Abstract
The intergenic region of spliced-leader (SL-IR) genes from 105 Trypanosoma cruzi I (Tc I) infected biological samples, culture isolates and stocks from 11 endemic countries, from Argentina to the USA were characterised, allowing identification of 76 genotypes with 54 polymorphic sites from 123 aligned sequences. On the basis of the microsatellite motif proposed by Herrera et al. (2007) to define four haplotypes in Colombia, we could classify these genotypes into four distinct Tc I SL-IR groups, three corresponding to the former haplotypes Ia (11 genotypes), Ib (11 genotypes) and Id (35 genotypes); and one novel group, Ie (19 genotypes). Genotypes harboring the Tc Ic motif were not detected in our study. Tc Ia was associated with domestic cycles in southern and northern South America and sylvatic cycles in Central and North America. Tc Ib was found in all transmission cycles from Colombia. Tc Id was identified in all transmission cycles from Argentina and Colombia, including Chagas cardiomyopathy patients, sylvatic Brazilian samples and human cases from French Guiana, Panama and Venezuela. Tc Ie gathered five samples from domestic Triatoma infestans from northern Argentina, nine samples from wild Mepraia spinolai and Mepraia gajardoi and two chagasic patients from Chile and one from a Bolivian patient with chagasic reactivation. Mixed infections by Tc Ia + Tc Id, Tc Ia + Tc Ie and Tc Id + Tc Ie were detected in vector faeces and isolates from human and vector samples. In addition, Tc Ia and Tc Id were identified in different tissues from a heart transplanted Chagas cardiomyopathy patient with reactivation, denoting histotropism. Trypanosoma cruzi I SL-IR genotypes from parasites infecting Triatoma gerstaeckeri and Didelphis virginiana from USA, T. infestans from Paraguay, Rhodnius nasutus and Rhodnius neglectus from Brazil and M. spinolai and M. gajardoi from Chile are to our knowledge described for the first time.
Keywords: Trypanosoma cruziI, PCR, Spliced-leader intergenic region, Discrete typing unit, Chagas disease, Genotype
1. Introduction
Trypanosoma cruzi (Tc), the etiological agent of Chagas disease, affects approximately 15 million people from different endemic regions in America (Moncayo and Silveira, 2009). This parasitic disease shows a variable clinical course, which ranges from acute, asymptomatic cases to serious chronic stages characterised by low parasitaemia and involving cardiac and/or gastrointestinal disorders (WHO, 2002). One of the possible causes of such clinical variability has been attributed to the high genetic diversity and multiclonality of natural populations of T. cruzi (Macedo and Pena, 1998). This diversity was extensively demonstrated by different research groups using different biological, biochemical and molecular strategies targeting different genetic markers, allowing identification of six discrete typing units (DTUs) among the globally characterised T. cruzi isolates, designated as Tc I and Tc IIa to Tc IIe (Brisse et al., 2000; Miles et al., 2009). A satellite symposium held in Buzios, Brazil in August 2009 established a new nomenclature for these DTUs, namely Tc I, Tc II (formerly Tc IIb), Tc III (formerly Tc IIc), Tc IV (formerly Tc IIa), Tc V (formerly Tc IId) and Tc VI (formerly Tc IIe) (Zingales et al., 2009). Trypanosoma cruzi I is one of the parental T. cruzi phylogenetic lineages proposed by all experts in the field using different molecular-based strategies (reviewed in Sturm and Campbell, 2010). Further, it has been recently proposed that T. cruzi is synonymous with two different species, Tc I and Tc II (Tomazi et al., 2009).
Trypanosoma cruzi I populations are predominant in the Andean region, in Central America and in Mexico (Cortez et al., 2006; Brenière et al., 2007; Samudio et al., 2007; Llewellyn et al., 2009). They were first recognized in the sylvatic transmission cycle, mainly in marsupials and sylvatic triatomines (Jansen et al., 1999; Yeo et al., 2005; Ceballos et al., 2006) and are associated with human infection predominantly in northern South America, detected in cardiomyopathy patients fro m Venezuela (Añez et al., 2004) and Colombia (Cuervo et al., 2002; Triana et al., 2006; Salazar et al., 2006; Herrera et al., 2007) and recently in cardiomyopathy patients from Argentina (Burgos et al., in press). Tc I strains typically have less DNA content, fewer and smaller chromosomes (Vargas et al., 2004; Lewis et al., 2009). A remarkable intra-DTU variability has been observed, leading to a proposal of associations between internal structuring and transmission cycles (Diosque et al., 2003; Herrera et al., 2007; O´Connor et al., 2007; Brito et al., 2008; Mejía-Jaramillo et al., 2009; Llewellyn et al., 2009). However, knowledge of internal clustering within Tc I is still in its preliminary phase and there is not yet consensus on classification.
On the basis of single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) within a motif at the intergenic spacers of the multicopy spliced-leader gene (SL-IR), Herrera et al. (2007) proposed four Tc I haplotypes in Colombia. These haplotypes were recently designated as Ia–Id by Falla et al. (2009). As part of a wider project involving the characterisation of T. cruzi DTUs in different scenarios of Chagas disease (Marcet et al., 2006; Burgos et al., 2007, 2008; Cardinal et al., 2008; Bisio et al., 2009) we aimed to identify Tc I SL-IR based genotypes from natural populations infecting triatomines and patients with Chagas disease, as well as to type Tc I culture isolates from different triatomine species, animal reservoirs and humans, collected from different regions of America.
2. Materials and methods
2.1. Study samples
A total of 105 Tc I samples were included in the study. Ninety-five were obtained from different biological and geographical sources, including 70 culture stocks from triatomine bugs, animal reservoirs and human samples obtained in endemic areas of Argentina, Brazil, Chile, Colombia, Mexico, Panama and the USA; five faecal samples from Triatoma infestans from Paraguay and one faecal sample from Rhodnius neglectus from Brazil; seven homogenized abdomen samples from Triatoma gerstaeckeri from the USA (Table 1), and 12 clinical samples (seven peripheral blood, three cardiac biopsy, one skin biopsy and one CSF samples) from patients with Chagas disease from Argentina, Bolivia, French Guiana and Venezuela (Table 2). In addition, reference stocks from 10 laboratory Tc I strains, namely TCC (Basombrío et al., 1982), Sylvio X10 (Postan et al., 1983), G (Yoshida, 1983) and its clone D11 (Verbisck et al., 1998), EP (Torrealba A.B., 1970. Observaciones sobre el Comportamiento en Ratón Blanco de una Cepa Humana de Trypanosoma cruzi. Pregrade Thesis. Universidad Central de Venezuela. Caracas, Venezuela), CA-I CL-72 (ATCC, USA) and its clone K98 (Gonzalez Cappa et al., 1999), MIRANDA CL78 (ATCC, USA), 13379 cl7 (Tibayrenc and Miles, 1983) and SE 9V (Macina et al., 1987) were included (Table 3).
Table 1.
Description of Trypanosoma cruzi I Spliced Leader Intergenic Region (SL-IR) groups obtained directly from faeces, vectors tissues and culture stocks from vectors, reservoirs and human samples.
