Abstract
Bacterial spores are specialized cells that are exceptionally resistant to environmental stress. Spores convert back to actively growing cells, a process called germination, upon nutrient detection. The most common, initial step in the germination process is the recognition of small molecule germinants by germination (Ger) receptors. Ger receptors are inner-membrane heterocomplexes formed by three distinct protein products, the A-, B-, and C-subunits. In this review, we discuss and contrast published reports on representative Ger receptors from different Bacilli and Clostridia. We also present evidence for unrecognized germination pathways independent of Ger receptors. We further emphasize the function of L-alanine as a universal germinant. We also comment on biochemical aspects of germinant recognition and interaction between Ger receptor proteins. We propose that there are six general strategies used by Bacilli and Clostridia to integrate multiple germination signals. The use of different germinant recognition strategies results in germination response flexibility. Consequently, sporulating bacterial species that use the same biomolecules as germination signals can have different germination profiles. Finally, we discuss future directions for understanding the function of Ger receptors.
The germination (Ger) receptor family
Endospore-forming bacteria produce some of the most potent toxins known and are important pathogens in hospital-borne infections (Clostridium difficile) (Surowiec et al., 2006), food contamination (Bacillus cereus, Clostridium botulinum,)(Dembek et al., 2007; Ehling-Schulz et al., 2004), wound infestation (Clostridium perfringens, Clostridium tetani) (Edlich et al., 2003; Finsterer and Hess, 2007), and bioterrorism (Bacillus anthracis) (Mock and Fouet, 2001).
Bacilli and Clostridia spores form in response to unfavorable environmental conditions and can withstand extremes of heat, radiation, and chemical agents (Piggot and Hilbert, 2004). The spore's durability is even more remarkable considering that dormant spores revert back to actively growing cells almost immediately after nutrients return to the environment (Moir, 2006). The intrinsic resistance and the ability to remain dormant for long periods make spores the perfect delivery vehicle for infectious diseases.
Spores transform into replicative bacteria by a complex process called germination. Germination is triggered by environmental stimuli that are then transduced into a series of interrelated degradation events. This ultimately results in the loss of typical dormant spore properties (Moir et al., 2002). The mechanism of spore germination has been reviewed in detail elsewhere (Moir, 2006; Moir et al., 2002; Setlow, 2003).
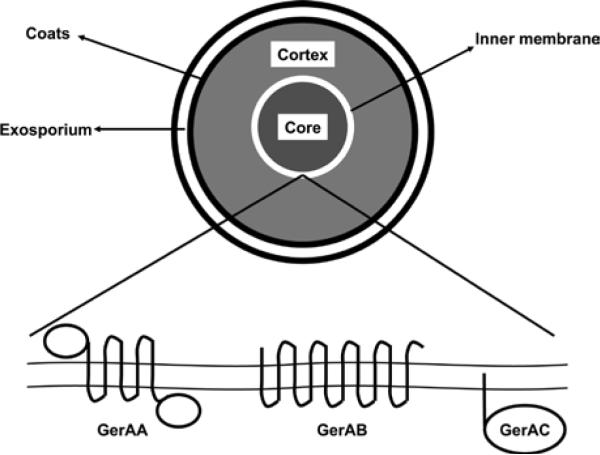
In most cases, Ger receptor activation is the first committed step in the germination process (Moir, 2006; Moir et al., 2002). Ger receptors are encoded, in general, as tricistronic operons containing three protein-coding genes, the A-, B-, and C-subunits (Figure 1) (Moir et al., 2002; Setlow, 2003). However, some Ger receptor subunits are encoded as orphan monocistronic genes (Peck and Robert, 2009) and yet other ger receptor operons encode duplicated subunit genes (Broussolle et al., 2002; Christie and Lowe, 2007).
Figure 1.
Model of the quintessential germination receptor GerA from B. subtilis. Suggested topology and localization of the GerAA, GerAB, and GerAC proteins are shown. All other Ger receptors are believed to fold in a similar fashion (adapted from Moir et al., 2002).
The A-subunit protein of Ger receptors consist of five or six predicted membrane-spanning domains, as well as large N- and C-terminal hydrophilic domains (Setlow, 2003). A-subunit proteins share significant homology to SpoVAF, a late-sporulation protein with no known function (Setlow, 2003). Intriguingly, Ger receptors have been shown to interact with proteins from the spoVA operon (Venkata Ramana Vepachedu, 2007). Whether these interactions are relevant to spore germination remains to be elucidated.
The B-subunit protein is predicted to be an integral membrane protein with 10 to 12 transmembrane helices (Moir et al., 2002). Genetic evidence suggests that the B-subunit may be involved in germinant recognition (Christie and Lowe, 2007; Paidhungat and Setlow, 1999) (vide infra). B-subunit proteins share homology with a superfamily of transmembrane amino-acid permeases (Jack et al., 2000).
The C-subunit protein, a hydrophilic gene product, is predicted to have a pre-lipoprotein signal sequence, suggesting that C-subunit proteins are anchored to the outer surface of the membrane via an N-terminally attached lipid moiety (Igarashi and Setlow, 2005). Indeed, mutational analysis have shown that lipidation of C-subunits are essential for germination (Igarashi and Setlow, 2005). C-subunit proteins show no significant homology to other protein families.
The organization of ger receptor operons suggests that each receptor is a heterocomplex formed by A-, B- and C-subunit proteins (Igarashi and Setlow, 2005). Indeed, deletion of any of the three subunits compromises the function of the encoded receptor (Moir et al., 2002). Genetic evidence has also shown that A-subunits interact with their corresponding B-subunits (Paidhungat and Setlow, 1999). Similarly, C-subunits seem to interact with the A- and B-subunits encoded in their genetic locus and, in some cases, with the A- and B-subunits of other receptors (Igarashi and Setlow, 2005). In addition to intra-subunit interactions, different Ger receptor complexes are believed to interact with each other. The cooperative nature of germinant recognition suggests that Ger receptors can form homo- and heterocomplexes, but the exact composition of these complexes remains to be determined (Atluri et al., 2006). Although genetic methods can be used to infer protein-protein contacts, no biochemical evidence for physical interactions between the subunits has been obtained.
Although activation of ger receptor operons occurs early in the sporulation process, their gene products are expressed at low levels. Indeed, it has been estimated that there are only a few dozen molecules of Ger receptors in a spore (Paidhungat and Setlow, 2001). Interestingly, overexpression of Ger receptors can increase spore germination rates significantly (Cabrera-Martinez et al., 2003). Hence, Ger receptors expression does not seem to be optimized for maximal spore germination. It is possible that this is an evolutionary strategy to ensure that germination only occurs when sufficient nutrients are present to sustain vegetative cell growth.
Representative Ger receptors in Bacilli and Clostridia
Genes for Ger receptor homologues have been found in most Bacilli and Clostridia. The number of tricistronic ger receptor operons vary from one in C. perfringens (Paredes-Sabja et al., 2008) to seven in B. anthracis (Fisher and Hanna, 2005). However, a number of ger receptor operons encode truncated subunit proteins and are probably inactive. Ger receptors from different species that detect the same biomolecules are more closely related than Ger receptors from the same species that recognize structurally different compounds.
