Abstract
In studies of extinction risk, it is often insufficient to conclude that species with narrow ranges or small clutch sizes require prioritized protection. To improve conservation outcomes, we also need to know which threats interact with these traits to endanger some species but not others. In this study, we integrated the spatial patterns of key threats to Australian amphibians with species' ecological/life-history traits to both predict declining species and identify their likely threats. In addition to confirming the importance of previously identified traits (e.g. narrow range size), we find that extrinsic threats (primarily the disease chytridiomycosis and invasive mosquitofish) are equally important and interact with intrinsic traits (primarily ecological group) to create guild-specific pathways to decline in our model system. Integrating the spatial patterns of extrinsic threats in extinction risk analyses will improve our ability to detect and manage endangered species in the future, particularly where data deficiency is a problem.
Keywords: amphibian declines, extinction risk, threats, Batrachochytrium dendrobatidis, Random Forests, classification tree
1. Introduction
Particular vertebrate life-history and ecological traits correlate with their risk of decline and extinction [1–4]. For example, for amphibians (and several other groups) small geographical range size has been consistently identified as the most important predisposing factor to decline [5–7]. Other factors, including clutch size, body size and ecological niche, have also been identified as potentially important [8–11].
Despite particular traits predisposing species or groups of species to decline, external factors are necessary as proximate agents of decline in most cases [1,12]. Few external threats in isolation, however, are widespread enough to explain global or continental vertebrate declines and in many regions multiple threats may operate in concert [10,13]. Furthermore, the effects of a threat can be expected to vary both spatially, according to its distribution and magnitude, and across species because unique suites of intrinsic traits of species respond differently to different threatening processes (e.g. habitat specialists are particularly vulnerable to habitat destruction; [14]). This may (typically) result in complex, context-dependent and/or indirect pathways to decline [2,15]. For these reasons, it has been recently stressed that considering both intrinsic traits and extrinsic threats together is important for describing the complete picture of extinction risk [1,16,17].
More practically, identifying and mitigating threats may also be the most direct and efficient management action available for slowing or reversing contemporary species declines. In studies of extinction risk, it is therefore likely to be insufficient or impractical to generically conclude that species with narrow ranges or small clutch sizes require prioritized protection (e.g. [5]). To improve conservation outcomes and the allocation of scarce conservation resources, we also need to know which threats interact with these traits to endanger some species but not others in order to identify how to prioritize and how to protect.
The Global Amphibian Assessment (GAA) identified several important threats for amphibians globally, the most important being habitat loss and species overexploitation (e.g. collection for the pet trade). A third category grouped species declining owing to ‘enigmatic’ (unknown) causes [10]. With the discovery of chytridiomycosis [18,19], many of these enigmatic declines have been attributed to the effects of this pandemic disease [13]. Nevertheless, in contrast to the progress that has been made in addressing habitat degradation in recent quantitative studies of extinction risk in amphibians (e.g. [20]), little attention has been given to spatially explicit aspects of the threat posed by chytridiomycosis. While this is almost certainly due to a lack of suitable or available data, it is an omission that could clearly hinder management at regional scales. Murray et al. [21], for example, show that the spatial distribution of chytridiomycosis is a better correlate of decline than range size in Australian frogs (cf. [7]), while Bielby et al. [22] show that the intrinsic traits of infected species correlate with whether they have suffered rapid declines or not as a result of infection. There is thus a clear need to better integrate host life history and ecology with the pathogen's distribution/environmental requirements for improved risk analysis.
In addition, other significant threats may also be present locally, exemplifying the appropriateness of regional studies over global studies in cases where there is a degradation of information resolution as spatial scale increases [17]. For example, invasive fish have been implicated in the decline of some Australian frog species [23,24]. Given the diversity of known threats and the array of ecological and life-history traits that predispose species to decline, untangling the complex interactions that result in specific pathways to decline or extinction is a challenging task [2,15,22,25] but one that is important for predicting further declines and, more importantly, improving our ability to do something about them [26–28].
Here we present an analysis that tackles inclusively the range of key threats and life-history traits previously shown or suspected to be associated with declines in our model system, Australian amphibians. We present a novel integration of the intrinsic traits that make some species susceptible to decline or extinction (life history, ecology) with spatial models of the multiple key threats (disease, invasive species, habitat destruction) that endanger them. In addition to powerful prediction of species declines, we employ flexible decision tree-based models to derive species-specific information about their likely threats. This identifies and prioritizes paths for action, so starting to close the gap between prediction and prevention of species declines.
2. Methods
(a). Tree-based modelling approach
We used Random Forests (RF), an extension of classification-tree analysis that has emerged from the field of machine learning [29], for our analyses. Forests are combinations of hundreds or thousands of trees that combine predictions to improve classification accuracy and stability and reduce the effects of bias and correlation among variables [31]. Unlike some machine-learning classifiers, RF is designed to provide accurate predictions as well as useful information about the underlying data. Only recently appearing in the ecological literature, RF frequently outperforms traditional statistical approaches for classification [32], and it has been suggested that tree-based methods may be more appropriate in extinction risk studies in which a species-by-species result is desirable ([26,33]; see also electronic supplementary material, appendix S1 and [25]). Discussion of the algorithm, associated metrics and uses of RF in ecology is provided by Cutler et al. [31]. We used the ‘randomForests’ package implemented in R [34] to run models and the ‘cforest’ function in package ‘party’ to obtain unbiased variable importance estimates to corroborate variable selection [35].
