Abstract
Detecting regions of the human genome that are, or have been, influenced by natural selection remains an important goal for geneticists. Many methods are used to infer selection, but there is a general reliance on an accurate understanding of how mutation and recombination events are distributed, and the well-known link between these processes and their evolutionary transience introduces uncertainty into inferences. Here, we present and apply two new, independent approaches; one based on single nucleotide polymorphisms (SNPs) that exploits geographical patterns in how humans lost variability as we colonized the world, the other based on the relationship between microsatellite repeat number and heterozygosity. We show that the two methods give concordant results. Of these, the SNP-based method is both widely applicable and detects selection over a well-defined time interval, the last 50 000 years. Analysis of all human genes by their Gene Ontology codes reveals how accelerated and decelerated loss of variability are both preferentially associated with immune genes. Applied to 168 immune genes used as the focus of a previous study, we show that members of the same gene family tend to yield similar indices of selection, even when located on different chromosomes. We hope our approach will provide a useful tool with which to infer where selection has acted to shape the human genome.
Keywords: natural selection, genetic diversity, humans, microsatellite, balancing selection, immune genes
1. Introduction
One hundred and fifty years after Darwin published ‘The Origin’, literally millions of single nucleotide polymorphisms (SNPs) [1–3] finally provide the tools that should allow us to analyse in detail how natural selection has acted on, and continues to shape the human genome. Various approaches have been explored [4], including the study of linkage disequilibrium blocks [5], detection of SNP clusters, testing for an excess of SNPs with one very common allele [6], discovery of unusually large or small genetic distances between populations [7] and, within genes, inferences about the ratio of synonymous to non-synonymous substitutions [5]. Although these studies have told us much, they tend to focus on directional rather than balancing selection and to rely on poorly tested assumptions about where and at what rate recombination events and mutations occur, assumptions that are increasingly being challenged [8–11]. Where balancing selection has been tested for, it seems elusive [12], possibly because ‘the requirements for detection by means of SNP data alone will rarely be met’ [12], a notable exception being Andrés et al. [13]. This is potentially of concern because there is increasing evidence that heterozygote advantage may be common, particularly at immune loci, in both humans [14,15] and many other species [15–17].
An alternative, and we believe novel, approach to the detection of natural selection is suggested by humankind's unusual demographic history. Somewhat over 50 000 years ago, anatomically modern humans moved out of Africa to colonize the world [18–20]. As they did so, one or a series of population bottlenecks caused a dramatic loss of neutral genetic variability [18,21–23], manifest everywhere people have looked, from microsatellites [22] and SNPs [24] to morphological traits [25] and even commensal bacterial diversity [26]. The signature of this loss is a monotonic decline in neutral genetic variability with land-only distance from Africa [19,23]. Previous methods for inferring selection have tended either to ignore this trend completely or to treat it as a nuisance variable that has to be controlled [15]. However, the uniformity and ubiquity of the decline in variability itself provides a useful new null hypothesis. Deviations from the overall trend should be informative about the action of natural selection. For example, balancing selection maintains two or more lineages within a population, thereby creating regions of enhanced diversity [27]. During a population bottleneck, such regions should show reduced diversity loss, manifest as genomic regions in which the gradient of diversity against distance from Africa is close to zero. Similarly, positive selection acting on variants that helped early modern humans adapt to new environments will have accelerated the reduction of diversity and created steeper slopes. Finally, positive slopes might be generated wherever the non-African environment presented new challenges that were best met by multiallelic solutions, for example when humans encountered new classes of pathogens or parasites [15,28].
A pervasive problem with many tests for selection is the lack of independent verification. Most tests rely on assumptions about local recombination or mutation rates, if only neither have changed appreciably in the recent past. In practice, these assumptions are open to question. Point mutations appear to occur non-randomly, falling in clusters [9,10], and these clusters themselves correlate with local recombination rate [29], though this may reflect correlation of both with features such as local GC base composition [30]. Nonetheless, recombination hotspots can be both intense and highly localized [31] and appear to be evolutionarily transient [11,32]. Equally, the clustering of SNPs may reflect gene conversion events focused on existing polymorphisms [10,33], potentially creating a dynamic and constantly changing mutation landscape. The main method for detecting selection that directly bypasses these issues involves dn/dS ratios [34], the proportion of all nucleotide substitutions that cause changes at the level of the protein. However, being based on several/many mutations in coding regions, this method cannot be used meaningfully to infer current selection acting on a single variant allele, or selection acting on variants in non-coding regions.
Given the above uncertainties, it is desirable to compare two independent methods of inference. For a second test, we therefore turned to microsatellites. It is well-established that microsatellite heterozygosity is positively correlated with repeat number [35–37]. Consequently, the average relationship between heterozygosity and repeat number provides an expectation for how variable an ‘average’ microsatellite of a given repeat number should be [36]. Wherever a microsatellite lies near to a gene experiencing selection, this expectation will change. In regions affected by balancing selection, a microsatellite should carry greater heterozygosity than expected from the number of repeats it carries. Similarly, microsatellites in regions affected by strong directional selection will have lost variability through selective sweep effects, and should show less variability than expected for their length.
The two methods for detecting selection described above are essentially independent: the first looks at how heterozygosity varies across the world regardless of absolute levels, while the latter looks at patterns within a single population and focuses on absolute variability relative to an extrinsic relationship, the way microsatellite heterozygosity scales with repeat number. Here we cross-test these two methods using large, published human datasets and show that they yield concordant patterns. We then apply the more general SNP-based approach to show how immune genes in particular exhibit patterns consistent with both balancing and directional selection.
