Abstract
Carboxyl-terminal binding protein 1 (CtBP1) is a transcriptional co-repressor and metabolic sensory protein, which often represses tumor suppressor genes. Hence, we sought to determine if CtBP1 affects expression of the tumor suppressor Brca1 in head and neck tissue, as down-regulation of Brca1 begins at the early stages of head and neck squamous cell carcinomas (HNSCCs). We found that CtBP1 represses Brca1 transcription by binding to the E2F4 site of the Brca1 promoter. Additionally, the recruitment of CtBP1 to the Brca1 promoter is redox-dependent, i.e., increased at high NADH levels in hypoxic conditions. Further, immunostaining using a human HNSCC tissue array revealed that nuclear CtBP1 staining began to accumulate in hyperplasic lesions and HNSCCs, this staining correlated with Brca1 down-regulation in these lesions. Pharmacological disruption of CtBP1 binding to Brca1 promoter by the antioxidant Tempol, which reduces NADH levels, relieved CtBP1-mediated repression of Brca1, leading to increased DNA repair in HNSCC cells. Since tumor cells are generally hypoxic with increased NADH levels, the dynamic control of Brca1 by a "metabolic switch" found in this study not only provides an important link between tumor metabolism and tumor suppressor expression, but also suggests a potential chemo preventative or therapeutic strategy for HNSCC via blocking NADH-dependent CtBP1 activity at early stages of HNSCC carcinogenesis.
Keywords: Brca1, CtBP1, NADH, transcription, tumor suppressor, HNSCC
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide (Hunter et al., 2005). Despite advances in cancer biology and therapy, the 5-year survival for patients with HNSCC has remained 50% for the past 20 years (Forastiere et al., 2001). To date, the best-studied germline mutations leading to increased HNSCC susceptibility are in genes belonging to the Fanconi anemia/Brca (Fanc/Brca) pathway (Kutler et al., 2003). Patients with germline mutations in Fanc/Brca pathway genes have a high incidence of HNSCC at a young age (Kutler et al., 2003). Among Fanc/Brca family genes, some are frequently mutated while others are down-regulated in sporadic HNSCC (Marsit et al., 2004; Sparano et al., 2006; Weber et al., 2007; Wreesmann et al., 2007). Mice with epithelia-specific heterozygous knockout of Brca1 developed HNSCCs, indicating that Brca1 loss plays an important role in HNSCC tumorigenesis (Berton et al., 2003). Cells with absent or mutated alleles of Brca1 show many features characteristic of reduced genome stability, including impaired cell cycle checkpoints, reduced efficiency in homologous recombination, and defectiveDNA repair following genotoxic insults (Shen et al., 1998). Recently we found that a reduction in Brca1 protein occurs frequently in HNSCCs (Bornstein et al., 2009). Since mutations or promoter hyper-methylation of Brca1 was detected in only a small fraction of HNSCC patients (Marsit et al., 2004; Sparano et al., 2006), Brca1 down-regulation could occur at the transcriptional or post-transcriptional level in HNSCC. In fact, the Brca1 promoter is controlled by a complex and dynamic array of DNA binding proteins, transcriptional co-activators and co-repressors (Atlas et al., 2001; Baker et al., 2003; Bindra et al., 2005; Bindra & Glazer, 2006; De Siervi et al., 2010; Mueller & Roskelley, 2003; Thakur & Croce, 1999). In search for potential Brca1 promoter repression, in this study, we examined the effect of CtBP1 on Brca1 transcription. CtBP1 was initially recognized as an adenoviral E1A-binding protein and its over-activation, in combination with a mutant Ras, leads to tumorigenesis and metastasis, suggesting that CtBP1 plays a critical role in oncogenesis (Boyd et al., 1993; Grooteclaes & Frisch, 2000; Schaeper et al., 1995; Subramanian et al., 1989). The underlying molecular mechanisms of CtBP1 in oncogenesis could be linked to its function as a transcriptional co-repressor of multiple tumor suppressors, including PTEN, p16INK4a, and p15INK4b (Chinnadurai, 2009).
