Abstract
Purpose
Keratoconus (KTCN) is a non-inflammatory, usually bilateral disorder of the eye which results in the conical shape and the progressive thinning of the cornea. Several studies have suggested that genetic factors play a role in the etiology of the disease. Several loci were previously described as possible candidate regions for familial KTCN; however, no causative mutations in any genes have been identified for any of these loci. The purpose of this study was to evaluate role of the collagen genes collagen type IV, alpha-1 (COL4A1) and collagen type IV, alpha-2 (COL4A2) in KTCN in Ecuadorian families.
Methods
COL4A1 and COL4A2 in 15 Ecuadorian KTCN families were examined with polymerase chain reaction amplification, and direct sequencing of all exons, promoter and intron-exon junctions was performed.
Results
Screening of COL4A1 and COL4A2 revealed numerous alterations in coding and non-coding regions of both genes. We detected three missense substitutions in COL4A1: c.19G>C (Val7Leu), c.1663A>C (Thr555Pro), and c.4002A>C (Gln1334His). Five non-synonymous variants were identified in COL4A2: c.574G>T (Val192Phe), c.1550G>A (Arg517Lys), c.2048G>C (Gly683Ala), c.2102A>G (Lys701Arg), and c.2152C>T (Pro718Ser). None of the identified sequence variants completely segregated with the affected phenotype. The Gln1334His variant was possibly damaging to protein function and structure.
Conclusions
This is the first mutation screening of COL4A1 and COL4A2 genes in families with KTCN and linkage to a locus close to these genes. Analysis of COL4A1 and COL4A2 revealed no mutations indicating that other genes are involved in KTCN causation in Ecuadorian families.
Introduction
Keratoconus (KTCN, OMIM 148300) is a non-inflammatory, usually bilateral disorder of the eye, characterized by progressive thinning and protrusion of the central cornea which results in altered refractive powers and loss of visual acuity [1]. The prevalence of the disease is estimated to be 1 in 2,000 individuals, and is the most common ectatic disorder of the cornea [1]. KTCN afflicts males and females in all ethnic groups [1]. Signs and symptoms depend on the stage of disease, with the first signs usually appearing in the third decade of life [1,2]. The cause of KTCN is still unknown; both genetic and environmental factors seem to play a role in its etiology. Although most cases of KTCN are isolated, an association with many syndromes, such as Down syndrome [3], Ehlers-Danlos syndrome [4], and Leber congenital amaurosis [5] has been described. Furthermore, extensive studies have shown an association between KTCN and constant eye rubbing [6], contact lens wear [7], or atopy [8]. Usually, KTCN is a sporadic disorder, but positive family history has been observed in 6%–8% of cases [1]. An autosomal dominant inheritance pattern with reduced penetrance has been suggested in 90% of patients with familial KTCN [9,10].
Genomewide linkage analyses have indicated several loci involved in the etiology of familial KTCN at 16q22.3-q23.1 (KTCN2; OMIM 608932), 3p14-q13 (KTCN3; OMIM 608586), 2p24 (KTCN4; OMIM 609271), 1p36.23–36.21, 5q14.3-q21.1, 5q21.2, 5q32-q33, 8q13.1-q21.11, 9q34, 14q11.2, 14q24.3, 15q2.32, 15q22.33-q24.2, 17p13, and 20q12 [10-20]. However, no mutations in any genes at any of these loci have been associated with KTCN.
We have demonstrated an evidence of linkage to a novel locus at 13q32 [21]. Collagen type IV, alpha-1 (COL4A1; OMIM 120130) and collagen type IV, alpha-2 (COL4A2; OMIM 120090) are mapped in close proximity to that locus. The COL4A1 and COL4A2 genes are organized in a head-to-head conformation [22]. These gene pairs share a common promoter, and each gene is transcribed in opposite directions [23]. The COL4A1 gene is placed on the minus strand and consists of 52 exons, while the COL4A2 gene is on the opposite strand and consists of 48 exons. They encode two of six collagen type IV chains – α1 and α2 (1,669 and 1,712 amino acids, respectively) – forming a heterotrimeric protein molecule of collagen type IV (α1α1α2), which is found in the structure of the basement membrane (BM) [22,23]. Each chain contains three domains: an NH2-terminal 7S domain, a major collagenous domain with Gly-X-Y repeats (the X position is frequently occupied by proline, whereas the Y position is often occupied by 4-hydroxyproline) and a non-collagenous domain (NC1) at the COOH-terminus. Repetitions of the Gly-X-Y motif determine the formation of the triple-helical structure of collagen [22].
Collagens are the major protein components of the human cornea, and several types of collagen, including collagen type IV, have been identified [24]. Biochemical studies have revealed thinning of corneas from patients with KTCN, which may occur as a result of a reduced amount of total collagen proteins [25] and changes in collagen fibers orientation [26]. Moreover, a cornea affected by KTCN contains defects in BM and alterations in the BM composition [27]. The presence of collagen type IV in normal human cornea has remained unclear [28]. Results from expression arrays have shown an expression of COL4A1 in transplant-quality human donor corneas [29] and a downregulation of COL4A1 in keratoconus corneas [30]. Immunohistochemical studies have found collagen type IV α1/α2 chains in keratoconus corneas in large defect sites [28]. In light of these results, we recognize COL4A1 and COL4A2 as candidate genes for KTCN.
