Supplementary Table 1).
LPS, a pathogen-associated molecular pattern (PAMP) recognized by the pattern recognition receptor (PRR) TLR4, activates AP-1 and NF-κB-signaling and this culminates in the expression of inflammatory genes like those perturbed by TRIM5 KD10,11. Monocyte-derived dendritic cells (MDDC), macrophages (MDM) and THP-1 cells were challenged with LPS and induction of the AP-1- and NF-κB-dependent genes CXCL9, CXCL10, CCL8, IL-6, IL-8, and PTGS2 (COX2), was found to be attenuated by TRIM5 KD (Fig. 1e and f and Supplementary Figure 2d and e). These results demonstrate that TRIM5 activates MAPK- and NF-κB-dependent genes and makes a major contribution to LPS signaling and gene induction (Supplementary Figure 1f).
Given the contribution of TRIM5 to the production of inflammatory mediators by LPS, the effect of TRIM5 on the previously reported anti-HIV-1 activity of LPS12 was examined. Transduction of MDDC, MDM, or THP-1 macrophages by VSV G-pseudotyped HIV-1 was blocked by LPS, by other PAMPs, and by type 1 IFN (Supplementary Fig. 3a–c). TRIM5 mRNA increased 10-fold in response to these factors (Supplementary Fig. 3d and e) but this increase was not sufficient for the anti-HIV-1 state (Supplementary Fig. 3f and g). Nonetheless, TRIM5 KD rescued HIV-1 from LPS, though not from type 1 IFN, and the magnitude rescue correlated with the efficiency of TRIM5 KD (Fig. 1g and Supplementary Fig. 4a and b). These phenotypes were indistinguishable from those observed with KD of IRF3, a critical transcription factor that acts proximal to IFNβ10 (Fig. 1h and Supplementary Fig. 4c). In contrast, KD of STAT2, a factor that acts downstream of the type I IFN receptor, blocked the anti-HIV-1 activity of either LPS or type 1 IFN (Fig. 1i and Supplementary Fig. 4d).
Rescue from LPS appears to be independent of capsid-recognition by TRIM5 in that TRIM5 KD rescued SIVMAC, a retrovirus that differs greatly from HIV-1 in terms of its sensitivity to TRIM5-mediated restriction1,2, as well as two non-retroviruses, the rhabdovirus vesicular stomatitis virus and the paramyxovirus Newcastle disease virus (Fig. 1j and Supplementary Fig. 4e–i). Though TRIM5 is not sufficient to activate IFN-β (Fig. 1c), it promotes the first wave of innate immune signaling upstream of IFN-β and thereby contributes to the antiviral state established by LPS (Supplementary Fig. 1f).
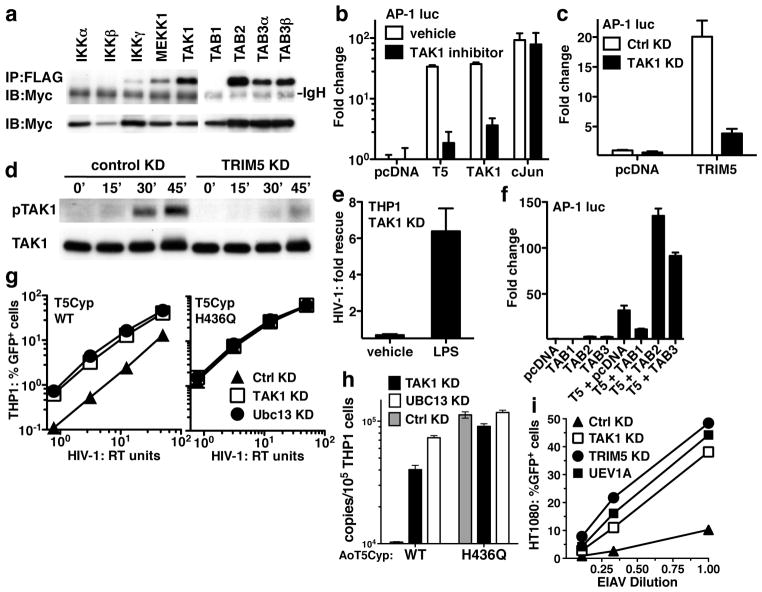