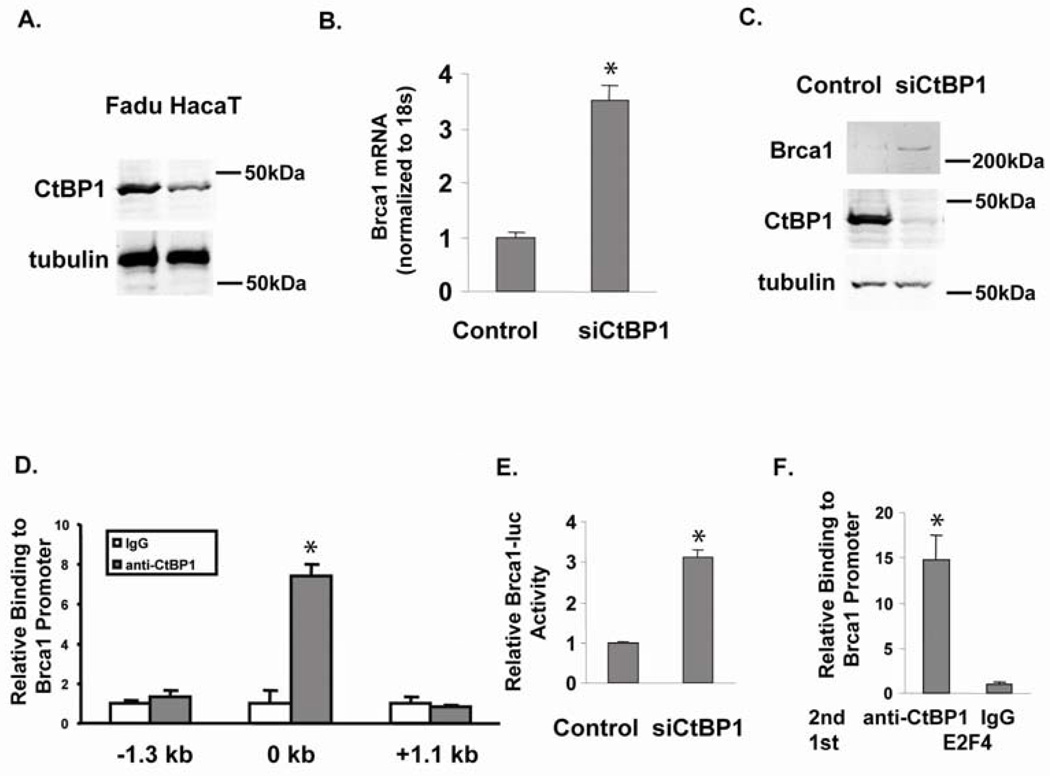
We examined CtBP1 protein levels in normal and cancer keratinocytes respectively (Fig. 1A). By western blotting, we found that CtBP1 protein levels in normal human keratinocyte HaCaT cells (Gift from Dr. Petra Boukamp) were about 1/3 of that in Fadu cells, an HNSCC cell line from a human hypopharyngeal carcinoma (from ATCC). After knocking down CtBP1 expression in Fadu cells using a CtBP1 specific siRNA (Zhang et al., 2003), we measured Brca1 mRNA by qRT-PCR; Brca1 mRNA levels increased 3 fold (Fig. 1B). Consistently, the increased Brca1 protein level correlated with the reduced CtBP1 protein level in siCtBP1 transfected Fadu cells (Fig. 1C). These data suggest that CtBP1 down-regulates Brca1 expression in HNSCC cells.
Figure 1.
CtBP1 represses Brca1 expression. (A) CtBP1 expression levels in the normal keratinocyte HaCaT cells and in Fadu SCC cells. Tubulin was used as a loading control and molecular markers are labeled. (B) CtBP1 knockdown in Fadu cells increased Brca1 mRNA; * p<0.05 vs. control cells. HNSCC Fadu cells were maintained in DMEM with 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. Fadu cell transfections were performed by Lipofecatmine 2000 in suspension, 5 × 104 Fadu cells were transfected with scrambled siRNA (Control) or siRNA targeting CtBP1 (siCtBP1) (Zhang et al., 2003). Cells were immediately transferred to 0.5 ml DMEM with serum and incubated at 37°C for 48 h. Total RNA was isolated using TRIzol (Invitrogen) and qRT-PCR was performed as previously described (Zhang et al., 2006). An 18S probe was used as an internal control. The relative RNA expression levels were determined by normalizing with internal controls, values were calculated using the comparative Ct method. Samples were assayed in triplicate for each experiment and at least two independent experiments were performed. Data are presented as mean + SEM from a representative experiment. (C) CtBP1 knockdown increases Brca1 protein in Fadu cells. Fadu, a human HNSCC cell line with a high CtBP1 level, was transfected with scrambled siRNA (Control) or siRNA to CtBP1 (siCtBP1) and its CtBP1 and Brca1 protein levels were measured by western blotting using antibodies from Santa Cruz. Tubulin was used as a loading control and molecular markers are labeled. (D) CtBP1 binding to the Brca1 regulatory element. Fadu cells were used for ChIP assay with an anti-CtBP1 antibody as described previously (Zhang et al., 2006). Primer sets encompassing −1.3 kb to +1.1 kb of the Brca1 promoter were used to q-PCR-amplify the ChIP sample. * p<0.05 vs. IgG. (E) CtBP1 represses Brca1 reporter, as siCtBP1 relieves repression of Brca1 reporter. The pGL4.26 Brca1 promoter luciferase reporter plasmid was generated by cloning a PCR-amplified 700 bp fragment of the Brca1 promoter spanning the TSS into the KpnI and BglII sites of pGL4.26 vector (Promega). Brca1 promoter–specific primers used were 5'-ggggtaccGACCTCTTCTTACGACTG-3' (forward) and 5'-gaagatctTTCCTGATCCTCAGCGC-3' (reverse). An empty renilla luciferase vector (pGL4.79) was use for normalization. Fadu cells were transfected with scrambled siRNA (Control) or siRNA to CtBP1 (siCtBP1) and the luciferase activity was measured (Zhang et al., 2002). (F) CtBP1 bound to E2F4 at the Brca1 promoter. Sequential ChIP using an anti-E2F4 antibody (Santa Cruz) followed by a CtBP1 antibody (Millipore) was performed in Fadu cells; p<0.02 vs. second ChIP using IgG. Primers surrounding the proximal promoter region of Brca1 were used to PCR-amplify the ChIP sample.