The purpose of this study was to screen COL4A1 and COL4A2 genes and determine whether sequence variants in these genes are involved in the causation of KTCN in Ecuadorian families.
Methods
Subjects
Twenty-three individuals from family KTCN-014, 25 affected individuals from other Ecuadorian families with KTCN, and 64 Ecuadorian control subjects were included in the study. The pedigrees of these families have been described elsewhere [21]. All individuals were examined in the Hospital Metropolitano in Quito, Ecuador, undergoing a complete ophthalmic evaluation as previously described [21]. The possible consequences of the study were explained and informed consent was obtained from all family members, according to the Declaration of Helsinki. Study protocol was approved by both the Institutional Review Board at Washington State University Spokane, Spokane, WA and Poznan University of Medical Sciences (Poland).
Sequencing analyses
Oligonucleotide primers were designed to amplify all coding sequences and intron-exon junctions, promoter, and UTRs of both COL4A1 and COL4A2 (Table 1). PCR amplifications were performed using Taq DNA Polymerase (Fermentas Inc., Glen Burnie, MD). PCR products were purified with ExoSAP-IT® (USB Corporation, Cleveland, OH) or Montage® PCR Filter Units (Millipore, Jaffrey, NH) and sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc. [ABI], Foster City, CA). Sequencing was visualized on an ABI PRISM® 3100 Genetic Analyzer (ABI) and a 3730xl DNA Analyzer (ABI). The DNA sequences of study subjects were compared with the reference sequences of COL4A1 and COL4A2 (GRCh37/hg19, GenBank accession numbers for the mRNA NM_001845.4 and NM_001846.2, respectively) using Sequencher® 4.1.4. Software (Gene Codes Corporation, Ann Arbor, MI).
Table 1. Primer sequences and annealing temperature used to PCR amplifications of COL4A1 and COL4A2 fragments.
| Name | Forward | Reverse | Annealing Temperature (°C) | Amplicon Size (bp) |
|---|---|---|---|---|
| COL4A1.1 |
CACCCTCCCCCTTTCTACTC |
GCCCAGAGAATGCACCTG |
59 |
837 |
| COL4A1.2 |
TTGGGCTGAGTAACACTTGG |
GCCTGGTTTGGCTTCATTTG |
58 |
459 |
| COL4A1.3–4 |
GGGCAACAGAATGAGACTCC |
TGTGAGCTGGGAGAGGAGAT |
66 |
477 |
| COL4A1.5–6 |
TGCTCTGTCTGCTTTGTGTG |
ACAAGCTGTGCTACTGGGTA |
60 |
698 |
| COL4A1.7–8 |
CCAACAAATGAAGGGTAGGG |
TGTGCCAAGTGTCTGAACG |
58 |
578 |
| COL4A1.9–10 |
CCTTTGCTTTGCCGTCTCTA |
TCATCATCCCTTTCCCACAG |
60 |
691 |
| COL4A1.11 |
GGAGATGGATTGGTATTGGT |
GACTAAGGGATGGATGAAAG |
58 |
451 |
| COL4A1.12 |
GGGACAAAGCTATTGCCTGA |
GACATTGATCCAAAGGTGGG |
58 |
239 |
| COL4A1.13 |
GCAGAGGCAAGGATGATTAG |
GGGGCTCGTATTTTATGGAC |
58 |
393 |
| COL4A1.14–15 |
CCCTGCCCTGCTTACATT |
GTCCCTACGAGCCTTTTCTG |
60 |
505 |
| COL4A1.16–17 |
TTAGTGGAGACGGGATTTCG |
AACTGCCTGCTTGTGTATGC |
60 |
725 |
| COL4A1.18 |
GATGGGACAAGTATCTGGGC |
CATCTCCTCTCCTTCCTCTC |
60 |
459 |
