Abstract
Leishmaniasis is a major public health problem and till date there are no effective vaccines available. The control strategy relies solely on chemotherapy of the infected people. However, the present repertoire of drugs is limited and increasing resistance towards them has posed a major concern. The first step in drug discovery is to identify a suitable drug target. The genome sequences of Leishmania major and Leishmania infantum has revealed immense amount of information and has given the opportunity to identify novel drug targets that are unique to these parasites. Utilization of this information in order to come up with a candidate drug molecule requires combining all the technology and using a multi-disciplinary approach, right from characterizing the target protein to high throughput screening of compounds. Leishmania belonging to the order kinetoplastidae emerges from the ancient eukaryotic lineages. They are quite diverse from their mammalian hosts and there are several cellular processes that we are getting to know of, which exist distinctly in these parasites. In this review, we discuss some of the metabolic pathways that are essential and could be used as potential drug targets in Leishmania.
Keywords: Leishmaniasis, Leishmania, Metabolic pathways, Drug targets
Introduction
Leishmaniasis is a group of parasitic diseases caused by at least 20 different species of the protozoan parasite Leishmania. It constitutes of a wide spectrum of diseases ranging in severity from simple cutaneous lesions to the usually fatal visceral form. The parasite is transmitted through the bite of sandfly to the mammalian hosts. The disease is endemic in 88 countries and affects as many as 12 million people around the globe with an incidence of 0.5 million cases of the visceral form of the disease and 1.5–2.0 million cases of the cutaneous form of the disease, causing extensive mortality and morbidity.
Since, effective vaccines against leishmaniasis are still under development, the current control measures rely solely on chemotherapy. Pentavalent antimonials are the standard first line of treatment but emergence of resistance towards them, has limited their usefulness. Alternative chemotherapeutic treatments with amphotericin B and its lipid formulation, miltefosine and paromomycin are available but their use is limited either due to toxicity or high cost of treatment. The current challenges in the chemotherapy include availability of very few drugs, emergence of resistance to the existing drugs, their toxicity and lack of cost-effectiveness. Therefore, it is of utmost importance to look for effective drugs and new drug targets for the treatment of leishmaniasis.
There seems to be a welcome change in terms of flow of funds for antiparasitic drug discovery. Some of the organisations like Institute of One World Health (IOWH), Drugs for Neglected Diseases Initiative (DNDi), Bill and Melinda Gates foundation have had a significant impact on working towards the drug development for tropical diseases. This has enabled technological advances in the field, which includes publicly funded sequencing of the genomes, thus fastening up the course towards drug development. A major breakthrough in the field is the availability of the complete genome sequence of various species of Leishmania like, L. major (Ivens et al. 2005), L. infantum and L. braziliensis. It has provided us with enormous information about the parasite genome for our better understanding of these organisms. With the availability and easily accessible genome sequences, the conventional method of drug target identification, which was done on the basis of the biochemical and physiological differences between the pathogen and host, is now changing. Microarrays and proteome analysis make use of the genome sequences and has allowed us to find genes unique to the parasite, some species-specific genes that can help us understand the pathogen better. Several bioinformatic approaches have also been proposed which will fasten the pace to come up with anti-leishmanial drugs (Myler 2008). The analysis of the complete genome sequence may significantly contribute in drug development by revealing the presence of novel enzymes and receptors. Furthermore, the comparison of the parasite genome with the human genome sequence will make the identification of genes, unique to the parasite, easier.
Identification of drug targets
One of the characteristic features in the process of drug development is target identification in a biological pathway. In theory, during identification of a target in a pathogen, it is important that the putative target should be either absent in the host or substantially different from the host homolog so that it can be exploited as a drug target. Trypanosomatids, phylogenetically, branch out quite early from the higher eukaryotes. In fact, their cell organization is significantly different from the mammalian cells and thus, it is possible to find targets that are unique to these pathogens. Secondly, the target selected should be absolutely necessary for the survival of the pathogen. It is also important to consider the stage of the life-cycle of the pathogen in which the target gene is expressed. It is crucial to look at the biochemical properties of the protein; it should have a small molecule binding pocket, so that specific inhibitors can be designed and if the target protein is an enzyme, its inhibition should lead to loss in cell viability. It is of high importance that the target selected should be assayable. Inexpensive and specific assay system should be available for high-throughput screening of molecules (Pink et al. 2005; Barrett et al. 1999).
Potential drug targets in Leishmania
Sterol biosynthetic pathway
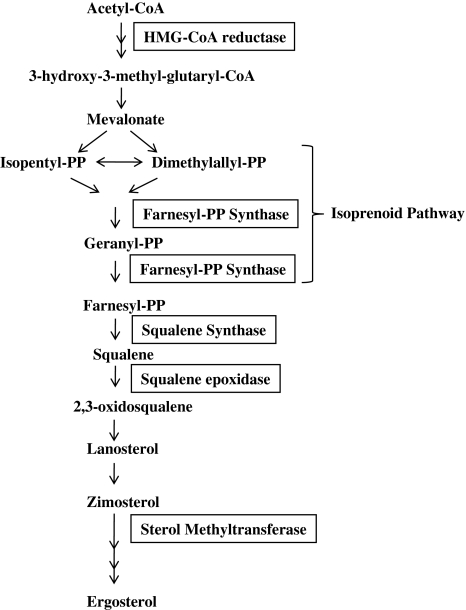
Sterols are important components of the cell membrane that are vital to cellular function and maintenance of cell structure. Unlike mammalian cells, which have cholesterol as the major membrane sterol, trypanosomatids synthesize ergosterol and other 24-methyl sterols that are required for their growth and viability. These sterols are absent from the mammalian cells. Therefore, the sterol biosynthetic pathway from Leishmania is considered to be an important drug target (Fig. 1).
Fig. 1.
Sterol biosynthetic pathway in Leishmania. The pathway shows the important steps and the enzymes involved in sterol biosynthesis. The final product in trypanosomatids is ergosterol as opposed to cholesterol in mammalian cells
One of the enzymes that is being studied deeply is squalene synthase (SQS) (EC 2.5.1.21) that catalyzes the first committed step of sterol synthesis by coupling two farnesyl molecules to form squalene. Zaragozic acids and quinuclidines are known to inhibit SQS. Two quinuclidine derivatives, ER-119884 and E5700 have been shown to be potent anti-Leishmania (Fernandes Rodrigues et al. 2008) and Trypanosoma (Urbina et al. 2004) agents. The inhibition of SQS by these compounds decreased the parasite’s endogenous sterol levels which had an antiproliferative effect on the parasite. In L. amazonensis, IC50 value of ER-119884 and E5700 for promastigotes was found to be 10 and 30 nM respectively (Fernandes Rodrigues et al. 2008). Squalene is converted to 2,3-oxidosqualene by the enzyme squalene epoxidase (EC 1.14.99.7). It converts squalene chain to tetracyclic sterol skeleton. Terbinafine, an allylamine, is known to inhibit squalene epoxidase. It has been shown that terbinafine inhibits the growth of promastigotes and intracellular amastigotes and leads to changes in structural organization in mitochondrion (Vannier-Santos et al. 1995). When it is used in combination with ketoconazole, another inhibitor of ergosterol biosynthesis, the effect is synergistic. Another class of inhibitors, bisphosphonates, inhibits the isoprenoid pathway that is catalyzed by the enzyme farnesyl diphosphate synthase (FPPS). They have been tested in vivo and in vitro against the protozoan parasites (Martin et al. 2001; Docampo and Moreno 2008). A potent inhibition of the cell growth and suppression of the activity of isolated enzymes (from L. major) was observed thus validating the isoprenoid pathway as a drug target.
Another important putative target in ergosterol biosynthesis is the enzyme Δ24,25-sterol methyltransferase (SMT) (EC 2.1.1.41). This enzyme is only present in trypanosomatids and absent from the human host. Therefore, this enzyme could be exploited as a potential drug target. Azasterols are known to inhibit SMT in case of Candida spp. (Ishida et al. 2009). The effect of azasterols has been studied on Leishmania and Trypanosoma species among the protozoan parasites. Several azasterols have been shown to have anti-proliferative effect with their IC50s in submicromolar to nanomolar range against L. amazonensis (Magaraci et al. 2003; Lorente et al. 2004), signifying that this step could be a potential chemotherapeutic target. In, L. amazonensis, they also led to disorganization of the mitochondrial membrane followed by intense swelling and loss of matrix contents (Vivas et al. 1996; Rodrigues et al. 2002). Interestingly, azasterols when used in combination with azoles act synergistically and are even more effective suggesting that inhibiting multiple steps of this pathway, that is, combination therapy may be used against the parasitic protozoa.