CtBP1 serves a key role in cellular regulation by binding to a variety of transcriptional repressors critical for development and tumorigenesis (Chinnadurai, 2002). Therefore, we assessed whether CtBP1 was recruited to the Brca1 gene to repress transcription. We performed chromatin immunoprecipitation (ChIP) to identify the CtBP1 binding sites at the Brca1 promoter region in Fadu cells, using an antibody against CtBP1 (Millipore) (Fig. 1D). Since CtBP1 typically binds near the TSS at promoters (Kim & Youn, 2009; Zhang et al., 2007; Zhang et al., 2006), we searched for possible binding sites in the Brca1 promoter regions within −1.3 kb to +1.1 kb of the transcriptional start site (TSS). PCR primer sets encompassing the above regions were used in q-PCR of ChIP DNA to identify CtBP1 binding sites, and ChIP using a normal IgG (Jackson Immuno Research) was used as a negative control (Fig. 1D). A CtBP1 binding site was found surrounding the TSS of the Brca1 promoter. CtBP1 binding was confirmed by an independent ChIP using a different anti-CtBP1 antibody (Santa Cruz) (data not shown). To examine if this CtBP1 binding site confers transcriptional repression to the Brca1 gene, we constructed a firefly luciferase reporter (pGL4.26, Promega) with a 700 bp region containing the CtBP1 binding site spanning the TSS at the Brca1 promoter and assayed the reporter activity with CtBP1 knockdown. Co-transfected empty Renilla luciferase reporter pGL4.79 (Promega) was used for normalization in the dual luciferase assay. Consistent with the increased Brca1 transcription observed with endogenous Brca1 genes when CtBP1 was abrogated in HNSCC Fadu cells, CtBP1 knockdown increased Brca1 reporter activity (Fig. 1E).
As a co-repressor, CtBP1 has been found in co-repressor complexes containing the CtBP1 binding adaptor protein CtIP and transcription factor E2F4 (Meloni et al., 1999). Therefore, we searched potential transcription factor binding sites around the above CtBP1 binding site, and found that E2F4 has been reported to bind in this region of the Brca1 promoter (Bindra & Glazer, 2006). We performed sequential ChIP using an antibody against E2F4 (Santa Cruz) for the first ChIP and an antibody against CtBP1 (Millipore) in the second ChIP. CtBP1 co-existed with E2F4 on the Brca1 promoter (Fig. 1F).
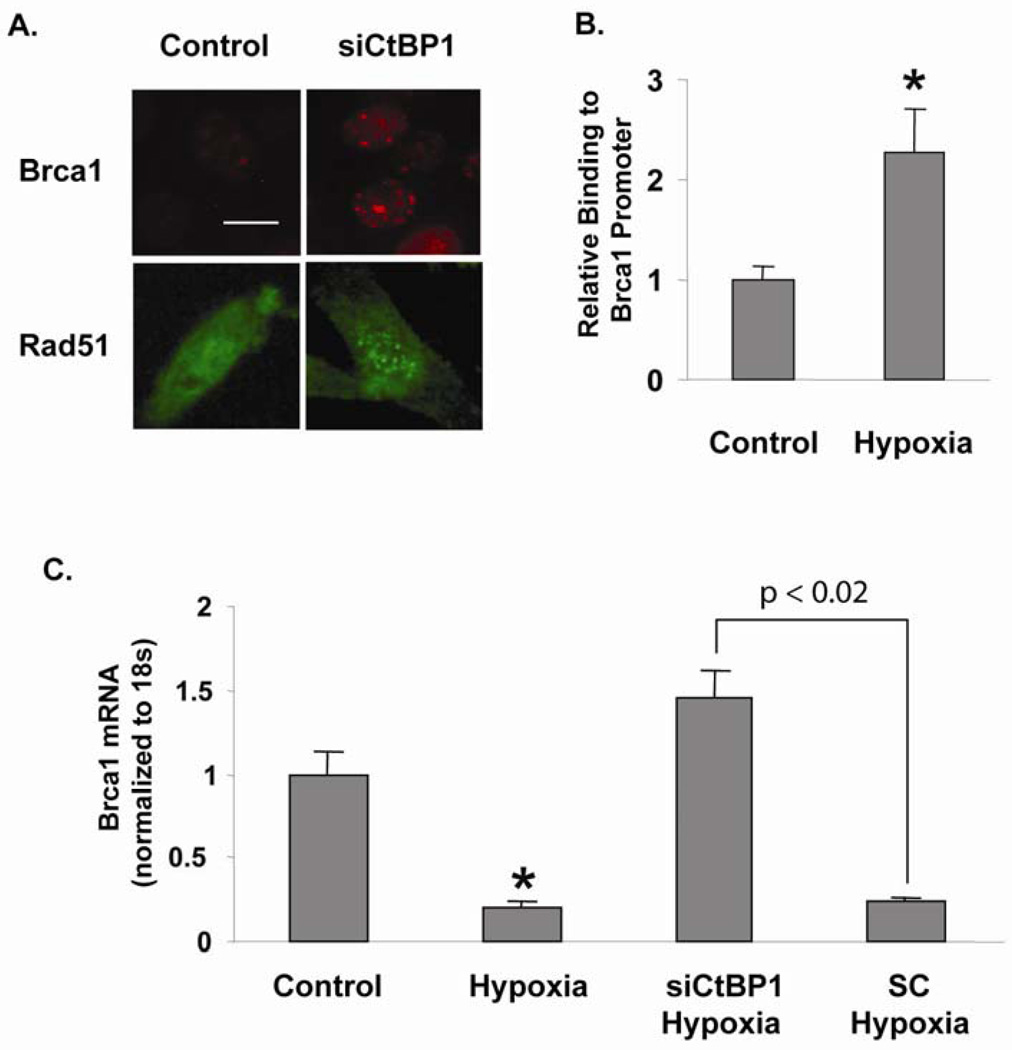
To further assess if restoration of Brca1 expression by CtBP1 knockdown in Fadu cells attenuates loss of Brca1-mediated DNA repair foci formation, we examined Brca1-mediated DNA repair foci formation by immunofluorescence staining using the Fadu and Fadu-siCtBP1 cells treated with mitomycin C (MMC, Sigma). Under normal conditions, Brca1 translocates to sites of MMC-induced DNA damage with other members of the Fanc/Brca pathway to form DNA repair nuclear foci (D'Andrea & Grompe, 2003). 24 h after 10 ng/ml MMC treatment, only about 10% of Fadu cells were able to form Brca1 foci, whereas Fadu cells with siCtBP1 added 48 h prior to MMC treatment (i.e., 72 h of CtBP1 knockdown) exhibited a 3-fold increase in the number of cells able to form MMC-induced DNA repair foci, from 13.5 ± 1.3 to 42.2 ± 2.9 per 100 cells (p<0.01) (Fig. 2A). Furthermore, foci-formation using a Rad51 antibody also revealed that the number of foci changed from 5.6 ± 3.0 to 17.5 ± 3.7 per 100 cells after CtBP1 knocking down (p<0.02) (Fig. 2A). These data suggest that CtBP1-mediated Brca1 repression abrogates Brca1 functions.