| COL4A1.19 |
GCTACCATTGCTGCTACTTCAC |
AATAGAAAGCGTGGGGAGAG |
62 |
447 |
| COL4A1.20 |
GTCACAACAGGCTTCAGGAG |
CCCAGGAGAGACATAAGGGT |
60 |
486 |
| COL4A1.21 |
CAGTGATGGTCTGGTTGGAT |
ATGCCAGGAGTCTCAGAGGT |
60 |
532 |
| COL4A1.22 |
TGGGTGGTGTGTGGTGATTA |
GAGAAGGGGCAAAACTCTGA |
60 |
516 |
| COL4A1.23 |
TTCCACCCATTAGCAGAGAG |
GCCAACACACCAAAGCAA |
60 |
304 |
| COL4A1.24 |
GTCCGTCTTGGGCATTTTAG |
ATTTGGGCTCTGTGGGTAAC |
60 |
431 |
| COL4A1.25 |
GTGCCCAAAGCCACACTATA |
TGTTCAGTTCCCCCAAATGC |
60 |
718 |
| COL4A1.26 |
CCTGGGAGGGTAGATGAAGT |
GAAAGGGAGGCACAAAAGG |
62 |
488 |
| COL4A1.27–28 |
AAGTGGAGAACACAGGCAGA |
TCTTCCCAACCAAACCCTAC |
56 |
636 |
| COL4A1.29 |
AGGTGCTGGAAGAGACAGCA |
GCTGAGGCTGAGAAACCATC |
60 |
678 |
| COL4A1.30 |
GCTTGAAAAGGGTTGAGCAG |
GGCCTCTAAGATTTGCATCG |
64 |
315 |
| COL4A1.31 |
CAGAGCCCCTACCGAGTATA |
CAGTGGGTGGGAGAAGAATC |
61 |
483 |
| COL4A1.32–33 |
CATTCAAGTTCCCAGTGTGG |
GCCTTCTGCTTGATGTTCCT |
60 |
653 |
| COL4A1.34 |
CTCATTTACCTGGGGTTGGA |
TATGGAGGACCCGATAACCC |
60 |
411 |
| COL4A1.35–36 |
TGTGCCTTTCCTGGGTTATC |
AATGTCATCCATCCCTGAGC |
64 |
594 |
| COL4A1.37 |
GGGGGATTCACGTTCTTGTA |
TCCCTGTGTGTTATGGCTCA |
58 |
364 |
| COL4A1.38–39 |
TGGCAGGTAGAAACCAGATG |
TGAAGATGGGAGACAGGACA |
61 |
641 |
| COL4A1.40 |
GACCTCAGGAAAACCAGGTG |
GTAGTTGCAGGGATGTGCAG |
60 |
359 |
| COL4A1.41 |
TGGTGGTTCTGAGCTGAAAG |
CATGTGTCTTGCAGGCATTG |
60 |
447 |
| COL4A1.42 |
TAAAGAGAAGGAGGGATCGG |
TCTTCACCAGAACCCACAAG |
60 |
673 |
| COL4A1.43 |
CCTGCCTCGATTTCTGTCTC |
TAGTGGGGATGTGGGAGTGT |
60 |
435 |
| COL4A1.44 |
CCACAAGGCACCATTTGTTC |
TACAAATTGGGCTGCCACAC |
60 |
376 |
| COL4A1.45 |
GGACCAAAAACAGTGCCCTA |
GAGCCTTGGGAAGTTTCTGA |
60 |
790 |
| COL4A1.46 |
CCAGAATGCAGTGGGAAGTT |
TTCCTGGGTTTTCTTCTGGA |
60 |
590 |
| COL4A1.47 |
ACAGCAAGAAACCAGGGAGA |
GGCTGCCTTTCAACAACATC |
60 |
591 |
| COL4A1.48 |
TGAAGGAGGTAGGCTGCTGT |
CGCAGTGTTTCACTCGCTAC |
60 |
516 |
| COL4A1.49 |
TGTTGTGAAAGACATTGCCC |
GCCCAGCCAACTGACTTTTA |
60 |
650 |
| COL4A1.50 |
AAAACCAACGGGGAGGTACT |
TAAGCAGCGAGATGCAGAGA |
59 |
407 |
| COL4A1.51 |
GGAAGCAGCCATTAGACGAT |
AAATCGTCTCGGTCATCTGC |
60 |
573 |
| COL4A1.52.1 |
TACCAGGTTGAGGCCTGATG |
ACCTCCTAGCACCCTTTGGT |
65 |
530 |
| COL4A1.52.2 |
GAAAACCAAAGGGTGCTAGG |
CCGAATGTGCTTACGTGTGA |
65 |
793 |
| COL4A1.52.3 |
CCTGGCTTGAAAAACAGCTC |
AATCACCCCCAGTCTGTGAC |
60 |
429 |
| COL4A2.1 |
TCGTGGGAAAGCTCAGATAC |
AGACAAAGCGAGTTTAGCGC |
60 |
1454 |
| COL4A2.2 |
GCTTCTGGAAGGGCCAAT |
GGGAAAGGGAGGAAGAGAGA |
60 |
587 |
| COL4A2.3 |
CCTCATCCTGCGCTAAACTC |
ACACTTTCCTGGCCTCTACG |
60 |
625 |
| COL4A2.4 |
ATTTCAGGGGTGGGAGAGAC |
CGGCCATCTAGGTTTGTGTG |
60 |
467 |
| COL4A2.5–6 |
TTCTTTCATCCCAACCCAGT |
TCCCCACGTGTTTTATGTCA |
59 |
663 |
| COL4A2.7 |
AGACAGAAGAAACCCCGACA |
TCTTGGGCGTCAACATACAG |
60 |
515 |
| COL4A2.8 |
TCAGAATAACCCCCATCAGC |
AACAGATCAGCCCTATCAGGAC |
60 |
568 |
| COL4A2.9–10 |
AGGTCCTGATAGGGCTGATCT |
TAACTGGCAGAGAGCTGGTG |
