Abstract
Excessive production of reactive oxygen species can lead to alteration of cellular functions responsible for many diseases including cardiovascular, neurodegenerative, cancer, and aging. Hydroxyl radical is a short-lived radical which is considered very aggressive due to its high reactivity towards biological molecules. In this study, a COumarin-NEutral Red (CONER) nanoprobe was developed for the detection of hydroxyl radical based on the ratiometric fluorescence signal between 7-hydroxy coumarin 3-carboxylic acid and neutral red dyes. Biocompatible poly-lactide-co-glycolide (PLGA) nanoparticles containing encapsulated neutral red were produced using a coumarin 3-carboxylic acid conjugated poly(sodium N-undecylenyl-Nε-lysinate) (C3C-poly-Nε-SUK) as moiety reactive to hydroxyl radicals. The response of the CONER nanoprobe was dependent on various parameters such as reaction time and nanoparticle concentration. The probe was selective for hydroxyl radicals as compared with other reactive oxygen species including O2•−, H2O2, 1O2, and OCl−1. Furthermore, the CONER nanoprobe was used to detect hydroxyl radicals in vitro using viable breast cancer cells exposed to oxidative stress. The results suggest that this nanoprobe represents a promising approach for the detection of hydroxyl radicals in biological systems.
INTRODUCTION
Aerobic organisms produce energy through oxidation of biological substrates in the presence of oxygen. Complete oxygen reduction takes place in mitochondria involving a series of redox reactions controlled by a complex enzymatic mechanism. Such reactions generate radical and non-radical reactive oxygen species, i.e. superoxide anion (O2•−), hydroxyl radical (OH•), singlet oxygen (1O2), and hydrogen peroxide (H2O2).1-3 The cells of aerobic organisms experience oxidative stress when an imbalance exists between free radical production and elimination of such species through reaction with various reducing agents, enzymes, and antioxidants.4-7 Reactive oxygen species are involved in many metabolic processes, including signal transduction, carcinogenesis, and inflammatory response. In addition, excessive production of radical species can lead to alteration of cellular functions responsible for cardiovascular diseases, neurodegenerative diseases, diabetes, cancer, joint diseases, as well as aging.8-13 During cellular oxidation processes, hydroxyl radicals can be produced by Fenton and Habber-Weiss reactions of hydrogen peroxide enabled by transition metals such as iron or copper.5,12 The hydroxyl radical has a short half-life, and is considered the most aggressive free radical primarily due to its high reactivity with many different biological species. Studies have shown that it can react with lipids, amino acids, proteins, DNA, and sugars at extremely high rates leading to cell damage and even cell death.14-16
Reactive oxygen species are detectable using various methods, including electron spin resonance (ESR), UV-visible, fluorescence, and luminescence spectroscopies.17-19 Fluorescence probes provide several advantages in comparison to other methods such as high specificity, localized information at the target site, and great versatility in possible detection schemes including fluorescence spectroscopy and in vitro fluorescence microscopy are several benefits of the fluorescent probes.20-22 In particular, coumarin 3-carboxylic acid (C3C) has been used as a fluorescent probe for detecting hydroxyl radicals.23 The non-fluorescent C3C molecule can react with a hydroxyl radical to undergo hydroxylation at position C7 of the coumarin structure, producing a highly fluorescent compound, i.e. 7-hydroxy coumarin 3-carboxylic acid (7-OH C3C). This compound exhibits a strong emission signal at 450 nm when excited at 400-410 nm.23-25 The chemical structure of C3C contains a carboxylic acid group in the C3 position that can be easily coupled with other molecules. Other coumarin derivatives such as the succinimidyl ester of C3C (SECCA), phospholipid – linked coumarins, and C3C – derivatized amino acids and peptides have been used for detection of hydroxyl radicals.26-28
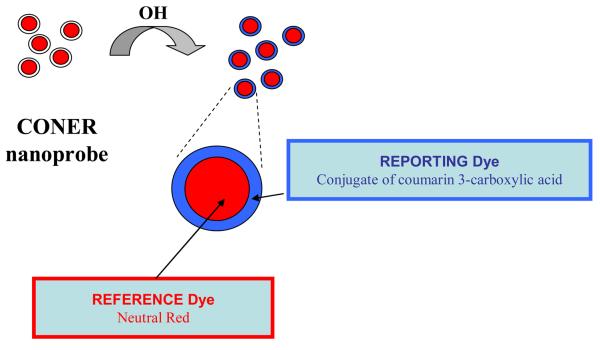
In developing a successful fluorescent probe for free radical detection, several aspects should be taken into consideration. For example, instrumental artifacts and sensitivity to environmental factors such as temperature and pH can alter the reliability of fluorescent probes. Furthermore, real time in vitro imaging becomes challenging if other molecules interfere with the probe within the cellular medium resulting in photobleaching or the generation of secondary radicals.18 Therefore, fluorescence ratiometric detection can be employed to reduce such limitations. The ratio of the two signals represented by the fluorescence intensity of a reporting molecule and the fluorescence intensity of a reference molecule can be used for detection of various radicals. In the recent literature, C3C-coupled polyacrylamide nanoparticles incorporating Texas Red as a reference dye were used as ratiometric nanoparticles, and hydroxyl radicals were detected based on the fluorescence intensity ratio between these two dyes.29 In the present manuscript, we propose a strategy where biocompatible CONER nanoprobes are developed and used as ratiometric fluorescent nanoprobes for hydroxyl radical detection according to the schematic illustrated in Figure 1.