| SL-IR group | Tc I sample | Reference | Geographical origin | Biological origin | Transmission Cycle |
|---|---|---|---|---|---|
| Tc Ia | TEV91Cl5 | 1 | Chaco, Argentina | Triatoma infestans (S) | Domestic |
| A-26b-2-motoH | 2 | Amamá, Santiago del Estero, Argentina | Canis familiaris (S) | ||
| SN5, SN8 | 3 | Guajira, Colombia | Rhodnius prolixus (S) | ||
| SN12 | 1 | Guajira, Colombia | Rhodnius prolixus (S) | ||
| HA | 4 | Casanare, Colombia | Human (S) | ||
| W3534 | 4 | Sucre, Colombia | Human (S) | ||
| SO8 | 4 | Sucre, Colombia | Rhodnius pallescens (S) | Sylvatic | |
| HD/Duran, PG/Gamma | 1 | Mexico | - (S) | ||
| TR/Triatoma | 1 | Mexico | Triatoma sp. (S) | ||
| USA1, 28, 104, 108, 111, 115, BG08011 | 1 | Texas, USA | Triatoma gerstaeckeri (A) | ||
| Flop2 | 5 | Florida, USA | Didelphis virginiana (S) | ||
| Tc Ib | Cas16 | 4 | Casanare, Colombia | Rhodnius prolixus (S) | Peridomestic |
| SP | 6 | Casanare, Colombia | Human (S) | Domestic | |
| STP3.3 | 4 | Tolima, Colombia | Rhodnius prolixus (S) | Sylvatic | |
| Sebas1, Sebas16, Fer1 | 7 | Depresión Momposina, Colombia | Rhodnius pallescens (S) | ||
| Mg6 | 7 | Magdalena, Colombia | Panstrongylus geniculatus (S) | ||
| Mg11 | 7 | Magdalena, Colombia | Rhodnius pallescens (S) | ||
| Coy10 | 1 | Tolima, Colombia | Rhodnius colombiensis (S) | ||
| Necocli2, 3, 4, 5 | 1 | Antioquía, Colombia | Rhodnius pallescens (S) | ||
| Tc Id | Ov1, Ov17 | 4 | Sucre, Colombia | Panstrongylus geniculatus (S) | Peridomestic |
| V81, V92 | 1 | Gran Chaco, Paraguay | Triatoma infestans (F) | ||
| V195 | 1 | Eastern region, Paraguay | Triatoma infestans (F) | Domestic | |
| V305 cl3, cl4, cl6, cl7 a | 1 | Eastern region, Paraguay | Triatoma infestans (F) | ||
| SN3 | 3 | Guajira, Colombia | Rhodnius prolixus (S) | ||
| Cas1 | 1 | Casanare, Colombia | Rhodnius prolixus (S) | ||
| PALV2-1 Cl4 | 1 | Chaco, Argentina | Triatoma infestans (S) | ||
| PAV00Cl7 | 8 | Chaco, Argentina | Triatoma infestans (S) | ||
| CHAMICAL7, CHAMICALII | 2 | La Rioja, Argentina | Triatoma infestans (S) | ||
| COMA375, 665, 701, 729 | 2 | Santiago del Estero, Argentina | Didelphis albiventris (S) | Sylvatic | |
| RN1612 | 1 | Quirinópolis, Goiás, Brazil | Rhodnius neglectus (F) | ||
| RN1153 | 1 | Riberao Cascalhera, Mato Grosso, Brazil | Rhodnius neglectus (S) | ||
| RN01 | 9 | Ceara State, Brazil | Rhodnius nasustus (S) | ||
| LB53 | 4 | Sucre, Colombia | Triatoma dimidiata (S) | ||
| SO6 | 10 | Sucre, Colombia | Rhodnius pallescens (S) | ||
| PalDa | 11 | Chaco, Argentina | Didelphis albiventris (S) | ||
| Put4 | 4 | Putumayo, Colombia | Rhodnius robustus (S) | ||
| Mg1 | 7 | Magdalena, Colombia | Rhodnius pallescens (S) | ||
| Amp07 | 7 | Antioquía, Colombia | Panstrongylus geniculatus (S) | ||
| Cas15 | 4 | Casanare, Colombia | Rhodnius prolixus (S) | ||
| Cas18 | 7 | Casanare, Colombia | Didelphis marsupialis (S) | ||
| Cas10, Cas19 | 1 | Casanare, Colombia | Rhodnius prolixus (S) | ||
| Coy8 | 4 | Tolima, Colombia | Didelphis marsupialis (S) | ||
| Coy9, Coy12 | 1 | Tolima, Colombia | Rhodnius colombiensis (S) | ||
| Medellin | 1 | Medellín, Colombia | - (S) | Missing Data | |
| Tc Ie | PALV2-2 Cl5 | 8 | Chaco, Argentina | Triatoma infestans (S) | Domestic |
| PALV1 Cl1 | 1 | Chaco, Argentina | Triatoma infestans (S) | ||
| BAJO DI LUCA | 2 | La Rioja, Argentina | Triatoma infestans (S) | ||
| TALAVERDE | 2 | La Rioja, Argentina | Triatoma infestans (S) | ||
| BELGRANOIII | 2 | La Rioja, Argentina | Triatoma infestans (S) | ||
| LGN cl7 / cl8 a | 12 | IV Region, Chile | Human (S) | ||
| SpTi 1 cl6 / cl7 / cl8 a | 1 | Metropilitan Region, Chile | Mepraia spinolai (S) | Sylvatic | |
| Col 108 | 1 | Metropolitan Region, Chile | Mepraia spinolai (S) | ||
| Sp 31, Sp 104 | 12 | IV Region, Chile | Mepraia spinolai (S) | ||
| Sp 54 | 13 | IV Region, Chile | Mepraia spinolai (S) | ||
| Sp Guayacan | 1 | IV Region, Chile | Mepraia spinolai (S) | ||
| Sp 130 cl6 / cl8 a,b | 13 | IV Region, Chile | Mepraia spinolai (S) | ||
| Gaj 29 cl6 / cl7 / cl8 a | 1 | Arica, Chile | Mepraia gajardoi (S) | ||
| Tc Ia + Tc Id | V75 a,c | 1 | Gran Chaco, Paraguay | Triatoma infestans (F) | Peridomestic |
| Pan4 a,d | 1 | Arraiján, Panama | Human (S) | Domestic | |
| Tc Ia + Tc Ie | WT a,e | 14 | IV Region,Chile | Human (S) | Domestic |
| Tc Id + Tc Ie | Til70 a,f | 1 | Metropolitan Region, Chile | Mepraia spinolai (S) | Sylvatic |
Samples with SL-IR PCR products cloned into pGEM-T easy Vector (cl).
cl6 (Ie like), cl8 (Ie);
cl6, cl7 (Ia), cl1, cl2, cl5 (Id);
cl1, cl5 (Ia), cl4 (Id);
cl7 (Ia), cl6 (Ie);
cl6 (Id), cl7, cl8 (Ie); (S), Culture stock; (A), Homogenized abdomen sample; (F), Faecal sample.
References: 1, This work; 2, Cardinal et al., 2008; 3, Téllez-Meneses et al., 2008; 4, Salazar et al., 2006; 5, Roellig et al., 2008; 6, Falla et al., 2009; 7, Mejía-Jaramillo et al., 2009. 8, Duffy et al., 2009; 9, Dias et al., 2007; 10, Rojas et al., 2007; 11, Lewellyn et al., 2009; 12, Brenière et al., 1991; 13, Venegas et al., 1997; 14, Torres et al., 2004.
Table 2.
Description of Trypanosoma cruzi I Spliced Leader Intergenic region (SL-IR) groups identified directly from human samples.
| SL-IR group | Tc I human sample | Reference | Geographical origin | Biological origin | Clinical manifestation | Cycle |
|---|---|---|---|---|---|---|
| Tc Ia | FRA | 1 | La Plata, Buenos Aires, Argentina | Peripheral blood | Chronic Chagas Heart disease | Domestic |
| PA | 1 | Chaco, Argentina | Peripheral blood | |||
| TA | 1 | Primero de Mayo, Chaco, Argentina | Peripheral blood | |||
| PASCh | 1 | Chaco, Argentina | Skin Biopsy sample | |||
| Tc Id | VA | 1 | Metán, Salta, Argentina | Peripheral blood | Chronic Chagas Heart disease | Domestic |
| AY | 1 | Ledesma, Jujuy, Argentina | Heart Explant | |||
| PAECl18 | 1 | Chaco, Argentina | Heart Explant | |||
| CHA | 1 | Monte Grande, Buenos Aires, Argentina | Heart Explant | |||
| 2414 | 2 | Chichiriviche de La Costa, Venezuela | Peripheral blood | Acute/Oral transmission | Sylvatic | |
| VTH, VTL | 4 | French Guiana | Peripheral blood | |||
| Tc Ie | AS | 3 | Bolivia | Cerebrospinal Fluid | Chagas encephalytis/AIDS | Domestic |
References: 1, Burgos et al., in press; 2, Alarcón de Noya and Martínez, 2009; 3, Burgos et al., 2008; 4, This work.
Table 3.