To understand how TRIM5 activates AP-1 and NF-κB, 20 candidate proteins, selected based on signaling activity above the MAPK/NF-κB bifurcation in the LPS signaling pathway, were tested for the ability to immunoprecipitate with TRIM5. Strong signal was observed with TAK1, TAB2, and TAB3 (Fig. 2a and Supplementary Fig. 5a), all components of the TAK1 (MAP3K7) kinase complex that phosphorylates proximal MAPK and NF-κB kinases in response to LPS11. Like TRIM5, TAK1 potently activated AP-1 and modestly activated NF-κB (Fig 2b). 5Z-7-oxozeaenol, a TAK1-inhibitor, blocked AP-1 induction by TRIM5 or TAK-1 without effect on AP-1 induction by the downstream effector c-Jun (Fig 2b). TAK1 KD blocked AP-1 activation by TRIM5 (Fig 2c), but not by c-Jun (Fig. 2c and Supplementary Fig. 5c). TRIM5 KD blocked LPS-induced TAK1 autophosphorylation on threonine 187 (Fig. 2d), a post-translational modification required for TAK1 activation11. Like TRIM5 KD, TAK1 KD rescued HIV-1 from the LPS-induced antiviral state (Fig. 2e and Supplementary Fig. 5d), and either TAB2 or TAB3 acted synergistically with TRIM5 to activate AP-1 (Fig. 2f). These results indicate that TRIM5 and the TAK1 kinase complex cooperate to promote signal transduction, and given that TAK1 phosphorylates both IKKs and MKKs11, explains how TRIM5 activates both MAPK and NF-κB signaling pathways.
Figure 2. The TAK1 kinase complex interacts biochemically and functionally with TRIM5.
a, HEK-293T cells were co-transfected with Myc-tagged human TRIM5α and the indicated FLAG-tagged constructs. Shown are immunoblots with anti-Myc antibody after immunoprecipitation with anti-FLAG (upper panel), or of total cell lysate (bottom panel). b, c, and f, HEK-293 cells were transfected with the indicated pcDNA-based expression plasmids and an AP-1 luciferase reporter and show the effect of TAK1 inhibitor 5z-7-oxozeaenol (b) or TAK1 KD (c). TAK1 KD and control KD THP-1 macrophages were treated with LPS for the indicated times and immunoblotted with anti-TAK1 antibody (lower panel) or anti-phospho-TAK1 antibody (upper panel) (d), or, cells were treated 24 hrs with LPS or vehicle and challenged with an HIV-1 luciferase reporter virus (e). The results in (e) are reported as fold rescue due to TAK1 KD, with respect to control KD. g and h, THP-1 cells were transduced with lentiviral vectors encoding owl monkey TRIM5Cyp, either wild-type or the H436Q mutant. Pools of each were then transduced with lentiviral KD vectors targeting either TAK1, UBC13, or control, and challenged with an HIV-1-GFP reporter vector. Infectivity was monitored by FACS (g) or by PCR for synthesis of full-length viral cDNA (h). i, HT1080 cells were transfected with dsRNA oligonucleotides targeting TRIM5, TAK1, or UEV1A and challenged with EIAV-GFP reporter vector.