Figure 1.
Design of ratiometric CONER nanoprobe.
Novel molecular micelles containing lysine-C3C conjugates were used for synthesis of biocompatible poly-lactide-co-glycolide (PLGA) nanoparticles through emulsification solvent evaporation.30,31 The reference dye neutral red (NeR) dye was encapsulated inside the polymeric matrix which in turn promoted protection from side reactions with reactive species. Nanoparticles containing only the C3C moiety and nanoparticles containing only the NeR dye were also synthesized using emulsification solvent evaporation. Optimization of CONER nanoprobe response was performed. In addition, CONER nanoprobe was investigated in the presence of other reactive oxygen species such as superoxide anion radical, hydrogen peroxide, hypochlorite and singlet oxygen. The CONER nanoprobe was tested on MCF-7 breast tumor cells exposed to H2O2 – induced oxidative stress. In vitro detection of hydroxyl radicals was achieved by use of fluorescence microscopy.
EXPERIMENTAL SECTION
Materials
Poly (D,L-lactide-co-glycolide) (PLGA, lactide:glycolide 50:50, MW 40,000-75,000), undecylenic acid, ethyl acetate, trifluoroacetic acid (TFA), sodium carbonate, coumarin 3-carboxylic acid, Nε-Boc lysine, dimethyl sulfoxide (DMSO), anhydrous dimethyl formamide (DMF), sucrose, hydrogen peroxide, and neutral red (NeR) were purchased from Sigma-Aldrich (St. Louis, MO, USA). NeR was neutralized with NaOH in water, resulting into a water-insoluble red precipitate that was freeze-dried and used further in the nanoprobe preparation. Dibasic sodium phosphate was purchased from Mallinckrodt (Hazelwood, MO, USA). N-hydroxysuccinimide (NHS), N,N′-dicyclohexylcarbodiimide (DCC), and cuprous sulfate were purchased from Fluka (Milwaukee, WI, USA). Dichloromethane (DCM), sodium phosphate monobasic, isopropyl alcohol, tetrahydrofuran, and hexanes were purchased from EMD Chemicals Inc. (Gibbstown, NJ, USA). Sodium bicarbonate and sodium hydroxide were purchased from Fisher Scientific (Pittsburgh, PA, USA). Doubly-distilled deionized water was obtained from an ELGA PURELAB Ultra water polishing system (US Filter, Lowell, MA, USA). All reagents were of analytical grade and were used as received. Synthesis of coumarin functionalized molecular micelles is presented in Supporting Information.
Nanoparticle Synthesis
PLGA nanoparticles were synthesized by use of an emulsification solvent evaporation method. Briefly 0.5 mL of 1 mg/mL NeR in DCM was added to 0.5 mL DCM solution containing 50 mg of PLGA. An aqueous phase was prepared by dissolving C3C-poly-Nε-SUK in 5 mL water. The organic phase was added dropwise to the aqueous phase under stirring conditions using a homogenizer (model 398, Biospec Products, Inc., Racine, WI, USA), at 15,000 rpm for 2 minutes, resulting in a single o/w emulsion. The emulsion droplets were further reduced by sonication using a probe ultrasound processor (model VC750, Sonics and Materials Inc., Newton, CT, USA), operating at an amplitude intensity of 35 %, for 10 minutes. The solvent was evaporated using a rotary evaporator (Büchi rotovapor R-200, Brinkmann Instruments, Inc., Westbury, NY, USA). The nanoparticles were centrifuged at 10,000 rpm for 30 minutes, followed by washing by centrifugation in the same conditions. The nanoparticle suspension was stored at 4 °C until further use. Nanoparticles containing only C3C (blank nanoparticles) were prepared using C3C-poly-Nε-SUK and DCM as the replacement for NeR dye solution. For non-C3C-functionalized NeR-loaded nanoparticles, C3C-poly-Nε-SUK was replaced with a non-C3C functionalized Boc-protected poly-Nε-Boc-SUK molecular micelle.