59 |
551 |
| COL4A2.11 |
GCATCAGAAACCTCCATGC |
ACATTGGCCTCCCTACAACA |
59 |
556 |
| COL4A2.12 |
TCCAATCTCAGCTCCCACTC |
TGTCCTCACCTCCACCTTCT |
60 |
548 |
| COL4A2.13 |
GGAAACAACCCCACAGAAAC |
GGAGGACCCGGTTATGTTTT |
59 |
524 |
| COL4A2.14 |
GTAAACATCTGCCTGGAACG |
CTATGGACAAGGGGATGAGA |
58 |
469 |
| COL4A2.15 |
TGTCACTGCCTGTCCTCAGA |
CCCCAGTGCTAGATGTTCGT |
61 |
513 |
| COL4A2.16 |
ATTATTTCCCATCCCCACCT |
GCAAAAATGAGAGCCAAGGT |
59 |
473 |
| COL4A2.17 |
CCCAGTGTCTTCAACAACCA |
TGTCAGAGGCCGTGTATTTG |
59 |
505 |
| COL4A2.18 |
AGCACAGTCTCCTGGCATTC |
CAGGCAACATGAAGGTCTCC |
60 |
569 |
| COL4A2.19 |
TTCGAGCTTTGGACTCACCT |
CTGTGAAGGTGTCCAAAGCA |
60 |
521 |
| COL4A2.20 |
ACCCATCGGAGTTATTGACG |
TACAGGGCTTCAGCTTCCAT |
60 |
490 |
| COL4A2.21 |
CCTGCATCTGTGGTTGTCTC |
AAGTTCGCCTCCTCATCAAC |
59 |
609 |
| COL4A2.22 |
CCTCTGAATGTGGTCCCAGT |
AAAGTCCGCCTTGGGGTAT |
59 |
602 |
| COL4A2.23–24 |
ATCGCAGAAAGTGCTCCTTG |
ATGAGCAGCCTGTCCTATGC |
60 |
545 |
| COL4A2.25 |
TGGCACTAGGTTCCTGTTCA |
ACAGGAGAGGCTGCATGTTT |
59 |
553 |
| COL4A2.26 |
AAACATGCAGCCTCTCCTGT |
TTCTGACAAGAGGGGTTTGG |
60 |
492 |
| COL4A2.27 |
CCAGAATGGTAGCCGGTTT |
GCAAGACCAGTTTGTGCTGA |
60 |
318 |
| COL4A2.28 |
TAAGCCTGGAGGTGCTGTTT |
CCGAAACACCTGTCTCCTTT |
59 |
499 |
| COL4A2.29 |
GCGAAGGTTGTAGGTTCCAA |
TGCCAAGACAAACAGTGAGC |
60 |
708 |
| COL4A2.30 |
GAATAGACAAGGGCAGGAAGG |
CAGAGGATGAGCCGATGTCT |
60 |
581 |
| COL4A2.31 |
CACAGCCTCAACCTCCAGAT |
CAGGCAGGAGCAGTTTGTCT |
60 |
643 |
| COL4A2.32 |
TGCTCCTCTGCCTTTGTCTT |
TGTTGAGGCAGGGATAAAGC |
60 |
656 |
| COL4A2.33 |
TGGTCTCTCTCCAAGGCTTC |
ACCGAGGTTACTCAGGCATC |
59 |
442 |
| COL4A2.34 |
ACAGCACGTAGGACAGCAAA |
ACATCTGCATGGTGTCCAAG |
59 |
470 |
| COL4A2.35 |
GCTAAGCAAACCGCCTATGA |
ACAGGACTTTCCACTGGGACT |
60 |
416 |
| COL4A2.36 |
GGGAGTCCACAATTCAGAGC |
GACCCTTCGCTGTTTCTGAG |
59 |
629 |
| COL4A2.37 |
CCCATGCTTCTCTCCAATTC |
ATGCCTCTCTCCATTCCTGA |
60 |
446 |
| COL4A2.38 |
CTGCTGCTGCTTTCTGTGTT |
CCTGTGCTGCTATGTTGGTG |
60 |
626 |
| COL4A2.39 |
GTGCTGTCCCACACATGAAA |
AGTCCATTCAACCCAGCAAC |
61 |
510 |
| COL4A2.40 |
ATGGGCCTCGATCCTCTTAT |
AAACCAGCTCTTTCCTGCAC |
60 |
484 |
| COL4A2.41 |
CCCACCATGAGATGTTCCTT |
ATGACACAGGAGGAGCCATC |
60 |
427 |
| COL4A2.42–43 |
AGTCATTCCATGCCACAGAC |
TAAGCTCTCCATTCCCCAAG |
60 |
666 |
| COL4A2.44–45 |
CCCGTTAGTGTCTGGCTCAT |
AGGTGTTCTGCTGGGCATAG |
60 |
744 |
| COL4A2.46 |
GAAACTGCCCTGCACTCCT |
TAGATGGACCCTTCCGTCAG |
60 |
664 |
| COL4A2.47 |
CACTCCCTGGTGATCCAACT |
CCAACTACCCTTGTGCAGTG |
60 |
675 |
| COL4A2.48.1 |
GGATGCCTCATGTCCGTATT |
TACATGGGTGTGTGCGAAGT |
60 |
689 |
| COL4A2.48.2 |
CATCCAGCAGCAGCACTTAG |
AGGTCTCCACTTCTGCCTGA |
59 |
530 |
| COL4A2.48.3 | CCTGCTTTCTACGCCAATGT | CTGGTTGGGGTGTTTTCTGT | 60 | 573 |
In the table, Amplicon Size represents length of the PCR product in base pairs (bp) and Annealing Temperature represents the annealing temperature of the primers used for PCR amplifications.