The well-characterized restriction of HIV-1 by owl monkey TRIMCyp1,13 was exploited to determine if TAK1 contributes to TRIM5-mediated, capsid-specific restriction. Pools of THP-1 cells were selected for puromycin-resistance after transduction with a bicistronic lentiviral vector encoding owl monkey TRIM5Cyp13. As shown previously, these cells were resistant to infection with wild-type HIV-1, but not to the HIV-1/G89V capsid mutant, and the infectivity of wild-type HIV-1 was rescued by cyclosporine13 (Supplementary Fig. 5e). Control cells transduced with vector bearing TRIM5Cyp-H436Q, a mutant that does not bind HIV-1 capsid and does not restrict HIV-113, were infected with efficiency equal to that of cells transduced with the empty vector. THP-1 cells transduced with either wild-type or H436Q mutant TRIM5Cyp were then subjected to a second round of selection after transduction with miR30-based KD vectors targeting TAK1 or luciferase control and expressing hygromycin-resistance. The pools of puromycin/hygromycin-double resistant THP-1 cells were then challenged with HIV-1. TAK1 KD rescued HIV-1 transduction and nascent HIV-1 cDNA synthesis (Figs. 2g and h). This effect was specific to the cells with TRIM5Cyp-mediated restriction activity since TAK1 KD had no effect on HIV-1 transduction in the non-restrictive, H436Q control cells (Fig 2g).
The contribution of TAK1 to restriction of N-tropic MLV by human TRIM5α was examined using miR30-based KD vectors in THP-1, HeLa, and HT1080 cells. Inhibition of both N-tropic and B-tropic MLV infection by the TAK1 KD was observed, perhaps because, unlike HIV-1, infection with MLV is cell-cycle dependent14, and these viruses were sensitive to growth inhibitory effects of the KD. This precluded assessment of capsid-specific effects on reporter gene transduction, though nascent viral cDNA synthesized after infection of THP-1 cells was rescued by the TAK1 KD in an N-tropic MLV-specific manner (Supplementary Fig. 5g). Similar non-specific effects on MLV were observed after transfection of dsRNA oligonucleotides targeting TAK1. Like HIV-1, Equine infectious anemia virus (EIAV) is a lentivirus that infects non-dividing cells, but it differs from HIV-1 in that it is relatively sensitive to human TRIM5α-mediated restriction15. Transfection of dsRNAs targeting TAK1 rescued EIAV transduction almost to the same level as the TRIM5 KD (Fig. 2i). These results indicate that TAK1 contributes to capsid-specific restriction mediated by TRIM5.
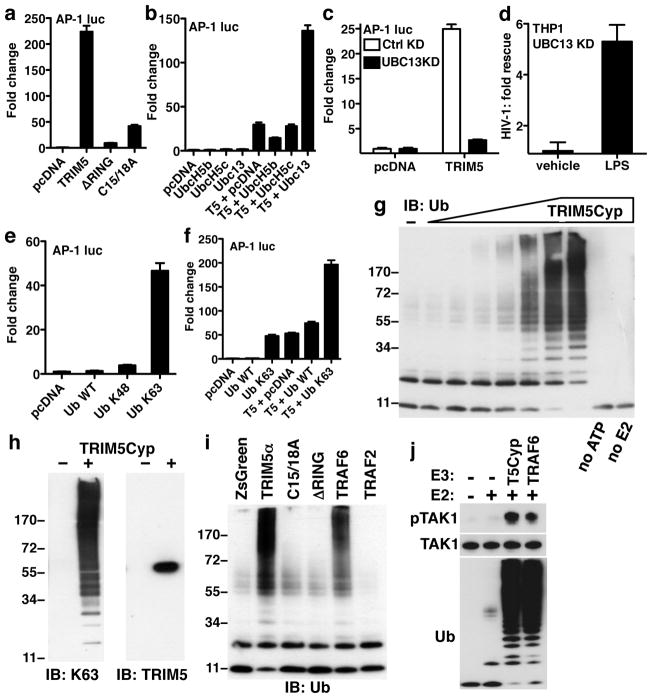
AP-1 induction by TRIM5 was impaired by mutants of the RING E3 ubiquitin (Ub)-ligase domain (Fig. 3a). This raised the question which of the many E2 Ub-conjugating enzymes might be relevant for TRIM5-mediated effects on signal transduction. Among candidate E2s, UBC13 synergized with TRIM5 to activate AP-1 (Fig. 3b). Interestingly, the TAK1 kinase complex is activated by the heterodimeric E2 UBC13/UEV1A11. KD of UBC13 or UEV1A severely blocked AP-1 activation by TRIM5 (Fig. 3c and Supplementary Fig. 6a–c), rescued HIV-1 from the LPS-induced antiviral state in THP-1 macrophages (Fig. 3d and Supplementary Fig. 6d), and rescued HIV-1 and EIAV from TRIM5-mediated restriction (Fig. 2g–i).