Nanoparticle Characterization
Nanoparticle size and morphology were investigated using transmission electron microscopy (TEM) (JEOL 100CX, JEOL USA Inc., Peabody, MA, USA) operating at 80kV. A drop of nanoparticle suspension was dried at room temperature on a carbon coated copper grid and negatively stained using a 2 % solution of uranyl acetate. Polydispersity index (PDI) was measured by use of dynamic light scattering (DLS) (Zetasizer NanoZS, Malvern Instruments Ltd., Malvern, UK). Nanoparticle zeta potential was measured by use of laser doppler anemometry (Zetasizer NanoZS, Malvern Instruments Ltd., Malvern, UK) using a capillary cell. The reported values represent the average of three different nanoparticle batches.
Dye Content
The NeR dye content (% w/w) was determined by difference between the known dye amount added to the formulation and the amount eliminated in the purification steps by centrifugation. The supernatant containing the excess dye was analyzed by the use of absorbance spectroscopy (UV-3101PC UV-Vis-near-IR scanning spectrometer, Shimadzu, Columbia, MD) using a reduced volume (1.4 mL) quartz cuvet. A calibration curve was constructed for NeR by representing the absorbance intensity at 530 nm as a function of NeR concentration (y = 1.1178×−0.0019, R2 = 0.9989). Similarly, a calibration curve was constructed for C3C by representing the absorbance intensity at 290 nm as a function of C3C concentration (y = 0.0111×−0.0258, R2 = 0.9927). The C3C coverage (% w/w) was calculated based on the C3C content of poly-Nε-SUK remaining attached on the surface of nanoparticles after purification.
Hydroxyl Radical Generation
Hydroxyl radicals were generated in phosphate buffer (pH=7.4) based on the reaction of cuprous sulfate and hydrogen peroxide in the presence of ascorbic acid, as described by equations (1) and (2).27 The reduction of Cu2+ takes place in the presence of ascorbic acid resulting into Cu+ that further reacts with hydrogen peroxide leading to the formation of hydroxyl radicals. The concentration of OH• was varied by varying the concentration of cuprous sulfate added to the reaction. For various experiments, the reaction was stopped at various time intervals using DMSO as OH• scavenger.
| (1) |
| (2) |
Fluorescence Spectroscopy
Steady-state fluorescence spectra were recorded using Fluorolog-3 spectrometer (model FL1073, Horiba Jobin Yvon, Edison, NJ) operated in the front face mode, at 25°C as provided by a temperature control chamber. The samples were prepared in phosphate buffer (pH 7.4), and a short path (0.4 cm2) cuvet was used. The excitation and emission slit widths were both set at 5 nm, respectively. The samples were excited at 410 nm, and the emission was collected from 420 to 700 nm. The blanks containing all reagents except nanoparticles were subtracted for each sample. All measurements were performed in triplicates.
In the experiments for nanoparticle reaction with hydroxyl radicals the reaction conditions were 0.07 mg/mL nanoparticles, 200 μM CuSO4, 20 mM H2O2 and 200 μM ascorbic acid that reacted for 5 minutes in a total volume of 500 mL. For the experiments investigating the effect of coumarin location, the reaction mixture contained 0.07 mg/mL nanoparticles, 200 μM CuSO4, 20 mM H2O2 and 200 μM ascorbic acid. The mixture was allowed to react for 5 minutes in a total volume of 500 mL before collecting the fluorescence spectra. In the experiments for the effect of nanoparticle concentration, various concentrations of nanoparticles (0.03, 0.07, 0.10, 0.14, 0.21 mg/mL nanoparticles) were reacted with 400 μM CuSO4, 20 mM H2O2 and 200 μM ascorbic acid for 5 min in a total volume of 500 mL. In the experiments for the effect of hydroxyl radical concentration, ratiometric nanoparticles (0.07mg/mL) were incubated with 20 mM H2O2, 200 μM ascorbic acid, and various concentrations of CuSO4 (10, 20, 30, 40, 50, 100, 200, 400, and 1000 μM respectively) for 5 min in a total volume 500 mL. For the selectivity experiments, the nanoparticles (0.07 mg/mL) were reacted with OH• (400 μM CuSO4, 20 mM H2O2 and 200 μM ascorbic acid), O2−• (200 μM KO2), H2O2 (20 mM), OCl− (200 μM NaOCl), and 1O2 (200 μM H2O2 + 200 μM NaOCl).