Haplotype analysis
PEDSTATS [31] was used to verify the structure of KTCN-014 family and identify potential Mendelian inconsistencies in the inheritance of single nucleotide polymorphisms (SNPs) in COL4A1 and COL4A2. For that region, to determine the full haplotypes inherited along with the substitutions occurring in affected individuals, a reconstruction of observed sequence variants was prepared using SimWalk2 [32,33]. Allele frequencies were set as equal. The location of genetic markers was determined on the basis of the Rutgers combined linkage-physical map of the human genome [34], either directly or by interpolation. Haplotype was generated with HaploPainter [35].
Statistical analysis for Gln1334His substitution
The difference in distribution of Gln1334His substitution between affected and unaffected individuals in family KTCN-014 was analyzed by Fisher's Exact Test for Count Data. Similarly, 25 affected individuals from the remaining KTCN families versus 64 Ecuadorian control individuals were compared using Fisher's Exact Test. The difference between the examined groups was considered significant if the value of probability (p) did not exceed 0.05.
Prediction of effect of amino acid substitutions on protein function
The potential impact of amino acid substitutions on the COL4A1 and COL4A2 proteins was examined using PolyPhen, SIFT, PMUT, PANTHER, and SNAP tools.
The PolyPhen tool predicts which missense substitution affects the structure and function of protein, and uses Position-Specific Independent Counts software to assign profile scores. These scores are the likelihood of the occurrence of a given amino acid at a specific position, compared to the likelihood of this amino acid occurring at any position (background frequency) [36].
The SIFT analytic tool, on the basis of gene sequences homology, evaluates conserved positions, and calculates a score for the amino acid change at a particular position. A score of <0.05 is considered as pathogenic and has a phenotypic effect on protein structure [37].
The PMUT calculates the pathological significance of non-synonymous amino acid substitution using neural networks (NN). NN output >0.5 is considered to be deleterious [38]. PANTHER estimates the likelihood of a particular amino acid’s change affecting protein function. On the basis of an alignment of evolutionarily related proteins, it generates the substitution Position-Specific Evolutionary Conservation (subPSEC). The subPSEC could achieve values from 0 (neutral) to about −10 (most likely to be deleterious). The value −3 is the cutoff point for functional significance, and corresponds to a Pdeleterious of 0.5. If the substitution occurs at a position not appearing in the multiple sequence alignment, a subPSEC score cannot be calculated and change is not likely to be pathogenic [39,40].
The SNAP tool predicts the functional consequences of exchanging amino acids using evolutionary conservation and structure/function relationships. The SNAP output shows prediction neutral or non-neutral, and the expected accuracy [41].
Results
Forty eight members of 15 Ecuadorian families and 64 Ecuadorian control subjects were included in the study. Twenty-three individuals from family KTCN-014, two affected individuals from each of the families KTCN-011, 015, 019, 020, 021, 024, 025, 030, 031, 034, and 035, and one patient from each of KTCN-05, 013, and 017 were examined.
COL4A1 and COL4A2 sequence analyses
Screening of COL4A1 (NM_001845.4) coding regions revealed 12 sequence variants, three of which were amino acid substitutions: c.19G>C (Val7Leu), c.1663A>C (Thr555Pro), and c.4002A>C (Gln1334His). We identified one novel synonymous change, c.3693G>A (Thr1231Thr), and eight previously reported sequence variants: c.432T>A (Ala144Ala), c.1257T>C (Pro419Pro), c.1815T>C (Pro605Pro), c.2130G>A (Pro710Pro), c.3183G>A (Gly1061Gly), c.3189A>T (Arg1063Arg), c.4470C>T (Ala1490Ala), and c.4800C>T (Ser1600Ser). In the 5′ untranslated region (5′ UTR), one novel sequence variant, c.84+124T>A, was identified. In the 3′ untranslated region (3′ UTR), two previously reported variants, c.*587C>A and c.*975A>C, were detected.
Sequencing analyses of COL4A2 (NM_001846.2) coding regions revealed 13 previously reported sequence variants, including five non-synonymous substitutions: c.574G>T (Val192Phe), c.1550G>A (Arg517Lys), c.2048G>C (Gly683Ala), c.2102A>G (Lys701Arg), and c.2152C>T (Pro718Ser), and eight synonymous substitutions: c.297G>A (Thr99Thr), c.1008C>T (Pro336Pro), c.1095G>A (Pro365Pro), c.1179C>T (Ile393Ile), c.1488G>A (Pro496Pro), c.4089G>A (Ala1363Ala), c.4290T>C (Phe1430Phe), c.4515A>G (Pro1505Pro). In the 5′ UTR, five known nucleotide changes, c.-277A>C, c.-232C>G, c.-215C>T, c.-203T>C, and c.-133A>G, were identified. In the 3′ UTR, eight previously reported sequence variants, c.*76T>C, c.*101_*102del2, c.*417C>G, c.*541C>T, c.*557A>G, c.*650T>C, c.*663T>C, and c.*727G>C were detected.