While RF is a powerful classifier and can effectively identify and characterize traits or threats that increase risk of decline across cases, it is difficult to trace an individual species' path through the forest and arrive simply at the cause(s) of its classification (since there are thousands of trees, each with only a subset of predictors). To aid interpretation and applicability of the RF results, we used a single-tree method (conditional inference tree, using the ctree function in R package party developed by Hothorn et al. [36]) for corroboration and to visualize and help trace the effects of the most influential predictors on the classification of individual species. We allowed the tree to show splits up to a significance level of p < 0.25 to provide added descriptive value [36].
(b). Decline as a response variable
We used the ‘Trend’ classification from the IUCN Red List (downloaded July 2009) to partition species into two response classes, ‘declining’ or ‘stable’. Species of ‘increasing’ trend were classed as stable for this analysis, and species of ‘unknown’ trend were excluded for model training, except for ‘extinct’ (or presumed extinct) species, for which trend was not listed but which have clearly declined. Species of unknown trend were then introduced as new cases and their trends predicted.
(c). Decline predictor variables
(i). Intrinsic traits
Intrinsic traits were taken or inferred from published sources and supplemented by expert opinion (electronic supplementary material, appendix S2). Species morphology was represented by body size (snout-vent length; SVL). Life-history traits were represented by the variables testes mass, clutch size and ova size. Ecological variables included: ecological group (terrestrial breeders, T; ephemeral pond breeders, E; permanent water associated, P; stream associated, S; or moist bog/soak associated, M) and larval mode (direct developing or free-swimming larvae). We also included categories of relative normal abundance (1 = low, 6 = high) and relative reproductive potential (a 1–5 ordinal score of the number of eggs typically produced per female per year). Geographical range size was represented by extent of occurrence (EOO), calculated in ArcGIS9.2 from species distribution maps developed during the GAA (downloaded June 2009).
Of the 223 Australian species listed by the IUCN (2009), 81 had a complete set of data for all intrinsic variables, although all variables in isolation contained values for a minimum of 58.7 per cent of species (mean 87.7% complete) (table 1). We used multiple imputation with predictive mean matching to replace missing data on continuous variables (see electronic supplementary material, appendix S3, for further details).
Table 1.
Summary of model variables, the number of species with data for each variable, the proportion of total species with data and the proportion of species for which data were imputed (using multiple imputation, MI, for continuous variables and RF for categorical variables) for each variable. ES is environmental suitability for the organism as modelled with species distribution models (see text).
| species with data | % (n = 223 species) | % imputed | |
|---|---|---|---|
| intrinsic variable | |||
| trend (dependent variable) | 207 | 92.8 | 0 (excluded) |
| range size (EOO) | 212 | 95.1 | 0 (excluded) |
| body size (SVL) | 223 | 100 | 0 |
| larval mode | 223 | 100 | 0 |
| ecological group | 223 | 100 | 0 |
| clutch size | 143 | 64.1 | 35.9 |
| ova size | 140 | 62.8 | 37.2 |
| testes mass | 166 | 74.4 | 25.6 |
| abundance category | 131 | 58.7 | 41.3 |
| reproductive category | 209 | 93.7 | 6.3 |
| extrinsic variable | |||
| ES for Bd | 215 | 96.4 | 0 (excluded) |
| ES for Gambusia | 215 | 96.4 | 0 (excluded) |
| ES for cane toad | 215 | 96.4 | 0 (excluded) |
| habitat degradation | 215 | 96.4 | 0 (excluded) |
(ii). Extrinsic threats
Extrinsic threats considered in this analysis have all been linked with amphibian declines in previous studies. We calculated threat levels by averaging spatial patterns of each threat across all available occurrence records for each species (approx. 300 000 records for all Australian species; [37], updated by D. Rosauer in 2009); see electronic supplementary material, appendix S1 and figure S1 for details. Habitat degradation was calculated as the mean VAST score (Vegetation Assets, States and Transitions as per [38]), a national gridded dataset coded as discrete states ranging from intact native (1) to total removal (5). Potential exposure to three invasive species was calculated as above but on spatial models of environmental suitability (ES) for each threat. Murray et al. [21] describe the procedure (for chytridiomycosis) in detail and further notes meriting this approach are provided in electronic supplementary material, appendix S1 and figure S1. Invasive species considered here included the pathogen Batrachochytrium dendrobatidis (Bd; cause of amphibian chytridiomycosis; [21]), the predatory cane toad (Rhinella marina; J. Elith 2009, unpublished data) and the predatory eastern mosquitofish (Gambusia holbrooki; K. A. Murray 2009, unpublished data). Our database of frog occurrences contained data on 215 described Australian species, which represents the number of species for which threats could be calculated. We did not impute missing data for threats owing to their idiosyncratic distributions with respect to amphibian species ranges but rather excluded these cases (n = 8 species; mainly representing newly described and data-deficient species, final n = 198 species for analysis).
3. Results
In the full RF model, the estimate of classification error rate (the out-of-bag or OOB estimate) was low at 12.1 per cent, meaning that the overall percentage of cases correctly classified was 87.9 per cent (174 of 198 species). Specificity (per cent of stable species correctly classified) was 91.3 per cent, while sensitivity (per cent of declining species correctly classified) was somewhat lower at 77.6 per cent (Cohen's κ = 0.68; 95% CI 0.56–0.80, where κ = 0 indicates discrimination not better than random, κ = 1 indicates perfect discrimination; see electronic supplementary material, appendix S4, for information on our modifications to the default RF settings for this analysis). Of 24 species misclassified, 11 were ‘incorrectly’ predicted to be stable and 13 were incorrectly predicted to be in decline. A further one (of 14) species of unknown trend was predicted to be in decline (table 2). Misclassified species contained multiple representatives of all four frog families occurring in the training data, suggesting that factors missing from the model were not strongly phylogenetically patterned.