2. Methods
SNP data were downloaded from http://hapmap.ncbi.nlm.nih.gov/, specifically HapMap phase II and III (5 February 2009 release) genotyped in the following population samples: Yoruba from Nigeria (YRI), Europeans from Utah (CEU), Lahuya from Kenya (LWK), Maasai from Kenya (MKK), Toscans from Italy (TSI), Han from China (CHB) and Japanese from Japan (JPT) [38]. Four other populations were excluded owing to their greater risk of mixed ancestry. Heterozygosity was estimated assuming two alleles in Hardy–Weinberg equilibrium. Distance from Africa was measured as the land-only route from Addis Ababa to the town of sampling/centre of sampling region [22]. CEU was taken as Paris, an intermediate western European location.
To determine the local slope of SNP heterozygosity against distance from Africa for any given point in the genome, a custom macro was written in Visual Basic. SNP data for the relevant chromosome were read into an array and stored as heterozygosities for each of the seven populations. Local slope was then calculated as the Pearson correlation coefficient of average heterozygosity against distance from Africa across the seven populations, average heterozygosity being based on all SNPs within a given distance of the focal location. A correlation coefficient was preferred to the actual slope because, with so few data points, steep but poorly supported slope values often arise by chance, while large correlation coefficients more often imply a well-defined relationship, regardless of whether the slope itself is steep (for a given set of SNPs, heterozygosity varies little among populations, so large outliers are unlikely). In all cases, we compared the results obtained using four different window sizes: ±10, ±25, ±50 and ±100 kb.
Microsatellite data were downloaded from the Centre d'Etude du Polymorphisme Humain (CEPH) website (http://www.cephb.fr/en/cephdb/) and are based on the data published by Dib et al. [39]. The location of each microsatellite on the human genome, build 36.6 (chosen for maximum compatibility across all datasets used), was determined through the sequence-tagged sites database, and expected heterozygosity calculated using the frequencies of alleles listed, assuming Hardy–Weinberg equilibrium. Wherever possible, we extracted the clone sequence and the primer sequences, with which we calculated the mean allele length converted to numbers of repeat units (= ‘length’), on the assumption of no insertions or deletions in the regions between the primer sites and the microsatellite. Finally, we calculated residual heterozygosity at each locus. Plotting heterozygosity against length yields the expected positive relationship. However, the variance in heterozygosity declines strongly with increasing repeat number, owing to the fact that while essentially all long microsatellites have high heterozygosity, short microsatellites can have almost any value. To reduce this bias, we therefore expressed the heterozygosity of each microsatellite as the standardized residual heterozygosity of all loci within 0.5 repeat units in length. Loci with extreme residuals (greater than 2.5 s.d.) were excluded, since these may include strongly aberrant loci with unusual features such as insertions or deletions in their flanking DNA.
A full list of all annotated human genes was downloaded from the Gene Ontology (GO) website (http://www.geneontology.org) on 10 March 2009 [40]. Locations on the human genome build 36.6 were verified and each locus stored as its unweighted mid-point location (i.e. we used the middle base rather than the middle exonic base), along with all associated GO codes. In addition, we also downloaded a list of 168 genes from a previous paper examining selection on immune genes [28], along with their locations. This list was used as a supplementary test of the association between selection and immune genes.
3. Results
(a). Microsatellite heterozygosity and single nucleotide polymorphism variability
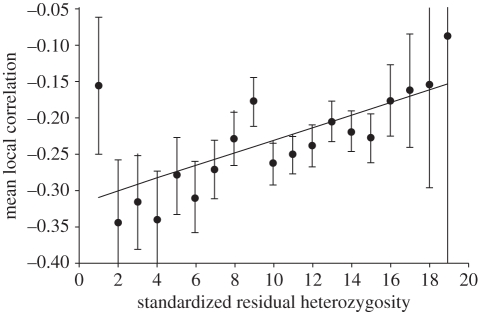
After excluding loci with extreme residual heterozygosity and where lack of sequence/primer information precluded inference of repeat number, data from a total of 4524 microsatellites were retained. Data were combined into 20 equal-width bins spanning the range of residual heterozygosity, standardized by subtracting the mean and dividing by the standard deviation, of −2.5 to 2.5. Within each bin, each microsatellite was placed at the centre of a symmetrical window (four sizes examined = ±10, ±25, ±50 and ±100 kb) and in each case the correlation coefficient of the relationship between SNP heterozygosity and distance from Africa was calculated based on the seven study populations. Figure 1 shows how the mean correlation coefficient varies across the 20 microsatellite bins for a window size of ±25 kb. A regression based on the data as shown is significant (r2 = 0.488, n = 19, p = 0.0009), but becomes appreciably stronger if the first data point, a major outlier, is removed (r2 = 0.812, n = 19, p = 3.3 × 10−7). The lowest bin is likely to be an outlier because very low heterozygosity can result from several processes other than selection, most obviously stabilization of the locus through internal point mutations [41]. The data point for the highest bin contained only a single locus and was omitted in both cases. Other window sizes yield substantially weaker associations, the narrowest window being non-significant and the two larger windows approaching significance (p ∼ 0.07 in both cases). In all cases, excluding the extreme bins yields stronger, more positive slopes. We believe our optimum window size lies at 25 kb because while smaller windows reflect well local conditions, they carry more statistical noise owing to the small number of SNPs included, and for larger windows the converse is true; with more reliable numbers of SNPs reducing stochastic noise but the larger regions tending to embrace more than one functional block.
Figure 1.
Relationship between residual expected microsatellite heterozygosity and the extent to which local heterozygosity was lost as humans colonized the world. Standardized residual heterozygosity is the standardized residual of the relationship between average repeat number and heterozygosity in Europeans, placed in 20 equal-width bins (bin 1 = −2.5 to –2.25 s.d. etc.). Bin 20 is omitted because it contained only one observation. Mean local correlation is the average correlation between local SNP heterozygosity (all SNPs within 25 kb of the microsatellite) and distance from Africa across seven worldwide populations. Error bars are ±1 standard error.