Figure 3. TRIM5 acts with UBC13/UEV1A to synthesize free K63-linked Ub chains that activate TAK1.
a –c, e and f, HEK-293 cells were transfected with an AP-1 luciferase reporter and the indicated pcDNA-based expression plasmids. Bars show mean +/− s.d. (n = 6). In c, HEK-293 cells had stable UBC13 KD or control KD. d, UBC13 KD or control KD THP-1 macrophages were treated for 24 hrs with LPS or vehicle and challenged with an HIV-1 luciferase reporter virus. Shown is the fold rescue due to TRIM5 KD, with respect to the control KD. g–j, Products of in vitro reactions with ATP, Ub, UBE1, UBC13/UEV1A, and the indicated E3 Ub ligases were revealed by immunoblot for total Ub (g, i, and j), K63-linked Ub chains (left panel of h), or TRIM5 (right panel of h). E3 ubiquitin ligases included purified owl monkey TRIM5Cyp (g, h, and j), or the indicated FLAG-tagged proteins immunoprecipitated from HEK-293T cells (i). j, In vitro Ub reactions like those in (i) were incubated with purified TAK1 kinase complex. Products were probed in immunoblot with the indicated antibodies.
The UBC13/UEV1A E2 heterodimer is notable in that it generates K63-linked Ub chains that are unlinked to substrates; these free Ub chains multimerize and activate the TAK1 kinase complex via the Ub binding components, TAB2 and TAB311. Ub in which all lysines except K63 are mutated to arginine (Ub K63) activated AP-1 and NF-κB (Fig. 3e and Supplementary Fig. 6e and f), and enhanced the ability of TRIM5 to activate AP-1 (Fig. 3f). K48-only Ub did not have these activities (Fig. 3e and f), nor did WT Ub, perhaps because of the dominance of competing Ub metabolic pathways and the tight regulation of K63 chains within cells16. These experiments indicate that the heterodimeric E2 UBC13/UEV1A and the K63-linked Ub chains that it produces play a role in TRIM5-mediated signaling.
Since TRIM5 interacted biochemically and functionally with TAK1, TAB2, TAB3, UBC13, UEV1A, and K63-Ub, the ability of TRIM5 to synthesize K63-linked Ub chains was assessed. Towards this end, a purification protocol was established that yielded 0.5 mg of soluble, full-length, owl monkey TRIM5Cyp from 1 liter of Sf9 culture (Supplementary Fig. 7). No procedure has been reported to date for the production of purified, full-length, recombinant TRIM5 protein17. Purified TRIM5Cyp was incubated with purified Ub, E1, and the E2 Ub-conjugases UBC13 and UEV1A, and reaction products were separated by SDS-PAGE. With increasing TRIM5Cyp concentration, monomeric Ub was progressively depleted and the yield of Ub chains increased (Fig. 3g and Supplementary Fig. 8a). Synthesis of Ub chains was ATP-dependent and required both UBC13 and UEV1A.
The Ub chains generated by TRIM5Cyp were detected with antibody specific for K63-linked Ub and immunoblot revealed TRIM5Cyp to be a monomer with no detectable incorporation into the Ub chains (Fig. 3h). To obtain an independent assessment of their identity, reaction products were isolated by PAGE and analyzed by matrix-assisted laser desorption/ionization and tandem mass spectrometry (Supplementary Fig. 8a and 9a–c). These methods identified peptides corresponding to K63-linked Ub and failed to detect conjugates with other Ub lysines or peptides corresponding to TRIM5Cyp, confirming that reaction products were free, unattached, K63 Ub chains. Additionally, synthesis of Ub chains was undetectable with a Ub mutant in which K63 was mutated to arginine (Supplementary Fig. 8b). Conversely, Ub was efficiently incorporated into chains when all lysines except K63 were mutated to arginine (Supplementary Fig. 8b), indicating that K63 was necessary and sufficient to form the Ub chains.