Description of Trypanosoma cruzi I Spliced Leader Intergenic region (SL-IR) groups identified from reference cultured stocks.
| SL-IR group | Tc I stock | Reference | Geographical origin | Biological origin |
|---|---|---|---|---|
| Tc Ia | TCC | 1 | Tulahuen, Chile | Human |
| K98 | 2 | San Luis, Argentina | ||
| CA-I CL72 | 3 | San Luis, Argentina | ||
| MIRANDA CL78 | 3 | Argentina | ||
| Tc Id | G | 4 | Amazon region, Brazil | Didelphis marsupialis |
| D11 | 5 | Amazon region, Brazil | ||
| EP | 6 | Cojedes, Venezuela | Human | |
| Sylvio X10 | 7 | Pará, Brazil | ||
| Tc Ie | 13379 cl7 | 8 | Santa Cruz, Bolivia | Human |
| SE 9V | 9 | Santiago del Estero, Argentina |
References: 1, Basombrío el al., 1982; 2, Gonzalez Cappa et al., 1999; 3, ATCC T. cruzi strains; 4, Yoshida, 1983; 5, Verbisck et al., 1998; 6, Torrealba A.B., 1970. Observaciones sobre el Comportamiento en Ratón Blanco de una Cepa Humana de Trypanosoma cruzi. Pregrade Thesis. Universidad Central de Venezuela. Caracas, Venezuela; 7, Postan et al., 1983; 8, Tibayrenc and Miles, 1983; 9, Macina et al., 1987.
Procedures related to clinical samples comply with the criteria of the Ethical Committees of the participating institutions, in agreement with the Declaration of Helsinski.
2.2. PCR amplification of SL-IR fragments from Tc I samples
Total DNA was extracted from the above mentioned samples (Tables 1, 2 and 3). In the case of culture isolates, a 350 bp fragment from Tc I SL-IR was amplified with primers TCC (5´-CCCCCTCCCAGGCCACACACTG-3´) and TC2 (5´-CCTGCAGGCACACGTGTGTG-3´) as previously reported (Burgos et al., 2007). In the case of clinical samples, a hemi-nested PCR was carried out using, in the first round, TC2 and UTCC (5´-CGTACCAATATAGTACAGAAACTG-3´) as antisense primer, which is located 125 bp downstream of TCC, thus amplifying a fragment of 475 bp; the second round was carried out using TC2 and TCC, as mentioned above. The 350 bp amplicons were purified using a QIAquick PCR purification kit (QIAgen®) and sequenced (Macrogen, Korea). In the case of some biological samples (Table 1), the PCR products were cloned into a pGEM-T easy Vector (Promega, USA) in order to reveal the existence of mixed Tc I SL-IR genotypes.
2.3. Sequence analysis of SL-IR amplicons
Electropherograms of forward and reverse sequences were edited using the program Chromas Version 1.45. A consensus of forward and reverse sequences (contig) was created. SL-IR sequences were aligned using the ClustalW algorithm (Thompson et al., 1994) and adjusted by eye. Nucleotide sites showing ambiguous base calls were resolved with the consensus genotype.
The haplotypes Ia, Ib, Ic and Id described by Falla et al. (2009) and Herrera et al. (2007) will be referred as SL-IR groups. Reference genotypes for each SL-IR group were obtained from the GenBank database: CGC (AM259467), FChC (AM259469), X380C (AM259472), EFC (AM259474), PALC (AM259473), P/217 clone 1 (EF576840), P/209cl1 clone 4 (EF576817) and P/209cl1 clone 7: EF576818.
2.4. Phylogenetic inferences
Phylogenetic analyses were performed using Maximum Parsimony (MP) and Bayesian Inference. Molecular data sets were first analyzed using TNT ver. 1.0 (Goloboff, P.A., Farris, J. S., Nixon, K., 2003. Tree Analysis Using New Technology Version 1.0 Available from the authors and from http://www.zmuc.dk/public/phylogeny) with a gap as the fifth state to account for the evolution by indels. Analysis was conducted using a heuristic search procedure: 100 random addition sequences plus Tree bisection reconnection (TBR) branch swapping, retaining 10 trees per replicate, keeping up to 1,000 trees and swapping the trees in memory. Clade stability was assessed by 10,000 parsimony bootstrap replications (Felsenstein, 1985). MrModeltest software v. 2.2 (Nylander,J.A.A., 2004. MrModelTest Evolutionary Biology Centre, Uppsala University. http://www.abc.se/~nylander) was used to infer the most appropriate model of molecular evolution based on the Akaike Information Criterion (AIC), as suggested by Posada and Buckley (2004). The best model selected for this data set was GTR + I + G. Bayesian phylogenetic analysis was performed using MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist, 2001). Two independent analyses were run using a random starting tree with three heated chains and one cold chain over 4,000,000 generations, with sampling every 100 generations. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University (USA) which is partially funded by Microsoft Corporation.
2.5. Identification of recombination break points and network analysis
The automated system for detection of recombination using a genetic algorithm (Kosakovsky Pond et al., 2006) known as Genetic Algorithm for Recombination Detection (GARD), available at http://www.datamonkey.org, was used to search for evidence of recombination break points in 53 selected sequence variants representative of the five SL-IR groups (larger alignments involved limitations in the number of sites for the small sample AIC (AICc) inference). The nucleotide-substitution model was selected automatically before being applied to the recombination-site analysis and the goodness-of-it was evaluated by the AICc (Sugiura, 1978).
Provided that recombination breakpoints were not detected, the pattern of relationships among SL-IR genotypes was investigated using Median-joining (MJ) networks. The network was constructed using the software Network 4.5.1.0 (Bandelt et al., 1999) available at http://www.fluxus-engineering.com. This method analyses recombination-free population data, combining features of Kruskal’s algorithm to find minimum spanning trees by favoring short connections, and Farris’s MP heuristic algorithm, which sequentially adds new vertices called “median vectors”. Indels and SNPs within positions 12–19 of the alignment were coded as three SNPs in order for indels to represent a single mutational event. Indels were double-weighted, as recommended by the Network 4.5.1.0 user guide. The MJ network was post-processed by MP calculation to eliminate non-parsimonious links and find the shortest (Steiner) tree (Polzin and Daneschmand, 2003).
2.6. Tc Ie specific PCR assay
Specific primers to amplify SL-IR group Tc Ie were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast): TcIe-Fw (5´-CACG TGTGTGTATGTATGTATGTGC- 3´) and TcIe-Rv (5´-ATCAGCGCCACAGAAAGTG- 3´). A PCR reaction was carried out at a final volume of 30 µl of reaction mix containing buffer 1X, 1.5 mM MgCl2, 250 µM of each dNTP, 0.2 µM of each primer and 0.75 U of Taq Polymerase (Promega). Amplification was carried out in a MJR PTC-100 thermocycler (MJ Research, Watertown, MA, USA) as follows: 40 cycles at 94 °C for 45 s, 65 °C for 45 s, 72 °C for 45 s and a final extension step at 72 °C for 10 min. PCR products were analysed by 2% agarose gel electrophoresis followed by ethidium bromide staining and u.v. visualization.
3. Results
3.1. Analysis of the SL-IR gene
The origins of the 105 characterised Tc I samples are described in Table 1 (tissue and faeces of vectors and cultured stocks originally obtained from vectors, reservoirs and humans), Table 2 (human tissue and blood samples) and Table 3 (reference cultured stocks). In nine samples, the electropherograms exhibited considerable nucleotide ambiguity, suggesting the existence of mixed SL-IR genotypes. Consequently, sequences from individual recombinant clones from the PCR products were obtained (27 sequences; see Table 1).