In vitro Detection of Hydroxyl Radicals
Human mammary MCF-7 tumor cells (HTB-22, American Tissue Culture Collection, ATCC, Manassas, VA) were grown to 90% confluence according to ATCC's instructions and used for in vitro detection of hydroxyl radicals. Specifically, the cells were incubated with nanoparticle suspension (0.07 mg/ mL cell suspension) in a 6-well plate at 10,000 cells/ well. After two hours of incubation, the cells were washed with growth media to eliminate the excess of nanoparticles. The cells were then exposed to H2O2-induced oxidative stress (400 μM) for 10, 20 and 40 minutes, and washed with phosphate buffer. Fluorescence images were taken before and after exposure, using a Leica DM RXA2 upright microscope (Leica Microsystems Inc., Bannockburn, IL) equipped with GFP and TRITC filter cubes, and an immersion 40X objective.
RESULTS AND DISCUSSION
Nanoparticle Characterization
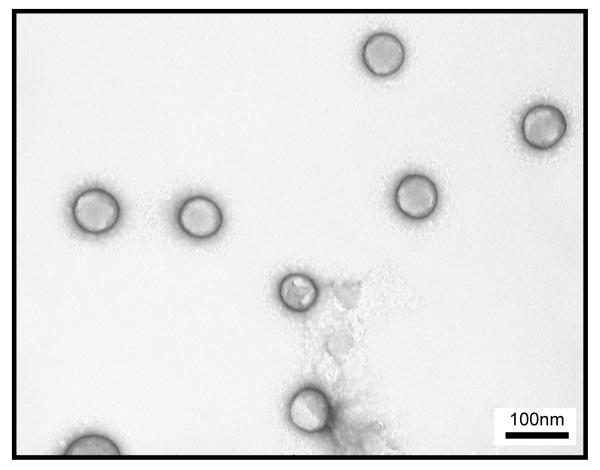
In this study, a CONER nanoprobe based on molecular micelle-modified PLGA nanoparticles was prepared by emulsification solvent evaporation using fluorescent molecular micelles (C3C-poly-Nε-SUK) as emulsifiers. Previously, PLGA nanoparticles were successfully synthesized in our laboratory using molecular micelles as emulsifiers, offering many advantages including small size, monodispersed suspension, and excellent stability.30 The molecular micelle-modified NeR loaded PLGA nanoparticles synthesized in this study had an average size of 65 ± 4 nm as measured by TEM. In addition, the nanoparticles were monodispersed as expressed by a low PDI value of 0.052 ± 0.012. A zeta potential of −52.77 ± 0.76 mV indicating high stability of nanoparticles in solution was afforded by the anionic molecular micelle present on the nanoparticle surface. Electron microscopy images showed spherical individual nanoparticles as shown in Figure 2.
Figure 2.
TEM micrograph of CONER nanoprobes with average diameter of 65 nm.
Molecular micelle – modified PLGA nanoparticles have been previously reported by our group for the encapsulation and delivery of hydrophobic molecules.31 In this study, NeR was neutralized with NaOH to produce a neutral and hydrophobic reference dye. NeR was then encapsulated in the PLGA nanoparticles using C3C-poly-Nε-SUK as emulsifier. NeR content was determined by UV-vis. Typical NeR content was 83.9 ± 0.8 % w/w while C3C content was 48.4 ± 0.9 % w/w as determined using the same approach.