Each Ger receptor can detect a unique germinant including amino acids, nucleosides, sugars, and cations. Hence, expressing multiple ger receptor operons allow spores to respond to structurally diverse compounds. Furthermore, each sporulating species encodes a unique complement of Ger receptors and responds to a different set of germinants. Interestingly, different Ger receptors can cooperate to recognize a single germinant or multiple germinants (Moir, 2006; Moir et al., 2002). Below, we discuss the information available for Ger receptors and their cognate germinants in different Bacilli and Clostridia.
Bacillus subtilis Ger receptors
B. subtilis spores contain three functional Ger receptors encoded by the gerA, gerB, and gerK operons (Paidhungat and Setlow, 2000). GerA is the only receptor in B. subtilis that is activated by a single germinant, L-alanine. GerA-mediated germination is strongly inhibited by the stereoisomer, D-alanine (Yasuda and Tochikubo, 1984). GerA is considered as the model Ger receptor since most of what is known about Ger receptor function has been derived from studies in this operon (McCann et al., 1996).
No single germinant has been identified for the GerB or GerK receptors (Paidhungat and Setlow, 2000). The GerK receptor seems to recognize glucose, while GerB seem to recognize amino acids and fructose. However, no cogerminant is sufficient to trigger germination alone (Atluri et al., 2006). GerB and GerK act cooperatively to trigger germination with either L-asparagine supplemented with glucose, fructose, and potassium ions (GFK) or L-alanine supplemented with GFK (Wax and Freese, 1968; Wax et al., 1967). It is important to note that glucose-activated GerK receptors also stimulates L-alanine germination mediated by GerA (Atluri et al., 2006). Furthermore, GerB can interact with GerA to trigger spore germination in response to appropriate nutrients (Atluri et al., 2006). Hence, all three functional receptors encoded by B. subtilis interact to modulate each other's activities.
Bacillus anthracis Ger receptors
Genome sequencing revealed six genomic (gerA, gerH, gerK, gerL, gerS, and gerY) and one plasmidic (gerX) ger receptor operons in B. anthracis (Read et al., 2003). These receptors interact in response to different germinant combinations to form four distinct amino acid and nucleoside dependent germination pathways (Fisher and Hanna, 2005).
Functional analysis revealed that the GerH receptor is activated by nucleosides (Weiner et al., 2003), while the GerS receptor is activated by aromatic amino acids (Ireland and Hanna, 2002a). The GerK receptor recognizes L-alanine and L-valine, while the GerL receptor recognizes L-alanine and L-methionine. Ligands for GerA and GerY have not been identified, and it is possible that they encode non-functional receptors (Fisher and Hanna, 2005). The ligand for the GerX receptor is also not known. Germination of B. anthracis spores appears to require the coordinated detection of at least two germinants, either a nucleoside and an amino acid, or two different amino acids (Fisher and Hanna, 2005).
Previous reports had suggested that the GerX, GerH, and to a lesser extent the GerS receptors, are important for the establishment of anthrax infection and disease progression (Guidi-Rontani et al., 1999; Ireland and Hanna, 2002b; Kang et al., 2005; Weiner and Hanna, 2003). However, recent data shows that as long as one of the genomically encoded receptors (GerH, GerK, GerL, or GerS) is present, B. anthracis spores remain fully virulent in an inhalational anthrax murine model (Carr et al., 2010). This suggests that none of the genomically encoded Ger receptors is absolutely required for infection and that their function is redundant. In contrast, when mice were infected subcutaneously with B. anthracis spores, the GerH receptor became the primary determinant for pathogenesis (Carr et al., 2010). This suggests that distinct germination pathways are activated under different cellular conditions in the host.
Bacillus cereus Ger receptors
Analysis of the B. cereus ATCC 14579 genome revealed the presence of seven putative Ger receptor operons, termed gerG, gerI, gerK, gerL, gerQ, gerR, and gerS (Ivanova et al., 2003). Wild-type B. cereus ATCC 14579 spores germinated strongly with L-alanine, L-cysteine, L-threonine, and L-glutamine (Hornstra et al., 2006). Mutational analysis shows that the GerR receptor influences all germination responses to single amino acids, except L-glutamine (Hornstra et al., 2006). Instead, the GerG receptor was shown to be required in L-glutamine-mediated germination. This suggests that GerG and GerR activation trigger different germination pathways.
Inosine is a known germinant of B. cereus spores (Barlass et al., 2002; Clements and Moir, 1998; Hornstra et al., 2006). Indeed, B. cereus ATCC 14579 spores are able to germinate with inosine as sole germinant (Hornstra et al., 2005). Interestingly, no single Ger receptor seemed to be essential for inosine-mediated germination. However, GerI, GerQ, and GerR mutants germinated at lower rates than wild-type spores when treated with inosine alone (Hornstra et al., 2006). This suggests that GerI, GerQ, and GerR are able to individually recognize inosine, but that their function is redundant. It is worth noticing that B. anthracis spores also use inosine as a germinant, but in B. anthracis, inosine must be supplemented with amino acids for germination initiation (Ireland and Hanna, 2002a).
The addition of inosine as a co-germinant with amino acids enhanced the germination response of B. cereus ATCC 14579 (Hornstra et al., 2006). The germination response to co-germinants was stronger with inosine/L-glutamine and inosine/L-phenylalanine. The combination of L-glutamine and inosine required the GerG and GerI receptors, while L-phenylalanine/inosine combinations required only the GerI receptor (Hornstra et al., 2006). This indicates that the GerI receptor interacts with L-phenylalanine and inosine and that both molecules contribute to germination.
In contrast to B. cereus ATCC 14579, which seems to use the GerR, GerI, or GerQ receptors redundantly to recognize inosine (Hornstra et al., 2006), B. cereus strain 569 strictly requires both the GerI and GerQ receptors to activate inosine-mediated germination (Barlass et al., 2002; Clements and Moir, 1998). Thus, while B. anthracis, B. cereus ATCC 14579 and B. cereus 569 spores recognize nucleoside germinants, each strain shows a different response to these compounds.
We recently determined that there are two distinct nucleoside-dependent germination pathways in B. cereus 569 (Abel-Santos and Dodatko, 2007): The nucleoside/alanine germination pathway and the inosine-only germination pathway. These two pathways recognize nucleoside differently. The nucleoside/alanine germination pathway seems to require only one active nucleoside receptor, either GerI or GerQ (Barlass et al., 2002; Clements and Moir, 1998). Interestingly, B. anthracis lacks the GerQ receptor, but can germinate with nucleosides/alanine by activating a GerI receptor analog (Weiner et al., 2003).
Analysis of structurally diverse nucleosides used as cogerminants with L-alanine suggested that, in the nucleoside/alanine pathway, germination-inducing activity is governed by the structural properties of the sugar rather than the base moieties of the nucleoside (Shibata et al., 1986). These results are consistent with previous spore germination experiments in B. cereus NCIB 8122 and T strains (Preston and Douthit, 1988; Senesi et al., 1991; Shibata et al., 1986; Yousten, 1975).