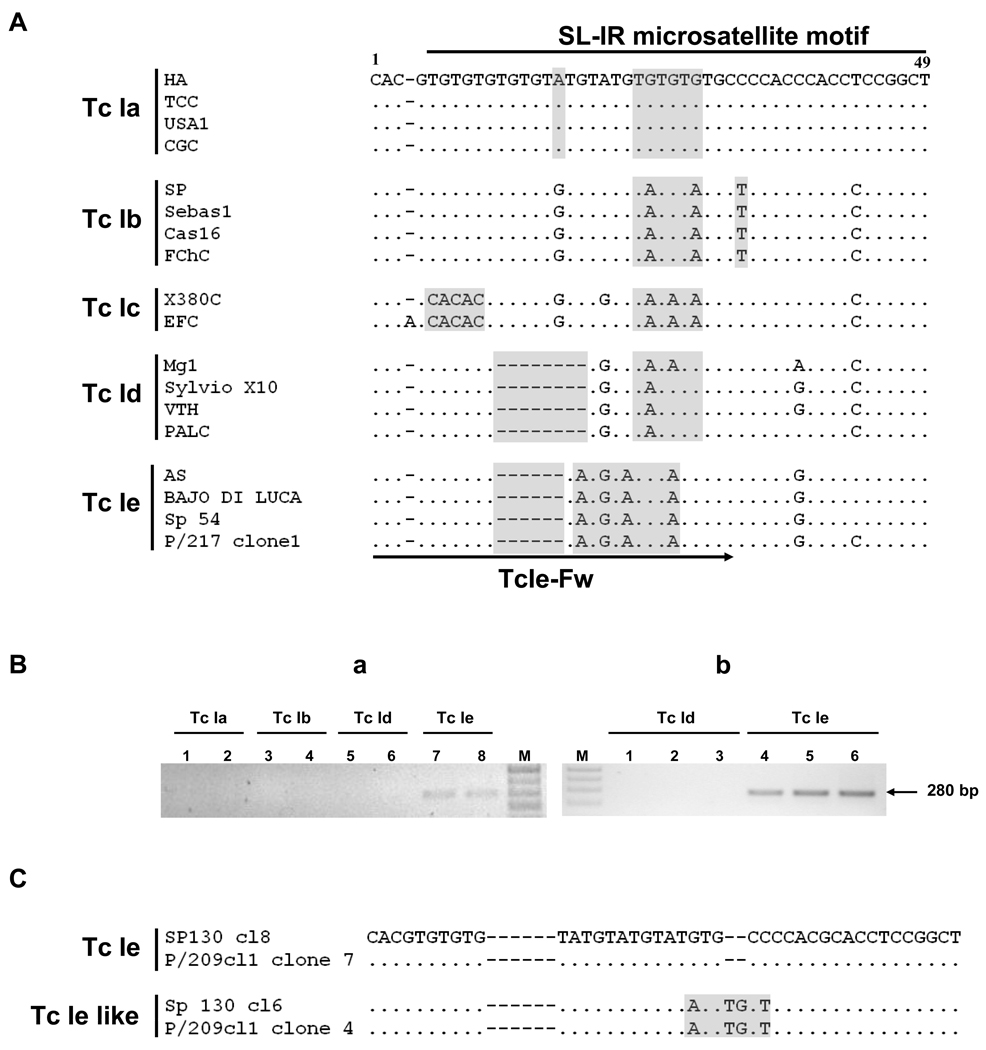
Sequencing of the SL-IR amplicons allowed identification of 76 genotypes with 54 polymorphic sites from the 123 obtained sequences. Alignment of the 44 bp microsatellite motif that defines the SL-IR groups (Fig. 1, sites 6–49) revealed that 11 genotypes belonged to groups Tc Ia, 11 to Tc Ib, 35 to Tc Id and none to Tc Ic. Interestingly, 19 genotypes revealed novel patterns of indels and adenine-guanine transitions.
Fig. 1.
Sequence analysis of spliced-leader intergenic region (SL-IR) amplicons and Trypanosoma cruzi SL-IR group Tc Ie-specific PCR assay. A) Alignment of a 49 bp region within the SL-IR of 12 out of the 123 analysed T. cruzi I (Tc I) sequences. The 44 bp microsatellite motif used to identify Tc I SL-IR groups is underlined. Boxed areas correspond to the nucleotide polymorphisms found among the SL-IR groups. Partial reference sequences reported in Herrera et al. (2007) and O´Connor et al. (2007) are also shown: CGC (AM259467), FChC (AM259469), X380C (AM259472), EFC (AM259474), PALC (AM259473) and P/217 clone 1 (EF576840). The specific sense primer for Tc Ie DNA amplification is indicated by an arrow. Gaps (−). Conserved sites (·). B) Gel electrophoresis of Tc Ie PCR amplicons using primers TcIe-Fw and TcIe-Rv from SL-IR groups a-b-d-e isolate representatives. a) Lanes: 1: TCC (Tc Ia), 2: Tev91Cl5 (Tc Ia), 3: Mg11 (Tc Ib), 4: Coy10 (Tc Ib), 5: G (Tc Id), 6: PAECl18 (Tc Id), 7: Palv2-2 Cl5 (Tc Ie), 8: AS (Tc Ie), M: 1 kb plus DNA ladder (Invitrogen). Lanes 1–8: 10 ng of DNA template. b) Lanes: 1–3: G (Tc Id); 4–6: Palv2-2 Cl5 (Tc Ie). Lanes: 1 and 4, 5 pg; 2 and 5, 50 pg; 3 and 6, 0.5 ng of DNA template, respectively. M: 1 kb plus DNA ladder. C) Tc Ie and Tc Ie like motifs found in different SL-IR PCR products cloned from the Chilean isolate Sp 130 and the reference parasite clone P/209cl1 (clone 4: EF576817, clone 7: EF576818, O´Connor et al., 2007) indicating intra-genomic variability.
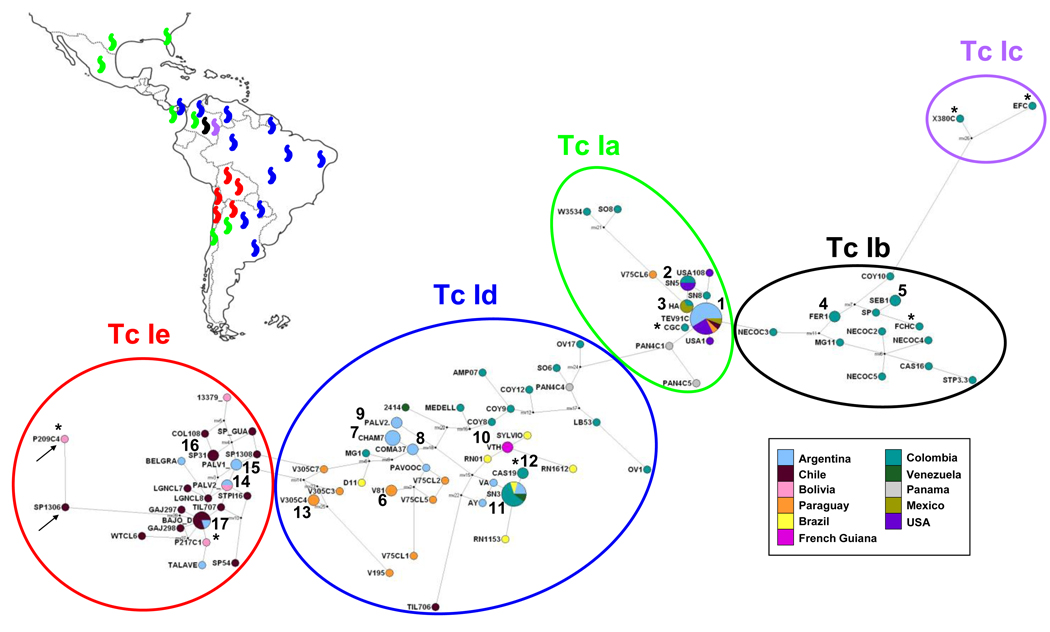
Phylogenetic trees constructed from the multiple sequence alignment using MP and Bayesian Inference methods showed neither clear clustering nor concordance between the derived topologies (data not shown). Thus, in order to analyze the intra-specific relationships among the SL-IR genotypes and, providing that recombination breakpoints among those were not detected (data not shown), a MJ network was constructed with the 123 SL-IR sequences and another seven available in GenBank were used as references. Genotypes belonging to SL-IR groups Tc Ia, Tc Ib, Tc Ic and Tc Id clustered in four groups. Moreover, the 19 novel genotypes were assigned to a different group, clustering with reference sequences from cloned strains P/217 clone1 and P/209cl1 clone4. On the basis of the microsatellite motif, these genotypes formed a distinct group, designated as Tc Ie (red oval in Fig. 2), with the exception of one genotype obtained from an isolate from Mepraia spinolai specimen (Sp 130) in the metropolitan region of Chile (cl6, Table 1). The latter harbors an insertion of two bases and a guanine-adenine and a cytosine-hymine transition with respect to the other Tc Ie genotypes. This region was identical to that present in reference P/209cl1 clone4 (Tc Ie like; Fig. 1C; arrows in Fig. 2).
Fig. 2.