Figure 2.
Redox-sensitive regulation of Brca1 by CtBP1 in HNSCC cells. (A) CtBP1 knockdown increases MMC-induced DNA repair foci formation. Brca1 foci increased from 13.5 ± 1.3 to 42.2 ± 2.9 per 100 cells (p<0.01) and Rad51 foci increased from 5.6 ± 3.0 to 17.5 ± 3.7 per 100 cells (p<0.02). Brca1 and Rad51 antibodies (Santa Cruz) were used to immunostain DNA repair foci. Scale bar = 2 µm. (B) NADH-dependent CtBP1 binding to the Brca1 regulatory element. Fadu cells were incubated in a hypoxia chamber (1% O2, 5% CO2, 94% N2) for 3 h. ChIP assays were performed using an anti-CtBP1 antibody (Millipore) and compared to samples without special treatment (Control). Primers surrounding the proximal promoter region of Brca1 were used to PCR-amplify the ChIP sample. ChIP of Fadu cells without special treatment was used for normalization. * p<0.05 vs. non-treated Fadu cells. (C) NADH-dependent regulation of Brca1 expression. Fadu cells were treated with hypoxia for 48 hr and Brca1 mRNA was assayed in comparison to untreated Fadu cells (Control). Furthermore, CtBP1 knockdown (siCtBP1) relieved hypoxia-mediated Brca1 repression, p<0.02 for Brca1 level in siCtBP1 treatment vs. the scrambled siRNA (SC) treatment of Fadu cells.
Different from other transcriptional co-repressors, CtBP1 protein is uniquely structured to sense changes in free nuclear NADH concentration (Fjeld et al., 2003; Zhang et al., 2002). Studies by us and others have demonstrated that changes in cellular redox potential alter the interaction of CtBP1 with DNA-binding transcription repressors (Barnes et al., 2003; Kim et al., 2005; Kim & Youn, 2009; Mirnezami et al., 2003; Zhang et al., 2002; Zhang et al., 2007; Zhang et al., 2006). Therefore, as a “foe” of multiple tumor suppressors, CtBP1 also provides a link between transcriptional regulation and the metabolic status of the cells. This link is especially important given the high NADH concentration associated with hypoxia and the glycolytic nature of solid tumors. To investigate whether CtBP1-mediated repression of Brca1 gene is sensitive to NADH levels, we used ChIP assays to examine CtBP1 occupancy on the Brca1 promoter under hypoxic condition. Our previous study has shown that hypoxia increases free cellular NADH levels, which affects CtBP1 activity without affecting its levels (Zhang et al., 2002; Zhang et al., 2006). Consequently, Fadu cells exposed to hypoxia (1% O2 for 3 h) showed a 2.5 fold increase in CtBP1 recruitment to the proximal region of Brca1 promoter (Fig. 2B). Consistent with CtBP1 binding, Brca1 mRNA levels were reduced in hypoxia (Fig. 2C). To further determine if hypoxia-mediated Brca1 reduction depends on endogenous CtBP1, we knocked down CtBP1 in Fadu cells with hypoxia treatment. siRNA knockdown of CtBP1 largely attenuated the repressive effect of hypoxia on Brca1 transcription (Fig. 2C). Therefore, recruitment of CtBP1 to repress Brca1 gene transcription clearly depends on NADH level. Brca1 transcription has been shown to be regulated by various environmental stimuli (Andres et al., 1998; Bindra et al., 2005; De Siervi et al., 2010). Our study demonstrates the unique redox regulation of Brca1 transcription via the NADH sensor CtBP1. CtBP1’s recruitment to the Brca1 promoter is up-regulated by hypoxia, supporting the proposed role of hypoxia and anaerobic glycolysis in promoting tumor formation through the down-regulation of tumor suppressors including Brca1.