(b). Single nucleotide polymorphism variability and GO codes
Using the ‘best’ bin size determined from the microsatellite analysis, 25 kb, we next analysed a list of 65508 genes and gene functions downloaded from the GO website. Multiple GO codes for the same gene (defined as having the same start and stop location) were treated as separate entries and gene location was taken as the mid-point of the gene. Having determined the local SNP slope at each locus, mean slopes were calculated for each GO code found, with qualifying codes having more than five different genes. To assess whether immune-related genes tend to have extreme slopes, suggesting selection, all retained GO codes (n = 1308) were classified blind by one of us (C.B. presented with an alphabetically ordered list of gene classes without any inferred selection coefficients) as to whether they were or were not directly linked to immune function. Examples include ‘defence against bacteria’, ‘positive regulation of chemokine biosynthetic process’ and ‘natural killer cell activation’ (for full list, see the electronic supplementary material, table S1). Attempts to use the GO coding system directly failed because key descriptors such as ‘immune response’, while capturing many relevant genes, also exclude many legitimate and important classes (e.g. ‘I-κB kinase/NF-κB cascade’) which would have to be added manually. After sorting by mean slope, the frequencies of immune genes were determined for each consecutive block of 100 codes (figure 2). The two highest bin counts are found in the highest and lowest mean slope classes, significantly more often than expected by chance (χ21 = 7.43, p = 0.006). The 24 GO codes associated with the strongest positive and negative slopes are listed in table 1. Note that the standard errors of GO codes with negative slopes tend to be appreciably lower than those of the top positive slopes, despite being based on similar numbers of genes, suggesting that the selective forces acting on genes in code classes that yield positive slopes are more heterogeneous. Finally, to get an idea of the level of non-independence, we also estimated the correlation between the slopes of adjacent genes, classified according to genomic separation (end of gene1, start of gene2) in 10 kb bins, finding a decline from r = 0.62 (genes less than 10 kb apart) down to r = 0.32 (genes separated by 190–200 kb), suggesting that only extremely close genes will have similar slopes owing to proximity alone.
Figure 2.
Distribution of genes with immune function GO codes with respect to the extent to which local heterozygosity was lost as humans colonized the world. A total of 94 GO codes out of 1308 with six or more representative genes were considered immune-related. After calculating the mean correlation between local SNP heterozygosity (all SNPs within 25 kb of the centre of the gene) and distance from Africa across seven worldwide populations for all genes and averaging within each GO code, the codes were ordered according to their mean slope and the number of immune genes in each block of 100 codes counted. Thus, the 100 codes that gave the most positive correlations had an average correlation of 0.17 and included 13 codes that were deemed immune-related.
Table 1.
Summary of immune-related gene classes lying in genomic regions where unusually high or low levels of genetic variability were lost as modern humans colonized the world from Africa. (GO code is the Gene Ontology code with its associated description of the gene class function. Slope is the average correlation coefficient between local SNP heterozygosity and distance from Africa across seven worldwide populations with standard error in parentheses. n is the number of occurrences of genes of that code. Codes above the line are in the 100 most negative slopes, indicative of purifying selections, while codes below the line are in the top 100 positive values, indicative of diversifying or balancing selection.)
| GO code | description of function | corr | n |
|---|---|---|---|
| 16032 | viral reproduction | −0.71 (0.13) | 6 |
| 19047 | provirus integration | −0.7 (0.08) | 8 |
| 30889 | negative regulation of B cell proliferation | −0.68 (0.12) | 7 |
| 33077 | T cell differentiation in the thymus | −0.65 (0.09) | 11 |
| 50830 | defence response to Gram-positive bacterium | −0.6 (0.11) | 14 |
| 43280 | positive regulation of caspase activity | −0.6 (0.15) | 6 |
| 50718 | positive regulation of interleukin-1 beta secretion | −0.6 (0.13) | 11 |
| 19059 | initiation of viral infection | −0.6 (0.11) | 11 |
| 42116 | macrophage activation | −0.53 (0.22) | 6 |
| 42098 | T cell proliferation | −0.52 (0.13) | 15 |
| 6956 | complement activation | −0.5 (0.16) | 7 |
| 16064 | immunoglobulin mediated immune response | 0.04 (0.27) | 11 |
| 45060 | negative thymic T cell selection | 0.04 (0.2) | 9 |
| 32755 | positive regulation of interleukin-6 production | 0.05 (0.29) | 8 |
| 6911 | phagocytosis, engulfment | 0.08 (0.29) | 8 |
| 1782 | B cell homeostasis | 0.09 (0.3) | 7 |
| 45089 | positive regulation of innate immune response | 0.11 (0.28) | 6 |
| 50778 | positive regulation of immune response | 0.11 (0.28) | 7 |
| 19885 | antigen processing and presentation via MHC class I | 0.12 (0.22) | 8 |
| 48535 | lymph node development | 0.13 (0.2) | 10 |
| 45410 | positive regulation of interleukin-6 biosynthetic process | 0.15 (0.27) | 6 |
| 2504 | antigen processing via MHC class II | 0.28 (0.15) | 15 |
| 46718 | entry of virus into host cell | 0.34 (0.25) | 6 |
| 45059 | positive thymic T cell selection | 0.44 (0.24) | 6 |
(c). Single nucleotide polymorphism variability around 168 immune-related genes
Slopes were determined for each of the 168 genes studied by Walsh et al. [28], plus the gene APCS, which does not appear in the main list, but is discussed in the text. We also included Walsh et al.'s positive control, beta haemoglobin (HBB). Results are summarised in table 2. Several trends are apparent. First, the six genes identified as putatively under selection (IL9, CAV2, FUT2, ABCC1, VAV3 and APCS) and the positive control, HBB, tend to yield strongly negative slopes (−0.947, −0.908, −0.98, 0.67, −0.31, −0.912, −0.94, respectively). Indeed, IL9 and FUT2, and other members of the CAV and VAV gene families, CAV1 and VAV2, yield four of the 12 most negative values found. ABCC1 and VAV3 are very big genes (approx. 0.2 and 0.4 Mb, respectively), and both contain regions outside the window used that give strongly negative slopes, though other ATP-binding cassette (ABC) genes are also positive (see below).