Screening of exon/intron junctions in COL4A1 and COL4A2 revealed numerous sequence variants in the surrounding non-coding sequences, 71 and 86, respectively, including single nucleotide changes, insertions, and deletions. All screening results are summarized in Table 2.
Table 2. Sequence variants found in COL4A1 and COL4A2 genes.
| |
|
|
|
|
Affected KTCN-014 (n=10) |
Unaffected KTCN-014 (n=11) |
Unknown KTCN-014 (n=2) |
All KTCN-014 (n=23) |
Other KTCN Families Affected (n=25) |
All (n=48) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | dbSNP refID | Chromosome Position | Allele Change | Residue Change | no. | % | no. | % | no. | % | no. | % | no. | % | no. | % | |
|
COL4A1 (NM_001845.4) | |||||||||||||||||
| - |
110959464 |
c.-90G>T |
- |
5 |
50 |
7 |
63.6 |
0 |
0 |
12 |
52.2 |
15 |
60 |
27 |
56.3 |
||
| 1 |
rs9515185 |
110959356 |
c.19G>C |
Val7Leu |
8 |
80 |
6 |
54.5 |
1 |
50 |
15 |
65.2 |
18 |
72 |
33 |
68.8 |
|
| |
- |
110959167 |
c.84+124T>A |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs75270666 |
110895200 |
c.85–119C>T |
- |
0 |
0 |
2 |
18.2 |
1 |
50 |
3 |
13.0 |
5 |
20 |
8 |
16.7 |
|
| |
rs41275106 |
110895150 |
c.85–69T>C |
- |
1 |
10 |
3 |
27.3 |
0 |
0 |
4 |
17.4 |
4 |
16 |
8 |
16.7 |
|
| |
rs9521650 |
110866265 |
c.234+8C>T |
- |
2 |
20 |
1 |
9.1 |
0 |
0 |
3 |
13.0 |
5 |
20 |
8 |
16.7 |
|
| |
rs3737328 |
110866065 |
c.279+64G>A |
- |
5 |
50 |
5 |
45.5 |
1 |
50 |
11 |
47.8 |
10 |
40 |
21 |
43.8 |
|
| 7 |
rs532625 |
110864225 |
c.432T>A |
Ala144Ala |
7 |
70 |
8 |
72.7 |
2 |
100 |
17 |
73.9 |
14 |
56 |
31 |
64.6 |
|
| |
rs71805366 |
110863985, 110863989 |
c.468+5_468+9del5 |
- |
3 |
30 |
2 |
18.2 |
0 |
0 |
5 |
21.7 |
4 |
16 |
9 |
18.8 |
|
| |
rs76574181 |
110862750 |
c.469–191C>T |
- |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
4 |
16 |
12 |
25.0 |
|
| |
rs2166208 |
110862686 |
c.469–127C>T |
- |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
4 |
16 |
12 |
25.0 |
|
| |
rs9521649 |
110862303 |
c.615+24C>T |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs2166207 |
110862268 |
c.615+59T>G |
- |
7 |
70 |
9 |
81.8 |
2 |
100 |
18 |
78.3 |
21 |
84 |
39 |
81.3 |
|
| |
rs645114 |
110861785 |
c.616–11G>C |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs7333204 |
110861671 |
c.651+68A>G |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs7332120 |
110861670 |
c.651+69C>T |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs10687642 |
110861652, 110861653 |
c.651+86_651+87ins2 |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs55833821 |
110861649 |
c.651+90C>G |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs35638294 |
110861620, 110861621 |
c.651+118_651+119ins4 |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs7333008 |
110861560 |
c.651+179A>G |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
16 |
64 |
26 |
54.2 |
|
| |
rs598893 |
110859743 |
c.780+7G>A |
- |
7 |
70 |
8 |
72.7 |
2 |
100 |
17 |
73.9 |
17 |
68 |
34 |
70.8 |
|
| |
rs598819 |
110859690 |
c.780+60T>C |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs9588116 |
110859069 |
c.808–7C>G |
- |
7 |
70 |
9 |
81.8 |
2 |
100 |
18 |
78.3 |
21 |
84 |
39 |
81.3 |
|
| |
rs67772891 |
110859326 |
c.781–88delT |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs677877 |
110857895 |
c.859–10T>C |
- |
7 |
70 |
5 |
45.5 |
2 |
100 |
14 |
60.9 |
17 |
68 |
31 |
64.6 |
|
| |
rs482757 |
110857823 |
c.903+18G>A |
- |
7 |
70 |
5 |
45.5 |
2 |
100 |
14 |
60.9 |
17 |
68 |
31 |
64.6 |
|
| |
rs665713 |
110857502 |
c.957+198T>C |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
19 |
76 |
42 |
87.5 |
|
| |
rs648735 |
110856180 |
c.958–226T>C |
- |
7 |
70 |
8 |
72.7 |
2 |
100 |
17 |
73.9 |
17 |
68 |
34 |
70.8 |
|
| |
rs648705 |