Glycolytic pathway
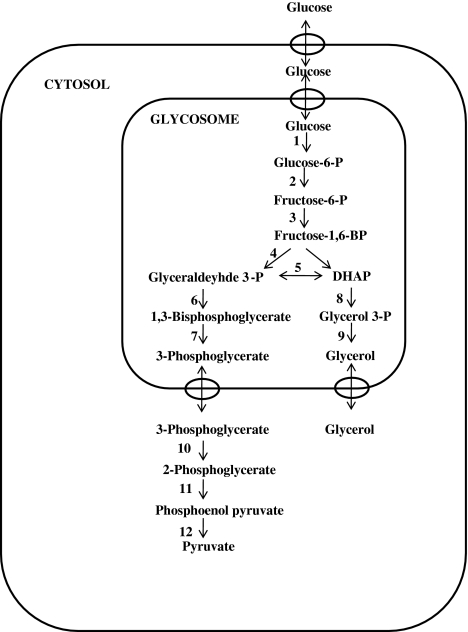
The energy metabolism of trypanosomatids solely depends on the carbon sources available in the host. Since the African trypanosomes lack a functional Krebs cycle, they use glycolysis as the only source of ATP generation (Opperdoes 1987). Seven of the glycolytic enzymes are compartmentalized in peroxisome-like organelles, glycosomes, which is a unique feature of trypanosomatids (Fig. 2). The unique compartmentalization of glycolytic enzymes in glycosomes in Leishmania and their large phylogenetic distance with the mammalian hosts provides them with unique features. These features can be exploited by designing specific inhibitors on the basis of the structure of the parasitic enzymes. Structure based drug designing is being employed to obtain compounds that bind to the enzymes with high affinity. The 3-D structures are available for some of the trypanosomatid enzymes: glyceraldehydes-3-phosphate dehydrogenase (EC 1.2.7.6) (Vellieux et al. 1993; Kim et al. 1995), triosephosphate isomerase (5.3.1.1) (Wierenga et al. 1991; Williams et al. 1999), phosphoglycerate kinase (EC 2.7.2.10) (Bernstein et al. 1997), pyruvate kinase (2.7.1.40) (Rigden et al. 1999), fructose-1,6-bisphosphate aldolase (EC 4.1.2.13) (Chudzik et al. 2000) and glycerol-3-phosphate dehydrogenase (EC 1.1.1.8) (Suresh et al. 2000). The differences in the 3-D structures of the parasite and host enzymes can be used to design specific inhibitors (Verlinde and Hol 1994).
Fig. 2.
Glycolytic pathway in trypanosomatids. The figure shows the glycolytic pathway as occurs in trypanosomatids. Seven of the glycolytic enzymes are compartmentalized in glycosomes. The enzymes involved in the pathway are (1) hexokinase (2) glucose-6-phosphate isomerase (3) phosphofructokinase (4) aldolase (5) triosephosphate isomerase (6) glyceraldehyde-3-phosphate dehydrogenase (7) phosphoglycerate kinase (8) glycerol-3-phosphate dehydrogenase (9) glycerol kinase (10) phosphoglycerate mutase (11) enolase (12) pyruvate kinase
Specific inhibitors have been designed for the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) based on its crystal structure. Adenosine analogs were synthesized as tight binding inhibitors that occupy the pocket on the enzyme that accommodates adenosyl moiety of NAD+ co-substrate. Adenosine was shown to be a poor inhibitor of the enzyme but its analog, with substitutions at the 2′ position of ribose and N6-position of adenosine (disubstituted analogs) resulted in enhanced inhibition of the enzyme. One of the analogs, N6-(1-naphthalenemethyl)-2′-(3-methoxybenzamido) adenosine inhibited growth of L. mexicana with IC50 of 0.28 μM (Aronov et al. 1999). This indicates that it is possible to block the energy production by synthesis of specific inhibitors of glycolytic pathway enzymes. Since glycolysis is the only source of energy for these parasites, it could serve as an excellent drug target.
Purine salvage pathway
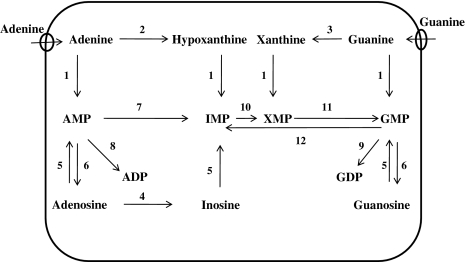
The parasitic protozoa, including Leishmania lack the enzymes to synthesize purine nucleotides de novo, therefore, they have to depend upon the purine salvage system to utilize purine bases from their mammalian hosts (Fig. 3). Purine bases are translocated through the parasite cell surface by nucleoside transporters.
Fig. 3.
Purine salvage pathway of Leishmania species. The enzymes involved in salvage of purines are (1) phosphoribosyltransferase (2) adenine deaminase (3) guanine deaminase (4) adenosine deaminase (5) nucleoside kinase (6) nucleotidase (7) AMP deaminase (8) AMP kinase (9) GMP kinase (10) IMP dehydrogenase (11) GMP synthetase (12) GMP reductase. AMP adenosine monophosphate; ADP adenosine diphosphate; IMP inosine monophosphate; XMP xanthine monophosphate; GMP guanosine monophosphate; GDP guanosine diphosphate
Nucleoside transporters
They are involved in transport of nucleosides across the membrane. Two specific transporters which have been well documented from Leishmania are LdNT1 (present in both promastigote and amastigotes) and LdNT2 (present in amastigotes) (Vasudevan et al. 1998; Carter et al. 2000). LdNT1 is responsible for transportation of adenosine and pyrimidine nucleosides and LdNT2 transports purine nucleosides (inosine and guanosine). LdNT1 and LdNT2 were first cloned from L. donovani by functional complementation of adenosine-pyrimidine transport deficient mutant, TUBA5 (tubercidin resistant) and inosine-purine transport deficient mutant FBD5 (Formycin resistant), respectively. Two very closely related genes encoding for pyrimidine nucleoside transporters LdNT1.1 and LdNT1.2 have been identified which show very high affinity towards pyrimidines. It has been seen that point mutations in LdNT1.1 and LdNT2 alter the substrate specificity and confer resistance towards drug (Vasudevan et al. 2001; Galazka et al. 2006). Two purine nucleobase transporters have also been identified from L. major, LmaNT3 and LmaNT4. LmaNT3 transports only bases hypoxanthine, xanthine, adenosine and guanine but not nucleosides whereas, LmaNT4 takes up only adenine at neutral pH but at acidic pH, it can take up hypoxanthine, guanine and xanthine as well (Ortiz et al. 2009). Its function is required for the optimal viability of the parasite inside the acidic phagolysosome of human macrophages. The parasitic transporters are different from the mammalian transporters in terms of their higher specificity towards the substrate. The fact that there are so many pathways for the uptake of purines; it is very difficult to target them with selective inhibitors. But, they will remain pharmacologically important as these transporters also uptake toxic nucleoside analogs which are inhibitory to the cell growth.
Purine Salvage enzymes
In Leishmania, the enzyme phosphoribosyltransferase (PRT), that converts dephosphorylated purines to nucleosides monophosphates, plays an important role in salvage of purines. Three PRTs have been identified and characterized from Leishmania (Glew et al. 1988) namely, adenine phosphoribosyl transferase (APRT) (EC 2.4.2.7), hypoxanthine-guanine phosphoribosyl transferase (HGPRT) (EC 2.4.2.8) and xanthine phosphoribosyl transferase (XPRT) (2.4.2.22). HGPRT converts hypoxanthine to inosine monophosphate and guanine to guanine monophosphate. Various inhibitors have been designed to target HGPRT due to its difference in substrate specificity with the host enzyme. The most common inhibitor used is allopurinol that is phosphorylated by HGPRT and incorporated into nucleic acid thus leading to selective death of the parasite (Marr 1983; Fish et al. 1985). Allopurinol has been shown to be effective against cutaneous (Martinez and Marr 1992) and visceral Leishmaniasis (Kager et al. 1981). Allopurinol when used with other anti-leishmanial drugs was found to be even more effective. Phthalic anhydride derivative (structural analogs of purine bases), TF1 and phthalimide derivative, TF2, have also been shown to be effective against Leishmania (Somoza et al. 1998). However, it has been found that PRTs are not essential for parasite’s survival. This is possible since there are various alternative purine salvage pathways present in Leishmania. Therefore, it is necessary to target more than one enzyme at a time to come up with an anti-leishmanial chemotherapy.