Table 2.
Predicted decliners (trend unknown) and misclassified cases from the pruned RF model. Species currently considered declining, which are predicted to be stable, may have lower risk than previously described or they may be undergoing declines owing to idiosyncratic or local extrinsic threats not shared by ecologically similar stable species. Species currently considered stable but predicted to be in decline may be at greater risk of decline than previously described or they may be otherwise relatively resistant to (or recovered from) specific threats causing declines in ecologically similar species. Node is the terminal node that each species falls in the (unpruned) conditional inference tree (figure 3).
| IUCN trend | predicted trend | species | p of decline | node |
|---|---|---|---|---|
| declining | stable | Adelotus brevis | 0.28 | 12 |
| Cophixalus neglectus | 0.12 | 10 | ||
| Geocrinia alba | 0.17 | 11 | ||
| Heleioporus australiacus | 0.37 | 12 | ||
| Litoria dentata | 0.28 | 13 | ||
| Litoria raniformis | 0.31 | 12 | ||
| Litoria spenceri | 0.37 | 4 | ||
| Litoria verreauxii | 0.16 | 12 | ||
| Pseudophryne australis | 0.28 | 12 | ||
| Pseudophryne bibronii | 0.46 | 4 | ||
| Pseudophryne semimarmorata | 0.04 | 11 | ||
| stable | declining | Cophixalus saxatilis | 0.56 | 10 |
| Geocrinia rosea | 0.54 | 4 | ||
| Litoria citropa | 0.79 | 4 | ||
| Litoria jervisiensis | 0.56 | 12 | ||
| Litoria nannotis | 0.89 | 4 | ||
| Litoria phyllochroa | 0.72 | 4 | ||
| Litoria revelata | 0.55 | 12 | ||
| Litoria tyleri | 0.71 | 13 | ||
| Mixophyes fasciolatus | 0.58 | 12 | ||
| Mixophyes schevilli | 0.51 | 10 | ||
| Pseudophryne raveni | 0.62 | 12 | ||
| Spicospina flammocaerulea | 0.79 | 4 | ||
| Taudactylus liemi | 0.97 | 4 | ||
| unknown | declining | Litoria jungguy | 0.74 | 4 |
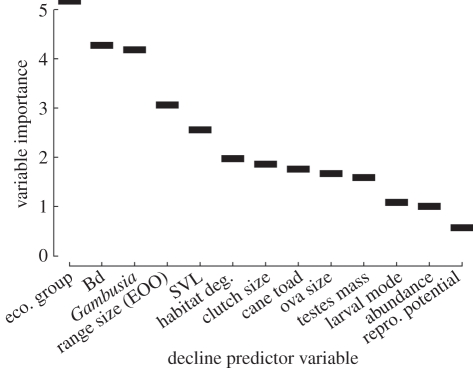
Both intrinsic and extrinsic factors were identified as important predictors of decline (figure 1). Ecological group and ES for Bd and Gambusia were unambiguously the most important variables (confirmed in cforest), followed by range size (EOO; also confirmed in cforest). Body size (SVL) and ES for cane toads were ranked next in RF, but cforest ranked all of the remaining variables including habitat degradation as being of lower, roughly equal importance (data not shown).
Figure 1.
Variable importance plot from the full RF model. To assess importance of each variable: after growing the kth tree, the values of the target variable among all out-of-bag (OOB) cases are randomly permuted and the OOB cases are run down the tree. The decrease in the number of votes for the correct class (fading confidence in the prediction) owing to permuting is averaged over the forest [29]. For variable selection, importance was corroborated with cforest in package party [30].
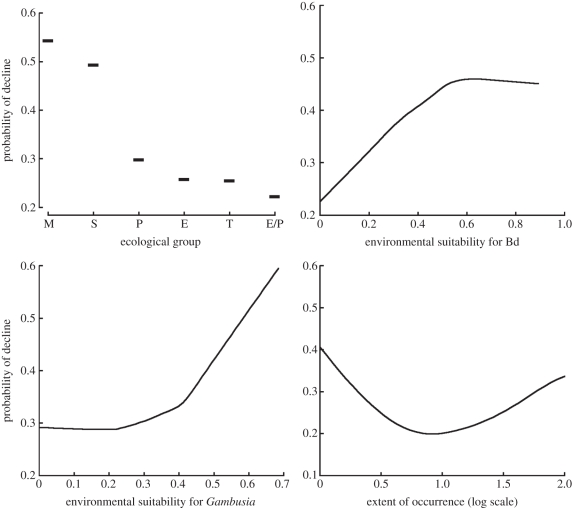
Continuous predictor variables were known a priori to be inter-correlated (electronic supplementary material, figure S2), so to improve interpretation of the effects of the most important variables (as indicated by cforest), a second RF was run in which only the four most important variables were included ([29,34]; classification accuracy was similar in this second model (data not shown), indicating no loss of predictive power via pruning). Partial dependence plotting [39], which is a common visualization tool for showing the effects of a small number of predictors on the ‘blackbox’ classification of the response variable, shows the probability of decline response (transformed from the logit scale) for the four most important variables (figure 2). The partial dependence plots suggest that species that are stream or moist bog/soak dwelling and experiencing environments highly suited to Bd and Gambusia are most likely to be declining. Probability of decline also increased for species with small range sizes (EOO; figure 2).