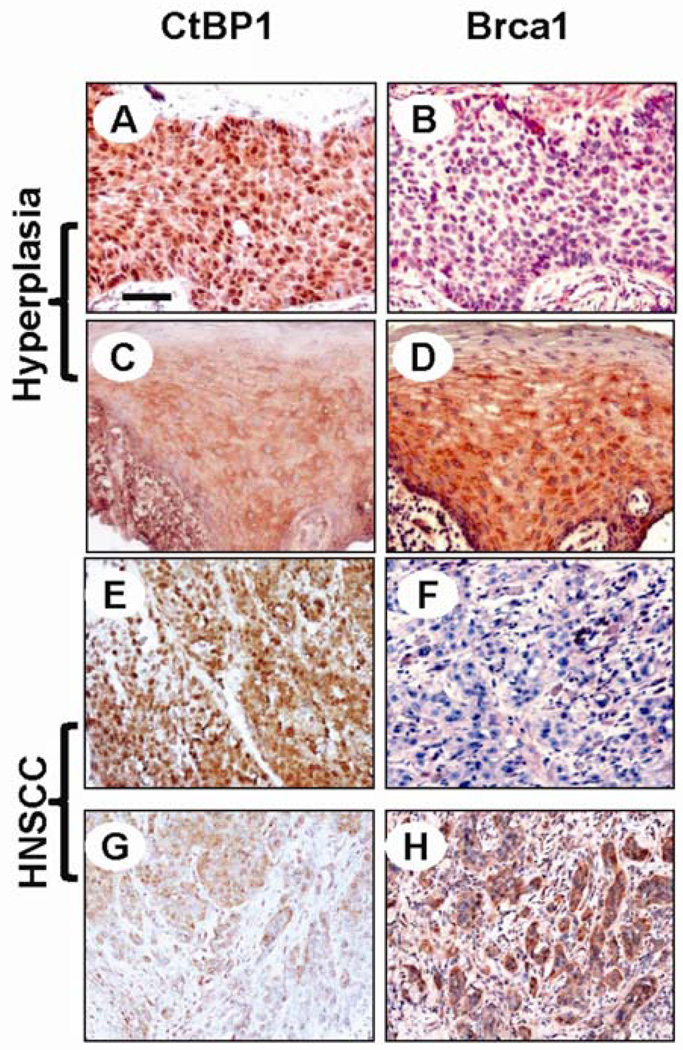
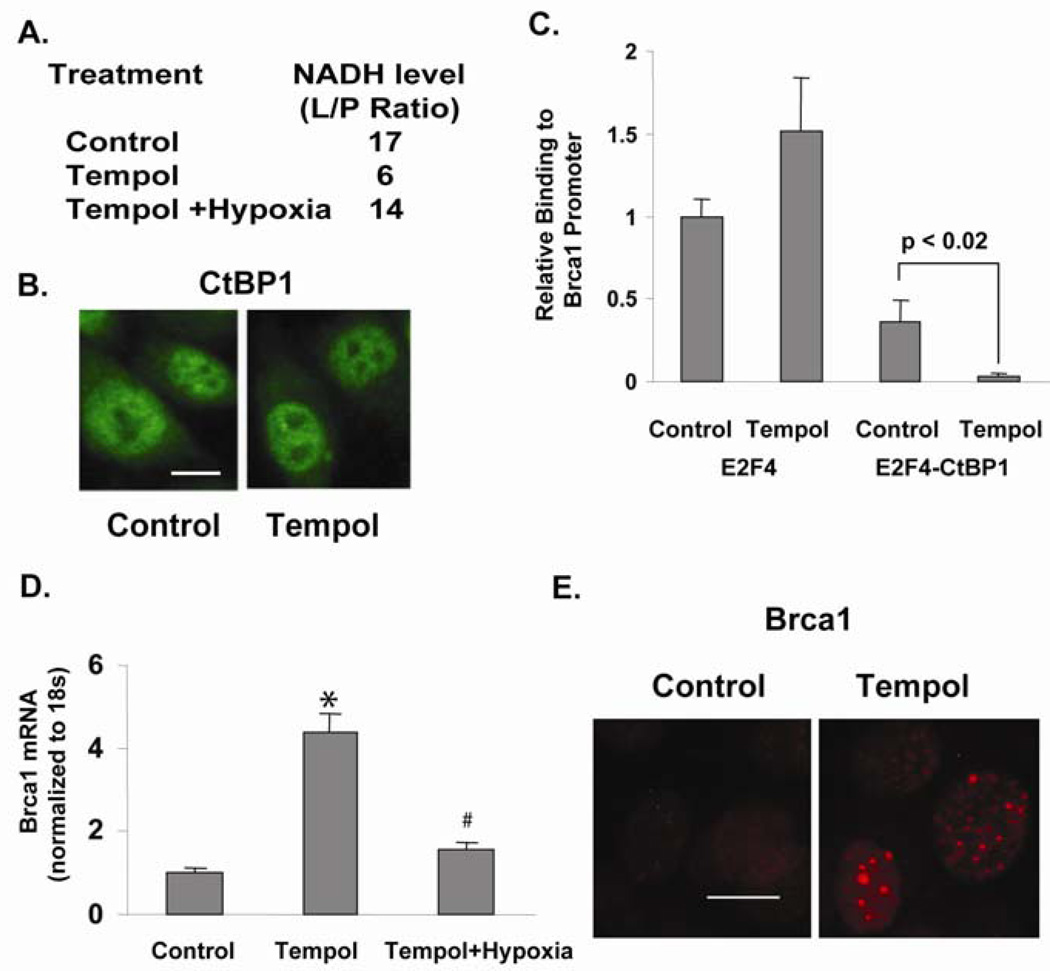
Our data suggest that under normal conditions, Brca1 expression in keratinocytes is not repressed by CtBP1 due to low levels of CtBP1 and NADH. However, during HNSCC carcinogenesis, increased levels of CtBP1 and NADH facilitate CtBP1-mediated repression of Brca1. To determine at which stage CtBP1 begins to be over-expressed in vivo during HNSCC carcinogenesis, we used immunohistochemistry (IHC) to examine CtBP1 expression on an HNSCC tissue array containing HNSCCs, hyperplastic mucosal tissues, and non-cancer controls (Biomax). Among 20 cases of hyperplastic lesions and 54 cases of HNSCCs, we found nuclear CtBP1 staining in 45% hyperplastic lesions and 80% HNSCCs (Table 1 and Fig. 3) but not in non-cancer head and neck tissues (data not shown). As we previously reported (Bornstein et al., 2009), down-regulation of Brca1 began in hyperplastic lesions of HNSCC patients (Table 1 and Fig. 3). Interestingly, CtBP1 nuclear staining correlated with Brca1 down-regulation in hyperplastic and HNSCC cases (Table 1 and Fig. 3). These data strongly support CtBP1 repression of Brca1 expression in vivo. Further, our data suggest that CtBP1 up-regulation begins in the early stages of HNSCC carcinogenesis, hence blocking CtBP1 activity could be a potential chemopreventive approach. Tempol, an antioxidant exhibiting a chemoprevention effect in several studies (Erker et al., 2005; Mitchell et al., 2003; Schubert et al., 2004; Zhang et al., 2008) and functioning as a topical radioprotector in a Phase I clinical study (Metz et al., 2004), is a stable free radical that down-regulates NADH by converting nitroxide to the corresponding hydroxylamine (Iannone et al., 1990; Krishna et al., 1992). Therefore, we investigated whether CtBP1 repression of Brca1 can be relieved by Tempol. Since free cellular NADH/NAD=1/700, the conversion of NAD to NADH mainly affects the free cellular NADH level, i.e., the decreased cellular NADH/NAD ratio measured by