Median-joining network for Trypanosoma cruzi I (Tc I) spliced-leader intergenic region (SL-IR) groups. SL-IR groups are indicated by ovals and geographical distribution is colour-coded. Circles represent genotypes and their size is proportional to the number of sequences harboring a given genotype: Tc Ia. 1: T ev91Cl5, TCC, A-26b-2-motoH, USA111, USA115, BG08011, Flop2, FRA, PA, TA, PASCh, K98, CA-I CL72, MIRANDA CL78, V75 cl7, WT cl7, HD/Duran. 2: SN5, SN12, USA28, USA104. 3: HA, PG/Gamma, TR/Triatoma. Tc Ib. 4: Fer1, Mg6. 5: Sebas1, Sebas16. Tc Id. 6: V81, V92. 7: CHAMICAL7, CHAMICALII, COMA665, COMA729. 8: COMA375, COMA701. 9: PALV2-1 Cl4, PalDa. 10: VTH, VTL. 11: SN3, G, PAECl18, CHA, Put4, Cas1, Cas10, Cas15, Cas18, EP. 12: Cas19, PALC*. 13. V305 cl4, V305 cl6. Tc Ie. 14: PALV2-2 Cl5, AS. 15: PALV1 Cl1, SE 9V. 16: SP 31, SP 104. 17: BAJO DI LUCA, Til70 cl8, Gaj 29 cl6, SpTi1 cl7, SpTi1 cl8. Black points represent hypothetical genotypes (median vectors). Reference genotypes reported in Herrera et al. (2007) and O´Connor et al. (2007) are indicated by an asterisk (*): CGC (Ia), FChC (Ib), X380C and EFC (Ic), PALC (Id), P/217 clone 1 and P/209cl1 clone 4 (Ie). Genotypes harboring a Tc Ie like sequence are indicated by arrows.
A median vector appeared connecting the Tc Ib genotype Coy10 and the reference Tc Ic genotypes X380C and EFC. Tc Ib and Tc Ia appeared connected by the genotype Necocli3 from Colombia, whose motif shared adenine-guanine transitions with both groups. Tc Ia and Tc Id were connected by the genotype Pan4 cl1 from Panama, which exhibited the microsatellite motif typical of the Tc Ia group modified by an additional 2 bp deletion. Finally, Tc Id and Tc Ie appeared connected by a group of sequences originally obtained from sample V305 from Paraguay.
3.2. Characterisation of mixed genotypes
In some biological samples and isolates we detected multiple SL-IR genotypes belonging to different groups. Indeed, mixed infections by Tc Ia + Tc Id were identified in faecal samples of a T. infestans bug from Paraguay (V75, Table 1) and in a culture stock derived from a human blood sample from Panama (Pan4, Table 1).
Furthermore, mixed groups were detected in cultured stocks isolated from the bloodstream of a patient with chronic Chagas disease from the IV region of Chile (Tc Ia + Tc Ie (WT), Table 1) and from one M. spinolai specimen captured at the metropolitan region of Chile (Tc Id + Tc Ie (Til70), Table 1).
In addition, genotypes belonging to Tc Ia and Tc Id were identified from natural parasite populations from an Argentinean heart transplanted Chagas cardiomyopathy patient undergoing reactivation. Tc Ia was detected in blood (PA) and skin chagoma (PASCh) specimens, while Tc Id was found in the heart explant (PAECl18) (Table 2), suggesting Tc I group histotropism.
3.3. Geographical distribution of SL-IR genotypes
Genotypes presenting the microsatellite motif Tc Ia were found in wild triatomine species from Colombia (SO8), Mexico (HD/Duran, PG/Gamma, TR/Triatoma) and USA (USA1, USA28, USA104, USA105, USA108, USA111, BG08011), and in one wild Didelphis virginiana specimen captured in Florida, USA (Flop2) (Table 1). Interestingly, Tc Ia was also identified in different types of clinical samples obtained from patients with Chagas disease from Argentina (FRA, PA, TA and PASCh) (Table 2) and cultured stocks originally isolated from patients from Colombia, Chile and Panama (HA, W3534, WT cl7, Pan4 cl1, Pan4 cl5) (Table 1) as well as in a dog (A-26b-2-motoH) and a T. infestans bug from Argentina (Tev91Cl5), and domestic Rhodnius prolixus from Colombia (SN5, SN8 and SN12) (Table 1). The Chilean strain TCC and the Argentinean clone K98 (originally isolated from patients with Chagas disease) and both ATCC strains (CA-I CL-72 and MIRANDA CL-78) also belonged to this group (Table 3).
Genotypes harboring the microsatellite motif Tc Ib were only found in Colombian populations of T. cruzi from a peridomestic R. prolixus (Cas16) and in one cultured stock originally isolated from a patient (SP). It was also found in samples belonging to wild species of Rhodnius spp. and Panstrongylus geniculatus from Colombia (STP3.3, Sebas1, Sebas16, Mg6, Mg11, Fer1, Coy10, Necocli2, Necocli3, Necocli4 and Necocli5) (Table 1).
More than 46% of the genotypes harbored the microsatellite motif typical of SL-IR group Tc Id. It was identified from Tc I populations infecting: a) vectors from: i) domestic cycles from Argentina, Paraguay (T. infestans) and Colombia (R. prolixus) (Table 1); ii) peridomestic cycles from Colombia (P. geniculatus) and Paraguay (T. infestans) (Table 1); iii) sylvatic cycles from Brazil (R. neglectus and Rhodnius nasutus), Colombia (Triatoma dimidiata, Rhodnius pallescens, Rhodnius robustus, Rhodnius colombiensis, R. prolixus and P. geniculatus) and Chile (M. spinolai) (Table 1); b) wild reservoirs from Argentina (Didelphis albiventris) and Colombia (Didelphis marsupialis) (Table 1) and c) human samples from Argentina, French Guiana, Panama and Venezuela (VA, AY, PAECl18, CHA, VTH, VTL, Pan4 cl4 and 2414) (Tables 1 and 2). Moreover, the Brazilian (Sylvio X10, G and its clone D11) and Venezuelan (EP) reference stocks belonged to this group (Table 3).
Genotypes presenting the motif Tc Ie were identified in domestic T. infestans samples from northern Argentina (PALV2-2 Cl5, PALV1 Cl1, BAJO DI LUCA, TALAVERDE, BELGRANOIII), in wild M. spinolai (SpTi 1, Col 108, Sp 31, Sp 104, Sp 54, Sp Guayacan, Sp130 cl6, Sp 130 cl8, Til70 cl7, Til70 cl8), wild Mepraia gajardoi (Gaj 29) and patients with chronic Chagas disease (LGN, WT cl6) from northern and central endemic areas of Chile (Table 1), and in the CSF specimen of a Bolivian patient with encephalitis due to AIDS (AS, Table 2). Moreover, it was identified from laboratory strains, such as SE 9V from Santiago del Estero, Argentina and 13379 cl7 from Santa Cruz, Bolivia (Table 3).
3.4. Tc Ie-specific PCR assay
The finding of the SL-IR group Tc Ie motivated us to design a specific oligonucleotide as sense primer for a PCR assay aiming to provide a rapid typing tool (underlined tract in Fig. 1A). An antisense primer was designed, located 238 bp downstream of the 44 bp microsatellite motif to amplify a differential Tc Ie fragment of 280 bp (Fig. 1B). The specificity of the PCR procedure was confirmed with DNA samples from SL-IR groups Ia, Ib and Id, whereas all of the 17 Tc Ie specimens amplified the specific PCR product (examples in Fig. 1Ba). Moreover, the PCR analysis sensitivity was tested using serial dilutions of PalV2-2 Cl5 DNA as template (Fig. 1Bb), allowing a detection limit of 1.5 pg of DNA in the reaction tube, which is equivalent to approximately 10 parasite cells.