Human TRIM5α, produced by transfection of 293T cells and enriched by immunoprecipitation, catalyzed the synthesis of free K63 Ub chains like those of TRIM5Cyp, in a RING domain-dependent manner (Fig. 3i and Supplementary Fig. 8c and d). It had at least as much activity as TRAF6 (Fig. 3i), an E3 Ub ligase previously reported to synthesize unattached K63 chains that activate TAK111. TRAF2, a close paralogue of TRAF6 that does not interact with UBC1318 lacked activity (Fig. 3i).
Free K63-linked Ub chains generated by TRAF6 result in TAK1 autophosphorylation on threonine 18711, a modification required for TAK1 activation. To test the effect of K63-linked Ub chains generated by TRIM5 on TAK1 activation, the essential components of a TAK1 kinase complex, TAK1, TAB1, and TAB211, were purified and combined (Supplementary Fig. 10). This complex was then incubated with Ub, UBC13/UEV1A, and either TRAF6 or purified owl monkey TRIM5Cyp. TAK1 phosphorylation was observed in response to the K63-linked Ub chains synthesized by either TRAF6 or by TRIM5Cyp (Fig. 3j). Kinase activity required the TAK1-associated TAB1, the Ub receptor TAB2, and UBC13/UEV1A (Fig. 3j). These experiments show that, like TRAF611, TRIM5 synthesizes free K63-linked Ub chains which activate TAK1 autophosphorylation.
If TRIM5 were a PRR specific for the retroviral capsid lattice, infection with retroviruses would activate signaling, the magnitude of which would correlate with TRIM5 avidity for the capsid of the challenge virus. To determine if this is the case, myeloid cells were challenged with pairs of retroviruses that differ with respect to TRIM5 avidity for the capsid3,4 and the subsequent induction of NFκB and MAPK-dependent genes was assessed. VSV G-pseudotyped N-tropic and B-tropic MLV vectors, normalized for exogenous reverse transcriptase activity and for titer on non-restrictive MDTF cells19, were used to challenge THP-1 macrophages. The MOI of the nonrestricted B-tropic MLV on cycling THP-1 cells was 0.1. mRNA was harvested from the THP-1 cells and processed by qRT-PCR. Greater induction of PTGS2, CXCL10, CCL8, and IL6 mRNA was observed after challenge with N-MLV than with B-MLV (Fig. 4a and b), in correlation with the higher avidity of human TRIM5α for the capsid of N-tropic MLV than for the capsid of B-tropic MLV3. TRIM5 KD suppressed the higher inflammatory gene induction by N-MLV indicating its dependence upon endogenous TRIM5 (Fig. 4b). Similar differential induction of PTGS2, CXCL10, CCL8, and IL6 mRNAs by N-tropic and B-tropic MLV was observed after challenge of MDDCs or MDMs (Supplementary Fig. 11).
Figure 4. Retrovirus capsid sensing by TRIM5.
THP-1 cells (a and b), MDDCs (c and d), or owl monkey kidney cells (e and f), were challenged with matched pairs of VSV G-pseudotyped particles bearing retrovirion capsids that are restricted by the TRIM5 orthologue endogenous to that cell type (black bars), or unrestricted (white bars), or VSV G–derived particles that are devoid of capsid (gray bars). Restricted capsids were from N-tropic MLV (a–d) or HIV-1 (e and f). Unrestricted capsids were B-tropic MLV (a and b), N/B-tropic MLV (c and d) or SIVMAC239 (e and f). Particles bore viral genomes in a, b, e, and f, but not in c and d. mRNA was harvested for qRT-PCR (a–c and e) and reported as fold change versus media control. Protein in the supernatant was quantitated (d and f). Bars show means +/− s.d. (n = 3), and are representative of at least 3 independent experiments. g, Immunoblots with the indicated antibodies of products from in vitro time-course with ATP, Ub, UBE1 (E1), UBC13/UEV1A (E2), and purified owl monkey TRIM5Cyp, with or without assembled HIV-1 capsid-A14C/E45C, and with or without competitive inhibitor MeIle4CsA. (h) Schematic showing entry of an HIV-1 virion core27 (courtesy of Pornillos and Yeager) into the target cell cytoplasm where it induces dimeric TRIM5 to form a hexameric lattice25 with increased E3 Ub ligase activity. With UBC13/UEV1A, TRIM5 synthesizes free K63 Ub chains that are recognized by TAB2, which multimerizes and activates the TAK1 kinase complex.