Figure 2.
Partial dependence plots showing the decline response as a function of each variable included in the pruned RF model. For the ecological group, M = moist bog/soak associated, S = stream associated, P = permanent water body associated, T = terrestrial, E = ephemeral pond breeder.
(a). Single-tree visualization
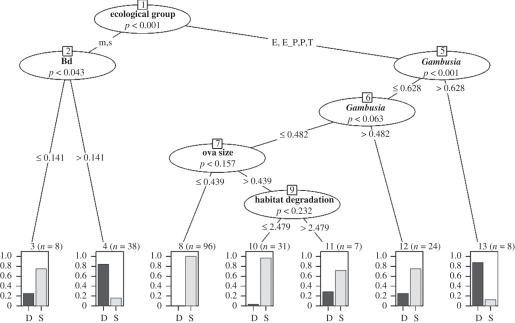
The most important primary split in the conditional inference tree was the ecological group; of 50 declining species, 34 (68.0%) were in the stream or moist bog/soak ecological groups (figure 3). On this branch, species experiencing environments suitable for Bd (ES > 0.14; node 4) were at the maximal risk of decline (84.2% of 38 species declining), while just two of eight species with ES for (Bd < 0.14 node 3) are declining. In the other ecological groups, just 16 (9.6%) of 166 species are declining. For these groups, the tree split on high ES for Gambusia (greater than 0.63), with seven of eight species experiencing declines (node 13), compared with just nine (5.7%) of 158 species below this threshold. Non-significant splits (at p > 0.05; [36]) are presented for descriptive value only. Species' membership in the terminal nodes is shown in electronic supplementary material, appendix S5 and table 2.
Figure 3.
Conditional inference tree on the full dataset (n = 212 species, comprising the 198 training cases used in RF and 14 cases with predicted trends) using all available variables (note: only three variables are retained in the tree at p < 0.05). Inner nodes (ovals) indicate which variables were used for splitting (threshold values on the line). Terminal nodes are numbered for reference in the text. n is the number of species falling in each terminal node; bars express the proportion of species in the node that are declining (D) or stable (S). A list of species with their RF class predictions and their membership in the terminal nodes of the tree is shown in electronic supplementary material, appendix S5. For ecological group, M = moist bog/soak associated, S = stream associated, P = permanent water body associated, T = terrestrial, E = ephemeral pond breeder.
4. Discussion
(a). A global perspective
Small geographical range size has previously been identified as the most important factor predisposing amphibians to decline, both in Australia and globally [5,7,20]. Other factors, including clutch size, body size and ecological niche, are also considered important [8–11]. Although in agreement about the importance of these traits, our results go beyond previous results by highlighting the way multiple extrinsic threats may operate across specific ecological groups. Targeting species with small ranges for conservation as prescribed by Sodhi et al. [20] and Cooper et al. [5] is certainly important; however, it is the threats themselves that must be addressed to achieve better conservation outcomes.
We demonstrate that extrinsic factors interact with intrinsic species traits to create multiple pathways to decline in Australian amphibians. Previous large-scale analyses have modelled the decline or extinction risk of amphibians from intrinsic traits only, both regionally [8,11] and globally [5]; some extrinsic threats have also been considered at a national (e.g. cane toads, feral pigs and habitat destruction; [7]) or global (e.g. climate, human density, habitat loss; [20,22]) scale. Our analysis substantially extends previous work by directly integrating spatially varying threats from the numerous, currently recognized key threatening processes (chytridiomycosis, Gambusia, R. marina, habitat destruction) with intrinsic traits of amphibian species that may be used to resolve context or ‘guild’-dependent amphibian declines.
The characterization of complex nonlinear responses and the presence of interaction effects among numerous inter-correlated intrinsic and extrinsic predictor variables underpin the novelty of our results and the utility of this non-parametric approach for studying trends in decline amidst multiple guilds and threats [15,26,40]. Parametric approaches can be expected to perform less well under these circumstances [2,25,33]. Because tree-based methods quantitatively depict relationships of the predictors to the response variable over subgroups, which can re-appear in numerous branches in the tree as required, they are highly suited to identifying interactions and describing ‘pathways’ to a particular outcome, in our case decline. While this is not intended to replace the processes of traditional, labour-intensive, descriptive, single-species-based risk assessments, in many ways, our analysis quantitatively and efficiently mimics this approach but at a fraction of the cost and effort. The results can then be used to better inform and direct future intensive/expensive efforts to improve cost-efficiency.
(b). Important risk factors for Australian amphibians
As depicted in the variable importance plots, the analysis highlighted four variables considered most important for the classification prediction of decline: ecological group, ES for Bd, ES for Gambusia and range size (EOO). Variables outside the top four were generally indistinguishable in their order of importance (as measured with cforest). Taken together with the partial dependence plots, risk of decline increased with association with stream and moist bog/soak environments, increased ES for Bd and Gambusia and smaller range sizes. Drivers of decline are thus clearly threat and intrinsic trait guild-specific. This was confirmed in the single-tree analysis, with species inhabiting streams or moist bog/soak environments suitable for Bd being at particularly high risk of decline. Species in the other groups inhabiting areas highly suitable for Gambusia were also at risk. In terms of the number of species at risk (compare nodes 4 and 13), the combined analyses suggested that chytridiomycosis is the most important single threat for declining amphibians in Australia.