4. Discussion
The SL gene is a tandemly arrayed multi-copy gene encoding a mini-exon of 39 nucleotides of length which is added to the 5’ end of mRNAs (Campbell et al., 2003). The sequence analysis of the SL-IR, a flanking region between SL repeats, has been widely used to discriminate Tc I from the rest of the DTUs, using the different size fragments obtained by PCR amplification (Souto et al., 1996; Brisse et al., 2001). Four groups, named as haplotypes, have been defined within Colombian Tc I (Herrera et al., 2007; Falla et al., 2009), based on a microsatellite motif present at the SL-IR. An additional group (Tc Ie) with a different motif is described in this study. The term “haplotype” is generally used to describe a set of markers, SNPs or alleles inherited as a unit in a haploid genome. Since T. cruzi is considered a diploid organism, the occurrence of sequence variability between allelic copies in a cloned strain implies the existence of at least two different haplotypes and in the case of a multicopy SL-IR gene may also imply polymorphic copies in a tandem array. In this context, we have designated as genotypes those PCR products that shared more than 99% sequence identity and as groups of genotypes those sharing a highly similar 44 bp microsatellite motif and a closer location within the MJ Network, which have been classified as Tc Ia, Tc Ib, Tc Ic, Tc Id and Tc Ie groups following the nomenclature of Falla et al. (2009).
Trypanosoma cruzi I SL-IR sequences have been characterised from wild and domestic animal reservoirs such as D. marsupialis, Canis familiaris (Herrera et al., 2007, 2009; Falla et al., 2009); D. albiventris, Procyon lotor and Dasyprocta aguti (O´Connor et al., 2007), as well as from a variety of sylvatic and domestic vectors such as T. infestans, Triatoma pallidipennis, Triatoma longipennis, Triatoma picturata, Triatoma barberi and other Triatoma spp. (O´Connor et al., 2007); T. dimidiata, Triatoma venosa and R. prolixus (Herrera et al., 2007), and more recently from sylvatic R. pallescens, P. geniculatus and Eratyrus cuspidatus specimens (Mejía-Jaramillo et al., 2009) and R. colombiensis (Falla et al., 2009). In the present study, SL-IR sequences from Tc I infecting wild T. gerstaeckeri and D. virginiana from the USA, T. infestans from Paraguay, R. nasutus and R. neglectus from northeastern Brazil and M. spinolai and M. gajardoi from Chile are described, to our knowledge, for the first time.
Furthermore, we identified Tc I SL-IR genotypes from clinical specimens, such as in a) peripheral blood samples from i) patients from French Guiana presenting acute cardiomyopathy who had probably acquired the infection through oral contamination with Attalea spp. juice (Christine Aznar, unpublished results), ii) a Venezuelan acute Chagas disease case due to a recent oral outbreak in Chichiriviche that affected 51 people at a school and caused three deaths (Alarcón de Noya and Martínez, 2009); b) three cardiac explant sections, four peripheral blood samples and one skin biopsy sample from patients with end-stage Chagas heart disease and with clinical reactivation after heart transplantation (Burgos et al., in press); c) a CSF sample of a Bolivian patient with Chagas encephalitis due to AIDS (Burgos et al., 2008).
Most of the sequences exhibited a low percentage of nucleotide ambiguity, in agreement with previous studies by O´Connor et al. (2007) who detected more than 96% similarity among SL-IR repeats in two strains of Tc I, thus validating the use of a consensus sequence as a genetic marker for population genetics.
In contrast to the topology proposed by Herrera et al. (2009), working with 31 sequences from Colombia, the phylogenetic trees from the multiple alignment of our genotypes using MP and Bayesian Inference methods did not render a clear clustering (data not shown). However, the MJ network-based analysis showed that the genotypes harboring the same Tc Ie motifs were more closely related. This analysis was possible given that no recombination break points with statistical support were found.
Distribution of genotypes harboring the microsatellite motif Tc Ia was in agreement with findings by O´Connor et al. (2007) who gathered SL-IR sequences from Mexico and Argentina and with more recent findings by Llewellyn et al. (2009) using a 48-marker panel of polymorphic microsatellite loci, who reported a close resemblance between domestic Tc I populations from the northern region of South America and sylvatic populations from Central and North America. In addition, the analysis of human tissue specimens allowed identification of Tc Ia genotypes associated with Chagas disease in the Southern Cone of America. Interestingly, two genotypes belonging to Tc Ia (Fig. 2) were SO8 from a sylvatic R. pallescens vector and W3534 from a Chagas disease patient, both from Sucre, Colombia, suggesting a connection between both parasite transmission cycles in this endemic region.
Genotypes harboring the motif Tc Ib were only found in Colombian populations of T. cruzi circulating in sylvatic, peridomestic and domestic cycles, extending previous findings by Falla et al. (2009) and Herrera et al. (2009) in peridomestic and domestic cycles.
Genotypes with the motif Tc Ic, which have been described in only two isolates from Boyacá, Colombia (X380C and EFC - Herrera et al., 2007), were not detected in our study, despite our having analyzed 36 Colombian stocks. Given that the differential motif distinguishing Tc Ic includes an insertion of six (X380C) or seven (EFC) nucleotides that maps within the sequence of the forward primer used by Herrera et al. (2007) for SL-IR PCR, we cannot discard the possibility that this motif could be an artifact which has arisen during amplification or sequencing procedures. Further studies on a larger sample size are needed to assess the existence and relevance of Tc Ic (Herrera et al., 2009).
Genotypes presenting the motif Tc Id were detected in 47 out of the 105 analyzed samples. They were identified from Tc I populations infecting T. infestans and D. albiventris specimens from the Argentine Chaco (Table 1), suggesting a connection between both domestic and sylvatic cycles (Cardinal et al., 2008). In Colombia, wild animal reservoirs and triatomine species, peridomestic P. geniculatus and domestic R. prolixus specimens were infected by Tc Id parasites. In addition, other genotypes with the Tc Id motif were detected in central Brazil, Paraguay, Venezuela and French Guiana.
Our study revealed genotypes sharing another microsatellite motif, defining a new group designated as Tc Ie. The MJ network gathered Tc Ie sequences from Bolivian, Argentinean and Chilean samples, five from cultured stocks from faeces of domestic T. infestans bugs collected in the northern and southwestern sections of the Argentine Chaco (Lauricella et al., 2005), eight from cultured stocks from faeces of sylvatic M. spinolai, one from M. gajardoi and two from chronic Chagas disease cases from Chile, and one from the CSF sample of a Bolivian patient with Chagas encephalitis due to AIDS. Moreover, it was identified from laboratory strains, such as SE 9V from Santiago del Estero, Argentina and 13379 cl7 from Santa Cruz, Bolivia. These samples present a 44 bp motif identical to that present in SL-IR sequences of four Bolivian DTU I strains (three from patients with Chagas disease and one from T. infestans) and one Brazilian isolate from D. aguti available in GenBank, clustered in the Pan-American group of isolates by O´Connor et al. (2007). Therefore, genotypes with the Tc Ie motif could be associated with strains circulating in the domestic cycles of Argentina and Bolivia and in the sylvatic cycle in Chile, represented by M. spinolai and M. gajardoi vectors.
Recently, Falla et al. (2009) proposed a PCR-based procedure in order to identify SL-IR groups Tc Ia, Tc Ib and Tc Id using different primers. As a complement to this strategy, we have designed a Tc Ie-specific PCR procedure to detect genotypes belonging to this group, avoiding the need for amplicon sequencing.
Recent data from samples of the Chaco Region showed the occurrence of multiple ambiguous sites in the sequences obtained from the PCR products of cloned strains, indicating intra-genomic variability and suggesting incomplete concerted evolution of the multicopy tandem array (Tomasini et al., unpublished data). Interestingly, in our study, one culture isolate from M. spinolai (SP 130) revealed two different Tc Ie genotypes, one of those showing the typical SL-IR microsatellite motif of the Tc Ie group, and the other one, a motif identical to that of the reference sequence P/209cl1 clone4, which was also clustered in the Panamerican group by O´Connor et al. (2007) (Tc Ie like). Both genotypes were identified from samples that also presented typical Tc Ie genotypes, indicating that, at least for P/209cl1 clone4, they were distinct SL-IR copies of the same parasite genome. This could also be the case for mixed genotypes belonging to Tc Ia + Tc Id, Tc Ia + Tc Ie and Tc Id + Tc Ie in other culture isolates and biological specimens, but these findings may be the result of polyclonal infections.
Interestingly, we have identified genotypes Tc Ia and Tc Id at different body sites of a patient with end-stage Chagas Heart disease who underwent heart transplantation presenting skin reactivation due to immunosuppressive therapy. Indeed, Tc Ia populations from blood (PA) and skin (PASCh) samples depicted almost identical Restriction Fragment Length Polymorphism (RFLP)-PCR based mini-circle signatures, different from those profiled from the Tc Id cardiac population (PAECl18) (Burgos et al., in press). This finding pointed to differential tissue tropism of Tc I populations belonging to different SL-IR groups.