lactate/pyruvate ratio (L/P ratio) indicates a decrease in free NADH concentration (Williamson et al., 1967). Treating Fadu cells with 0.1 mM Tempol (Sigma) for 16 h caused a 3 fold decrease of cellular NADH level (Fig. 4A). Concomitantly, CtBP1 recruitment to the Brca1 promoter was decreased (Fig. 4C), even though CtBP1 level and localization remained unchanged during the Tempol treatment (Fig. 4B). Consequently, Brca1 expression was increased by Tempol treatment of HNSCC Fadu cells (Fig. 4D). qRT-PCR assay showed that Brca1 mRNA levels in Fadu cells increased 3–4 fold by Tempol treatment. Since Tempol is an anti-oxidant, its effect on Brca1 expression may be contributed from its effect on NADH and/or anti-oxidant effects. To determine the contribution of NADH levels to the effect of Tempol, we induced hypoxia in Tempol-treated cells and examined Brca1 levels. Hypoxia treatment largely attenuated the effect of Tempol on Brca1 expression (Fig. 4D). Since oxidation is suppressed during acute hypoxia (Wu et al., 2007), the attenuated Tempol effect on Brca1 expression suggests that Tempol-mediated CtBP1 repression on the Brca1 gene depends, at least in part, on its effect on reducing NADH levels. To determine whether restoration of Brca1 expression by Tempol treatment enhances Brca1 function, we assayed Brca1-mediated DNA repair foci formation by immunofluorescence staining (Fig. 4E). Similar to CtBP1 knockdown, Tempol treatment of Fadu cells increased the number of MMC-induced Brca1-DNA repair foci from 13.5 ± 1.3 to 40.1 ± 1.4 per 100 cells (p<0.01). Therefore, NADH dependence of CtBP1-mediated repression of tumor suppressors provides a perfect targeting strategy: NADH-blockade may represent chemopreventative or therapeutic approaches against HNSCC via its ability to increase Brca1-mediated DNA damage repair thus enhancing genome stability. This study instigates future investigation of the long-term effects of Tempol on chemoprevention of cancer types in which CtBP1 plays an important role.
Table 1.