110856153 |
c.958–199T>G |
- |
7 |
70 |
8 |
72.7 |
2 |
100 |
17 |
73.9 |
17 |
68 |
34 |
70.8 |
|
| |
rs7327728 |
110856094 |
c.958–140T>A |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs648263 |
110856085 |
c.958–131T>C |
- |
7 |
70 |
8 |
72.7 |
2 |
100 |
17 |
73.9 |
17 |
68 |
34 |
70.8 |
|
| |
- |
110855997 |
c.958–43delT |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
- |
110853032 |
c.1085–205A>T |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
22 |
88 |
45 |
93.8 |
|
| |
rs995223 |
110851036 |
c.1121–58A>G |
- |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
17 |
68 |
27 |
56.3 |
|
| |
rs496916 |
110851014 |
c.1121–36C>G |
- |
10 |
100 |
7 |
63.6 |
1 |
50 |
18 |
78.3 |
8 |
32 |
26 |
54.2 |
|
| 21 |
rs995224 |
110850842 |
c.1257T>C |
Pro419Pro |
3 |
30 |
6 |
54.5 |
1 |
50 |
10 |
43.5 |
17 |
68 |
27 |
56.3 |
|
| |
rs683309 |
110850770 |
c.1285+44A>G |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs9588112 |
110847566 |
c.1286–101G>A |
- |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
4 |
16 |
12 |
25.0 |
|
| |
rs505050 |
110847227 |
c.1381+143C>A |
- |
10 |
100 |
10 |
90.9 |
1 |
50 |
21 |
91.3 |
21 |
84 |
42 |
87.5 |
|
| |
rs9521643 |
110847217 |
c.1381+153T>C |
- |
4 |
40 |
6 |
54.5 |
1 |
50 |
11 |
47.8 |
18 |
72 |
29 |
60.4 |
|
| |
rs2241966 |
110847190 |
c.1381+180A>G |
- |
4 |
40 |
6 |
54.5 |
1 |
50 |
11 |
47.8 |
18 |
72 |
29 |
60.4 |
|
| |
rs685884 |
110845314 |
c.1382–54C>T |
- |
9 |
90 |
11 |
100.0 |
2 |
100 |
22 |
95.7 |
21 |
84 |
43 |
89.6 |
|
| |
rs2241967 |
110844721 |
c.1466–90G>A |
- |
8 |
80 |
7 |
63.6 |
1 |
50 |
16 |
69.6 |
11 |
44 |
27 |
56.3 |
|
| 25 |
rs536174 |
110839550 |
c.1663A>C |
Thr555Pro |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs9521638 |
110839428 |
c.1728+57T>C |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
19 |
76 |
34 |
70.8 |
|
| 26 |
rs61749897 |
110838814 |
c.1815T>C |
Pro605Pro |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
4 |
16 |
12 |
25.0 |
|
| |
rs2305080 |
110838703 |
c.1897+29A>G |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
19 |
76 |
34 |
70.8 |
|
| |
rs565470 |
110838646 |
c.1897+86T>C |
- |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
25 |
100 |
46 |
95.8 |
|
| |
rs72654112 |
110835460 |
c.1991–16G>A |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
4 |
16 |
4 |
8.3 |
|
| |
rs7329411 |
110835195 |
c.2095+145G>T |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
19 |
76 |
34 |
70.8 |
|
| 29 |
rs16975492 |
110833702 |
c.2130G>A |
Pro710Pro |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
rs16975491 |
110833564 |
c.2193+75G>A |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
rs10492497 |
110831866 |
c.2194–98A>G |
- |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
6 |
24 |
14 |
29.2 |
|
| |
rs2131939 |
110831837 |
c.2194–69C>T |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
4 |
16 |
4 |
8.3 |
|
| |
rs503053 |
110831451 |
c.2345–68A>G |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
- |
110830612 |
c.2626–34T>C |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs2305081 |
110830090 |
c.2716+99C>T |
- |
5 |
50 |
4 |
36.4 |
1 |
50 |
10 |
43.5 |
9 |
36 |
19 |
39.6 |
|
| |
rs1562173 |
110828922 |
c.2968+51C>T |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
rs1975514 |
110828891 |
c.2969–31A>G |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| 37 |
rs874204 |
110827580 |
c.3183G>A |
Gly1061Gly |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| 37 |
rs874203 |
110827574 |
c.3189A>T |
Arg1063Arg |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
- |
110826231 |
c.3505+16C>T |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs17517598 |
110825264 |
c.3506–147C>A |