Our study is thus in strong agreement with Bielby et al. [22], who identified the intrinsic traits that made Bd-positive species susceptible to rapid declines (small range, small clutch size, partially aquatic species) and warned against the dangers of equating the simple presence of Bd with declines amidst other complex, potentially interactive intrinsic factors. Indeed, approximately two-thirds of Bd-positive species in Australia are currently considered stable [41]. Unlike Bielby et al. [22], however, our analysis directly and spatially incorporated the potential threat posed by Bd to all species [21] in the context of numerous threats, thereby allowing our model to construct other pathways to decline for particular subgroups of species among a number of potential threats that clearly impact species guilds in different ways.
(c). Conservation implications: predictions, misclassification and drivers of decline
Our analysis not only provides insight into the intrinsic factors that make species susceptible to declines for a given threat, but also supplies a species-specific tool with which to assess those threats, predict declines and respond with more informed conservation decisions [27,28,33]. We review our findings with reference to the specifics of our study system, highlighting where our approach offers insight that is broadly relevant to endangered species and protected area management, including IUCN Red List assessments and the treatment of data-deficient species.
The effects of Bd on amphibians are inherently non-random with respect to the species affected [13,42], and the role of Bd in many of the 32 declining stream or moist bog/soak species in the high-Bd group (node 4) in this study has been reasonably well documented (e.g. Pseudophryne corroborree, Taudactylus acutirostris, Taudactylus eungellensis, Litoria (Nyctimistes) dayi, Litoria genimaculata, Litoria leseuerii, Litoria pearsoniana, Litoria rheocola; [13,18,43]). Nevertheless, less than 50 per cent of species in this group have yielded positive test results for the disease to date [41]. Some of these species can no longer be located and/or have been declared extinct (Rheobatrachus silus, Rheobatrachus vitellinus, Taudactulus diurnus, Taudactulus rheophilus, Litoria nyakalensis), strengthening the inference that Bd has been the proximate cause of their extirpations [13,18,44] as asserted for a confirmed Bd-positive, ecologically similar species (T. acutirostris; [45]). Others in this group should be prioritized for disease testing, highlighting the utility of our predictive results for identifying threats as well as future actions.
Misclassification in this framework can also provide important insights. In classification problems, the binary response is normally assumed to be known without error. If this is the case here, misclassified declining species (predicted to be stable) could flag idiosyncratic or unknown causes of decline (e.g. climate change), which could help focus future investigations. For misclassified stable species (predicted declining), misclassification could indicate that some species have been able to evade the threats causing declines in ecologically similar species (e.g. resistance to chytridiomycosis). This group could thus be instructive for identifying factors that allow some species to mediate threats while other similar species cannot. Conversely, species may be identified that should be classified as in decline based on our model but are not listed as such (see also [32,46]). Similarly, species that ‘should’ be stable but are believed to be in decline could represent species potentially at lower risk than previously assessed.
In the high-Bd terminal node discussed above, for example, almost every species was predicted by RF to be in decline, whereas several are currently classed as stable (e.g. Geocrinia rosea, Litoria phyllochroa, Litoria citropa, Litoria nannotis, Taudacytlus liemi, Spicospina flammocaerulea). These species represent potentially unrecognized decliners, and their membership in this node identifies Bd as a potential threat. Indeed, the former four species have been reported with Bd [41] and L. nannotis has certainly undergone dramatic declines such that all may require re-assessment of their trends and/or further disease investigation to elucidate how they may be mediating or escaping this threat (e.g. disease resistance). Similarly, RF predicted a species of unknown trend to be in decline (Litoria jungguy) given its ecological similarity to other declining species. When run down the single tree, L. jungguy falls in the high-Bd node, immediately identifying chytridiomycosis as a likely threat. Predicting trends in data-deficient species is an important task for conservation and for Red Listing. The two species in this node that were incorrectly predicted by RF to be stable (Litoria spenceri, Pseudophryne bibronii) represent equivocal cases; while the single tree indicated that Bd is a potential threat, RF suggested these species also shared traits with ecologically similar but stable species. Their declines may thus be due to local factors not captured by the model (e.g., other invasive species, climate change). Further surveys are nevertheless required to resolve the true status of misclassified species, prioritizing where possible those species that have the most influence in the model.
Our analysis implicated G. holbrooki as an agent in the decline of at least six amphibian species (Litoria aurea, Litoria cooloolensis, Litoria olongburesnsis, Litoria freycineti and Crinia tinnula) and RF predicted another one stable species (Litoria tyleri) to be a decliner, probably owing to the strong association of this threat with ecologically similar declining species (node 13). With the exception of Litoria brevipalmata (a highly unusual, extreme ephemeral pond breeding species), there is evidence from case studies that Gambusia can affect these species via predation of their eggs or tadpoles [23,47,48]. In conjunction with the current focus on the effects of habitat degradation and fragmentation in many species in this group, further Gambusia research and management intervention are strongly justified. The model does not, however, imply that other threats cannot be operating in conjunction; amidst this group, Litoria dentata was uniquely but incorrectly predicted by RF to be stable, representing a species that may be misclassified by the IUCN or otherwise declining owing to a more complex combination of factors. Similarly, L. aurea is known to be impacted by Bd in some populations [49] and its Bd exposure score (0.46) is consistent with declines occurring in species in other ecological groups.