Correlation between CtBP1 over-expression and Brca1 down-regulation in hyperplastic lesions and HNSCCs.
| Number of Cases/Total Cases | |||
|---|---|---|---|
| % of CtBP1 Over-expression | Hyperplastic mucosa | 9/20 (45%) | |
| HNSCC | 43/54 (80%) | ||
| CtBP1(+) | CtBP1(−) | ||
| % of Brca1 Down-regulation in CtBP1 (+) or (−) cases | Hyperplastic Mucosa | 5/9 (55.5%)* | 0/11(0%) |
| HNSCC | 37/43(86%)** | 3/11(27%) | |
p<0.01;
p<0.001, between CtBP1 (+) and CtBP1 (−) groups.
Figure 3.
Correlation between CtBP1 up-regulation and Brca1 down-regulation in a human HNSCC tissue array (Biomax HN801). Immunohistochemical staining was performed using antibodies against Brca1 (Santa Cruz) and CtBP1 (Millipore) to stain consecutive tissue sections as we previously described (Bornstein et al., 2009). Sections were counterstained with hematoxylin. Evaluation of CtBP1 and Brca1 staining of human HNSCC samples was performed by 2 independent investigators using methods described previously (Bornstein et al., 2009). Note that a hyperplastic lesion with CtBP1 nuclear staining (A) showed negative staining for Brca1 in the consecutive section (B). In contrast, a hyperplastic lesion with CtBP1 staining in the cytoplasm but little in the nucleus (C) exhibited positive Brca1 staining (D). The same correlation was also observed in HNSCC lesions, in which predominant nuclear CtBP1 staining in (E) barely had Brca1 staining in (F). In contrast, in section (G), tumor epithelial cells showed either cytoplasmic CtBP1 staining (upper areas) or no CtBP1 staining (lower areas); the consecutive section in (H) showed positive Brca1 staining. The scale bar in the first panel represents 40 µm for panels A-F and 80 µm for panels G and H.
Figure 4.
Tempol affects Brca1 transcription. (A) Tempol (0.1 mM for 16 h) decreases free NADH reflected by a decreased cellular lactate/pyruvate (L/P) ratio in Fadu cells. The lactate and pyruvate concentrations of Fadu cells were measured by colorimetric assays and used to calculate the free cellular NADH/NAD ratio (Williamson et al., 1967). Since free NAD is in greater excess than free NADH, the conversion of NAD to NADH mainly affects NADH level. Therefore, the L/P ratio indicates the free NADH level (Williamson et al., 1967). Hypoxia attenuated the NADH decrease induced by Tempol treatment. (B) CtBP1 level and localization stay unchanged during Tempol treatment of Fadu cells. Scale bar=2 µm. (C) Tempol treatment decreased CtBP1 recruitment to E2F4 (E2F4-CtBP1) at the Brca1 promoter. Sequential ChIP using an E2F4 antibody (Santa Cruz; E2F4 single ChIP: E2F4) followed by anti-CtBP1 (double ChIP: E2F4-CtBP1) was performed in Fadu cells treated with Tempol; p<0.02 vs. non-treated Fadu cells (Control). Primers surrounding the proximal promoter region of Brca1 were used to PCR-amplify the ChIP sample. E2F4 single ChIP of non-treated Fadu cells was used for normalization. (D) Tempol increases Brca1 expression in Fadu cells measured by qRT-PCR; * p<0.05 vs. non-treated Fadu cells (Control). Furthermore, hypoxia attenuated the stimulatory effect of Brca1 by Tempol; # p<0.02 vs. Tempol-treated Fadu cells (Tempol). (E) Tempol increases Brca1 foci formation from 13.5 ± 1.3 to 40.2 ± 1.4 per 100 cells (p<0.01). Scale bar = 2 µm.
In summary, our current analysis revealed a direct transcriptional repression of CtBP1 on Brca1 expression in head and neck epithelial cells, which is also dependent on cellular NADH levels. We also report that CtBP1 is over-expressed in human HNSCC and its over-expression correlated with Brca1 down-regulation. The unique NADH-dependence of CtBP1’s action not only highlights the importance of hypoxia and anaerobic glycolysis in promoting tumor formation through activation of CtBP1, but also reveals a potential chemo preventative/therapeutic approach for HNSCC, which warrants future studies to test this approach in HNSCC animal models prior to clinical trials. It will be interesting to explore if the mechanism of Tempol action found in this study can be applied to other anti-oxidants for their chemoprevention effects. Moreover, since Brca1 is a tumor suppressor in multiple organs/tissues, it will be interesting to assess if CtBP1-mediated Brca1 repression contributes to cancer in multiple cancer types as the chemoprevention potential suggested by our current study could have broader applications.
Acknowledgments
This work was supported by grants from the NIH, R01DE15953 (to X. J. W.) and R01CA115468 (to Q.Z.). We thank Dr. Petra Boukamp for providing the normal human keratinocytes HaCaT cells and Dr. James Mitchell for helpful discussions.