Table 2.
Summary of inferred recent selection acting on 168 immune genes listed in Walsh et al. (Genes are listed by their official abbreviations and are listed in alphabetical order along with their location specified as chromosome (‘C’) and location in Megabases (‘Loc’). For each gene we calculated the Pearson's correlation coefficient, r, between local SNP heterozygosity (all SNPs within 25 kb of the centre of the gene) and distance from Africa across seven worldwide populations (‘corr’). CCL3L1 did not yield enough neighbouring SNPs for a meaningful correlation to be calculated. We also calculated correlations for APCS (correlation = −0.912) and the positive control, HBB (correlation = −0.94). Taking microsatellite locations (figure 1) as representative of random locations across the genome, the mean correlation coefficient is −0.236 (n = 4524). n.a., not applicable.)
| gene | Loc | C | corr | gene | Loc | C | corr | gene | Loc | C | corr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABCB1 | 87.1 | 7 | 0.39 | F11R | 159.3 | 1 | 0.63 | IL1F9 | 113.5 | 2 | −0.95 |
| ABCC1 | 16.0 | 16 | 0.68 | FACL6 | 131.3 | 5 | 0.52 | IL1R1 | 102.2 | 2 | 0.66 |
| ABCD3 | 94.7 | 1 | 0.69 | FCER1A | 157.5 | 1 | −0.28 | IL1R2 | 102.0 | 2 | −0.23 |
| ABCG2 | 89.3 | 4 | 0.83 | FCER1G | 159.5 | 1 | 0.92 | IL1RL1 | 102.3 | 2 | −0.76 |
| AGT | 228.9 | 1 | 0.91 | FCGR2A | 159.7 | 1 | 0.40 | IL1RL1LG | 10.8 | 19 | 0.74 |
| AIM2 | 157.3 | 1 | −0.33 | FCGR2B | 159.9 | 1 | −0.69 | IL1RL2 | 102.2 | 2 | 0.89 |
| APOBEC3G | 37.8 | 22 | −0.43 | FCGR3A | 159.8 | 1 | −0.28 | IL1RN | 113.6 | 2 | −0.23 |
| CAV1 | 116.0 | 7 | −0.98 | FCGR3B | 159.9 | 1 | −0.58 | IL21R | 27.3 | 16 | −0.67 |
| CAV2 | 115.9 | 7 | −0.85 | FLOT2 | 24.2 | 17 | 0.47 | IL3 | 131.4 | 5 | 0.39 |
| CAV3 | 8.8 | 3 | −0.76 | FUT2 | 53.9 | 19 | −0.97 | IL4 | 132.0 | 5 | −0.52 |
| CCL1 | 29.7 | 17 | −0.79 | FY | 157.4 | 1 | −0.79 | IL4R | 27.3 | 16 | −0.99 |
| CCL2 | 29.6 | 17 | −0.37 | FYN | 112.2 | 6 | −0.30 | IL5 | 131.9 | 5 | −0.04 |
| CCL3 | 29.7 | 17 | 0.61 | GC | 72.8 | 4 | −0.19 | IL6 | 22.7 | 7 | −0.59 |
| CCL3L1 | 31.3 | 17 | n.a. | HP | 70.7 | 16 | 0.54 | IL8 | 74.8 | 4 | 0.24 |
| CCL5 | 31.3 | 17 | −0.09 | HSPA4 | 132.4 | 5 | −0.48 | IL9 | 135.3 | 5 | −0.88 |
| CCL7 | 56.0 | 17 | 0.02 | HSPA9B | 137.9 | 5 | −0.61 | ILF3 | 10.6 | 19 | 0.86 |
| CCL8 | 31.4 | 17 | 0.77 | ICAM1 | 10.3 | 19 | −0.32 | IRF1 | 131.9 | 5 | −0.67 |
| CCL11 | 29.6 | 17 | 0.50 | ICAM2 | 59.4 | 17 | −0.88 | ITK | 156.6 | 5 | 0.14 |
| CCL13 | 56.0 | 17 | −0.73 | ICAM3 | 10.3 | 19 | 0.23 | ITLN1 | 159.1 | 1 | −0.55 |
| CCL14 | 31.4 | 17 | −0.55 | ICAM4 | 10.3 | 19 | 0.64 | ITLN2 | 195.2 | 1 | −0.88 |
| CCL16 | 31.4 | 17 | −0.72 | ICAM5 | 10.3 | 19 | 0.65 | LCK | 32.6 | 1 | −0.88 |
| CCL17 | 31.6 | 16 | 0.83 | IFI16 | 157.3 | 1 | −0.18 | LCP2 | 169.6 | 5 | −0.68 |
| CCL18 | 31.2 | 17 | 0.69 | IFIX | 157.2 | 1 | −0.90 | LMAN1 | 55.2 | 18 | −0.07 |
| CCL22 | 29.6 | 16 | −0.47 | IFNA1 | 21.4 | 9 | 0.43 | LY9 | 159.0 | 1 | 0.83 |
| CCL23 | 29.7 | 17 | −0.72 | IFNA10 | 21.2 | 9 | 0.95 | LYN | 57.0 | 8 | −0.83 |
| CCNT1 | 47.4 | 12 | 0.50 | IFNA13 | 21.4 | 9 | 0.31 | MAL | 95.1 | 2 | 0.78 |
| CCR1 | 46.2 | 3 | −0.91 | IFNA14 | 21.2 | 9 | 0.88 | MBL2 | 54.2 | 10 | −0.68 |
| CCR3 | 46.3 | 3 | 0.58 | IFNA16 | 21.2 | 9 | 0.88 | MMP28 | 31.1 | 17 | −0.71 |
| CCR9 | 45.9 | 3 | −0.47 | IFNA17 | 21.2 | 9 | 0.78 | MNDA | 157.1 | 1 | 0.22 |
| CD14 | 140.0 | 5 | −0.54 | IFNA2 | 21.4 | 9 | 0.81 | NCL | 232.0 | 2 | 0.81 |
| CD244 | 159.1 | 1 | 0.77 | IFNA21 | 21.2 | 9 | −0.39 | NFATC1 | 75.3 | 18 | −0.32 |
| CD28 | 204.3 | 2 | −0.35 | IFNA4 | 21.2 | 9 | 0.73 | NOS2A | 23.1 | 17 | −0.67 |
| CD4 | 6.8 | 12 | 0.75 | IFNA5 | 21.3 | 9 | 0.09 | PF4 | 75.1 | 4 | −0.95 |