The utility of the single tree for interpreting the RF results degrades below the major splits described above. The additional splits in the tree were not significant (at p = 0.05), so there is reduced confidence in tracing individual species to the terminal nodes on this branch (nodes 8, 10, 11 and 12; if the additional splits were collapsed, all of these species would fall in node 6, which would become terminal). As the sister of node 13, species falling in these groups can be expected to be predicted as mainly stable. The classification by RF reflects this; considering the remaining nine declining species on this branch, RF incorrectly predicted all but one to be stable (Litoria castanea was a correctly predicted decliner). These misclassified species represent cases that may be declining owing to additional factors not captured in the model (or, less likely, species that may have been misclassified as declining by IUCN). Litoria castanea's membership in node 12 identifies Gambusia as a potential threat, although in light of the partial dependence plots, its classification by RF is likely to be influenced by multiple factors, with a Bd score (greater than 0.15) consistent with declines in other ecological groups and a very high degree of habitat degradation. In contrast, RF predicted that several stable species on this branch should be in decline (Cophixalus saxatilis, Litoria jervisiensis, Litoria revelata, Mixophyes fasciolatus, Mixophyes shevilli, Pseudophryne raveni). It is difficult to identify from the single tree what factors influenced the RF classification but a review of the partial dependence plots from RF and the variable values for these species is informative (electronic supplementary material, appendix S5). For example, all but one of these species had high Bd scores (greater than 0.5) and all but two had high Gambusia scores (greater than 0.5).
Our study thus provides quantitative information that could influence the allocation of conservation resources at a national scale. While habitat degradation remains a major contemporary conservation problem, prescriptively gazetting species with small distributional ranges in protected areas will in no way alleviate the key threatening processes of disease and invasive species impacting amphibians in Australia and, by extension, many other parts of the world. Without striving to better map and mitigate threats, thereby improving a species' ‘potential for successful recovery’ [27], declining amphibian species may continue to fall through the conservation prioritization safety net. We expect the same problems to exist for other declining taxa and geographical regions. The approach we employ is adaptable and could significantly improve the detection and, more importantly, the management of endangered species into the future, particularly where data deficiency precludes the traditional (albeit labour intensive) species-by-species approach.
Acknowledgements
K.A.M. thanks H. Hines and K. McDonald for expert knowledge on species' ecology and life-history traits, J. Elith for unpublished data, B. Murray for abundance data, D. Rowe for occurrence data, D. Segan, M. Watts and C. Klein for GIS wisdom and spatial data and R. Wilson and H. Possingham for lab space and morning teas. We also thank Prof. Andy Purvis and two anonymous referees whose comments improved the final manuscript. K.A.M. was supported by an Australian Postgraduate Award, an Australian Biosecurity CRC professional development award and a Wildlife Preservation Society of Australia student research award.
References
- 1.Cardillo M., Mace G. M., Gittleman J. L., Jones K. E., Bielby J., Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441–1448 10.1098/rspb.2008.0179 (doi:10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson A. D., Hamilton M. J., Boyer A. G., Brown J. H., Ceballos G. 2009. Multiple ecological pathways to extinction in mammals. Proc. Natl Acad. Sci. 106, 10 702–10 705 10.1073/pnas.0901956106 (doi:10.1073/pnas.0901956106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher D. O., Owens I. P. F. 2004. The comparative method in conservation biology. Trends Ecol. Evol. 19, 391–398 10.1016/j.tree.2004.05.004 (doi:10.1016/j.tree.2004.05.004) [DOI] [PubMed] [Google Scholar]
- 4.Olden J. D., Poff N. L., Bestgen K. R. 2008. Trait synergisms and the rarity, extirpation, and extinction risk of desert fishes. Ecology 89, 847–856 10.1890/06-1864.1 (doi:10.1890/06-1864.1) [DOI] [PubMed] [Google Scholar]
- 5.Cooper N., Bielby J., Thomas G. H., Purvis A. 2008. Macroecology and extinction risk correlates of frogs. Global Ecol. Biogeogr. 17, 211–221 10.1111/j.1466-8238.2007.00355.x (doi:10.1111/j.1466-8238.2007.00355.x) [DOI] [Google Scholar]
- 6.Gaston K. J., Fuller R. A. 2009. The sizes of species' geographic ranges. J. Appl. Ecol. 46, 1–9 10.1111/j.1365-2664.2008.01596.x (doi:10.1111/j.1365-2664.2008.01596.x) [DOI] [Google Scholar]
- 7.Murray B. R., Hose G. C. 2005. Life-history and ecological correlates of decline and extinction in the endemic Australian frog fauna. Austral Ecol. 30, 564–571 10.1111/j.1442-9993.2005.01471.x (doi:10.1111/j.1442-9993.2005.01471.x) [DOI] [Google Scholar]