GPI biosynthetic pathway as drug target
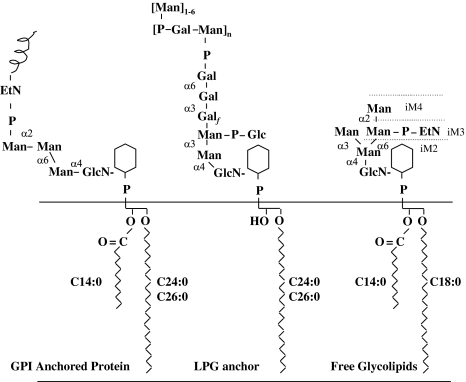
Glycosylphosphatidylinositol (GPI) glycolipids are major cell surface constituents in the Leishmania parasites that act as anchor to various cell surface glycoproteins. The cell surface of the promastigotes is coated by glycocalyx which consists of GPI anchored glycoproteins, GPI-anchored lipophosphoglycan (LPG) and a family of free GPIs, called as glycoinositolphospholipids (GIPLs), in high densities (Fig. 4). They protect the parasite from the alternate complement pathway and external hydrolases. LPG is essential for the infectivity of L. major promastigotes in both the mammalian and insect hosts (Spath et al. 2000; Sacks et al. 2000). The core glycan unit of all protein linked GPIs is conserved and has the structure (Ethanolamine-P-Manα1-2Manα1-6Manα1-4GlcNH2), which is linked to the 6-position of d-myo-inositol ring of phosphatidylinositol (PI) (Ferguson et al. 1999). GPI anchors are synthesized in a step-wise manner in endoplasmic reticulum membrane and then attached to the nascent proteins. These GPI anchored proteins are then transferred to the plasma membrane via the secretory pathway. GPI biosynthesis is essential for bloodstream form T. brucei parasites and has been validated as a chemotherapeutic target (Ferguson 2000).
Fig. 4.
Structure of the major GPI classes synthesized by L. mexicana promastigotes. These glycolipids act as anchors for major cell surface proteins (GPI anchored proteins), surface lipophosphoglycans (LPGs) and free glycolipids (GIPLs). M2–4 denotes GPIs with the structures Man2–4GlcN-PI; EP denotes the presence of an EtN-P cap or side chain; and the prefix i denotes the presence of an α1-3-linked mannose in the core structure
The first step in GPI biosynthesis is the formation of GlcNAc-PI, by the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to PI. The reaction is catalyzed by the protein complex, GPI-N-acetylglucosaminyltransferase (GPI-GnT). Then, GlcNAc-PI is de-N-acetylated to form GlcN-PI, catalyzed by GlcNAc-PI-de-N-acetylase. This is an essential step for all GPI biosynthetic pathways. Trypanosomal enzyme has been shown to be zinc metalloenzyme (Urbaniak et al. 2005). The enzymes from T. brucei (Sharma et al. 1997) and L. major (Smith et al. 1997) showed narrow substrate specificity than the human homolog in vitro. Substrate specificity was determined using substrate analogs of GlcNAc-PI. The substrate analog GlcNR-PI where R is an acyl group larger than propionyl group were not de-N-acetylated by the enzyme but when this group was substituted with benzoyl group, the analog was shown to inhibit the parasitic enzyme and not the human counterpart. The difference in the substrate specificities of the two enzymes has been exploited and two synthetic GlcNAc-PI analogs, GlcNCONH2-β-PI and GlcNCONH2-(2-O-octyl)-PI were shown to be T. brucei specific suicide inhibitors (Smith et al. 2001).
Differences between the parasitic and mammalian GPI biosynthetic pathways occur from GlcN-PI onwards, including the timing of inositol acylation and deacylation. The next step in GPI biosynthesis in case of parasites is mannosylation of GlcN-PI which is followed by inositol acylation, whereas, in yeast and human, inositol acylation is a pre-requisite for mannosylation. The trimannosyl core of GPI anchors is added stepwise and involves three distinct mannosyl transferases (MT). Mannosyltransferase III (MTIII) has been found to be substrate specific. There is a difference in the structure of the substrates for the mammalian and the trypanosomal MTIII which suggests that species-specific inhibition can be achieved by synthesizing small molecule inhibitors (Urbaniak et al. 2008). The mannose donor for all GPI-mannosyl transferases is dolichol-P-mannose (Dol-P-Man). Dol-P-Man is synthesized by the enzyme Dol-P-Man synthase which can be inhibited by the addition of lipopeptide antibiotic amphomycin which forms a complex with Dol-P, and thereby inhibiting GPI biosynthesis. The enzymes MT require divalent cations for their function and can be inhibited in vitro by metal chelators like EDTA. GlcN-PI analog, GlcN-(2-O-hexadecyl)PI inhibits specifically the first mannosyltransferase in T. brucei in a competitive manner.
The next step is the acylation of inositol. In T. brucei, it can be inhibited in vivo as well as in vitro using a serine esterase inhibitor phenylmethylsulphonyl fluoride (PMSF) leading to accumulation of the Man3-GlcN-PI intermediate in T. brucei. This inhibition of trypanosomal inositol acylation was also observed in vitro with either GlcN-(2-O-methyl)-PI or GlcN-(2-O-octyl)-PI (Smith et al. 1999). The final step of synthesis of GPI core glycan structure, involves transfer of ethanolamine phosphate to 6th position of the third mannose. Since, PMSF inhibits the inositol acylation of the intermediate Man3-GlcN-PI to form Man3-GlcN-(acyl)-PI in T. brucei, the addition of ethanolamine phosphate is prevented (Guther et al. 1994). The mammalian enzyme is not inhibited by PMSF. This selective inhibition implies significant differences in the structure of the enzymes which can be exploited for designing specific inhibitors.
After the completion of GPI anchor synthesis, lipid remodeling takes place which involves exchange of fatty acid components with other fatty acid of the lipid moiety or the whole lipid component. T. brucei contains myristate as the major lipid component in its GPI-anchored variable surface glycoproteins. The major product of fatty acid biosynthesis in T. brucei is myristate which is required to be incorporated into GPI anchors and therefore, this pathway can be targeted in T. brucei. Thiolactomycin, an inhibitor of fatty acid synthesis leads to death of parasites. Myristate specific remodeling has been reported for Leishmania GPIs also. Incorporation of myristate analogs into the GPI anchor is toxic to trypanosomes. Therefore, the enzymes of fatty acid remodeling are possible targets for anti-trypanosome chemotherapy. The mature GPI precursor is transferred to the protein by transamidation reaction by a multimeric protein complex, where, COOH terminal signal peptide is replaced by the formation of amide linkage to amino group of ethanolamine phosphate linked to the third mannose of GPI anchor. The protein components differ in different species, suggesting that these differences between the parasite and mammalian enzymes can be exploited for drug designing.
Protein kinases as drug targets
Cyclin dependent kinases (CDKs) are known to play a crucial role in cell division. They have been found to be abnormally regulated in cancer cells and have therefore drawn attention as drug targets. In Leishmania, the cdc-2 related kinase (CRK) family has attracted attention as potential drug targets. They are homologs of CDKs and are thought to be essential for cell cycle progression. Two putative CDKs in L. mexicana, LmexCRK1 and LmexCRK3 (Hassan et al. 2001) have been found to be essential to the promastigotes form of the parasite. Attempts to generate null mutants of CRK3 resulted in change in ploidy of the parasite (Hassan et al. 2001). CRK3 is found to be active throughout the life cycle in L. mexicana (Grant et al. 1998). CRK3 from L. major was able to complement a temperature sensitive cdc-2 mutant in S. pombe. Inhibitors of CRK3, inhibited the growth and replication of L. donovani amastigotes in peritoneal macrophages. These compounds also led to aberrant DNA content and abnormal morphology of the cells as determined by the flow cytometry (Grant et al. 2004).