| CD48 | 158.9 | 1 | −0.72 | IFNA6 | 21.3 | 9 | 0.35 | PF4V1 | 74.9 | 4 | 0.10 |
| CD58 | 116.9 | 1 | 0.40 | IFNA7 | 21.2 | 9 | 0.72 | PHB | 44.8 | 17 | 0.76 |
| CD84 | 158.8 | 1 | −0.85 | IFNA8 | 21.4 | 9 | −0.83 | PPBP | 75.1 | 4 | −0.96 |
| CRP | 157.9 | 1 | −0.92 | IFNAR1 | 33.6 | 21 | −0.84 | PPIA | 44.8 | 7 | −0.86 |
| CSF2 | 131.4 | 5 | −0.52 | IFNAR2 | 33.5 | 21 | 0.26 | PTPRC | 196.9 | 1 | −0.38 |
| CX3CL1 | 56.0 | 16 | −0.17 | IFNB1 | 21.1 | 9 | −0.52 | PVRL4 | 159.3 | 1 | 0.91 |
| CXCL1 | 75.0 | 4 | 0.90 | IFNG | 66.8 | 12 | −0.52 | RNPC2 | 33.8 | 20 | −0.84 |
| CXCL2 | 77.2 | 4 | 0.64 | IFNGR2 | 33.7 | 21 | 0.34 | SLAMF1 | 158.9 | 1 | −0.68 |
| CXCL3 | 77.2 | 4 | −0.50 | IFNW1 | 21.1 | 9 | 0.75 | SLAMF6 | 158.7 | 1 | 0.72 |
| CXCL5 | 78.7 | 4 | −0.97 | IGSF4B | 157.4 | 1 | −0.95 | SLAMF7 | 159.0 | 1 | −0.61 |
| CXCL6 | 75.2 | 4 | −0.81 | IGSF8 | 158.3 | 1 | 0.19 | SLAMF8 | 158.1 | 1 | −0.91 |
| CXCL9 | 75.1 | 4 | 0.42 | IGSF9 | 158.2 | 1 | 0.26 | SLAMF9 | 158.2 | 1 | −0.92 |
| CXCL10 | 75.1 | 4 | −0.47 | IL10RB | 33.6 | 21 | −0.76 | SLC11A1 | 219.0 | 2 | −0.87 |
| CXCL11 | 74.9 | 4 | −0.67 | IL13 | 132.0 | 5 | −0.55 | SLPI | 43.3 | 20 | −0.85 |
| CXCL13 | 77.1 | 4 | −0.48 | IL18R1 | 102.4 | 2 | 0.16 | SPBPBP | 75.1 | 4 | −0.72 |
| DEFA1 | 6.8 | 8 | 0.32 | IL18RAP | 102.4 | 2 | 0.20 | STOM | 123.2 | 9 | −0.67 |
| DEFA3 | 6.9 | 8 | 0.83 | IL1A | 113.3 | 2 | −0.60 | STOML1 | 72.1 | 15 | 0.81 |
| DEFA4 | 6.8 | 8 | 0.85 | IL1B | 113.3 | 2 | 0.60 | SYK | 92.7 | 9 | −0.40 |
| DEFA5 | 6.9 | 8 | 0.81 | IL1F10 | 113.5 | 2 | −0.95 | TGFB1 | 46.5 | 19 | −0.67 |
| DEFA6 | 6.8 | 8 | 0.91 | IL1F5 | 113.5 | 2 | −0.92 | THY1 | 118.8 | 11 | −0.90 |
| DEFB1 | 6.7 | 8 | −0.52 | IL1F6 | 113.5 | 2 | −0.76 | VAV1 | 6.8 | 19 | −0.91 |
| DEFT1 | 6.8 | 8 | 0.32 | IL1F7 | 113.4 | 2 | −0.86 | VAV2 | 135.7 | 9 | −0.97 |
| ETF1 | 137.9 | 5 | −0.82 | IL1F8 | 113.5 | 2 | −0.34 | VAV3 | 108.1 | 1 | −0.95 |
The second trend is for genes with similar names to yield similar slopes. A rigorous analysis is hampered both by non-independence owing to gene clustering and the fact that our understanding of function is insufficiently complete to group genes accurately by function. Some genes with similar names may have very different functions in terms of the precise role they play. Nonetheless, several groupings stand out. All three CAV and all three VAV genes have strongly negative values, despite lying on multiple chromosomes. Similarly, all four ABC genes, all five alpha defensin (DEFA) genes and 11 of 13 interferon alpha (IFNA) genes have positive/strongly positive slopes. Interestingly, although the DEFA genes all form a single cluster, DEFT1 lies within this cluster and has a negative slope, indicating that the generally positive slopes are not owing entirely to linkage disequilibrium. IFNA genes also form a cluster on chromosome 9, but the cluster is big enough (275 kb) to contain contrasting slopes and the two group members with negative slopes lie at either end.
4. Discussion
We show that microsatellites which are more heterozygous than expected for their repeat number tend to lie in genomic regions where SNP variability either fails to decline or actually increases with distance from Africa. Assays of regions around human genes reveal how key immune gene classes tend to show extreme SNP slopes, with antigen presentation genes having the most positive slopes and genes associated with defence against bacterial infection showing the most negative. Focusing on 168 immune genes studied previously [28], we find good agreement with the original conclusions in terms of genes experiencing directional selection, but also identify several candidate gene families that appear to be under balancing selection.