Reaction with Hydroxyl Radicals
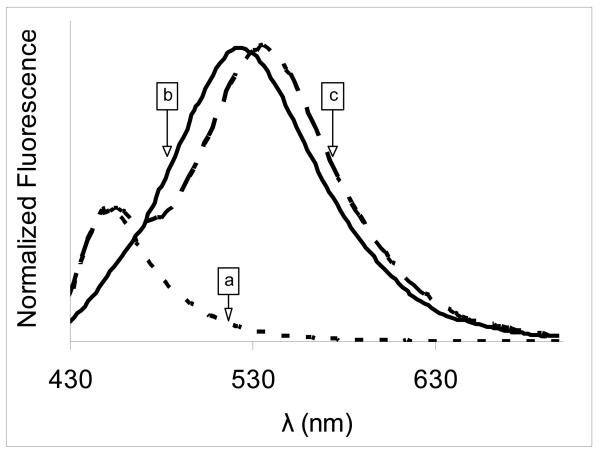
In contrast with variations of a single signal, ratiometric detection involved changes in the ratio of two signals. The first signal was given by a reporting dye that reacted with the molecule of interest, while the second signal was given by a reference dye that corrected for instrumental and environmental artifacts. Nanoparticles that contain both reference and reporting dyes were used in the ratiometric detection scheme. In the absence of hydroxyl radicals, the CONER ratiometric nanoprobe did not display a fluorescent signal at 450 nm (Figure S4, Supporting information). In contrast, the ratiometric nanoprobe exhibited two distinct signals, at 450 and 540 nm corresponding to reporting dye and reference dye, respectively. The reporting dye was 7-OH C3C-poly-Nε-SUK that resulted from the reaction of OH radicals with C3C-poly-Nε-SUK molecular micelle present on the nanoparticle surface. The product of such reaction presented a fluorescent signal at 450 nm when excited at 410 nm. At the same time, the reference dye, NeR, provided a separate fluorescent signal. The two signals obtained after the reaction with OH• can be observed in Figure 3, graph c.
Figure 3.
Normalized fluorescence of nanoparticles after reaction with OH• a) C3C-only nanoparticles; b) NeR-only nanoparticles; c) complete ratiometric nanoprobe .
C3C-only nanoparticles (NeR-free nanoparticles containing C3C-poly-Nε-SUK) exhibited a peak at 450 nm as a result of the reaction of OH• with C3C, while the fluorescence peak at 540 nm was absent (Figure 3, graph a). In contrast, NeR-only nanoparticles prepared using a non-functionalized Boc-protected poly-Nε-Boc-SUK molecular micelle as emulsifier had no C3C fluorescence (Figure 3, graph b). This confirmed that the two fluorescence peaks in the CONER probe spectrum were generated by the reference and the reporting dyes, respectively. In addition, the effect of coumarin location is discussed in Supporting Information. It was observed that the coumarin moiety present on the surface of nanoparticles was able to react with hydroxyl radicals. In contrast, the encapsulation of coumarin moiety into the polymer matrix produced no clear signal at 450 nm, suggesting that a polymer shielding effect took place.
Effect of Hydroxyl Radical Concentration
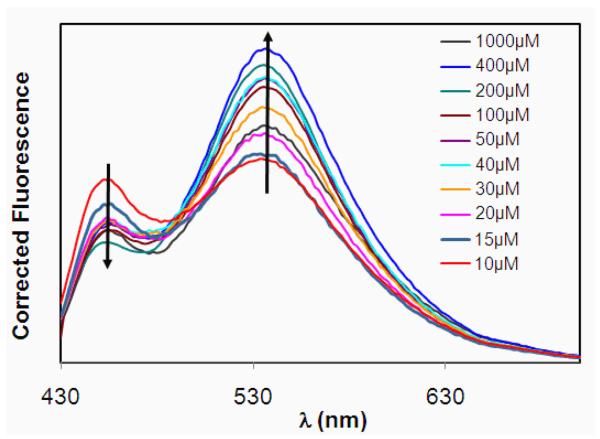
CONER nanoprobes were incubated with increasing concentrations of OH•. In general, if the reporting and reference dyes of a ratiometric probe are in close proximity, and there is an overlap between the fluorescence signal of the reporting dye and the absorbance of the reference dye, fluorescence resonance energy transfer (FRET) occurs. For such probes, the signal of the reporting dye decreases while the one of the reference dye increases. The reporting dye signal strictly depends on the reaction with OH•, and an increase in radical concentration will produce an increase in the reporting dye signal. The fluorescence spectra of CONER nanoprobe in the presence of different concentrations of OH• is shown in Figure 4. In this experiment, the concentration of OH• was changed by changing the concentration of CuSO4, as described in the experimental section.
Figure 4.
Fluorescence spectra of nanoparticles after reaction with OH• (at various CuSO4 concentrations).