In addition to the nucleoside/alanine germination pathway, B. cereus has also developed a mechanism that allows germination in presence of inosine as sole germinant. In contrast to the nucleoside/alanine germination pathway, both GerI and GerQ are absolutely required to activate the inosine-only germination pathway. The inosine-only germination pathway has been previously described in both B. cereus 569 and ATCC 14579 strains (Barlass et al., 2002; Clements and Moir, 1998; Hornstra et al., 2006).
Structure activity relationship of nucleoside agonists and antagonists of B. cereus spore germination have shown that the GerI receptor is constrained to recognize a single germinant (inosine) and is inhibited in a predictable pattern by structurally related nucleosides. On the other hand, the GerQ receptor is more promiscuous toward nucleoside agonists and is inhibited by nucleosides containing hydrophobic groups at the 6-position (Dodatko et al., 2009a).
Recent data from our laboratory suggest that when B. cereus 569 spores are exposed to inosine as sole germinant, the spores respond by releasing alanine and other amino acids into their environment (Dodatko et al., 2009b). Amino acid release during germination is dependent of both the GerI and GerQ receptors. Furthermore, the released amino acids are required for the completion of germination in the inosine-only pathway. Thus, the two nucleoside-mediated pathways are not isolated from each other, but are hierarchically organized. Inosine activates the first step in the germination process through the inosine-only pathway and the combined action of the added nucleoside and the released alanine completes the germination program through the nucleoside/alanine germination pathway.
A contrasting picture for the two germination pathways emerges. The GerI and GerQ receptors are differentially activated in B. cereus 569. In the presence of inosine only, both GerI and GerQ are necessary for germination activation. Upon inosine binding by the stringent GerI receptor, the spore releases alanine and other amino acids to complete the germination process. On the other hand, when both nucleoside and alanine are present in the medium, amino acid release is by-passed. In this case, the more promiscuous GerQ receptor seems to control the germination process.
Bacillus megaterium Ger receptors
Spores of B. megaterium QM B1551 germinate in response to glucose, L-proline, L-leucine, and inorganic salts (Christie and Lowe, 2007). In a recent study, it was shown that B. megaterium QM B1551 spore germination is mediated by the plasmid-borne tricistronic gerU operon (Christie and Lowe, 2007). The GerU receptor seems to be activated by L-leucine and glucose. Interestingly, a monocistronic B-subunit protein encoded on the same locus was shown to contribute to germinant specificity. This protein (GerVB) seems to interact with GerUA and GerUC to form a functional receptor for glucose, L-proline, and KBr (Christie and Lowe, 2007). Interestingly, both GerUB and GerVB are orthologues of the GerKB subunit from B. subtilis and all three proteins are involved in glucose-mediated germination (Irie et al., 1982). This suggests that these three B-subunit proteins recognize glucose even though different organisms encode them.
Clostridium botulinum Ger receptors
Spores of proteolytic C. botulinum show large variability in their germination response. Different strains germinated in the presence of L-alanine alone, amino acids/L-lactate, inosine, and/or AGFK (L-asparagine, glucose, fructose, and potassium ions) (Alberto, 2003). In contrast, germination of all non-proteolytic C. botulinum spores was triggered by defined by pairs of germinant combinations (L-alanine/L-lactate, L-serine/L-lactate or L-cysteine/L-lactate) (Plowman and Peck, 2002).
Analysis of three sequenced strains of proteolytic C. botulinum (ATCC 3502, ATCC 19397 and NCTC 2012) showed three well-conserved ger receptor operons. Interestingly, two additional B-subunit genes flank one of the ger receptor operons. Furthermore, ATCC 3502 and ATCC 19397 have an additional orphan B-subunit homolog (Peck and Robert, 2009). Two more proteolytic C. botulinum strains (Okra and Langeland) encode an additional tricistronic ger receptor operon. This extra operon is more closely related to Ger receptors from other Clostridia than to any C. botulinum Ger receptor (Peck and Robert, 2009). The differential expression of additional, divergent Ger receptor proteins may allow proteolytic C. botulinum strains to respond to different germinants and germination conditions (Peck and Robert, 2009).
Non-proteolytic C. botulinum showed a different Ger receptor organization than proteolytic C. botulinum strains (Peck and Robert, 2009). The three ger receptor operons present in all five strains of proteolytic C. botulinum were absent. Instead, non-proteolytic C. botulinum strains possess a single tricistronic ger receptor operon. This single ger receptor operon is more closely related to the extra Ger receptor found in the proteolytic Okra and Langeland strains (Peck and Robert, 2009). The presence of a single Ger receptor in non-proteolytic C. botulinum is consistent with the narrower range of germinants used b y these strains. However, it is not known if this ger receptor operon is actively transcribed during sporulation or if the encoded Ger receptor is active.
Clostridium perfringens Ger receptors
All C. perfringens spores tested use KCl and L-asparagine as a universal germination mixture (Paredes-Sabja et al., 2008). Additionally, C. perfringens strains carrying a chromosomal enterotoxin (cpe) gene can also germinate with KCl alone. In contrast, C. perfringens strains carrying a plasmid-borne cpe gene can germinate with L-alanine or L-valine, in addition to KCl/L-asparagine (Paredes-Sabja et al., 2008). These results are intriguing. Firstly, chromosomal enterotoxin (from SM101 strain) and plasmid enterotoxin (from F5603 strain) are 100% identical at the amino acid level. Secondly, Although cpe is expressed during sporulation, the enterotoxin remains in the mother cell and not in the spore (Harry et al., 2009). Thus, the function of the enterotoxin in modulating germination response of C. perfringens spores is not clear.
The unorthodox germination behavior of C. perfringens spores is further highlighted by the fact that all strains sequenced encode a single, identical GerK receptor. Thus, if strain-specific germination responses are dependent on GerK activation, this receptor must be able to recognize four structurally different molecules (KCl, L-asparagine, L-valine, and L-alanine). Furthermore, each C. perfringens strain uses only a subset of available co-germinants. Hence, the GerK receptor must be able to change substrate specificity between strains. Alternatively, some of the germinants could be recognized by unidentified germination receptors.
Germination response in Clostridium bifermentans and Clostridium sordellii
It has been previously reported that spores of C. bifermentans germinate in the presence of L-alanine, L-arginine, L-phenylalanine, and L-lactate, even though no Ger receptor has been identified in this species (Waites and Wyatt, 1971). This is similar to our recent findings that showed that C. sordellii, a close relative of C. bifermentans, needs a combination of a small amino acid, a basic amino acid, an aromatic amino acid, and bicarbonate to germinate (Ramirez and Abel-Santos, 2010). Even though germination rates are highest when spores are incubated with a mixture of L-alanine, L-phenylalanine, and L-arginine, germination also occurs when each amino acid is substituted with compounds with similar physical and chemical properties. Thus, C. sordellii spores seem to be quite promiscuous in the recognition of germinants (Ramirez and Abel-Santos, 2010).