MAP kinases
Mitogen-activated Protein (MAP) kinases are mediators of signal transduction and important regulators of cell differentiation and cell proliferation in eukaryotic cells. So far, ten MAP kinases have been identified in L. mexicana, of which LmxMKK, LmxMPK and LmxMPK9 (Wiese 1998; Wiese et al. 2003; Bengs et al. 2005) have been studied intensely. Null mutants of both LmxMKK and LmxMPK9 are viable both in amastigote and promastigote stages of the life cycle and therefore, neither of them can be exploited as drug targets. However, null mutants of LmxMPK, has the ability to infect peritoneal macrophages and differentiate into amastigotes, but are unable to proliferate within the parasitophorous vacuole. This phenotype was reverted back by the reintroduction of LmxMPK to the mutant (Wiese 1998). Therefore, LmxMPK is found to be essential for the growth of the more relevant amastigote form and can be used as a drug target.
Proteinases as drug targets
Proteinases are of four main types- cysteine, serine, aspartate and metallo-enzymes. The name is given on the basis of the residue present in the active site. In case of parasitic protozoa, the most identified and characterized are the cysteine proteinases (CPs), which are homologous to mammalian cathepsins. CPs have attracted attention as potential drug targets because of their role in host cell–parasite interaction, as putative virulence factor, and being structurally different from the mammalian homolog.
Analysis of L. major genome database revealed that there are genes encoding for as many as 65 CPs. CPs are further divided into different types. CPA and CPB are both cathepsin L-like proteinases in terms of amino acid sequence. CPC is cathepsin B-like proteinases. CPA, CPB and CPC are involved in host-parasite interaction as determined by gene replacement studies in L. mexicana. L. mexicana deficient in the multicopy CPB gene array (Δcpb) have reduced virulence with poor lesion growth in BALB/c mice (Alexander et al. 1998). A natural inhibitor of CP (ICP) from L. mexicana has been characterized and has been shown to be a potent inhibitor of CPB. BALB/c mice infected with mutants overexpressing ICP were able to resolve the infection faster (Bryson et al. 2009). In another study, T. brucei infected mice were treated with cysteine proteinase inhibitor, carbobenzoxy-phenylalanyl-alanine-diazomethyl ketone (Z-Phe-Ala-CHN2). It led to alteration in cell morphology and was lethal to the cultured parasites (Scory et al. 2007). These studies provide evidence of the therapeutic potential of the inhibitor of cysteine proteinases. In T. brucei, RNAi studies have shown a cathepsin B like protease tbcatB, as a key target (Mackey et al. 2004). Parasites containing single copy of tbcatb show enlargement of lysosome and decreased degradation of endocytosed host proteins (O’Brien et al. 2008). The CPs are also of interest because of their structural differences with the mammalian CPs. The protein tbcatB has been crystallized and its structure confirms an occluding loop which is important for substrate binding (Kerr et al. 2010). This loop creates a larger prime side pocket in active site cleft than is found in mammalian cathepsin B. The difference in the structure of the proteins may be utilized for design of inhibitors.
Folate biosynthesis
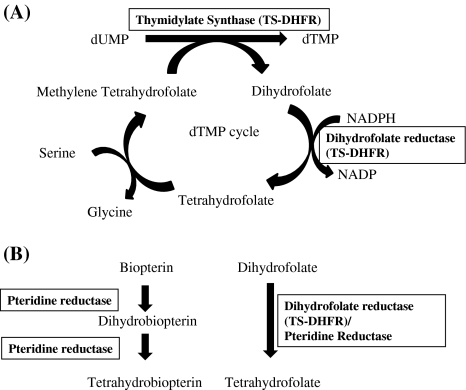
Folate pathway has been of interest as a drug target and has been used in anti-cancer and anti-malarial chemotherapy. Folates are important cofactors used in a variety of metabolic pathways like DNA and RNA synthesis and amino acid metabolism. Since they are essential for growth, the enzymes involved in their synthesis have been of interest as drug targets, particularly, thymidylate synthase (TS) (EC 2.1.1.45) and dihydrofolate reductase (DHFR) (EC 1.5.1.3) which is responsible for converting dihydrofolate to tetrahydrofolate, an important cofactor in the synthesis of thymine. TS and DHFR catalyze sequential reactions to synthesize dTMP (Fig. 5a). Both the enzymes have been studied intensively and used as targets for chemotherapy. Interestingly, TS and DHFR exist on a single polypeptide in case of trypanosomatids, with DHFR domain on amino terminus and TS domain on carboxy terminus. TS-DHFR has been characterized from L. major. The classic inhibitors of DHFR were found to be ineffective against Leishmania (Neal and Croft 1984). An answer to that came from the genetic analysis which showed amplification of the gene pteridine reductase (PTR1) (1.5.1.33) in some of the mutants in Leishmania which were resistant to methotrexate (an inhibitor of DHFR-TS) (Nare et al. 1997). PTR1 can reduce both pterins and folates (Fig. 5b) and is much less susceptible to inhibition by anti-folates, as revealed by structural studies, targeted against DHFR. Therefore, it can act as a bypass system for DHFR-TS. A number of compounds have been screened against PTR1 in L. major (Hardy et al. 1997). Several compounds were found to be inhibiting both DHFR-TS and PTR1. However, only four such compounds were identified that inhibited both the enzymes and the growth of the parasite potently. This indicates that an inhibitor is required that targets both the enzymes simultaneously or two compounds that can be used in combination specifically inhibiting both the enzymes.
Fig. 5.
Folate synthesis pathway. a The figure shows the synthesis of dTMP by the enzyme TS which uses methylene tetrahydrofolate and converts it into dihydrofolate. It is then converted back to tetrahydrofolate by the enzyme DHFR. b The schematic diagram shows that the enzyme pteridine reductase (PTR1) can reduce both pterins and folates
Glyoxalase system
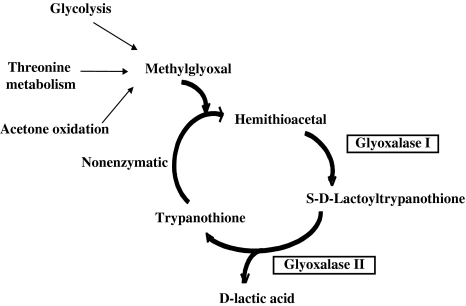
The Glyoxalase system functions to detoxify the cell by removal of toxic and mutagenic intermediate, methylglyoxal, which is mainly formed as a by-product of glycolysis. It is also formed during threonine catabolism and acetone oxidation (Cooper 1984; Vickers et al. 2004). The glyoxalase system comprises of two enzymes viz. glyoxalase I (lactoyl glutathione lyase) (EC 4.4.1.5) and Glyoxalase II (hydroxyacyl glutathione hydroxylase) (EC 3.1.2.6) and uses glutathione as a cofactor. However, trypanosomatids rely on a trypanothione dependent glyoxalase system (Fig. 6) which is unique to these parasites, where glyoxalase I catalyzes the formation of S-d-lactoyltrypanothione from hemithioacetal. Hemithioacetal is formed non-enzymatically from methylglyoxal and reduced trypanothione. S-d-lactoyltrypanothione is further hydrolysed to d-lactate regenerating trypanothione by glyoxalase II (Vickers et al. 2004).
Fig. 6.
Glyoxalase system in trypanosomatids. The glyoxalase system detoxifies the cell by removal of methylglyoxal which is formed as an intermediate during glycolysis, threonine oxidation and acetone oxidation. Methylglyoxal reacts non-enzymatically with trypanothione to form hemithioacetal. This hemithioacetal is used as the substrate by glyoxalase I forming S-d-lactoyltrypanothione. Glyoxalase II further hydrolyses S-d-lactoyltrypanothione to form d-lactate, regenerating trypanothione
Glyoxalase I has been characterized from L. donovani (Padmanabhan et al. 2005) and L. major (Vickers et al. 2004). The enzyme is highly substrate specific as it depends entirely on trypanothione as substrate, instead of glutathione as in their mammalian hosts. Superposition of L. donovani glyoxalase I with crystal structure of E. coli glyoxalase I showed that its substrate binding loop is smaller in comparison to glutathione utilizing enzymes and is devoid of positively charged residues, as observed for human and E. coli homolog (Padmanabhan et al. 2005). L. donovani glyoxalase I has been found to be an essential gene in the parasite (Chauhan and Madhubala 2009). Glyoxalase II was first identified from T. brucei. It is expressed in both procyclic and bloodstream forms of T. brucei (Irsch and Krauth-Siegel 2004). Glyoxalase II has also been characterized from L. donovani (Padmanabhan et al. 2006). Sequence comparison shows that it lacks the basic residues in its active site which are conserved in human homolog. These residues are important for binding to glutathione. The substrate for L. donovani Glyoxalase II is thioester of trypanothione, which is positively charged. Therefore, it cannot be accommodated in the active site of the human enzyme. This observation strongly suggests that the parasitic enzymes are highly substrate specific. Therefore, trypanothione-dependent glyoxalase pathway has drawn attention for additional biochemical and genetic investigation as a possible target for rational drug design.