The non-transcribed spacer is essential for the transcription of the mini-exon gene that becomes a substrate for the trans-splicing reaction in trypanosomatids, providing promoter elements that recruit transcription factors for the RNA polymerase (Saito et al., 1994; Hitchcock et al., 2007). In this way, sequence polymorphism of the intergenic spacer, as observed among the described genotypes, could alter DNA secondary structure responsible for this recruitment function, affecting the mRNA post-transcriptional process.
Finally, the remarkable genetic heterogeneity of Tc I should be taken into consideration for designing Tc I functional genomic projects, molecular diagnosis, trypanocidal drug trials and disease pathogenesis assays.
Acknowledgements
We thank Dr. Michael Yabsley (University of Georgia, Athens, USA) for providing Flop2 and BG08011 and Dr. Paula Marcet (Centers for Disease Control and Prevention, Atlanta, USA) for her collaboration in characterisation of USA samples. Sample 2414 was kindly provided by Dr. Belkisyolé Alarcón de Noya (Sección de Inmunología, Instituto de Medicina Tropical, Universidad Central de Venezuela, Venezuela). Financial support was obtained from: PICT 33955 from the Ministry of Science and Technology (FONCyT) [Argentina] and PIP 112-2008-01-02913 from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina to AGS; Colciencias 1115-04-14387, Proyecto de Sostenibilidad, University of Antioquia 2007–2008 [Colombia] to OTC; Proyecto 2020, Banco de la República de Colombia to AMJ and OTC; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Apoio à Pesquisa do Distrito Federal (FAP/DF), Brazil to CACC and RGG and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT)1085154 [Chile] to AS. The participation of REG and MVC was supported by National Institutes of Health (NIH) Research Grant #R01 TW05836 funded by the Fogarty International Center and the National Institute of Environmental Health Sciences (to Uriel Kitron and REG), and by grants from Universidad de Buenos Aires and FONCyT, Argentina to REG. MVC, REG, AGS are members of the CONICET Researcher’s Career.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcón de Noya B, Martínez J. II Tópicos de Actualidad: Transmisión oral de la Enfermedad de Chagas en Venezuela: un segundo brote escolar. Salus. 2009;13:9–10. [Google Scholar]
- Añez N, Crisante G, da Silva FM, Rojas A, Carrasco H, Umezawa ES, Stolf AM, Ramírez JL, Teixeira MM. Predominance of lineage I among Trypanosoma cruzi isolates from Venezuelan patients with different clinical profiles of acute Chagas Disease. Trop Med Int Health. 2004;9:1319–1326. doi: 10.1111/j.1365-3156.2004.01333.x. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Basombrío MA, Besuschio S, Cossio PM. Side effects of immunization with liver attenuated Trypanosoma cruzi in mice and rabbits. Infect Immun. 1982;36:342–350. doi: 10.1128/iai.36.1.342-350.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisio M, Cura C, Duffy T, Altcheh J, Giganti SO, Begher S, Scapellato P, Burgos JM, Levin MJ, Schreck R, Freilij H, Schijman AG. Trypanosoma cruzi discrete typing units in Chagas disease patients with HIV co-infection. Rev Biomed. 2009;20:166–178. [Google Scholar]
- Brenière SF, Braquemond P, Solari A, Agnèse JF, Tibayrenc M. An isoenzyme study of naturally occurring clones of Trypanosoma cruzi isolated from both sides of the West Andes highland. Trans R Soc Trop Med Hyg. 1991;85:62–66. doi: 10.1016/0035-9203(91)90160-z. [DOI] [PubMed] [Google Scholar]
- Brenière SF, Bosseno MF, Magallón-Gastelúm E, Castillo Ruvalcaba EG, Gutierrez MS, Montaño Luna EC, Basulto JT, Mathieu-Daudé F, Walter A, Lozano-Kasten F. Peridomestic colonization of Triatoma longipennis (Hemiptera, Reduviidae) and Triatoma barberi (Hemiptera, Reduviidae) in a rural community with active transmission of Trypanosoma cruzi in Jalisco state, Mexico. Acta Trop. 2007;101:249–257. doi: 10.1016/j.actatropica.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- Brisse S, Verhoef J, Tibayrenc M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol. 2001;31:1218–1226. doi: 10.1016/s0020-7519(01)00238-7. [DOI] [PubMed] [Google Scholar]
- Brito CM, Lima MM, Sarquis O, Pires MQ, Coutinho CF, Duarte R, Pacheco RS. Genetic polymorphism in Trypanosoma cruzi I isolated from Brazilian Northeast triatomines revealed by low-stringency single specific primer-polymerase chain reaction. Parasitol Res. 2008;103:1111–1117. doi: 10.1007/s00436-008-1102-5. [DOI] [PubMed] [Google Scholar]
- Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Burgos JM, Begher S, Valadares Silva H, Bisio M, Duffy T, Levin MJ, Macedo AM, Schijman AG. Case report: molecular identification of Trypanosoma cruzi I tropism for central nervous system in Chagas reactivation due to AIDS. Am. J. Trop. Med. Hyg. 2008;78:294–297. [PubMed] [Google Scholar]
- Burgos JM, Diez M, Vigliano C, Bisio M, Risso M, Duffy T, Cura C, Brusses B, Favaloro L, Leguizamon MS, Lucero RH, Laguens R, Levin MJ, Favaloro R, Schijman AG. Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin Infect Dis. doi: 10.1086/655680. in press. [DOI] [PubMed] [Google Scholar]
- Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, Kitron U, Gürtler RE, Schijman AG. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38:1533–1543. doi: 10.1016/j.ijpara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos LA, Cardinal MV, Vazquez-Prokopec GM, Lauricella MA, Orozco MM, Cortinas R, Schijman AG, Levin MJ, Kitron U, Gürtler RE. Long-term reduction of Trypanosoma cruzi infection in sylvatic mammals following deforestation and sustained vector surveillance in northwestern Argentina. Acta Trop. 2006;98:286–296. doi: 10.1016/j.actatropica.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MR, Pinho AP, Cuervo P, Alfaro F, Solano M, Xavier SC, D'Andrea PS, Fernandes O, Torrico F, Noireau F, Jansen AM. Trypanosoma cruzi (Kinetoplastida Trypanosomatidae): ecology of the transmission cycle in the wild environment of the Andean valley of Cochabamba, Bolivia. Exp Parasitol. 2006;114:305–313. doi: 10.1016/j.exppara.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Cuervo P, Cupolillo E, Segura I, Saravia N, Fernandes O. Genetic diversity of Colombian sylvatic Trypanosoma cruzi isolates revealed by the ribosomal DNA. Mem Inst Oswaldo Cruz. 2002;97:877–880. doi: 10.1590/s0074-02762002000600023. [DOI] [PubMed] [Google Scholar]
- Dias FB, Diotaiuti L, Romanha AJ, Bezerra CM, Machado EM. First report on the occurrence of Trypanosoma rangeli Tejera, 1920 in the state of Ceará, Brazil, in naturally infected triatomine Rhodnius nasutus Stål, 1859 (Hemiptera, Reduviidae, Triatominae) Mem Inst Oswaldo Cruz. 2007;102:643–645. doi: 10.1590/s0074-02762007005000057. [DOI] [PubMed] [Google Scholar]
- Diosque P, Barnabe C, Padilla A, Marco J, Cardozo R, Cimino R, Nasserd J, Tibayrenc M, Basombrıo M. Multilocus enzyme electrophoresis analysis of Trypanosoma cruzi isolates from a geographically restricted endemic area for Chagas' disease in Argentina. Int J Parasitol. 2003;33:997–1003. doi: 10.1016/s0020-7519(03)00139-5. [DOI] [PubMed] [Google Scholar]
- Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, Levin MJ, Favaloro RR, Freilij H, Schijman AG. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in chagas disease patients. PLoS Negl Trop Dis. 2009;3:e419. doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez Cappa SM, Mirkin GA, Solana ME, Tekiel VS. Trypanosoma cruzi pathology. Strain dependent? Medicina (B Aires) 1999;59:69–74. [PubMed] [Google Scholar]