- 8.Hero J. M., Williams S. E., Magnusson W. E. 2005. Ecological traits of declining amphibians in upland areas of eastern Australia. J. Zool. 267, 221–232 10.1017/S0952836905007296 (doi:10.1017/S0952836905007296) [DOI] [Google Scholar]
- 9.Lips K. R., Reeve J. D., Witters L. R. 2003. Ecological traits predicting amphibian population declines in Central America. Conserv. Biol. 17, 1078–1088 10.1046/j.1523-1739.2003.01623.x (doi:10.1046/j.1523-1739.2003.01623.x) [DOI] [Google Scholar]
- 10.Stuart S. N., Chanson J. S., Cox N. A., Young B. E., Rodrigues A. S. L., Fischman D. L., Waller R. W. 2004. Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786 10.1126/science.1103538 (doi:10.1126/science.1103538) [DOI] [PubMed] [Google Scholar]
- 11.Williams S. E., Hero J. M. 1998. Rainforest frogs of the Australian wet tropics: guild classification and the ecological similarity of declining species. Proc. R. Soc. Lond. B 265, 597–602 10.1098/rspb.1998.0336 (doi:10.1098/rspb.1998.0336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies R. G., et al. 2006. Human impacts and the global distribution of extinction risk. Proc. R. Soc. B 273, 2127–2133 10.1098/rspb.2006.3551 (doi:10.1098/rspb.2006.3551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skerratt L. F., Berger L., Speare R., Cashins S., McDonald K. R., Phillott A. D., Hines H. B., Kenyon N. 2007. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth 4, 125–134 10.1007/s10393-007-0093-5 (doi:10.1007/s10393-007-0093-5) [DOI] [Google Scholar]
- 14.Norris K., Harper N. 2004. Extinction processes in hot spots of avian biodiversity and the targeting of pre-emptive conservation action. Proc. R. Soc. Lond. B 271, 123–130 10.1098/rspb.2003.2576 (doi:10.1098/rspb.2003.2576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaustein A. R., Kiesecker J. M. 2002. Complexity in conservation: lessons from the global decline of amphibian populations. Ecol. Lett. 5, 597–608 10.1046/j.1461-0248.2002.00352.x (doi:10.1046/j.1461-0248.2002.00352.x) [DOI] [Google Scholar]
- 16.Fisher D. O., Blomberg S. P., Owens I. P. F. 2003. Extrinsic versus intrinsic factors in the decline and extinction of Australian marsupials. Proc. R. Soc. Lond. B 270, 1801–1808 10.1098/rspb.2003.2447 (doi:10.1098/rspb.2003.2447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purvis A., Cardillo M., Grenyer R., Collen B. 2005. Correlates of extinction risk: phylogeny, biology, threat and scale. Phylogen. Conserv. 8, 295–316 10.1017/CB09780511614927.013 (doi:10.1017/CB09780511614927.013) [DOI] [Google Scholar]
- 18.Berger L., et al. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc. Natl Acad. Sci. USA 95, 9031–9036 10.1073/pnas.95.15.9031 (doi:10.1073/pnas.95.15.9031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longcore J. E., Pessier A. P., Nichols D. K. 1999. Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia 91, 219–227 10.2307/3761366 (doi:10.2307/3761366) [DOI] [Google Scholar]
- 20.Sodhi N. S., Bickford D., Diesmos A. C., Lee T. M., Koh L. P., Brook B. W., Sekercioglu C. H., Bradshaw C. J. A. 2008. Measuring the meltdown: drivers of global amphibian extinction and decline. PLoS ONE 3, e1636. 10.1371/journal.pone.0001636 (doi:10.1371/journal.pone.0001636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray K. A., Retallick R. W. R., Puschendorf R., Skerratt L. F., Rosauer D., McCallum H., Berger L., Speare R., VanDerWal J. In press Assessing spatial patterns of disease risk to biodiversity: implications for the management of the amphibian pathogen, Batrachochytrium dendrobatidis. J. Appl. Ecol. [Google Scholar]
- 22.Bielby J., Cooper N., Cunningham A. A., Garner T. W. J., Purvis A. 2008. Predicting susceptibility to future declines in the world's frogs. Conserv. Lett. 1, 82–90 10.1111/j.1755-263X.2008.00015.x (doi:10.1111/j.1755-263X.2008.00015.x) [DOI] [Google Scholar]
- 23.Gillespie G., Hero M. 1999. Potential impacts of introduced fish and fish translocations on Australian amphibians. In Declines and disappearances of Australian frogs (ed. Campbell A.), pp. 131–144 Canberra, Australia: Environment Australia [Google Scholar]
- 24.Reynolds S. J. 2009. Impact of the introduced poeciliid Gambusia holbrooki on amphibians in southwestern Australia. Copeia 2009, 296–302 10.1643/CH-08-101 (doi:10.1643/CH-08-101) [DOI] [Google Scholar]
- 25.Bielby J., Cardillo M., Cooper N., Purvis A. 2010. Modelling extinction risk in multispecies data sets: phylogenetically independent contrasts versus decision trees. Biodivers. Conserv. 19, 113–127 10.1007/s10531-009-9709-0 (doi:10.1007/s10531-009-9709-0) [DOI] [Google Scholar]
- 26.Jones M. J., Fielding A., Sullivan M. 2006. Analysing extinction risk in parrots using decision trees. Biodivers. Conserv. 15, 1993–2007 10.1007/s10531-005-4316-1 (doi:10.1007/s10531-005-4316-1) [DOI] [Google Scholar]