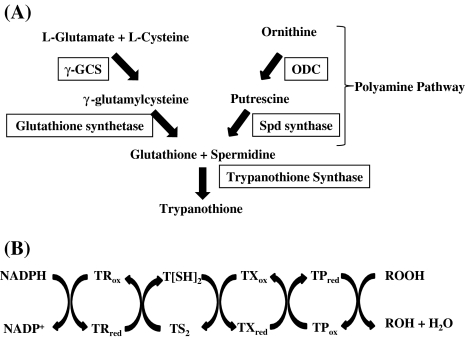
Trypanothione pathway
As discussed above, trypanothione (bis-(glutathionyl) spermidine) is a key molecule against oxidative stress in Trypanosoma and Leishmania. It is not only unique to the parasite but is also crucial in maintaining the cellular redox potential and thus is essential for parasite survival. Trypanothione synthesis is catalyzed by two enzymes, namely, trypanothione synthetase (TS) (EC 6.3.1.9) and trypanothione reductase (TR) (EC 1.8.1.12). TS catalyzes trypanothione synthesis from two molecules of glutathione and spermidine (Fig. 7a). Trypanothione is then maintained in its reduced form by the enzyme TR in the presence of NADPH (Fairlamb et al. 1985). Reduced trypanothione in turn reduces tryparedoxin (TX) followed by reduction of tryparedoxin recycling enzyme tryparedoxin peroxidase (TP) (EC 1.11.1.15) (Fig. 7b). Since, this is the only pathway that is crucially involved in regulating oxidative stress in these parasites; therefore, trypanothione pathway has become the focus of anti-trypanosomatid drug discovery. Both TR and TP have been shown to be important drug targets. Null mutants of TR show attenuated infectivity and decreased capacity to survive within intracellular macrophages (Dumas et al. 1997). TR is similar in sequence and structure to its human counterpart, glutathione reductase. However, the active site of TR shows five non conservative changes in its active site, giving it overall negative charge which can accommodate trypanothione disulfide and gives the enzyme its specificity towards its substrate (Zhang et al. 1996). This difference in substrate specificities has allowed synthesis of specific inhibitors against the parasite. Many lead compounds that inhibit TR have been identified including, polyamine derivatives, tricyclics and aminodiphenyl sulphides (Werbovetz 2000). Trypanothione pathway thus, provides a promising drug target in trypanosomatids.
Fig. 7.
Trypanothione pathway. a The figure shows synthesis of trypanothione from glutathione and spermidine in trypanosomatids. γ-GCS: γ-glutamylcysteine synthetase; ODC ornithine decarboxylase. b Trypanothione peroxidase pathway. Trypanothione is maintained in its reduced form by the enzyme TR which uses NADPH. The reduced trypanothione, T[SH]2 further reduces TX followed by reduction of the enzyme TP. TP catalyses final reduction of peroxides to water or alcohols
Topoisomerases as drug targets
DNA topoisomerases are ubiquitous enzymes that play an important role in many essential processes like DNA replication, transcription, recombination and repair. They are broadly classified as type I and type II topoisomerases that cleave single stranded and double stranded DNA, respectively. DNA topoisomerases have been used as chemotherapeutic targets for anti-bacterial and anti-parasitic diseases. Type I topoisomerase (EC 5.99.1.2) have been characterized from L. donovani and T. cruzi. The enzyme was found to be independent of ATP (Das et al. 2004a). L. donovani topoisomerase I was found to be present in both kinetoplast and nucleus (Das et al. 2004b). Inhibitors of type I topoisomerase include anti-leishmanial compounds such as sodium stibogluconate and urea stibamine. Camptothecin, a plant alkaloid and a known inhibitor of eukaryotic topoisomerase I, was found to be inhibitory to T. brucei, T. cruzi and L. donovani. Analogs of camptothecin have also been screened against trypanosomes and the structural motifs have been identified which specifically inhibit the parasitic topoisomerase I with more potency (Bodley et al. 1995). Topoisomerase II (EC 5.99.1.3) has been identified from T. brucei (Strauss and Wang 1990), T. cruzi (Fragoso and Goldenberg 1992) and L. donovani (Das et al. 2001). Interestingly, the topoisomerase II from T. brucei and L. donovani were found to contain both ATP dependent and independent activities. Topoisomerase II inhibitor, 9-anilinoacridine (used as anti-tumor agent) and other acridine derivatives inhibited Leishmania and Trypanosoma strongly (Figgitt et al. 1992). Dihydrobetulinic acid (DHBA), a derivative of betulinic acid (which is a pentacyclic triterpenoid) has been reported to be active against both Topoisomerase I and topoisomerase II from L. donovani (Chowdhury et al. 2003). Three isoflavanoids, 8-prenylmucronulatol, lyasperin H and smiranicin have been found to be anti-leishmanial and this activity has been correlated to kDNA linearization and inhibition of topoisomerase II (Salem and Werbovetz 2005). However, their use is limited due to their low cell toxicity. The structure of L. donovani topoisomerase I bound to nicked DNA captured as a vanadate complex has been elucidated (Davies et al. 2006). The structural analysis of these enzymes will give us an insight into their catalytic mechanisms and will also enable us to design specific inhibitors against Leishmania.
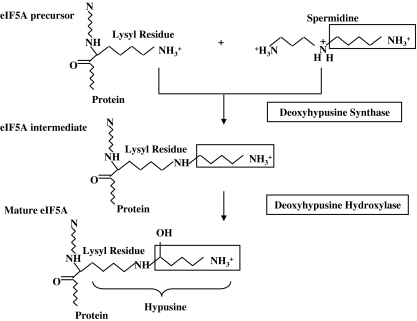
Hypusine pathway
Hypusine (Nε-(4-amino-2-hydroxybutyl) lysine), an unusual amino acid derived from the polyamine spermidine, is present in all the eukaryotes. Its name is derived from its two structural components, hydroxyputrescine and lysine (Shiba et al. 1971). It is synthesized as a result of post-translational modification occurring exclusively on one cellular protein, eukaryotic initiation factor 5A (eIF5A). It is formed in two enzymatic steps (Park et al. 1982) (Fig. 8). The first step is catalyzed by the enzyme deoxyhypusine synthase (DHS) (EC 2.5.1.46) which catalyses the NAD+ dependent transfer of the 4-aminobutyl moiety of spermidine to a specific lysine residue of the eIF5A precursor protein to form an intermediate, deoxyhypusine (Chen and Dou 1988; Murphey and Gerner 1987). This intermediate is subsequently hydroxylated by the enzyme deoxyhypusine hydroxylase (DOHH) (EC 1.14.99.29) (Abbruzzese et al. 1986) which completes the synthesis of hypusine and maturation of eIF5A.
Fig. 8.
Hypusine biosynthetic pathway. Hypusine is formed on the protein eIF5A by the subsequent action of enzymes DHS and DOHH
Hypusination of eIF5A is necessary for its function and cell viability. Disruption of two eIF5A genes (TIF51A and TIF51B) (Schnier et al. 1991) and DHS gene (Park et al. 1998) from Saccharomyces cerevisiae produces a lethal phenotype. Inhibitors of hypusine biosynthetic enzymes, DHS (spermidine analogs) and DOHH (metal chelators) have been shown to exert anti-proliferative effects in mammalian cells including cancer cell lines (Park et al. 1994; Nishimura et al. 2005). Recently, we have shown that hypusine biosynthesis occurs in Leishmania donovani. Interestingly, Leishmania has two genes containing DHS domains viz., DHS-like gene (DHSL20) and DHS34 gene, of which only DHS34 protein was found to contain functional activity in vitro. Null mutants of DHS34 gene could not be generated implying that the gene is essential to the parasite’s survival (Chawla et al. 2010). Structural modeling of DHS34 protein showed that one of the NAD+ binding domains lies in a 225-amino acid long insertion present in DHS34 protein as compared to the human homolog. The recombinant DHS34 remained uninhibited by most potent inhibitors of the human enzyme like GC7 and other spermidine analogs, indicating that there is a difference between the spermidine binding sites of both the enzymes which could be exploited to design inhibitors.