Retroviral cDNA activates innate immune signaling under some conditions20. Restriction by human TRIM5α results in N-MLV cDNA levels that are an order of magnitude lower than for B-MLV cDNA21 so the experiments described above might underestimate the effect of N-MLV capsid on TRIM5-mediated signaling. Therefore, MDDCs were challenged with matched pairs of virus-like particles (VLPs) devoid of the viral genome that serves as the reverse transcription template. VLPs bearing N-MLV capsid activated CXCL9, CXCL10, IFIT1, and IFIT2 mRNAs from 5 to 55-fold over the levels in untreated MDDCs (Fig. 4c). Inflammatory gene induction was not detected with VLPs bearing the unrestricted NB-MLV capsid22 (Fig. 4c). As with the mRNA, soluble IL8, CCL5, CXCL9, and CXCL10 protein was differentially induced by N-MLV (Fig. 4d).
To determine if differential gene induction after retrovirus challenge was peculiar to N-tropic MLV, a similar experiment was performed with OMK, a kidney cell line from the owl monkey, Aotus trivirgatus. The TRIM5 orthologue in this species restricts HIV-1 but not SIV1. VSV G-pseudotyped HIV-1 and SIV vectors, normalized for exogenous reverse transcriptase activity and for titer on HeLa cells, were used to challenge OMK cells. The MOI of the unrestricted SIV on OMK cells was 0.3. Among the MAPK- and NF-κB-dependent gene products that were detectable in this species using human probes, transcriptional activation of PTGS2, IFIT1 and IFIT2 mRNAs, and secretion of IL8, CCL2, CCL4, and CXCL10 proteins, was higher after challenge with the restricted virus (Fig. 4e and f).
TRIM5 senses retrovirus capsids in the target cell cytoplasm (Fig 4a–f) and activates MAPK- and NF-κB-dependent transcription via the synthesis of TAK1-activating, K63-linked Ub chains (Fig. 3g–j). If these observations were linked functionally, interaction with capsid would be expected to stimulate the synthesis of Ub chains by TRIM5Cyp. Soluble, recombinant HIV-1 capsid or capsid hexamers generated by the oxidation of recombinant capsid bearing strategically-placed cysteine substitutions (A14C/E45C/W184A/M185A)23 had no effect on the synthesis of K63-linked Ub chains (data not shown). Current models of the HIV-1 capsid lattice are based on cylinders generated under high salt with either capsid or capsid-nucleocapsid fusion protein17,24; both preparations were generated but the high salt necessary to maintain capsid cylinders blocked E3 Ub ligase activity. Capsid cylinders were then assembled with A14C/E45C-substituted capsid protein in 1 M NaCl and the cysteines were oxidized. These oxidized cylinders were stable in the absence of salt (Supplementary Fig. 12a) and greatly stimulated the production of K63-linked Ub chains by TRIM5Cyp (Fig. 4g). No Ub-linked products were detected with anti-capsid (p24) or anti-TRIM5 antibodies, indicating that the reaction products were unattached Ub chains (Fig. 4g). Finally, two factors that disrupt the HIV-1 capsid-TRIM5Cyp interaction and block restriction activity - a non-immunosuppressive cyclosporine analogue1 or the TRIM5Cyp H436Q mutant protein13 - each eliminated the enhancement of E3 Ub ligase activity by the A14C/E45C capsid cylinders, without effect on the baseline activity in the absence of capsid (Fig. 4g and Supplementary Figs. 7d and 12b–d).