Previous methods for detecting the action of natural selection on the human genome have met with mixed success [4]. Apart from the obvious problem of false positives that applies to all genome-wide analyses, many of the other methods rely on identifying regions of the genome with unusual characteristics, such as high levels of linkage disequilibrium or SNP density. Such approaches could be powerful with a complete understanding of how recombination and mutation events occur, but as yet we do not have this. Instead, it seems that recombination and mutation events tend to cluster with each other [42], and that rates can vary over periods of evolutionary time as short as that which separates humans and chimpanzees [11,32,43]. Also, mutations may be more common near to microdeletions [44] or simply to each other [33]. Such uncertainties make the interpretation of the distribution of SNPs within any given population difficult. Methods based on finding SNPs with unusually high differences in allele frequency among populations potentially overcome these issues, but are in turn hampered by ascertainment bias, the phenomenon in which the discovery process may favour SNPs with unusually large allele frequency differences among populations [45,46], which would exacerbate the (already non-trivial) issue of false positives.
Our new method offers two potentially important advantages over other methods. First, by comparing levels of variability among global populations relative to a well-defined expectation, the strong linear decline with distance from Africa, many of the problems associated with not knowing how patterns of linkage disequilibrium and mutations came to be distributed are avoided. Second, although ascertainment bias has the clear potential to enrich for SNPs that give large Fst values, our method averages heterozygosity over many SNPs, reducing greatly the impact of one or a few unusual markers. A further aspect of our approach is that it detects selection over a well-defined time scale, specifically the period in which humans colonized the world from Africa, somewhat over 50 000 years. On the one hand, this means our method is inappropriate, for example, in detecting selection acting on humans before they left Africa. On the other hand, having a known period may allow substantial future refinement, for example by modelling the impact of recombination.
A further benefit of our method is that it detects several different forms of selection, including balancing selection. Balancing selection has previously proved difficult to detect [12], despite evidence that it affects a number of genomic regions [14,47]. The key issue is that the primary prediction of balancing selection, that of maintaining locally higher levels of heterozygosity [27], is difficult to distinguish within a population from other factors such as the presence of mutation hotspots [48–50]. However, when a population goes through a bottleneck and as a result suffers genome-wide loss of neutral diversity, those regions experiencing balancing selection should stand out as islands where diversity has been unusually retained. Our approach appears to show this, both through the fact that microsatellites with higher than expected variability for their repeat number lie in genomic regions where variability has not declined across the world, and through the fact that genes most known for balancing selection, those associated with antigen presentation, also lie in these areas.
Our approach remains somewhat crude. The analysis presented is based only on seven populations, three of which are in Africa, and using a point of origin for the decline of variability, Addis Ababa, which was chosen somewhat arbitrarily and which should probably be replaced by a location lying more in central southern Africa [19,23,25]. Use of more populations could help immensely, particularly the inclusion of populations from South America, the part of the world most distant from Africa. Another improvement involves ascertainment bias during the discovery process [46]. Although we believe that ascertainment bias impacts rather little on our analysis overall, there remains a concern that locally one or a few unusual SNPs could impact our analysis. Use of larger SNP datasets based on markers developed so as to minimize bias would help reduce this potential problem further. Arguably the biggest improvement is likely to be achieved through a more sophisticated statistical analysis. We currently treat all SNPs as equal and independent (in the sense that we do not recover phase), even though it is clear that recombination rates vary widely across the genome. Algorithms that reconstruct phase and estimate local recombination rates [32] have the potential to yield appreciably improved estimates of heterozygosity, based more on haplotype blocks than on individual SNPs. A further issue relates to gene classification. When analysing all genes together we were forced to use a pragmatic rule of counting many genes several times, one for each GO code attracted. While this should not bias our results in terms of creating consistently high or low slopes for immune genes, it is clearly sub optimal. More focused studies should, by their nature, be able to avoid this problem. For example, one might compare members of a gene family, some involved in immune function and some not.
Finally, it is worth considering how our method works in practice. Applied to a list of known, immune-related genes, we find that our method tends to yield strong negative slopes when applied to ‘hits’ generated by other tests. Strong negative slopes should indicate purifying selection, selection that has acted to accelerate the loss of diversity relative to neutral sites. This makes biological sense, in that the other tests are generally aimed at detecting patterns generated by this form of selection, and we can imagine that humans encountered many new pathogens as they moved into new areas and encountered new foods and new climates. However, we also find several gene clusters that yield strongly positive values, suggestive of balancing or diversifying selection. These include: ABC proteins, a group only recently recognized as being important in the immune system, but whose functions include regulation of antigen presentation [51]; defensins, specifically the DEFA group, involved with defence against bacteria and antitoxin activity [52]; and IFNA, a group of proteins with direct antiviral, antiproliferative and immunomodulatory properties [53]. Across all qualifying GO codes, we find that immune-related genes are over-represented both in genes yielding extremely high and extremely low slopes, suggesting that immune genes in general are more likely than average to be under selection.
In conclusion, we present a new method for detecting the action of natural selection on the human genome that exploits our unusual demographic history. By using as our null hypothesis the changes in diversity that are known to have occurred when humans moved out of Africa to colonize the world, we bypass many of the uncertainties that attach to other approaches. Our method appears effective in pinpointing immune-related genes as foci for natural selection, supporting the findings of other studies [13]. Future expansion of SNP datasets to embrace further populations and rigorous modelling to determine null distributions for our measure should increase its power.