Concluding remarks
Over the last decade, there has been a burst of activity towards the development of efficient drugs against the parasitic diseases including leishmaniasis. Target based drug discovery is being employed rather than the random screening of compounds, as the former approach produces more specific results. Many technological advances have been made, which has led to an extensive search for identification of novel drug targets. In the post-genomic era, a lot of proteins, enzymes, metabolites and pathways have been identified in trypanosomatids which are unique to these pathogens and could be exploited as drug targets. However, as outlined in the review, it is very critical to identify the suitable proteins or other metabolites to be used as drug targets. Several biochemical pathways have been discussed here which have been identified as targets for anti-leishmanial drug discovery. Glycolytic pathway, sterol biosynthesis and trypanothione pathway are quite distinct in Leishmania as compared to its mammalian host and therefore have been of interest for drug discovery. We have also identified a novel hypusine biosynthetic pathway in L. donovani. The enzyme DHS was found to be essential to the parasite and structurally different from the human homolog. The identified drug targets can then be used for high throughput screening with chemical libraries for which development of cheap and efficient assay system is required. Furthermore, refinement in the structure of the available compounds is required to synthesise more specific and effective inhibitors for these metabolic pathways. We have to combine all the technology and the information available with us to come up with more drug targets and eventually, with much needed new drugs to combat this deadly disease.
Acknowledgments
Rentala Madhubala is supported by a grant from Council of Scientific and Industrial Research (CSIR), India for the ongoing project related to hypusine biosynthesis. Bhavna Chawla is also supported by CSIR, India.
References
- Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization. J Biol Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- Alexander J, Coombs GH, Mottram JC. Leishmania mexicana cysteine proteinase-deficient mutants have attenuated virulence for mice and potentiate a Th1 response. J Immunol. 1998;161:6794–6801. [PubMed] [Google Scholar]
- Aronov AM, Suresh S, Buckner FS, Voorhis WC, Verlinde CL, Opperdoes FR, Hol WG, Gelb MH. Structure-based design of submicromolar, biologically active inhibitors of trypanosomatid glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1999;96:4273–4278. doi: 10.1073/pnas.96.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MP, Mottram JC, Coombs GH. Recent advances in identifying and validating drug targets in trypanosomes and leishmanias. Trends Microbiol. 1999;7:82–88. doi: 10.1016/S0966-842X(98)01433-4. [DOI] [PubMed] [Google Scholar]
- Bengs F, Scholz A, Kuhn D, Wiese M. LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol Microbiol. 2005;55:1606–1615. doi: 10.1111/j.1365-2958.2005.04498.x. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Michels PA, Hol WG. Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation. Nature. 1997;385:275–278. doi: 10.1038/385275a0. [DOI] [PubMed] [Google Scholar]
- Bodley AL, Wani MC, Wall ME, Shapiro TA. Antitrypanosomal activity of camptothecin analogs. Structure–activity correlations. Biochem Pharmacol. 1995;50:937–942. doi: 10.1016/0006-2952(95)00215-L. [DOI] [PubMed] [Google Scholar]
- Bryson K, Besteiro S, McGachy HA, Coombs GH, Mottram JC, Alexander J. Overexpression of the natural inhibitor of cysteine peptidases in Leishmania mexicana leads to reduced virulence and a Th1 response. Infect Immun. 2009;77:2971–2978. doi: 10.1128/IAI.00558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NS, Drew ME, Sanchez M, Vasudevan G, Landfear SM, Ullman B. Cloning of a novel inosine-guanosine transporter gene from Leishmania donovani by functional rescue of a transport-deficient mutant. J Biol Chem. 2000;275:20935–20941. doi: 10.1074/jbc.M002418200. [DOI] [PubMed] [Google Scholar]
- Chauhan SC, Madhubala R. Glyoxalase I gene deletion mutants of Leishmania donovani exhibit reduced methylglyoxal detoxification. PLoS One. 2009;4:e6805. doi: 10.1371/journal.pone.0006805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla B, Jhingran A, Singh S, Tyagi N, Park MH, Srinivasan N, Roberts SC, Madhubala R. Identification and characterization of a novel deoxyhypusine synthase in Leishmania donovani. J Biol Chem. 2010;285:453–463. doi: 10.1074/jbc.M109.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Dou QP. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 1988;229:325–328. doi: 10.1016/0014-5793(88)81149-9. [DOI] [PubMed] [Google Scholar]
- Chowdhury AR, Mandal S, Goswami A, Ghosh M, Mandal L, Chakraborty D, Ganguly A, Tripathi G, Mukhopadhyay S, Bandyopadhyay S, Majumder HK. Dihydrobetulinic acid induces apoptosis in Leishmania donovani by targeting DNA topoisomerase I and II: implications in antileishmanial therapy. Mol Med. 2003;9:26–36. [PMC free article] [PubMed] [Google Scholar]
- Chudzik DM, Michels PA, de WS, Hol WG. Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases. J Mol Biol. 2000;300:697–707. doi: 10.1006/jmbi.2000.3910. [DOI] [PubMed] [Google Scholar]
- Cooper RA. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol. 1984;38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- Das A, Dasgupta A, Sharma S, Ghosh M, Sengupta T, Bandopadhyay S, Majumder HK. Characterisation of the gene encoding type II DNA topoisomerase from Leishmania donovani: a key molecular target in antileishmanial therapy. Nucleic Acids Res. 2001;29:1844–1851. doi: 10.1093/nar/29.9.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Dasgupta A, Sengupta T, Majumder HK. Topoisomerases of kinetoplastid parasites as potential chemotherapeutic targets. Trends Parasitol. 2004;20:381–387. doi: 10.1016/j.pt.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Das BB, Sen N, Ganguly A, Majumder HK. Reconstitution and functional characterization of the unusual bi-subunit type I DNA topoisomerase from Leishmania donovani. FEBS Lett. 2004;565:81–88. doi: 10.1016/j.febslet.2004.03.078. [DOI] [PubMed] [Google Scholar]
- Davies DR, Mushtaq A, Interthal H, Champoux JJ, Hol WG. The structure of the transition state of the heterodimeric topoisomerase I of Leishmania donovani as a vanadate complex with nicked DNA. J Mol Biol. 2006;357:1202–1210. doi: 10.1016/j.jmb.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Docampo R, Moreno SN. The acidocalcisome as a target for chemotherapeutic agents in protozoan parasites. Curr Pharm Des. 2008;14:882–888. doi: 10.2174/138161208784041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, Olivier M, Papadopoulou B. Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J. 1997;16:2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- Ferguson MA. Glycosylphosphatidylinositol biosynthesis validated as a drug target for African sleeping sickness. Proc Natl Acad Sci USA. 2000;97:10673–10675. doi: 10.1073/pnas.97.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MA, Brimacombe JS, Brown JR, Crossman A, Dix A, Field RA, Guther ML, Milne KG, Sharma DK, Smith TK. The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim Biophys Acta. 1999;1455:327–340. doi: 10.1016/s0925-4439(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Fernandes Rodrigues JC, Concepcion JL, Rodrigues C, Caldera A, Urbina JA, de Souza W. In vitro activities of ER-119884 and E5700, two potent squalene synthase inhibitors, against Leishmania amazonensis: antiproliferative, biochemical, and ultrastructural effects. Antimicrob Agents Chemother. 2008;52:4098–4114. doi: 10.1128/AAC.01616-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figgitt D, Denny W, Chavalitshewinkoon P, Wilairat P, Ralph R. In vitro study of anticancer acridines as potential antitrypanosomal and antimalarial agents. Antimicrob Agents Chemother. 1992;36:1644–1647. doi: 10.1128/aac.36.8.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish WR, Marr JJ, Berens RL, Looker DL, Nelson DJ, LaFon SW, Balber AE. Inosine analogs as chemotherapeutic agents for African trypanosomes: metabolism in trypanosomes and efficacy in tissue culture. Antimicrob Agents Chemother. 1985;27:33–36. doi: 10.1128/aac.27.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso SP, Goldenberg S. Cloning and characterization of the gene encoding Trypanosoma cruzi DNA topoisomerase II. Mol Biochem Parasitol. 1992;55:127–134. doi: 10.1016/0166-6851(92)90133-5. [DOI] [PubMed] [Google Scholar]