- Herrera C, Bargues MD, Fajardo A, Montilla M, Triana O, Vallejo GA, Guhl F. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions in Colombia. Infect Genet Evol. 2007;7:535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Herrera C, Guhl F, Falla A, Fajardo A, Montilla M, Vallejo GA, Bargues MD. Genetic variability and phylogenetic relationships within Trypanosoma cruzi I isolated in Colombia based on miniexon gene sequences. J Parasitol Res. 2009 doi: 10.1155/2009/897364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock RA, Thomas S, Campbell DA, Sturm NR. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 2007;7:44. doi: 10.1186/1471-2180-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck P, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jansen AM, Santos de Pinho AP, Lisboa CV, Cupolillo E, Mangia RH, Fernandes O. The sylvatic cycle of Trypanosoma cruzi: a still unsolved puzzle. Mem Inst Oswaldo Cruz. 1999;94:203–204. doi: 10.1590/s0074-02761999000700030. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Lauricella MA, Stariolo RL, Riarte AR, Segura EL, Gürtler RE. Distribution and pathogenicity of Trypanosoma cruzi isolated from peridomestic populations of Triatoma infestans and Triatoma guasayana from rural western Argentina. Mem Inst Oswaldo Cruz. 2005;100:123–129. doi: 10.1590/s0074-02762005000200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ, Miles MA. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol. 2009;39:1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, Torrico F, Diosque P, Valente V, Valente SA, Gaunt MW. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5:e1000410. doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo AM, Pena SD. Genetic Variability of Trypanosoma cruzi: Implications for the Pathogenesis of Chagas Disease. Parasitol Today. 1998;14:119–124. doi: 10.1016/s0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- Macina RA, Arauzo S, Reyes MB, Sanchez DO, Basombrio MA, Montamat EE, Solari A, Frasch AC. Trypanosoma cruzi isolates from Argentina and Chile grouped with the aid of DNA probes. Mol Biochem Parasitol. 1987;25:45–53. doi: 10.1016/0166-6851(87)90017-x. [DOI] [PubMed] [Google Scholar]
- Marcet PL, Duffy T, Cardinal MV, Lauricella MA, Levin MJ, Kitron U, Gürtler RE, Schijman AG. PCR-based screening and typing of Trypanosoma cruzi lineages directly from faecal samples of triatomine bugs from northwestern Argentina. Parasitology. 2006;132:1–9. doi: 10.1017/S0031182005008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Jaramillo AM, Arboleda-Sánchez S, Rodríguez IB, Cura C, Salazar A, Del Mazo J, Triana-Chávez O, Schijman AG. Geographical clustering of Trypanosoma cruzi I groups from Colombia revealed by low-stringency single specific primer-PCR of the intergenic regions of spliced-leader genes. Parasitol Res. 2009;104:399–410. doi: 10.1007/s00436-008-1212-0. [DOI] [PubMed] [Google Scholar]
- Miles MA, Llewellyn MS, Lewis MD, Yeo M, Baleela R, Fitzpatrick S, Gaunt MW, Mauricio IL. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136:1509–1528. doi: 10.1017/S0031182009990977. [DOI] [PubMed] [Google Scholar]
- Moncayo A, Silveira AC. Current epidemiological trends for Chagas disease in Latin America and future challenges in epidemiology, surveillance and health policy. Mem Inst Oswaldo Cruz. 2009;104:17–30. doi: 10.1590/s0074-02762009000900005. [DOI] [PubMed] [Google Scholar]
- O´Connor O, Bosseno MF, Barnabé C, Douzery EJ, Brenière SF. Genetic clustering of Trypanosoma cruzi I lineage evidenced by intergenic miniexon gene sequencing. Infect Genet Evol. 2007;7:587–593. doi: 10.1016/j.meegid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Polzin T, Daneschmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003;31:12–20. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Postan M, Dvorak JA, McDaniel JP. Studies of Trypanosoma cruzi clones in inbred mice. I. A comparison of the course of infection of C3H/HEN- mice with two clones isolated from a common source. Am J Trop Med Hyg. 1983;32:497–506. doi: 10.4269/ajtmh.1983.32.497. [DOI] [PubMed] [Google Scholar]
- Rojas W, Caro MA, Lopera JG, Triana O, Dib JC, Bedoya G. Analysis of polymorphisms in the trypanothione reductase and cruzipain genes in Colombian strains of Trypanosoma cruzi. Biomedica. 2007;1:50–63. [PubMed] [Google Scholar]
- Roellig DM, Brown EL, Barnabé C, Tibayrenc M, Steurer FJ, Yabsley MJ. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg Infect Dis. 2008;14:1123–1125. doi: 10.3201/eid1407.080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito RM, Elgort MG, Campbell DA. A conserved upstream element is essential for transcription of the Leishmania tarentolae mini-exon gene. EMBO J. 1994;13:5460–5469. doi: 10.1002/j.1460-2075.1994.tb06881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar A, Schijman AG, Triana O. High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures. Acta Trop. 2006;100:110–118. doi: 10.1016/j.actatropica.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Samudio F, Ortega-Barría E, Saldaña A, Calzada J. Predominance of Trypanosoma cruzi I among Panamanian sylvatic isolates. Acta Trop. 2007;101:178–181. doi: 10.1016/j.actatropica.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- Sturm NR, Campbell DA. Alternative lifestyles: The population structure of Trypanosoma cruzi. Acta Trop. 2010;115:35–43. doi: 10.1016/j.actatropica.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Sugiura N. Further analysis of the data by Akaike's information criterion and the finite corrections. Comm. Statist. 1978;A7:13–26. [Google Scholar]
- Téllez-Meneses J, Mejía-Jaramillo AM, Triana-Chávez O. Biological characterization of Trypanosoma cruzi stocks from domestic and sylvatic vectors in Sierra Nevada of Santa Marta, Colombia. Acta Trop. 2008;108:26–34. doi: 10.1016/j.actatropica.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Miles MA. A genetic comparison between Brazilian and Bolivian zymodemes of Trypanosoma cruzi. Trans R Soc Trop Med Hyg. 1983;77:76–83. doi: 10.1016/0035-9203(83)90021-4. [DOI] [PubMed] [Google Scholar]
- Tomazi L, Kawashita SY, Pereira PM, Zingales B, Briones MR. Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic. Genet Mol Res. 2009;8:458–476. doi: 10.4238/vol8-2gmr591. [DOI] [PubMed] [Google Scholar]
- Torres JP, Ortiz S, Muñoz S, Solari A. Trypanosoma cruzi isolates from Chile are heterogeneous and composed of mixed populations when characterized by schizodeme and Southern analyses. Parasitology. 2004;128:161–168. doi: 10.1017/s0031182003004475. [DOI] [PubMed] [Google Scholar]
- Triana O, Ortiz S, Dujardin JC, Solari A. Trypanosoma cruzi: variability of stocks from Colombia determined by molecular karyotype and minicircle Southern blot analysis. Exp Parasitol. 2006;113:62–66. doi: 10.1016/j.exppara.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Vargas N, Pedroso A, Zingales B. Chromosomal polymorphism, gene synteny and genome size in T. cruzi I and T. cruzi II groups. Mol Biochem Parasitol. 2004;138:131–141. doi: 10.1016/j.molbiopara.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Venegas J, Ortiz S, Munoz S, Solari A. Molecular karyotype and schizodeme analyses of Trypanosoma cruzi stocks from Chilean triatomines. Parasitology. 1997;115:41–46. doi: 10.1017/s0031182097001066. [DOI] [PubMed] [Google Scholar]
- Verbisck NV, Da-Silva S, Mortara RA. Trypanosoma cruzi: amastigote polymorphism defined by monoclonal antibodies. Braz J Med Biol Res. 1998;31:1583–1591. doi: 10.1590/s0100-879x1998001200011. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Control of Chagas Disease. Second report of the WHO expert committee. WHO Tech. Rep. Ser. 2002;905:1–109. [PubMed]
- Yeo M, Acosta N, Llewellyn M, Sánchez H, Adamson S, Miles GA, López E, González N, Patterson JS, Gaunt MW, Rojas deAriasA, Miles MA. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Yoshida N. Surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi. Infect Immun. 1983;40:836–839. doi: 10.1128/iai.40.2.836-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]