C. sordellii is an opportunistic pathogen that causes toxic shock syndrome after gynecological procedures (Aldape et al., 2006). In this context, the fact that C. sordellii spores requires bicarbonate and prefer slightly acidic pH for germination is intriguing (Ramirez and Abel-Santos, 2010). Maximal germination is observed at a pH range where both carbonic acid and bicarbonate are present in equilibrium. On the other hand, at more basic pH (where the concentration of carbonic acid is insignificant) or more acidic pH (where the concentration of bicarbonate is insignificant), no germination is detected. Thus, it seems that both the carbonic acid and bicarbonate forms are required germinants. Since bicarbonate is present at high concentrations in the female reproductive tract and healthy females have a vaginal pH below 4.5 (Vishwakarma, 1962), all bicarbonate forms would be protonated to the carbonic acid form, and C. sordellii spores would not be able to germinate under these conditions. Thus, only when the vaginal pH is altered due to vaginosis will the bicarbonate/carbonic acid ratio be optimal for germination.
Germination response in Clostridium difficile
Although C. difficile encodes every other known spore-specific protein, Ger receptor genes are conspicuously absent (Sebaihia et al., 2006). However, C. difficile spores must be able to germinate in order to complete their life cycle. A recent article showed that C. difficile spores recognize glycine (an amino acid) and taurocholate (a bile salt) as germinants (Sorg and Sonenshein, 2008).
Taurocholate is a bile acid involved in the emulsification of fats (Carey and Small, 1970). Thus, taurocholate could trigger germination by either permeabilizing the spore membrane or activating a cognate receptor. Different lines of evidence suggest that C. difficile spores possess unknown taurocholate and glycine germination receptors. Firstly, glycine is a common amino acid with no known membrane disrupting activity. Hence, the function of glycine as a co-germinant suggests the presence of a glycine-specific germination receptor. The specificity of glycine recognition is further highlighted by the inability of other amino acids to substitute as co-activators of spore germination (Sorg and Sonenshein, 2008). Secondly, a membrane disruptive mechanism would be nonspecific. Hence, other bile salts should be equally able to activate spore germination. Instead, chenodeoxycholate, another bile salt with potential detergent properties, is a competitive inhibitor of taurocholate-mediated germination (Sorg and Sonenshein, 2009). In this case, chenodeoxycholate does not activate but hinders spore germination, suggesting that chenodeoxycholate competes with taurocholate for binding at a putative bile salt receptor.
Putative unrecognized germination receptors
The absence of ger receptor operons in the genome of C. difficile and the complex germination response of C. perfringens indicate that either some Ger receptors are too divergent to be recognized by standard bioinformatics tools or that a different set of proteins can also recognize biomolecules to trigger germination (Peck and Robert, 2009). Indeed, a novel spore germination mechanism has been recently reported in B. subtilis (Shah et al., 2008). In these cases, B. subtilis spores germination was triggered by peptidoglycan degradation products, even in the absence of all Ger receptors (Shah et al., 2008). Peptidoglycan mediated germination seem to be dependent on a well conserved eukaryotic-like serine/threonine protein kinase (PrkC) located in the spore inner membrane (Shah et al., 2008) . A search of genome sequences reveals the presence of putative prkC homologues in a number of different Bacilli and Clostridia (Peck and Robert, 2009). Detection of peptidoglycan fragments represents a novel germination mechanism that may allow spores to determine whether other microbes are actively growing in the environment. Whether PrkC-like proteins contribute to the germination of other sporulating bacteria spores has not been determined. It is expected that other Ger receptor independent germination pathways may still be discovered.
L-alanine as a germinant
L- Alanine is a universal amino acid that has been shown to be a common germinant for both Bacilli and Clostridia. L-alanine-mediated germination has been mostly characterized in B. subtilis spores (McCann et al., 1996). However, L-alanine can also induce germination in B. cereus (Barlass et al., 2002), B. anthracis (Fisher and Hanna, 2005), B. megaterium (Christie and Lowe, 2007), C. botulinum (Broussolle et al., 2002), C. sporogenes (Broussolle et al., 2002) C. perfringens (Paredes-Sabja et al., 2008), and C. sordellii (Ramirez and Abel-Santos, 2010). Indeed, L-alanine seems to be the most ubiquitous germinant for bacterial spores.
Although L-alanine is able to induce B. subtilis spore germination on its own, spore activation in other Bacilli and Clostridia requires supplementing L-alanine with other cogerminants. These co-germinants vary from other amino acids, sugars, nucleosides or salts. This suggests that alanine-mediated germination evolved from being a singular germination signal to form part of an integrated response with other germination pathways.
GerA in B. subtilis and GerL in B. anthracis are close homologues that have been shown to directly influence germination in response to L-alanine (Ireland and Hanna, 2002a; McCann et al., 1996). On the other hand, C. botulinum and C. perfringens spores also responds to L-alanine as germinant (Alberto, 2003; Broussolle et al., 2002; Paredes-Sabja et al., 2008), but their Ger receptors form a phylogenetically divergent clade from the GerA B. subtilis receptor (Chris Ross, personal communication).
The divergence between Bacilli and Clostridia alanine-responsive Ger receptors implies that their alanine-binding sites are dissimilar. Indeed, no consensus motif for alanine binding can be found between the two bacteria classes (Chris Ross, personal communication). Since L-alanine can activate unrelated Ger receptors, it suggest that L-alanine may be the most recent, common ancestral germinant. If this is the case, the ancestral L-alanine Ger receptor genes may have duplicated multiple times in different Bacilli and Clostridia. After duplication, Ger receptor domains responsible for L-alanine binding would have mutated to create receptors with different specificity. Alternatively, L-alanine sensitivity could have arisen multiple times in the history of this receptor family because of the abundance of L-alanine in different ecological niches.
Binding site of germinants
Even though many Bacilli and Clostridia use similar metabolites as germinants, the mechanism of germinant-receptor recognition and the specific role of inter-subunit interactions remain to be elucidated. A standing question in the field is the location of the binding site for germinants in the Ger receptor complex. Mutation in the primary sequence of the Ger receptor could lead to changes in binding site environment and consequently, changes in germinant selectivity. Gain-of-function mutations that change the specificity of germinant recognition were mapped to either the A- or B-subunit of the corresponding Ger receptor, suggesting that both the A- and B-subunit are involved in germinant recognition (Atluri et al., 2006; Paidhungat and Setlow, 1999).
Recently Christie et al. has shown that by using alternate B-subunits (GerUB and GerVB) to form the B. megaterium Ger receptor, the specificity of germinant recognition can be changed (Christie and Lowe, 2007). Furthermore, point mutations in the GerVB protein from B. megaterium reduced substrate affinity and specificity. These mutations correspond with amino acid changes within transmembrane domains 9 and 10 and the connecting outer loop (Christie et al., 2008). Since only mutations in the B-subunit were found to affect germinant recognition, it suggests that the C-terminus of the B-subunits may be involved in the formation of a binding pocket, or affect the conformation of nearby functionally important regions (Christie and Lowe, 2008).