The experiments presented here demonstrate that TRIM5 is a multifunctional component of the innate immune system. In addition to functioning as a retroviral capsid-specific restriction factor, TRIM5 synthesizes K63 Ub chains that activate TAK1 and inflammatory transcription, most probably via multimerization of the TAK1-associated Ub-binding protein TAB211 (Fig. 4h). This activity was greatly increased by the hexameric capsid lattice, a molecular signature of HIV-1 and other retroviruses. TRIM5, then, satisfies criteria for a bona fide PRR10. Interestingly, TRIM5 spontaneously forms an hexagonal lattice that is complementary to the capsid lattice2525 but the efficiency of TRIM5 lattice formation is greatly stimulated by the capsid hexamer25 (Fig. 4h). Little is known about how the innate immune system detects retroviruses26 and the discovery that TRIM5 acts as a PRR is an important step towards filling this critical gap. The cellular factors required for TRIM5 E3 activity and inflammatory gene induction, UBC13, UEV1A and TAK1, also promoted capsid-specific restriction activity, indicating that the multiple functions of TRIM5 are mechanistically linked. Identification of relevant TAK1 substrates will inform future attempts to pinpoint the mechanism of restriction.
METHODS SUMMARY
Plasmids, cells and viruses
These methods were previously described1,13,19 or are detailed in the Supplementary Material.
Recombinant Protein
Production of full-length, soluble, TRIM5Cyp is described in the supplement. CA A14C E45C was produced and assembled into tubes as described23,24.
Microarray
Illumina HumanHT-12 V3.0 expression beadchips were probed with RNA from TRIM5 KD THP-1 cells. Data set and methods are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession number GSE25041.
Supplementary Material
Acknowledgments
We thank D. Baltimore, M. J. Birrer, J. Brojatsch, A. Cimarelli, A. DeIaco, S. Elledge, M. Emerman, W. Ferlin, D. Garcin, S. Ghosh, O. Haller, T. Hatziioannou, J. Hiscott, A. Iwasaki, D. Kolakofsky, M. Kosco-Vilbois, H. Malik, R. Medzhitov, M. R. Neagu, G. Napolitani, P. Palese, D. Pinschewer, O. Pornillos, L. Roux, O. Schwartz, M. Strubin, V. Studer, W. Sundquist, G. Towers, D. Trono, J. Tschopp, M. Yeager, M. Zufferey, and the Functional Genomics Center (Zürich), for ideas, technical assistance, and reagents. This work was supported by NIH grant RO1AI59159 to J.L., NIH grant R21AI087467 to W.M., Swiss National Science Foundation grant 3100A0-128655 to J.L. and 3100A0-122342 to M.G.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions T.P., S.H., J. G., C. R., C. S., M. P., W. M., M. G., and J.L. designed the experiments; T.P., S.H., D. M., S. Z., J. G., J. La., C. R., M. P., A. B., P.D.U. and L.C. performed the experiments. All authors contributed to the assembly and writing of the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 2.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 3.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthoux L, et al. As(2)O(3) enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J Virol. 2003;77:3167–3180. doi: 10.1128/JVI.77.5.3167-3180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi M, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 7.Tareen SU, Emerman M. Human Trim5alpha has additional activities that are uncoupled from retroviral capsid recognition. Virology. 2011;409:113–120. doi: 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. Journal of Virology. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host & Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med. 1989;169:1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neagu MR, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berthoux L, Sebastian S, Sokolskaja E, Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci U S A. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langelier CR, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. Journal of Virology. 2006;80:2855–2862. doi: 10.1128/JVI.80.6.2855-2862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci U S A. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulm JW, Perron M, Sodroski J, RM Complex determinants within the Moloney murine leukemia virus capsid modulate susceptibility of the virus to Fv1 and Ref1-mediated restriction. Virology. 2007;363:245–255. doi: 10.1016/j.virol.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 23.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 25.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5{alpha} protein. Proc Natl Acad Sci U S A. 2010;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medzhitov R, Littman D. HIV immunology needs a new direction. Nature. 2008;455:591. doi: 10.1038/455591a. [DOI] [PubMed] [Google Scholar]
- 27.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.