In the presence of OH•, it was expected that the C3C moiety present on the surface of nanoparticles would react with the radicals, resulting in formation of 7-OH-C3C. The fluorescence intensity of 7-OH-C3C depends on various factors: OH• concentration, C3C concentration, and Förster resonance energy transferred to NeR. The observed FRET phenomenon was favored by the spectral overlap and short spatial distance between the reporting and reference dyes. If the OH• concentration is constant and C3C concentration increased (nanoparticle concentration increased), then both 7-OH-C3C and NeR peaks increased (Figure S2, Supporting information). This was observed primarily because both C3C and NeR concentrations increased (both were incorporated into the same nanoparticle system) and the effect of energy was not observed. If the C3C and NeR concentrations were held constant (nanoparticle concentration was constant), then the fluorescence intensity of 7-OH-C3C decreased with an increase in concentration of OH•, primarily because of energy transfer to NeR. Therefore, the NeR peak increased, and 7-OH-C3C decreased. In the absence of NeR, 7-OH-C3C increased with an increase of OH• concentration. In Figure S4 (Supporting information), the fluorescence spectra of blank nanoparticles containing only C3C are represented for two OH• concentrations. Since the NeR was absent in this nanoprobe, energy transfer was also absent, and consequently the fluorescence intensity of 7-OH-C3C increased with an increase in OH• concentration. Furthermore, the I450/I528 was calculated as the ratio between C3C fluorescence intensity at 450 nm and NeR fluorescence intensity at 528 nm, and represented as a function of OH• concentration (Supporting Information, Figure S6). Using the linear region of the graph, a limit of detection of 0.73 μM OH• was calculated.
Reaction with Other Radicals
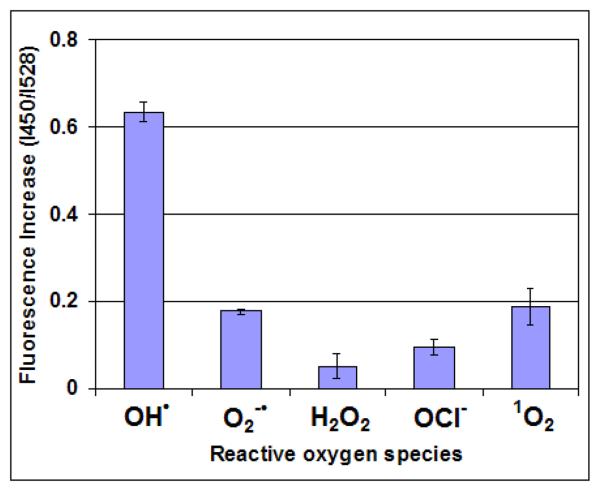
The selectivity of CONER nanoprobes for OH• is important in particular for radical detection in biological samples. Under steady state conditions, mitochondrial ROS concentrations are in the nanomolar range.32 However, under conditions of oxidative stress, ROS concentrations increase to dangerous levels. For example, hydrogen peroxide produces cell damage at 10 μM.5 In the present studies, the CONER probes were exposed to various radical and non-radical reactive species such as superoxide anion (O2•−), hydrogen peroxide (H2O2), singlet oxygen (1O2), and hypochlorite anion (OCl−). The reactions took place in phosphate buffer using the same nanoparticle concentration, reaction volume and reaction time. The ratio between C3C fluorescence intensity at 450 nm and NeR fluorescence intensity at 528 nm (I450/I528) is shown in Figure 5 for each investigated radical.
Figure 5.
Fluorescence spectra of nanoparticles after reaction with various radicals.
The I450/I528 ratio was 0.63 ± 0.03 for OH radicals. Three to four times smaller ratios were obtained for singlet oxygen (0.18 ± 0.04) and superoxide anion (0.17 ± 0.00). The smallest ratio was observed for hypochlorite anion (0.09 ± 0.01), representing a ratio seven times smaller than that of hydroxyl radicals. These results suggest that our CONER probe was more selective for OH• than for other reactive oxygen species.
In vitro Detection of Hydroxyl Radicals
In comparison with other reactive oxygen species, hydroxyl radical is a short-lived and highly reactive with numerous biological molecules including lipids, proteins and DNA. Therefore, a selective and sensitive detection method of OH• constitutes a vital technique for the diagnostics and understanding of OH• interaction with cells and its role in diseases. In order to verify the ability of CONER nanoprobe to react with hydroxyl radicals generated in a biological environment, live breast tumor cells were incubated with CONER nanoprobe over various periods of time.