Interaction between Ger receptors
Different metabolites are able to act as germinants in vitro, individually or in combinations (Fisher and Hanna, 2005; Hornstra et al., 2006; Paredes-Sabja et al., 2008; Wheeldon et al., 2008). In most cases, multiple Ger receptors appear to work in concert to induce the germination cascade (Christie and Lowe, 2007). In vivo, spores are probably exposed to numerous, simultaneous germination stimuli. Since Ger receptors seem to interact synergistically (Atluri et al., 2006; Wolgamott and Durham, 1971), it is possible that multiple interlocking pathways would increase the binding affinity of each cooperating receptor for its cognate ligand. As an example, the GerA receptor in B. subtilis functions independently to trigger spore germination in response to L-alanine (McCann et al., 1996). However, the germination profile of B. subtilis spores encoding mutant receptors show that Ger receptors can influence each other's activation (Atluri et al., 2006).
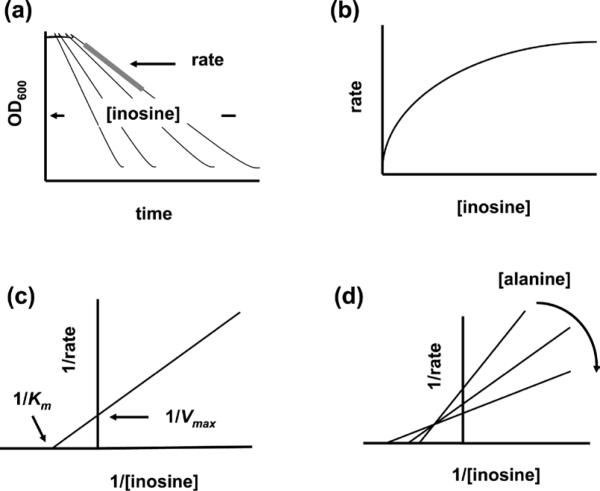
To understand the molecular determinants of Ger receptor interactions and their effect on germinant recognition, we recently applied kinetic methods to study bacterial spore germination (Abel-Santos and Dodatko, 2007; Akoachere et al., 2007; Ramirez and Abel-Santos, 2010). Spore germination can be analyzed by fitting optical density (OD) decreases to the Michaelis-Menten equation (Figure 2a) (Akoachere et al., 2007). The kinetic parameters obtained allow calculating apparent binding affinities (Km) of spores for the different co-germinants and maximum rates of spore germination (Vmax). In these instances, Km refers to the concentration of substrate required to reach half of the maximal germination rate. These parameters can be used, in turn, to propose mechanisms of germination, including potential interactions between germination receptors. Furthermore, by comparing apparent Km values to germinant concentrations in vivo, models for spore-germinant complex distribution can be proposed, and rate-limiting steps for the germination process can be inferred. Thus, kinetic analysis can yield information on spore activation even if the identities of the germination receptors are not known (Ramirez and Abel-Santos, 2010).
Figure 2.
Idealized B. anthracis germination kinetics analysis. (a) B. anthracis spores were treated with increasing amounts of inosine at a fixed L-alanine concentration (Akoachere et al., 2007). Germination rates were calculated from the change in optical density over time (grey bar). (b) Plotting germination rate vs. inosine concentration gave a hyperbolic curve indicating saturation of specific receptor sites. (c) Double reciprocal plots were used to calculate inosine's apparent binding constant (Km) and the maximal apparent rate (Vmax). (d) This process was repeated at different fix L-alanine concentrations to obtain a family of double reciprocal plots. Vmax increased with increasing L-alanine concentrations indicating that inosine and alanine bind to separate sites (Segel, 1993). The spore's affinity for inosine also increased with increasing L-alanine concentration revealing cooperativity between their receptors. The intersection of the curve family at a single point strongly supports the notion that both germinants are present simultaneously on their receptor(s).
Titrations of the germination rates of B. anthracis spores with either L-alanine or inosine yielded hyperbolas indicating saturation of germinant binding to specific receptor sites (Figure 2b) (Akoachere et al., 2007). The fact that the apparent germination Vmax increased with increasing germinant concentrations indicated that inosine and alanine bind to separate sites. Concomitantly, the affinity of either germinant increased when the complementary germinant is bound at its specific site, revealing remarkable cooperativity between their receptors (Figure 2d). Moreover, the intersection of the curve family at a single point strongly supports the notion that both germinants are present simultaneously on their receptor(s) to achieve the synergy in both velocity and affinity (Akoachere et al., 2007).
Using this type of analysis, we postulated that B. anthracis germination in the presence of L-alanine and inosine is consistent with an ordered bireactant system where inosine is required to bind first to activate spores. The spore-inosine complex then rapidly binds L-alanine to trigger germination (Akoachere et al., 2007). The proposed mechanistic information was used to design inhibitors that prevented B. anthracis spore germination in vitro and protected macrophages from anthrax cytotoxicity (Akoachere et al., 2007).
In contrast to the linear Michaelis-Menten kinetics observed in B. anthracis, inosine germination response in B. cereus 569 spores shows cooperative behavior (Abel-Santos and Dodatko, 2007). Double reciprocal and Hill plots of B. cereus germination kinetics suggest the presence of two distinct, strongly interacting inosine binding sites. This data is consistent with the existence of two different germination receptors (GerI and GerQ) that are required to respond to inosine as sole germinant (Barlass et al., 2002; Clements and Moir, 1998).
We have recently extended germination kinetic analysis to Clostridia. Even though germination receptors in C. sordellii spores have not been characterized, titrations of germination rates with NaHCO3 and L-arginine resulted in hyperbolas that could be analyzed using Michaelis-Menten approaches. In contrast, binding of L-phenylalanine and L-alanine by C. sordellii spores showed strong cooperative behavior, suggesting that putative L-phenylalanine and L-alanine receptors engage in protein-protein interactions that affect their corresponding binding sites (Ramirez and Abel-Santos, 2010).
Our recent results in the kinetic analysis of C. difficile spore germination suggested the presence of distinct receptors for the co-germinants, taurocholate and glycine (Ramirez et al.). Furthermore, taurocholate, and glycine bind to C. difficile spores through a complex mechanism, where both receptor homo- and heterocomplexes seem to be formed. The kinetic data also points to an ordered sequential progression of binding where the putative taurocholate receptor must be activated first, before recognition of glycine can take place (Ramirez et al.).
Strategies for spore germination
As seen above, Ger receptors can be differentially activated by environmental stimuli. The differences in responses can be described in terms of Boolean logic, where Ger receptors act as logical gates (Gregg, 1998). The responses observed so far can be divided into six distinct groups according to the type of signal integration used to trigger spore germination.
The first germination strategy represents the simplest case where a single compound triggers germination by activating its cognate Ger receptor (Figure 3a). This strategy is typified by the GerA receptor of B. subtilis that induces germination in the presence of L-alanine alone (McCann et al., 1996).