- 27.Marsh H., Dennis A., Hines H., Kutt A., McDonald K., Weber E., Williams S., Winter J. 2007. Optimizing allocation of management resources for wildlife. Conserv. Biol. 21, 387–399 10.1111/j.1523-1739.2006.00589.x (doi:10.1111/j.1523-1739.2006.00589.x) [DOI] [PubMed] [Google Scholar]
- 28.Wilson K. A., et al. 2007. Conserving biodiversity efficiently: what to do, where, and when. PLoS Biol. 5, 1850–1861 10.1371/journal.pbio.0050223 (doi:10.1371/journal.pbio.0050223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breiman L. 2001. Random forests. Berkeley, CA: University of California [Google Scholar]
- 30.Strobl C., Hothorn T., Zeileis A. 2009. Party on! A new, conditional variable-importance measure for random forests available in the party package. R J. 1, 14–17 [Google Scholar]
- 31.Cutler D. R., Edwards T. C., Beard K. H., Cutler A., Hess K. T., Gibson J., Lawler J. J. 2007. Random forests for classification in ecology. Ecology 88, 2783–2792 10.1890/07-0539.1 (doi:10.1890/07-0539.1) [DOI] [PubMed] [Google Scholar]
- 32.Maindonald J., Braun J. 2007. Cambridge series in statistical and probabilistic mathematics. In Data analysis and graphics using R: an example based approach. New York, NY: Cambridge University Press [Google Scholar]
- 33.Sullivan M. S., Jones M. J., Lee D. C., Marsden S. J., Fielding A. H., Young E. V. 2006. A comparison of predictive methods in extinction risk studies: contrasts and decision trees. Biodivers. Conserv. 15, 1977–1991 10.1007/s10531-005-4315-2 (doi:10.1007/s10531-005-4315-2) [DOI] [Google Scholar]
- 34.Liaw A., Wiener M. 2002. The randomforest package. R News 2, 18–22 [Google Scholar]
- 35.Strobl C., Boulesteix A. L., Zeileis A., Hothorn T. 2007. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinform. 8, 25. 10.1186/1471-2105-8-25 (doi:10.1186/1471-2105-8-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hothorn T., Homik k., Zeileis A. 2006. Unbiased recursive partitioning: a conditional inference framework. J. Comput. Graph. Stat. 15, 651–674 10.1198/106186006X133933 (doi:10.1198/106186006X133933) [DOI] [Google Scholar]
- 37.Slatyer C., Rosauer D., Lemckert F. 2007. An assessment of endemism and species richness patterns in the Australian Anura. J. Biogeogr. 34, 583–596 10.1111/j.1365-2699.2006.01647.x (doi:10.1111/j.1365-2699.2006.01647.x) [DOI] [Google Scholar]
- 38.Thackway R., Lesslie R. 2006. Reporting vegetation condition using the vegetation assets, states and transitions (VAST) framework. Ecol. Manag. Restor. 7, S53–S62 10.1111/j.1442-8903.2006.00292.x (doi:10.1111/j.1442-8903.2006.00292.x) [DOI] [Google Scholar]
- 39.Hastie T. J., Tibshirani R. J., Friedman J. H. 2001. The elements of statistical learning: data mining, inference and prediction, Springer series in statistics New York, NY: Springer [Google Scholar]
- 40.Beebee T. J. C., Griffiths R. A. 2005. The amphibian decline crisis: a watershed for conservation biology? Biol. Conserv. 125, 271–285 10.1016/j.biocon.2005.04.009 (doi:10.1016/j.biocon.2005.04.009) [DOI] [Google Scholar]
- 41.Murray K. A., et al. 2010. The distribution and host range of the pandemic disease chytridiomycosis in Australia spanning surveys from 1956 to 2007. Ecology 91, 1557. 10.1890/09-1608.1 (doi:10.1890/09-1608.1) [DOI] [Google Scholar]
- 42.Smith K. G., Lips K. R., Chase J. M. 2009. Selecting for extinction: nonrandom disease-associated extinction homogenizes amphibian biotas. Ecol. Lett. 12, 1069–1078 10.1111/j.1461-0248.2009.01363.x (doi:10.1111/j.1461-0248.2009.01363.x) [DOI] [PubMed] [Google Scholar]
- 43.Murray K. A., Skerratt L. F., Speare R., McCallum H. 2009. Impact and dynamics of disease in species threatened by the amphibian chytrid fungus, Batrachochytrium dendrobatidis. Conserv. Biol. 23, 1242–1252 10.1111/j.1523-1739.2009.01211.x (doi:10.1111/j.1523-1739.2009.01211.x) [DOI] [PubMed] [Google Scholar]
- 44.Daszak P., Cunningham A., Hyatt A. 2000. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287, 443–449 10.1126/science.287.5452.443 (doi:10.1126/science.287.5452.443) [DOI] [PubMed] [Google Scholar]
- 45.Schloegel L. M., Hero J. M., Berger L., Speare R., McDonald K., Daszak P. 2006. The decline of the sharp-snouted day frog (Taudactylus acutirostris): the first documented case of extinction by infection in a free-ranging wildlife species? EcoHealth 3, 35–40 10.1007/s10393-005-0012-6 (doi:10.1007/s10393-005-0012-6) [DOI] [Google Scholar]
- 46.Cardillo M., Mace G. M., Gittleman J. L., Purvis A. 2006. Latent extinction risk and the future battlegrounds of mammal conservation. Proc. Natl Acad. Sci. USA 103, 4157–4161 10.1073/pnas.0510541103 (doi:10.1073/pnas.0510541103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer E., Hero J. M., Shoo L., Lewis B. 2006. National recovery plan for the wallum sedgefrog and other wallum-dependent frog species. Canberra, Australia: Queensland Parks and Wildlife Service [Google Scholar]
- 48.NSW National Parks and Wildlife Service 2003. NSW threat abatement plan. predation by Gambusia holbrooki—the plague minnow. Hurstville, NSW: NPWS [Google Scholar]
- 49.Berger L. 2001. Diseases in Australian frogs. Townsville: James Cook University [Google Scholar]