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- Andres JL, Fan S, Turkel GJ, Wang JA, Twu NF, Yuan RQ, Lamszus K, Goldberg ID, Rosen EM. Oncogene. 1998;16:2229–2241. doi: 10.1038/sj.onc.1201752. [DOI] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Mueller CR. Oncogene. 2001;20:7110–7114. doi: 10.1038/sj.onc.1204890. [DOI] [PubMed] [Google Scholar]
- Baker KM, Wei G, Schaffner AE, Ostrowski MC. J Biol Chem. 2003;278:17876–17884. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. Nat Struct Biol. 2003;10:622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- Berton TR, Matsumoto T, Page A, Conti CJ, Deng CX, Jorcano JL, Johnson DG. Oncogene. 2003;22:5415–5426. doi: 10.1038/sj.onc.1206825. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Cancer Biol Ther. 2006;5:1400–1407. doi: 10.4161/cbt.5.10.3454. [DOI] [PubMed] [Google Scholar]
- Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, Deng C, Lu SL, Wang XJ. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. Embo J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- De Siervi A, De Luca P, Byun JS, Di LJ, Fufa T, Haggerty CM, Vazquez E, Moiola C, Longo DL, Gardner K. Cancer Res. 2010;70:532–542. doi: 10.1158/0008-5472.CAN-09-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker L, Schubert R, Yakushiji H, Barlow C, Larson D, Mitchell JB, Wynshaw-Boris A. Hum Mol Genet. 2005;14:1699–1708. doi: 10.1093/hmg/ddi181. [DOI] [PubMed] [Google Scholar]
- Fjeld CC, Birdsong WT, Goodman RH. Proc Natl Acad Sci U S A. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiere A, Koch W, Trotti A, Sidransky D. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Grooteclaes ML, Frisch SM. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- Hunter KD, Parkinson EK, Harrison PR. Nat Rev Cancer. 2005;5:127–135. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- Iannone A, Tomasi A, Vannini V, Swartz HM. Biochim Biophys Acta. 1990;1034:285–289. doi: 10.1016/0304-4165(90)90052-x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Cho EJ, Kim ST, Youn HD. Nat Struct Mol Biol. 2005;12:423–428. doi: 10.1038/nsmb924. [DOI] [PubMed] [Google Scholar]
- Kim JH, Youn HD. Cell Death Differ. 2009;16:584–592. doi: 10.1038/cdd.2008.186. [DOI] [PubMed] [Google Scholar]
- Krishna MC, Grahame DA, Samuni A, Mitchell JB, Russo A. Proc Natl Acad Sci U S A. 1992;89:5537–5541. doi: 10.1073/pnas.89.12.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP, Singh B. Arch Otolaryngol Head Neck Surg. 2003;129:106–112. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- Meloni AR, Smith EJ, Nevins JR. Proc Natl Acad Sci U S A. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz JM, Smith D, Mick R, Lustig R, Mitchell J, Cherakuri M, Glatstein E, Hahn SM. Clin Cancer Res. 2004;10:6411–6417. doi: 10.1158/1078-0432.CCR-04-0658. [DOI] [PubMed] [Google Scholar]
- Mirnezami AH, Campbell SJ, Darley M, Primrose JN, Johnson PW, Blaydes JP. Curr Biol. 2003;13:1234–1239. doi: 10.1016/s0960-9822(03)00454-8. [DOI] [PubMed] [Google Scholar]
- Mitchell JB, Xavier S, DeLuca AM, Sowers AL, Cook JA, Krishna MC, Hahn SM, Russo A. Free Radic Biol Med. 2003;34:93–102. doi: 10.1016/s0891-5849(02)01193-0. [DOI] [PubMed] [Google Scholar]
- Mueller CR, Roskelley CD. Breast Cancer Res. 2003;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci U S A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert R, Erker L, Barlow C, Yakushiji H, Larson D, Russo A, Mitchell JB, Wynshaw-Boris A. Hum Mol Genet. 2004;13:1793–1802. doi: 10.1093/hmg/ddh189. [DOI] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- Sparano A, Quesnelle KM, Kumar MS, Wang Y, Sylvester AJ, Feldman M, Sewell DA, Weinstein GS, Brose MS. Laryngoscope. 2006;116:735–741. doi: 10.1097/01.mlg.0000205141.54471.7f. [DOI] [PubMed] [Google Scholar]
- Subramanian T, La Regina M, Chinnadurai G. Oncogene. 1989;4:415–420. [PubMed] [Google Scholar]
- Thakur S, Croce CM. J Biol Chem. 1999;274:8837–8843. doi: 10.1074/jbc.274.13.8837. [DOI] [PubMed] [Google Scholar]
- Weber F, Xu Y, Zhang L, Patocs A, Shen L, Platzer P, Eng C. Jama. 2007;297:187–195. doi: 10.1001/jama.297.2.187. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreesmann VB, Estilo C, Eisele DW, Singh B, Wang SJ. ORL J Otorhinolaryngol Relat Spec. 2007;69:218–225. doi: 10.1159/000101542. [DOI] [PubMed] [Google Scholar]
- Wu W, Platoshyn O, Firth AL, Yuan JX. Am J Physiol Lung Cell Mol Physiol. 2007;293:L952–L959. doi: 10.1152/ajplung.00203.2007. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- Zhang QS, Eaton L, Snyder ER, Houghtaling S, Mitchell JB, Finegold M, Van Waes C, Grompe M. Cancer Res. 2008;68:1601–1608. doi: 10.1158/0008-5472.CAN-07-5186. [DOI] [PubMed] [Google Scholar]