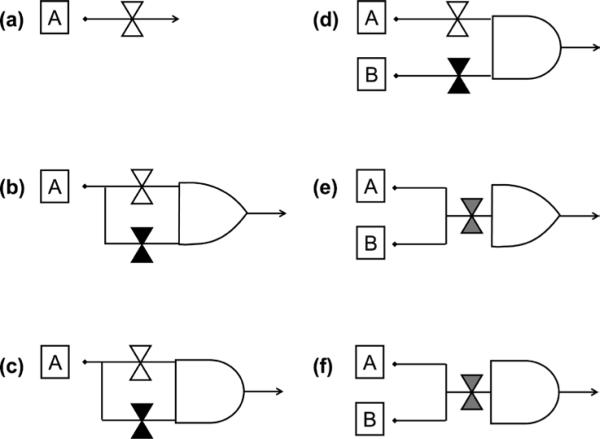
Figure 3.
Strategies for spore germination. Boxed letters represent germinants. White and black hourglasses represent Ger receptors that recognize a single germinant. Grey hourglasses represent Ger receptors that recognize multiple germinants. Arrows represent germination. (a) A single germinant triggers germination by activating its cognate Ger receptor. (b) A single germinant can trigger germination through multiple independent Ger receptors. (c) A single germinant must activate different Ger receptors simultaneously. (d) Multiple germinants must interact individually with their cognate receptors. (e) A single Ger receptor can be independently activated by different germinants. (f) A single Ger receptor must interact with multiple germinants to be activated.
The second germination strategy acts as an OR logical gate. This is a parallel process where a single germinant can trigger germination through multiple independent Ger receptors. In this case, activation of any single receptor is sufficient for germination (Figure 3b). This strategy is exemplified by the purine-mediated germination response of Bacillus cereus ATCC 14579, where three different receptors (GerI, GerQ, and GerR) redundantly activate germination in the presence of inosine (Hornstra et al., 2006).
The third germination strategy acts as an AND logical operator where a single germinant activates multiple Ger receptors simultaneously (Figure 3c). Thus, activation of all receptors is necessary before germination starts. This strategy is exemplified by B. cereus 569, where both GerI and GerQ must be activated by inosine to trigger germination (Barlass et al., 2002; Clements and Moir, 1998).
The fourth strategy acts as an integrator where multiple germinants must interact individually with their cognate receptors. In this case, each receptor recognizes its own cognate germinant, but more than one germinant must be present to activate germination (Figure 3d). This strategy is exemplified by the germination response of B. anthracis Sterne, where at least two different germinants must be detected for germination to occur (Fisher and Hanna, 2005).
In the fifth germination strategy, multiple germinants interact with a single receptor, but any one of the cognate germinants is sufficient to trigger germination (Figure 3e). This strategy is exemplified by B. megaterium spores that can germinate with either glucose, L-leucine, or KBr using the GerU receptor (Christie and Lowe, 2007).
The final strategy represents another AND logical operator where multiple germinants are required to activate a single Ger receptor. However, more than one germinant must be present to trigger germination (Figure 3f). This strategy is exemplified by the alanine/valine germination response of C. perfringens where both amino acids are used as germinant, but only a single gerK operon is present in the genome (Paredes-Sabja et al., 2008).
Interestingly, a single species could utilize different germination strategies depending on the combination of germinant and Ger receptors used. For example, in B. cereus ATCC 14579, GerI, GerQ, and GerR independently recognize inosine as a germinant. On the other hand, GerI alone is required to recognize both inosine and L-phenylalanine as signals for germination.
It is possible to speculate on the evolutionary history leading to the development of the different germination strategies. The most ancient strategy was probably the activation a single Ger receptor by its cognate germinant (Figure 3a). Alternatively, the most ancient Ger receptor could have been a complex with a promiscuous recognition site (Figure 3e).
A duplication event, followed by genetic drift would have allowed development of the OR logical gate for germination (Figure 3b). Mutations could then have disrupted protein-protein interactions between same complexes in favor of inter-receptor contact. This would have allow the development of the AND gate for germination (Figure 3c). Further mutations in the binding site of one of the duplicated receptors would have allowed changes in germinant selectivity (Figure 3d). Similar binding site mutation would have allowed the development of an AND gate (Figure 3f) from a single receptor that recognize multiple germinants (Figure 3e). Although compelling, more information is needed to understand the development of germination strategies.
Future directions
In this article, we have reviewed the current knowledge of the Ger receptor family from Bacilli and Clostridia. Ger receptor activation represents the first essential step in the establishment of infection by sporulating bacteria and therefore is of great importance for both clinical settings and biodefense. Recent work has made clear that Ger receptors interact to form intricate pathways for germination signal integration. Furthermore, new research has started to unravel the mechanism of germinant recognition. However, many questions remain unanswered. Ger receptor function has been mostly determined by genetic methods. In contrast, biochemical analysis of Ger receptor has been lacking. One of the main barriers for Ger receptor characterization is the difficulties of overexpressing and purifying integral membrane protein complexes (Tarmo Roosild, personal communication). New methods for recombinant membrane protein expression could lead to purification of individual subunits and reconstitution of Ger receptor complexes in vitro (Roosild et al., 2006). Alternatively, in vitro transcription/translation systems could allow expressing complete ger receptor operons in vitro that could then be purified as intact receptor complexes (Liguori et al., 2007). Once Ger receptors have been reconstituted in vitro, biochemical characterization will allow determining receptor stoichiometry, thermodynamics of receptor/ germinant interactions, and the mapping of binding sites and subunit protein-protein contacts.
Recombinant expression of Ger receptors may be complicated by the requirement for most Ger receptor to be formed by complexes of A-, B- and C- subunits. A further hurdle may arise if different Ger receptors organize into higher order arrays in the inner membrane. This type of higher order organization has yet to be confirmed, but may foster cooperation between individual Ger receptors (Atluri et al., 2006).
Even if Ger receptors prove refractive to protein expression and purification, biochemical characterization could still be achieved. Germination agonists and antagonists have been used to characterize the binding sites of B. cereus nucleoside receptors (Dodatko et al., 2009a). This approach could be extended to map germinant/Ger receptor interactions in other complexes. Furthermore, crosslinking groups could be used to identify Ger receptor domains that contact germinant molecules.
Future work should also focus on determining the molecular determinants underpinning the function of Ger receptors. Currently, it is not known how germinant selectivity is achieved, which other proteins interact with Ger receptors to transduce the initial germinant binding into spore structure germination, what signaling pathways are triggered upon Ger receptor activation, what are the specific functions of the A-, B-, and C- subunits, and how different germination signals are integrated in vivo.
Although germination is an all-or-nothing process for each individual spore, subpopulations of spores appear to have different germination thresholds (Ghosh and Setlow, 2009a). This might reflect a stochastic process due to the low expression levels of Ger receptor proteins. Indeed, spores that can only germinate when multiple Ger receptors are activated simultaneously have been isolated in B. subtilis, B. megaterium and B. cereus (Ghosh and Setlow, 2009a; Ghosh and Setlow, 2009b). Future work should elucidate whether differences in Ger receptors levels could account for these superdormant spores.