References
- 1.Brookes A. J. 1999. The essence of SNPs. Gene 234, 177–186 10.1016/S0378-1119(99)00219-X (doi:10.1016/S0378-1119(99)00219-X) [DOI] [PubMed] [Google Scholar]
- 2.Hinds D. A., Stuve L. L., Nilsen G. B., Halperin E., Eskin E., Balinger D. G., Frazer K. A., Cox D. R. 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 10.1126/science.1105436 (doi:10.1126/science.1105436) [DOI] [PubMed] [Google Scholar]
- 3.The International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–853 10.1038/nature06258 (doi:10.1038/nature06258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oleksyk T. K., Smith M. W., O'Brien S. J. 2010. Genome-wide scans for footprints of natural selection. Phil. Trans. R. Soc. B 365, 185–205 10.1098/rstb.2009.0219 (doi:10.1098/rstb.2009.0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabeti P. C., et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–919 10.1038/nature06250 (doi:10.1038/nature06250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sethupathy P., Giang H., Plotkin J. B., Hannenhalli S. 2008. Genome-wide analysis of natural selection on human cis-elements. PLoS ONE 3, e3137. 10.1371/journal.pone.0003137 (doi:10.1371/journal.pone.0003137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreiro L. B., Laval G., Quach H., Patin E., Quintana-Murci L. 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40, 340–345 10.1038/ng.78 (doi:10.1038/ng.78) [DOI] [PubMed] [Google Scholar]
- 8.Amos W. 2010. Heterozygosity and mutation rate: evidence for an interaction and its implications. BioEssays 32, 82–90 10.1002/bies.200900108 (doi:10.1002/bies.200900108) [DOI] [PubMed] [Google Scholar]
- 9.Drake J. W. 2007. Too many mutants with multiple mutations. Crit. Rev. Biochem. Mol. Biol. 42, 247–258 10.1080/10409230701495631 (doi:10.1080/10409230701495631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninio J. 1996. Gene conversion as a focusing mechanism for correlated mutations: a hypothesis. Mol. Gen. Genet. 251, 503–508 10.1007/BF02173638 (doi:10.1007/BF02173638) [DOI] [PubMed] [Google Scholar]
- 11.Ptak S., Hinds D. A., Koehler K., Nickel B., Patil N., Ballinger D. G., Przeworski M., Frazer K. A., Pääbo S. 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37, 429–434 10.1038/ng1529 (doi:10.1038/ng1529) [DOI] [PubMed] [Google Scholar]
- 12.Bubb K. L., et al. 2006. Scan of human genome reveals no new loci under ancient balancing selection. Genetics 173, 2165–2177 10.1534/genetics.106.055715 (doi:10.1534/genetics.106.055715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrés A. M., et al. 2009. Targets of balancing selection in the human genome. Mol. Biol. Evol. 26, 2755–2764 10.1093/molbev/msp190 (doi:10.1093/molbev/msp190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyons E. J., et al. 2009. Homozygosity and risk of childhood death due to invasive bacterial disease. BMC Med. Genet. 10, 55. 10.1186/1471-2350-10-55 (doi:10.1186/1471-2350-10-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prugnolle F., Manica A., Charpentier M., Guégan J. F., Guernier V., Balloux F. 2005. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027 10.1016/j.cub.2005.04.050 (doi:10.1016/j.cub.2005.04.050) [DOI] [PubMed] [Google Scholar]
- 16.Acevedo-Whitehouse K., Spraker T. R., Lyons E., Melin S. R., Gulland F., DeLong R. L., Amos W. 2006. Contrasting effects of heterozygosity on survival and hookworm resistance in California sealion pups. Mol. Ecol. 15, 1973–1982 10.1111/j.1365-294X.2006.02903.x (doi:10.1111/j.1365-294X.2006.02903.x) [DOI] [PubMed] [Google Scholar]
- 17.Reid J. M., Arcese P., Keller L. F. 2003. Inbreeding depresses immune response in song sparrows (Melospiza melodia): direct and inter-generational effects. Proc. R. Soc. Lond. B 270, 2151–2157 10.1098/rspb.2003.2480 (doi:10.1098/rspb.2003.2480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harpending H., Rogers A. 2000. Genetic perspectives on human origins and differentiation. Ann. Rev. Genomics Hum. Genet. 1, 361–385 10.1146/annurev.genom.1.1.361 (doi:10.1146/annurev.genom.1.1.361) [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Prugnolle F., Manica A., Balloux F. 2006. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 79, 230–237 10.1086/505436 (doi:10.1086/505436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macaulay V., et al. 2005. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308, 1034–1036 10.1126/science.1109792 (doi:10.1126/science.1109792) [DOI] [PubMed] [Google Scholar]
- 21.Amos W., Hoffman J. I. 2010. Evidence that two main bottleneck events shaped modern human genetic diversity. Proc. R. Soc. B 277, 131–137 10.1098/rspb.2009.1473 (doi:10.1098/rspb.2009.1473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prugnolle F., Manica A., Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160 10.1016/j.cub.2005.02.038 (doi:10.1016/j.cub.2005.02.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., Cavalli-Sforza L. L. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15 942–15 947 10.1073/pnas.0507611102 (doi:10.1073/pnas.0507611102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J. Z., et al. 2008. Worldwide human relationships inferred from genomewide patterns of variation. Science 319, 1100–1104 10.1126/science.1153717 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 25.Manica A., Amos W., Balloux F. 2007. The effect of ancient population bottlenecks on human phenotypes. Nature 448, 346–348 10.1038/nature05951 (doi:10.1038/nature05951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linz B., et al. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918 10.1038/nature05562 (doi:10.1038/nature05562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlesworth B., Nordberg M., Charlesworth D. 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. Camb. 70, 155–174 10.1017/S0016672397002954 (doi:10.1017/S0016672397002954) [DOI] [PubMed] [Google Scholar]