- Galazka J, Carter NS, Bekhouche S, Arastu-Kapur S, Ullman B. Point mutations within the LdNT2 nucleoside transporter gene from Leishmania donovani confer drug resistance and transport deficiency. Int J Biochem Cell Biol. 2006;38:1221–1229. doi: 10.1016/j.biocel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Glew RH, Saha AK, Das S, Remaley AT. Biochemistry of the Leishmania species. Microbiol Rev. 1988;52:412–432. doi: 10.1128/mr.52.4.412-432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KM, Hassan P, Anderson JS, Mottram JC. The crk3 gene of Leishmania mexicana encodes a stage-regulated cdc2-related histone H1 kinase that associates with p12. J Biol Chem. 1998;273:10153–10159. doi: 10.1074/jbc.273.17.10153. [DOI] [PubMed] [Google Scholar]
- Grant KM, Dunion MH, Yardley V, Skaltsounis AL, Marko D, Eisenbrand G, Croft SL, Meijer L, Mottram JC. Inhibitors of Leishmania mexicana CRK3 cyclin-dependent kinase: chemical library screen and antileishmanial activity. Antimicrob Agents Chemother. 2004;48:3033–3042. doi: 10.1128/AAC.48.8.3033-3042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guther ML, Masterson WJ, Ferguson MA. The effects of phenylmethylsulfonyl fluoride on inositol-acylation and fatty acid remodeling in African trypanosomes. J Biol Chem. 1994;269:18694–18701. [PubMed] [Google Scholar]
- Hardy LW, Matthews W, Nare B, Beverley SM. Biochemical and genetic tests for inhibitors of Leishmania pteridine pathways. Exp Parasitol. 1997;87:157–169. doi: 10.1006/expr.1997.4207. [DOI] [PubMed] [Google Scholar]
- Hassan P, Fergusson D, Grant KM, Mottram JC. The CRK3 protein kinase is essential for cell cycle progression of Leishmania mexicana. Mol Biochem Parasitol. 2001;113:189–198. doi: 10.1016/S0166-6851(01)00220-1. [DOI] [PubMed] [Google Scholar]
- Irsch T, Krauth-Siegel RL. Glyoxalase II of African trypanosomes is trypanothione-dependent. J Biol Chem. 2004;279:22209–22217. doi: 10.1074/jbc.M401240200. [DOI] [PubMed] [Google Scholar]
- Ishida K, Rodrigues JC, Ribeiro MD, Vila TV, de SW, Urbina JA, Nakamura CV, Rozental S. Growth inhibition and ultrastructural alterations induced by Delta24(25)-sterol methyltransferase inhibitors in Candida spp. isolates, including non-albicans organisms. BMC Microbiol. 2009;9:74. doi: 10.1186/1471-2180-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De GJ, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O’neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kager PA, Rees PH, Wellde BT, Hockmeyer WT, Lyerly WH. Allopurinol in the treatment of visceral leishmaniasis. Trans R Soc Trop Med Hyg. 1981;75:556–559. doi: 10.1016/0035-9203(81)90198-X. [DOI] [PubMed] [Google Scholar]
- Kerr ID, Wu P, Marion-Tsukamaki R, Mackey ZB, Brinen LS. Crystal Structures of TbCatB and rhodesain, potential chemotherapeutic targets and major cysteine proteases of Trypanosoma brucei. PLoS Negl Trop Dis. 2010;4:e701. doi: 10.1371/journal.pntd.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Feil IK, Verlinde CL, Petra PH, Hol WG. Crystal structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Leishmania mexicana: implications for structure-based drug design and a new position for the inorganic phosphate binding site. Biochemistry. 1995;34:14975–14986. doi: 10.1021/bi00046a004. [DOI] [PubMed] [Google Scholar]
- Lorente SO, Rodrigues JC, Jimenez JC, Joyce-Menekse M, Rodrigues C, Croft SL, Yardley V, Luca-Fradley K, Ruiz-Perez LM, Urbina J, de Souza W, Gonzalez PD, Gilbert IH. Novel azasterols as potential agents for treatment of leishmaniasis and trypanosomiasis. Antimicrob Agents Chemother. 2004;48:2937–2950. doi: 10.1128/AAC.48.8.2937-2950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey ZB, O’Brien TC, Greenbaum DC, Blank RB, McKerrow JH. A cathepsin B-like protease is required for host protein degradation in Trypanosoma brucei. J Biol Chem. 2004;279:48426–48433. doi: 10.1074/jbc.M402470200. [DOI] [PubMed] [Google Scholar]
- Magaraci F, Jimenez CJ, Rodrigues C, Rodrigues JC, Braga MV, Yardley V, Luca-Fradley K, Croft SL, de Souza W, Ruiz-Perez LM, Urbina J, Gonzalez PD, Gilbert IH. Azasterols as inhibitors of sterol 24-methyltransferase in Leishmania species and Trypanosoma cruzi. J Med Chem. 2003;46:4714–4727. doi: 10.1021/jm021114j. [DOI] [PubMed] [Google Scholar]
- Marr JJ. Pyrazolopyrimidine metabolism in Leishmania and trypanosomes: significant differences between host and parasite. J Cell Biochem. 1983;22:187–196. doi: 10.1002/jcb.240220307. [DOI] [PubMed] [Google Scholar]
- Martin MB, Grimley JS, Lewis JC, Heath HT, III, Bailey BN, Kendrick H, Yardley V, Caldera A, Lira R, Urbina JA, Moreno SN, Docampo R, Croft SL, Oldfield E. Bisphosphonates inhibit the growth of Trypanosoma brucei, Trypanosoma cruzi, Leishmania donovani, Toxoplasma gondii, and Plasmodium falciparum: a potential route to chemotherapy. J Med Chem. 2001;44:909–916. doi: 10.1021/jm0002578. [DOI] [PubMed] [Google Scholar]
- Martinez S, Marr JJ. Allopurinol in the treatment of American cutaneous leishmaniasis. N Engl J Med. 1992;326:741–744. doi: 10.1056/NEJM199203123261105. [DOI] [PubMed] [Google Scholar]
- Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J Biol Chem. 1987;262:15033–15036. [PubMed] [Google Scholar]
- Myler PJ. Searching the Tritryp genomes for drug targets. Adv Exp Med Biol. 2008;625:133–140. doi: 10.1007/978-0-387-77570-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nare B, Luba J, Hardy LW, Beverley S. New approaches to Leishmania chemotherapy: pteridine reductase 1 (PTR1) as a target and modulator of antifolate sensitivity. Parasitology. 1997;114(Suppl):S101–S110. [PubMed] [Google Scholar]
- Neal RA, Croft SL. An in vitro system for determining the activity of compounds against the intracellular amastigote form of Leishmania donovani. J Antimicrob Chemother. 1984;14:463–475. doi: 10.1093/jac/14.5.463. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K. Independent roles of eIF5A and polyamines in cell proliferation. Biochem J. 2005;385:779–785. doi: 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TC, Mackey ZB, Fetter RD, Choe Y, O’Donoghue AJ, Zhou M, Craik CS, Caffrey CR, McKerrow JH. A parasite cysteine protease is key to host protein degradation and iron acquisition. J Biol Chem. 2008;283:28934–28943. doi: 10.1074/jbc.M805824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes FR. Compartmentation of carbohydrate metabolism in trypanosomes. Annu Rev Microbiol. 1987;41:127–151. doi: 10.1146/annurev.mi.41.100187.001015. [DOI] [PubMed] [Google Scholar]
- Ortiz D, Sanchez MA, Koch HP, Larsson HP, Landfear SM. An acid-activated nucleobase transporter from Leishmania major. J Biol Chem. 2009;284:16164–16169. doi: 10.1074/jbc.M109.006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan PK, Mukherjee A, Singh S, Chattopadhyaya S, Gowri VS, Myler PJ, Srinivasan N, Madhubala R. Glyoxalase I from Leishmania donovani: a potential target for anti-parasite drug. Biochem Biophys Res Commun. 2005;337:1237–1248. doi: 10.1016/j.bbrc.2005.09.179. [DOI] [PubMed] [Google Scholar]
- Padmanabhan PK, Mukherjee A, Madhubala R. Characterization of the gene encoding glyoxalase II from Leishmania donovani: a potential target for anti-parasite drugs. Biochem J. 2006;393:227–234. doi: 10.1042/BJ20050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. The biosynthesis of protein-bound hypusine (N epsilon-(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon-(4-aminobutyl)lysine) J Biol Chem. 1982;257:7217–7222. [PubMed] [Google Scholar]