Ger receptors are naturally occurring small molecule binders. Thus, understanding how Ger receptors detect their cognate germinant, could lead to engineering of novel binding activities. Coupled with the stability of dormant spore and the ease for detecting germination, spores containing modified Ger receptors have the potential to be used as biodetectors of metabolic products or synthetic compounds.
Finally, unknown germination mechanisms may also play an important role in Bacilli and Clostridia (Shah et al., 2008). It would be interesting to determine if other Ger-independent germination pathways exist and whether these novel germination mechanisms synergize with Ger receptors to complete Bacilli and Clostridia life cycle.
Acknowledgements
The authors want to thank Prof. Eduardo Robleto for helpful discussions. E Abel-Santos was supported by USDA grant number 2010-65119-20603. C Ross was supported by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
References
- Abel-Santos E, Dodatko T. Differential nucleoside recognition during Bacillus cereus 569 (ATCC 10876) spore germination. New J Chem. 2007;31:748–755. [Google Scholar]
- Akoachere M, Squires RC, Nour AM, Angelov L, Brojatsch J, Abel-Santos EV. Identification of an in vivo inhibitor of Bacillus anthracis Sterne spore germination. J Biol Chem. 2007;282:12112–12118. doi: 10.1074/jbc.M611432200. [DOI] [PubMed] [Google Scholar]
- Alberto FB, Mason V, Carlin DR, Peck F, M.W. Variability in spore germination response by strains of proteolytic Clostridium botulinum types A, B and F. Letters in Applied Microbiology. 2003;36:41–45. doi: 10.1046/j.1472-765x.2003.01260.x. [DOI] [PubMed] [Google Scholar]
- Aldape MJ, Bryant AE, Stevens DL. Clostridium sordellii infection: Epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin Infec Dis. 2006;43:1436–1446. doi: 10.1086/508866. [DOI] [PubMed] [Google Scholar]
- Atluri S, Ragkousi K, Cortezzo DE, Setlow P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol. 2006;188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlass PJ, Houston CW, Clements MO, Moir A. Germination of Bacillus cereus spores in response to L-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology. 2002;148:2089–2095. doi: 10.1099/00221287-148-7-2089. [DOI] [PubMed] [Google Scholar]
- Broussolle V, Alberto F, Shearman CA, Mason DR, Botella L, Nguyen-The C, Peck MW, Carlin F. Molecular and physiological characterisation of spore germination in Clostridium botulinum and C. sporogenes. Anaerobe. 2002;8:89–100. [Google Scholar]
- Cabrera-Martinez R-M, Tovar-Rojo F, Vepachedu VR, Setlow P. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J Bacteriol. 2003;185:2457–2464. doi: 10.1128/JB.185.8.2457-2464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MC, Small DM. The characteristics of mixed micellar solutions with particular reference to bile. American Journal of Medicine. 1970;49:590–608. doi: 10.1016/s0002-9343(70)80127-9. [DOI] [PubMed] [Google Scholar]
- Carr KA, Lybarger SR, Anderson EC, Janes BK, Hanna PC. The role of Bacillus anthracis germinant receptors in germination and virulence. Molecular Microbiology. 2010;75:365–375. doi: 10.1111/j.1365-2958.2009.06972.x. [DOI] [PubMed] [Google Scholar]
- Christie G, Lazarevska M, Lowe CR. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J Bacteriol. 2008;190:2014–2022. doi: 10.1128/JB.01687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Lowe CR. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. Journal of Bacteriology. 2007;189:4375–4383. doi: 10.1128/JB.00110-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Lowe CR. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J Bacteriol. 2008;190:8009–8017. doi: 10.1128/JB.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements MO, Moir A. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J Bacteriol. 1998;180:6729–6735. doi: 10.1128/jb.180.24.6729-6735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek ZF, Smith LA, Rusnak JM. Botulism: cause, effects, diagnosis, clinical and laboratory identification, and treatment modalities. Disaster Med Public Health Prep. 2007;1:122–134. doi: 10.1097/DMP.0b013e318158c5fd. [DOI] [PubMed] [Google Scholar]
- Dodatko T, Akoachere M, Jimenez N, Alvarez Z, Abel-Santos E. Dissecting interactions between nucleosides and germination receptors in Bacillus cereus 569 spores. Microbiology. 2009a;156:1244–1255. doi: 10.1099/mic.0.030270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodatko T, Akoachere M, Muehlbauer SM, Helfrich F, Howerton A, Ross C, Wysock V, Brojatsch J, Abel-Santos E. Bacillus cereus spores release alanine that synergizes with inosine to promote germination. PLoS ONE. 2009b;4:e6398. doi: 10.1371/journal.pone.0006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich RF, Hill LG, Mahler CA, Cox MJ, Becker DG, Horowitz JH, Nichter LS, Martin ML, Lineweaver WC. Management and prevention of tetanus. J Long Term Eff Med Implants. 2003;13:139–154. [PubMed] [Google Scholar]
- Ehling-Schulz M, Fricker M, Scherer S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Molecular Nutrition & Food Research. 2004;48:479–487. doi: 10.1002/mnfr.200400055. [DOI] [PubMed] [Google Scholar]
- Finsterer J, Hess B. Neuromuscular and central nervous system manifestations of Clostridium perfringens infections. Infection. 2007;35:396–405. doi: 10.1007/s15010-007-6345-z. [DOI] [PubMed] [Google Scholar]
- Fisher N, Hanna P. Characterization of Bacillus anthracis germinant receptors in vitro. J Bacteriol. 2005;187:8055–8062. doi: 10.1128/JB.187.23.8055-8062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Setlow P. Isolation and characterization of superdormant spores of Bacillus species. J Bacteriol. 2009a;191:1787–1797. doi: 10.1128/JB.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Setlow P. The preparation, germination properties and stability of superdormant spores of Bacillus cereus. Journal of Applied Microbiology. 2009b;108:582–590. doi: 10.1111/j.1365-2672.2009.04442.x. [DOI] [PubMed] [Google Scholar]
- Gregg JR. Ones and Zeros: Understanding Boolean Algebra, Digital Circuits, and the Logic of Sets. Wiley-IEEE Press; 1998. [Google Scholar]
- Guidi-Rontani C, Pereira Y, Ruffie S, Sirard JC, Weber-Levy M, Mock M. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol Microbiol. 1999;33:407–414. doi: 10.1046/j.1365-2958.1999.01485.x. [DOI] [PubMed] [Google Scholar]
- Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J Bacteriol. 2009;191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstra LM, de Vries YP, de Vos WM, Abee T, Wells-Bennik MHJ. gerR, a novel ger operon involved in L-alanine- and inosine-initiated germination of Bacillus cereus ATCC 14579. Appl Environ Microbiol. 2005;71:774–781. doi: 10.1128/AEM.71.2.774-781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstra LM, de Vries YP, Wells-Bennik MH, de Vos WM, Abee T. Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl Environ Microbiol. 2006;72:44–53. doi: 10.1128/AEM.72.1.44-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Setlow P. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J Bacteriol. 2005;187:2513–2518. doi: 10.1128/JB.187.7.2513-2518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland JA, Hanna PC. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis DeltaSterne endospores: gerS mediates responses to aromatic ring structures. J Bacteriol. 2002a;184:1296–1303. doi: 10.1128/JB.184.5.1296-1303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland JA, Hanna PC. Macrophage-enhanced germination of Bacillus anthracis endospores requires GerS. Infect Immun. 2002b;70:5870–5872. doi: 10.1128/IAI.70.10.5870-5872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie R, Okamoto T, Fujita Y. A germination mutant of Bacillus subtilis deficient in response to glucose. J Gen Appl Microbiol. 1982;28:345–354. [Google Scholar]
- Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- Kang TJ, Fenton MJ, Weiner MA, Hibbs S, Basu S, Baillie L, Cross AS. Murine macrophages kill the vegetative form of Bacillus anthracis. Infect Immun. 2005;73:7495–7501. doi: 10.1128/IAI.73.11.7495-7501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori L, Marques B, Villegas-Mendez A, Rothe R, Lenormand J-L. Production of membrane proteins using cell-ree expression systems. Expert Review of Proteomics. 2007;4:79–90. doi: 10.1586/14789450.4.1.79. [DOI] [PubMed] [Google Scholar]
- McCann KP, Robinson C, Sammons RL, Smith DA, Corfe BM. Alanine germination receptors of Bacillus subtilis. Letters in Applied Microbiology. 1996;23:290–294. doi: 10.1111/j.1472-765x.1996.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Mock M, Fouet A. Anthrax. Annual Review of Microbiology. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Moir A. How do spores germinate? Journal of Applied Microbiology. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Moir A, Corfe BM, Behravan J. Spore germination. Cell Mol Life Sci. 2002;59:403–409. doi: 10.1007/s00018-002-8432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant D-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Setlow P. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Setlow P. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol. 2001;183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D, Torres JA, Setlow P, Sarker MR. Clostridium perfringens spore germination: characterization of germinants and their receptors. J Bacteriol. 2008;190:1190–1201. doi: 10.1128/JB.01748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck MW, Robert KP. In Advances in Microbial Physiology. Vol. 320. Academic Press; 2009. Biology and genomic analysis of Clostridium botulinum; pp. 183–265. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Current Opinion in Microbiology. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Plowman J, Peck MW. Use of a novel method to characterize the response of spores of nonproteolytic Clostridium botulinum types B, E and F to a wide range of germinants and conditions. Journal of Applied Microbiology. 2002;92:681–694. [Google Scholar]
- Preston RA, Douthit HA. Functional relationships between L- and D-alanine, inosine and NH4Cl during germination of spores of Bacillus cereus T. J Gen Microbiol. 1988;134:3001–3010. doi: 10.1099/00221287-134-11-3001. [DOI] [PubMed] [Google Scholar]
- Ramirez N, Abel-Santos E. Requirements for germination of Clostridium sordellii spores in vitro. J Bacteriol. 2010;192:418–425. doi: 10.1128/JB.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez N, Liggins M, Abel-Santos E. Kinetic evidence for the presence of putative germination receptors in C. difficile spores. data not published. doi: 10.1128/JB.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Roosild T, Vega M, Castronovo S, Choe S. Characterization of the family of Mistic homologues. BMC Structural Biology. 2006;6:10. doi: 10.1186/1472-6807-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature Genetics. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Segel IH. Behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley Classics Library Edition Wiley Interscience Publication; 1993. Enzyme kinetics. [Google Scholar]
- Senesi S, Cercignani G, Freer G, Batoni G, Barnini S, Ota F. Structural and stereospecific requirements for the nucleoside-triggered germination of Bacillus cereus spores. J Gen Microbiol. 1991;137:399–404. doi: 10.1099/00221287-137-2-399. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki M-H, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Ohnishi N, Takeda K, Fukunaga H, Shimamura K, Yasunobu E, Tani I, Hashimoto T. Germination of Bacillus cereus spores induced by purine ribosides and their analogs: effects of modification of base and sugar moieties of purine nucleosides on germination-inducing activity. Can J Microbiol. 1986;32:186–189. doi: 10.1139/m86-038. [DOI] [PubMed] [Google Scholar]
- Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiec D, Kuyumjian AG, Wynd MA, Cicogna CE. Past, present, and future therapies for Clostridium difficile-associated disease. Ann Pharmacother. 2006;40:2155–2163. doi: 10.1345/aph.1H332. [DOI] [PubMed] [Google Scholar]
- Venkata Ramana Vepachedu PS. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiology Letters. 2007;274:42–47. doi: 10.1111/j.1574-6968.2007.00807.x. [DOI] [PubMed] [Google Scholar]
- Vishwakarma P. The pH and bicarbonate-ion content of the oviduct and uterine fluids. Fertil Steril. 1962;13:481–485. doi: 10.1016/s0015-0282(16)34633-7. [DOI] [PubMed] [Google Scholar]
- Waites WM, Wyatt LR. Germination of spores of Clostridium bifermentans by certain amino acids, lactate and pyruvate in the presence of sodium or potassium Ions. J Gen Microbiol. 1971;67:215–222. doi: 10.1099/00221287-67-2-215. [DOI] [PubMed] [Google Scholar]
- Wax R, Freese E. Initiation of the germination of Bacillus subtilis spores by a combination of compounds in place of L-alanine. Journal of Bacteriology. 1968;95:433–438. doi: 10.1128/jb.95.2.433-438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wax R, Freese E, Cashel M. Separation of two functional roles of L-alanine in the initiation of Bacillus subtilis spore germination. J Bacteriol. 1967;94:522–529. doi: 10.1128/jb.94.3.522-529.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MA, Hanna PC. Macrophage-mediated germination of Bacillus anthracis endospores requires the gerH operon. Infect Immun. 2003;71:3954–3959. doi: 10.1128/IAI.71.7.3954-3959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MA, Read TD, Hanna PC. Identification and characterization of the gerH operon of Bacillus anthracis endospores: a differential role for purine nucleosides in germination. J Bacteriol. 2003;185:1462–1464. doi: 10.1128/JB.185.4.1462-1464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeldon LJ, Worthington T, Hilton AC, Elliott TSJ, Lambert PA. Physical and chemical factors influencing the germination of Clostridium difficile spores. J Appl Microbiol. 2008;105:2223–2230. doi: 10.1111/j.1365-2672.2008.03965.x. [DOI] [PubMed] [Google Scholar]
- Wolgamott GD, Durham NN. Initiation of spore germination in Bacillus cereus: a proposed allosteric receptor. Can J Microbiol. 1971;17:1043–1048. doi: 10.1139/m71-165. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Tochikubo K. Relation between D-glucose and L- and D-alanine in the initiation of germination of Bacillus subtilis spores. Microbiol Immunol. 1984;28:197–207. doi: 10.1111/j.1348-0421.1984.tb00671.x. [DOI] [PubMed] [Google Scholar]
- Yousten AA. Germination of Bacillus cereus endospores: a proposed role for heat shock and nucleosides. Can J Microbiol. 1975;21:1192–1197. doi: 10.1139/m75-178. [DOI] [PubMed] [Google Scholar]