- 28.Walsh E. C., et al. 2006. Searching for signals of evolutionary selection in 168 genes related to immune function. Hum. Genet. 119, 92–102 10.1007/s00439-005-0090-0 (doi:10.1007/s00439-005-0090-0) [DOI] [PubMed] [Google Scholar]
- 29.Nachman M. W. 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17, 481–485 10.1016/S0168-9525(01)02409-X (doi:10.1016/S0168-9525(01)02409-X) [DOI] [PubMed] [Google Scholar]
- 30.Spencer C. C. A., Deloukas P., Hunt S., Mullikin J., Myers S., Silverman B., Donnelly P., Bentley D., McVean G. 2006. The influence of recombination on human genetic diversity. PLoS Genet. 2, e148. 10.1371/journal.pgen.0020148 (doi:10.1371/journal.pgen.0020148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffreys A. J., Neumann R., Panayi M., Myers S., Donelly P. 2005. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 37, 601–606 10.1038/ng1565 (doi:10.1038/ng1565) [DOI] [PubMed] [Google Scholar]
- 32.Winckler W., et al. 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308, 107–111 10.1126/science.1105322 (doi:10.1126/science.1105322) [DOI] [PubMed] [Google Scholar]
- 33.Amos W. 2010. Even small SNP clusters are non-randomly distributed: is this evidence of mutational non-independence? Proc. R. Soc. B 277, 1443–1449 10.1098/rspb.2009.1757 (doi:10.1098/rspb.2009.1757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen R., et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3, e170. 10.1371/journal.pbio.0030170 (doi:10.1371/journal.pbio.0030170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brinkmann B., Klintschar M., Neuhuber F., Hühne J., Rolf B. 1998. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62, 1408–1415 10.1086/301869 (doi:10.1086/301869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber J. L. 1990. Informativeness of human (dC-dA)n. (dG-dT)n polymorphisms. Genomics 7, 524–530 10.1016/0888-7543(90)90195-Z (doi:10.1016/0888-7543(90)90195-Z) [DOI] [PubMed] [Google Scholar]
- 37.Weber J. L., Wong C. 1993. Mutation of human short tandem repeats. Hum. Mol. Genet. 2, 1123–1128 10.1093/hmg/2.8.1123 (doi:10.1093/hmg/2.8.1123) [DOI] [PubMed] [Google Scholar]
- 38.The International HapMap 3 Consortium 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 10.1038/nature09298 (doi:10.1038/nature09298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dib C., et al. 1996. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380, 152–154 10.1038/380152a0 (doi:10.1038/380152a0) [DOI] [PubMed] [Google Scholar]
- 40.The Gene Ontology Consortium 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 10.1038/75556 (doi:10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin L., Macaubas C., Hallmayer J., Kimura A., Mignot E. 1996. Mutation rate varies among alleles at a microsatellite locus: phylogenetic evidence. Proc. Natl Acad. Sci. USA 93, 15 285–15 288 10.1073/pnas.93.26.15285 (doi:10.1073/pnas.93.26.15285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lercher M. J., Hurst L. D. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18, 337–340 10.1016/S0168-9525(02)02669-0 (doi:10.1016/S0168-9525(02)02669-0) [DOI] [PubMed] [Google Scholar]
- 43.Jeffreys A. J., Neumann R. 2009. The rise and fall of a human recombination hotspot. Nat. Genet. 41, 625–629 10.1038/ng.346 (doi:10.1038/ng.346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian D., et al. 2008. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455, 105–109 10.1038/nature07175 (doi:10.1038/nature07175) [DOI] [PubMed] [Google Scholar]
- 45.Akey J. M., Zhang K., Xiong M., Jin L. 2003. The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequeilibrium. Mol. Biol. Evol. 20, 232–242 10.1093/molbev/msg032 (doi:10.1093/molbev/msg032) [DOI] [PubMed] [Google Scholar]
- 46.Wakeley J., Nielsen R., Liu-Cordero S. N., Ardlie K. 2001. The discovery of single-nucleotide polymorphisms: and inferences about human demographic history. Am. J. Hum. Genet. 69, 1332–1347 10.1086/324521 (doi:10.1086/324521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollox E. J., Armour J. A. L. 2008. Directional and balancing selection in human beta-defensins. BMC Evol. Biol. 8, 113. 10.1186/1471-2148-8-113 (doi:10.1186/1471-2148-8-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffreys A. J., May C. A. 2004. Intense and highly localized gene conversion activity in human meiotic crossover hotspots. Nat. Genet. 36, 151–156 10.1038/ng1287 (doi:10.1038/ng1287) [DOI] [PubMed] [Google Scholar]
- 49.Rogozin I. B., Pavlov Y. I. 2003. Theoretical analysis of mutation hotspots and their sequence context specificity. Mut. Res. 544, 65–85 10.1016/S1383-5742(03)00032-2 (doi:10.1016/S1383-5742(03)00032-2) [DOI] [PubMed] [Google Scholar]
- 50.Tenaillon M. I., Austerlitz F., Tenaillon O. 2008. Apparent mutational hotspots and long distance linkage disequilibrium resulting from a bottleneck. J. Evol. Biol. 21, 541–550 10.1111/j.1420-9101.2007.01490.x (doi:10.1111/j.1420-9101.2007.01490.x) [DOI] [PubMed] [Google Scholar]
- 51.Van de Ven R., Scheffer G. L., Scheper R. J., de Gruijl T. D. 2009. The ABC of dendritic cell development and function. Trends Immunol. 30, 421–429 10.1016/j.it.2009.06.004 (doi:10.1016/j.it.2009.06.004) [DOI] [PubMed] [Google Scholar]
- 52.Menendez A., Finlay B. B. 2007. Defensins in the immunology of bacterial infections. Curr. Opin. Immunol. 19, 385–391 10.1016/j.coi.2007.06.008 (doi:10.1016/j.coi.2007.06.008) [DOI] [PubMed] [Google Scholar]
- 53.Sozzani S., Bosisio D., Scarsi M., Tincani A. 2010. Type I interferons in systemic immunity. Autoimmunity 43, 196–203 10.3109/08916930903510872 (doi:10.3109/08916930903510872) [DOI] [PubMed] [Google Scholar]