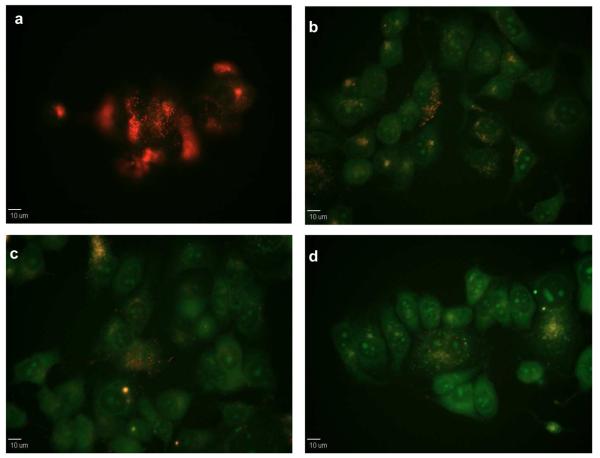
Oxidative stress was further induced using hydrogen peroxide. Hydroxyl radicals were likely generated by reaction of hydrogen peroxide with transition metals present in the cellular environment, following a route similar to the one described in the experimental section.4,5 The fluorescence signal was observed by fluorescence microscopy at different incubation times as shown in Figure 6. Since C3C was not fluorescent under the experimental conditions, the cells appeared initially red due to the NeR fluorescent signal. As the OH• became available in the cell environment after H2O2 exposure, the cells progressively started fluorescing green after 10, 20 and 40 minutes. Green fluorescence is strongly determined by NeR since the NeR fluorescence shifted from red to green due to the cellular environment. However, green fluorescence also increased in time due to more radicals reacting with C3C and increasing energy transfer towards NeR. Therefore, it can be inferred that the increase in green fluorescence correlates to an increase in OH• concentration.
Figure 6.
Fluorescence micrographs of CONER probe response in live cells exposed to H2O2 – induced oxidative stress a) before the addition of H2O2 and at b) t=10, c) t=20, and d) t= 40 minutes after the addition of H2O2.
For control experiments (Supporting Information), the cells were exposed to oxidative stress conditions in the absence of nanoparticles. Under these conditions, the cells displayed a weak green autofluorescence that was significantly lower (by visual comparison of identically processed images) than the fluorescence observed for cells incubated with CONER probes. In addition, the cells were incubated with nanoparticles containing only C3C moiety as well as nanoparticles containing only NeR. The fluorescence of 7-OH-C3C was not observed during in vitro experiments using the GFP cube. Therefore, in the absence of NeR there was no green fluorescence. In contrast, when only NeR was present, green fluorescence was observed, and the red fluorescence remained unchanged before and after exposure to oxidative stress. The imaging experiments suggested that the presence of both C3C and NeR were required in order to obtain a change in the fluorescence of cells exposed to oxidative stress.
CONCLUSION
The detection of reactive oxygen species is critical to the understanding of the mechanism of hydroxyl radical, one of the most reactive radicals involved in oxidative stress. In this study, biocompatible CONER nanoprobe (C3C-poly-Nε-SUK – modified NeR – loaded PLGA nanoparticles) was successfully developed and used to detect hydroxyl radicals by fluorescence ratiometric detection conferred by a 7-OH C3C reporting dye and a NeR reference dye contained within the same nanoparticle system. The ratiometric nanoparticles were sensitive and selective for hydroxyl radicals as compared to other reactive oxygen species such as superoxide anion (O2•−), hydrogen peroxide (H2O2), singlet oxygen (1O2), and hypochlorite (OCl−). In summary, the CONER probe successfully detected hydroxyl radicals in viable cells exposed to oxidative stress allowing them to be a great analytical tool for in vitro studies.