- Park MH, Wolff EC, Lee YB, Folk JE. Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J Biol Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- Park MH, Joe YA, Kang KR. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- Pink R, Hudson A, Mouries MA, Bendig M. Opportunities and challenges in antiparasitic drug discovery. Nat Rev Drug Discov. 2005;4:727–740. doi: 10.1038/nrd1824. [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Phillips SE, Michels PA, Fothergill-Gilmore LA. The structure of pyruvate kinase from Leishmania mexicana reveals details of the allosteric transition and unusual effector specificity. J Mol Biol. 1999;291:615–635. doi: 10.1006/jmbi.1999.2918. [DOI] [PubMed] [Google Scholar]
- Rodrigues JC, Attias M, Rodriguez C, Urbina JA, Souza W. Ultrastructural and biochemical alterations induced by 22,26-azasterol, a delta(24(25))-sterol methyltransferase inhibitor, on promastigote and amastigote forms of Leishmania amazonensis. Antimicrob Agents Chemother. 2002;46:487–499. doi: 10.1128/AAC.46.2.487-499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DL, Modi G, Rowton E, Spath G, Epstein L, Turco SJ, Beverley SM. The role of phosphoglycans in Leishmania–sand fly interactions. Proc Natl Acad Sci USA. 2000;97:406–411. doi: 10.1073/pnas.97.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MM, Werbovetz KA. Antiprotozoal compounds from Psorothamnus polydenius. J Nat Prod. 2005;68:108–111. doi: 10.1021/np049682k. [DOI] [PubMed] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scory S, Stierhof YD, Caffrey CR, Steverding D. The cysteine proteinase inhibitor Z-Phe-Ala-CHN2 alters cell morphology and cell division activity of Trypanosoma brucei bloodstream forms in vivo. Kinetoplastid Biol Dis. 2007;6:2. doi: 10.1186/1475-9292-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Smith TK, Crossman A, Brimacombe JS, Ferguson MA. Substrate specificity of the N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase of glycosylphosphatidylinositol membrane anchor biosynthesis in African trypanosomes and human cells. Biochem J. 1997;328(Pt 1):171–177. doi: 10.1042/bj3280171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Mizote H, Kaneko T, Nakajima T, Kakimoto Y. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim Biophys Acta. 1971;244:523–531. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- Smith TK, Sharma DK, Crossman A, Dix A, Brimacombe JS, Ferguson MA. Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J. 1997;16:6667–6675. doi: 10.1093/emboj/16.22.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Sharma DK, Crossman A, Brimacombe JS, Ferguson MA. Selective inhibitors of the glycosylphosphatidylinositol biosynthetic pathway of Trypanosoma brucei. EMBO J. 1999;18:5922–5930. doi: 10.1093/emboj/18.21.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Crossman A, Borissow CN, Paterson MJ, Dix A, Brimacombe JS, Ferguson MA. Specificity of GlcNAc-PI de-N-acetylase of GPI biosynthesis and synthesis of parasite-specific suicide substrate inhibitors. EMBO J. 2001;20:3322–3332. doi: 10.1093/emboj/20.13.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza JR, Skillman AG, Jr, Munagala NR, Oshiro CM, Knegtel RM, Mpoke S, Fletterick RJ, Kuntz ID, Wang CC. Rational design of novel antimicrobials: blocking purine salvage in a parasitic protozoan. Biochemistry. 1998;37:5344–5348. doi: 10.1021/bi973095z. [DOI] [PubMed] [Google Scholar]
- Spath GF, Epstein L, Leader B, Singer SM, Avila HA, Turco SJ, Beverley SM. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc Natl Acad Sci USA. 2000;97:9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss PR, Wang JC. The TOP2 gene of Trypanosoma brucei: a single-copy gene that shares extensive homology with other TOP2 genes encoding eukaryotic DNA topoisomerase II. Mol Biochem Parasitol. 1990;38:141–150. doi: 10.1016/0166-6851(90)90214-7. [DOI] [PubMed] [Google Scholar]
- Suresh S, Turley S, Opperdoes FR, Michels PA, Hol WG. A potential target enzyme for trypanocidal drugs revealed by the crystal structure of NAD-dependent glycerol-3-phosphate dehydrogenase from Leishmania mexicana. Structure. 2000;8:541–552. doi: 10.1016/S0969-2126(00)00135-0. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Crossman A, Chang T, Smith TK, Aalten DM, Ferguson MA. The N-acetyl-d-glucosaminylphosphatidylinositol De-N-acetylase of glycosylphosphatidylinositol biosynthesis is a zinc metalloenzyme. J Biol Chem. 2005;280:22831–22838. doi: 10.1074/jbc.M502402200. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Yashunsky DV, Crossman A, Nikolaev AV, Ferguson MA. Probing enzymes late in the trypanosomal glycosylphosphatidylinositol biosynthetic pathway with synthetic glycosylphosphatidylinositol analogues. ACS Chem Biol. 2008;3:625–634. doi: 10.1021/cb800143w. [DOI] [PubMed] [Google Scholar]
- Urbina JA, Concepcion JL, Caldera A, Payares G, Sanoja C, Otomo T, Hiyoshi H. In vitro and in vivo activities of E5700 and ER-119884, two novel orally active squalene synthase inhibitors, against Trypanosoma cruzi. Antimicrob Agents Chemother. 2004;48:2379–2387. doi: 10.1128/AAC.48.7.2379-2387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier-Santos MA, Urbina JA, Martiny A, Neves A, de SW. Alterations induced by the antifungal compounds ketoconazole and terbinafine in Leishmania. J Eukaryot Microbiol. 1995;42:337–346. doi: 10.1111/j.1550-7408.1995.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Vasudevan G, Carter NS, Drew ME, Beverley SM, Sanchez MA, Seyfang A, Ullman B, Landfear SM. Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc Natl Acad Sci USA. 1998;95:9873–9878. doi: 10.1073/pnas.95.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan G, Ullman B, Landfear SM. Point mutations in a nucleoside transporter gene from Leishmania donovani confer drug resistance and alter substrate selectivity. Proc Natl Acad Sci USA. 2001;98:6092–6097. doi: 10.1073/pnas.101537298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellieux FM, Hajdu J, Verlinde CL, Groendijk H, Read RJ, Greenhough TJ, Campbell JW, Kalk KH, Littlechild JA, Watson HC, et al. Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data. Proc Natl Acad Sci USA. 1993;90:2355–2359. doi: 10.1073/pnas.90.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinde CL, Hol WG. Structure-based drug design: progress, results and challenges. Structure. 1994;2:577–587. doi: 10.1016/S0969-2126(00)00060-5. [DOI] [PubMed] [Google Scholar]
- Vickers TJ, Greig N, Fairlamb AH. A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major. Proc Natl Acad Sci USA. 2004;101:13186–13191. doi: 10.1073/pnas.0402918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas J, Urbina JA, de SW. Ultrastructural alterations in Trypanosoma (Schizotrypanum) cruzi induced by Delta(24(25)) sterol methyl transferase inhibitors and their combinations with ketoconazole. Int J Antimicrob Agents. 1996;7:235–240. doi: 10.1016/S0924-8579(96)00325-1. [DOI] [PubMed] [Google Scholar]
- Werbovetz KA. Target-based drug discovery for malaria, leishmaniasis, and trypanosomiasis. Curr Med Chem. 2000;7:835–860. doi: 10.2174/0929867003374615. [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Noble ME, Vriend G, Nauche S, Hol WG. Refined 1.83 Å structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. J Mol Biol. 1991;220:995–1015. doi: 10.1016/0022-2836(91)90368-G. [DOI] [PubMed] [Google Scholar]
- Wiese M. A mitogen-activated protein (MAP) kinase homologue of Leishmania mexicana is essential for parasite survival in the infected host. EMBO J. 1998;17:2619–2628. doi: 10.1093/emboj/17.9.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese M, Kuhn D, Grunfelder CG. Protein kinase involved in flagellar-length control. Eukaryot Cell. 2003;2:769–777. doi: 10.1128/EC.2.4.769-777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Zeelen JP, Neubauer G, Vriend G, Backmann J, Michels PA, Lambeir AM, Wierenga RK. Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein Eng. 1999;12:243–250. doi: 10.1093/protein/12.3.243. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bond CS, Bailey S, Cunningham ML, Fairlamb AH, Hunter WN. The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 Å resolution. Protein Sci. 1996;5:52–61. doi: 10.1002/pro.5560050107. [DOI] [PMC free article] [PubMed] [Google Scholar]