Supplementary Material
Acknowledgments
The authors thank the National Science Foundation and the National Institutes of Health for the financial support. G.M. Ganea thanks Karen Mcdonough for her assistance in the cell culture studies.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McLennan HR, Esposti MD. Journal of Bioenergetics and Biomembranes. 2000;32:153–162. doi: 10.1023/a:1005507913372. [DOI] [PubMed] [Google Scholar]
- 2.Naoi M, Maruyama W, Shamoto-Nagai M, Yi H, Akao Y, Tanaka M. Molecular Neurobiology. 2005;31:81–93. doi: 10.1385/MN:31:1-3:081. [DOI] [PubMed] [Google Scholar]
- 3.Nohl H, Kozlov AV, Gille L, Staniek K. Biochemical Society Transactions. 2003;31:1308–1311. doi: 10.1042/bst0311308. [DOI] [PubMed] [Google Scholar]
- 4.Sorg O. Comptes Rendus Biologies. 2004;327:649–662. doi: 10.1016/j.crvi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Kohen R, Nyska A. Toxicologic Pathology. 2002;30:620–650. doi: 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- 6.Jones DP. Antioxidants & Redox Signaling. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 7.Arora S, Chopra SC, Gupta C. Journal of Anaesthesiology, Clinical Pharmacology. 2003;19:15–32. [Google Scholar]
- 8.Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Clinical Chemistry. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 9.Molavi B, Mehta Jawahar L. Current Opinion in Cardiology. 2004;19:488–93. doi: 10.1097/01.hco.0000133657.77024.bd. [DOI] [PubMed] [Google Scholar]
- 10.Passi S, Gianni G, Cocchi M. Progress in Nutrition. 2006;8:241–256. [Google Scholar]
- 11.Manea A, Constantinescu E, Popov D, Raicu M. Journal of Cellular and Molecular Medicine. 2004;8:117–126. doi: 10.1111/j.1582-4934.2004.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goetz ME, Luch A. Cancer Letters. 2008;266:73–83. doi: 10.1016/j.canlet.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 13.Afonso V, Champy R, Mitrovic D, Collin P, Lomri A. Joint, Bone, Spine. 2007;74:324–329. doi: 10.1016/j.jbspin.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget E-P, Ravanat J-L, Sauvaigo S. Mutation Research, Fundamental and Molecular Mechanisms of Mutagenesis. 1999;424:9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 2nd edition Oxford University Press; 1989. pp. 1–81. [Google Scholar]
- 16.Allen DA, Harwood SM, Varagunam M, Raftery MJ, Yaqoob MM. FASEB Journal. 2003;17:908–910. doi: 10.1096/fj.02-0130fje. [DOI] [PubMed] [Google Scholar]
- 17.Vieira AJSC, Telo JP, Dias RMB. Methods in Enzymology. 1999;300:194–201. doi: 10.1016/s0076-6879(99)00126-3. [DOI] [PubMed] [Google Scholar]
- 18.Bartosz G. Clinica Chimica Acta. 2006;368:53–76. doi: 10.1016/j.cca.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Busserolles J, Rock E, Rayssiguier Y, Mazur A. Current Topics in Biophysics. 2002;26:157–162. [Google Scholar]
- 20.Gomes A, Fernandes E, Lima JLFC. Journal of Biochemical and Biophysical Methods. 2005;65:45–80. doi: 10.1016/j.jbbm.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Soh N, Imato T. Current Bioactive Compounds. 2006;2:409–430. [Google Scholar]
- 22.Soh N. Analytical and Bioanalytical Chemistry. 2006;386:532–543. doi: 10.1007/s00216-006-0366-9. [DOI] [PubMed] [Google Scholar]
- 23.Manevich Y, Held KD, Biaglow JE. Radiation Research. 1997;148:580–591. [PubMed] [Google Scholar]
- 24.Martin-Aragon S, Benedi J, Villar A. Phytotherapy Research. 1996;10:S75–S78. [Google Scholar]
- 25.Newton GL, Milligan JR. Radiation Physics and Chemistry. 2006;75:473–478. [Google Scholar]
- 26.Makrigiorgos GM, Baranowska-Kortylewicz J, Bump E, Sahu SK, Berman RM, Kassis AI. International Journal of Radiation Biology. 1993;63:445–58. doi: 10.1080/09553009314550601. [DOI] [PubMed] [Google Scholar]
- 27.Soh N, Makihara K, Ariyoshi T, Seto D, Maki T, Nakajima H, Nakano K, Imato T. Analytical Sciences. 2008;24:293–296. doi: 10.2116/analsci.24.293. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh SC, Auzenne E, Farquhar D, Klostergaard J. Bioconjugate Chemistry. 2007:731–735. doi: 10.1021/bc060285q. [DOI] [PubMed] [Google Scholar]
- 29.King M, Kopelman R. Sensors and Actuators, B: Chemical. 2003;B90:76–81. [Google Scholar]
- 30.Ganea GM, Sabliov CM, Ishola AO, Fakayode SO, Warner IM. Journal of Nanoscience and Nanotechnology. 2008;8:280–292. [PubMed] [Google Scholar]
- 31.Ganea GM, Fakayode SO, Losso JN, van Nostrum CF, Sabliov CM, Warner IM. Nanotechnology. 2010;21:285104. doi: 10.1088/0957-4484/21/28/285104. [DOI] [PubMed] [Google Scholar]
- 32.Cadenas E, Davies KJA. Free Radical Biology & Medicine. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.