- |
1 |
10 |
1 |
9.1 |
0 |
0 |
2 |
8.7 |
4 |
16 |
6 |
12.5 |
|
| |
rs2289799 |
110824974 |
c.3556+93G>C |
- |
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
17 |
68 |
32 |
66.7 |
|
| |
rs2275845 |
110823178 |
c.3557–99C>T |
- |
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
6 |
24 |
14 |
29.2 |
|
| 42 |
- |
110822943 |
c.3693G>A |
Thr1231Thr |
0 |
0 |
2 |
18.2 |
1 |
50 |
3 |
13.0 |
7 |
28 |
10 |
20.8 |
|
| |
- |
110822653 |
c.3742+231C>T |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
2 |
8 |
2 |
4.2 |
|
| |
rs589985 |
110819586 |
c.3877–9C>T |
- |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
24 |
96 |
45 |
93.8 |
|
| |
rs1778817 |
110819460 |
c.3949+45C>T |
- |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
24 |
96 |
45 |
93.8 |
|
| |
rs652572 |
110819457 |
c.3949+48T>C |
- |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
24 |
96 |
45 |
93.8 |
|
| |
rs1213026 |
110819362 |
c.3949+143T>C |
- |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
24 |
96 |
45 |
93.8 |
|
| |
- |
110818760:110818763 |
c.3950–110_3950–113del4 |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| 45 |
rs3742207 |
110818598 |
c.4002A>C |
Gln1334His |
8 |
80 |
4 |
36.4 |
1 |
50 |
13 |
56.5 |
10 |
40 |
23 |
47.9 |
|
| |
rs1816884 |
110817171 |
c.4150+38C>G |
- |
4 |
40 |
6 |
54.5 |
2 |
100 |
12 |
52.2 |
14 |
56 |
26 |
54.2 |
|
| |
rs2298241 |
110816097 |
c.4151–189C>G |
- |
2 |
20 |
6 |
54.5 |
0 |
0 |
8 |
34.8 |
5 |
20 |
13 |
27.1 |
|
| |
rs2298240 |
110815673 |
c.4249+137G>C |
- |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
2 |
8 |
2 |
4.2 |
|
| |
rs16975424 |
110814923 |
c.4250–134T>C |
- |
1 |
10 |
5 |
45.5 |
0 |
0 |
6 |
26.1 |
5 |
20 |
11 |
22.9 |
|
| 49 |
rs1133219 |
110813709 |
c.4470C>T |
Ala1490Ala |
3 |
30 |
5 |
45.5 |
2 |
100 |
10 |
43.5 |
10 |
40 |
20 |
41.7 |
|
| |
rs2275843 |
110813532 |
c.4640+7C>T |
- |
2 |
20 |
6 |
54.5 |
0 |
0 |
8 |
34.8 |
5 |
20 |
13 |
27.1 |
|
| |
- |
110813531 |
c.4640+8G>A |
- |
4 |
40 |
2 |
18.2 |
0 |
0 |
6 |
26.1 |
0 |
0 |
6 |
12.5 |
|
| |
rs2275842 |
110813523 |
c.4640+16G>A |
- |
2 |
20 |
6 |
54.5 |
0 |
0 |
8 |
34.8 |
5 |
20 |
13 |
27.1 |
|
| |
rs617111 |
110807776 |
c.4641–32G>A |
- |
4 |
40 |
2 |
18.2 |
0 |
0 |
6 |
26.1 |
0 |
0 |
6 |
12.5 |
|
| |
rs681884 |
110805062 |
c.4756–209C>T |
- |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
24 |
96 |
47 |
97.9 |
|
| |
- |
110804970 |
c.4756–117G>C |
- |
1 |
10 |
1 |
9.1 |
0 |
0 |
2 |
8.7 |
0 |
0 |
2 |
4.2 |
|
| 51 |
rs650724 |
110804809 |
c.4800C>T |
Ser1600Ser |
2 |
20 |
4 |
36.4 |
0 |
0 |
6 |
26.1 |
11 |
44 |
17 |
35.4 |
|
| |
rs13260 |
110802123 |
c.*587C>A |
- |
2 |
20 |
4 |
36.4 |
0 |
0 |
6 |
26.1 |
11 |
44 |
17 |
35.4 |
|
| |
rs28362515 |
110801735 |
c.*975A>C |
- |
1 |
10 |
1 |
9.1 |
0 |
0 |
2 |
8.7 |
0 |
0 |
2 |
4.2 |
|
|
COL4A2 (NM_001846.2) | |||||||||||||||||
| |
rs7989823 |
110959643 |
c.-277A>C |
|
7 |
70 |
9 |
81.8 |
0 |
0 |
16 |
69.6 |
25 |
100 |
41 |
85.4 |
|
| |
rs7990009 |
110959688 |
c.-232C>G |
|
5 |
50 |
7 |
63.6 |
0 |
0 |
12 |
52.2 |
17 |
68 |
29 |
60.4 |
|
| |
rs7990017 |
110959705 |
c.-215C>T |
|
7 |
70 |
9 |
81.8 |
0 |
0 |
16 |
69.6 |
23 |
92 |
39 |
81.3 |
|
| |
rs7991332 |
110959717 |
c.-203T>C |
|
5 |
50 |
7 |
63.6 |
0 |
0 |
12 |
52.2 |
15 |
60 |
27 |
56.3 |
|
| |
rs35466678 |
110959787 |
c.-133A>G |
|
3 |
30 |
3 |
27.3 |
0 |
0 |
6 |
26.1 |
14 |
56 |
20 |
41.7 |
|
| |
rs7327528 |
110960044 |
c.-44–163G>C |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
4 |
16 |
4 |
8.3 |
|
| |
rs76536922 |
110960164 |
c.-44–43C>T |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
4 |
16 |
4 |
8.3 |
|
| |
rs4773143 |
110960685 |
c.99+215T>C |
|
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
21 |
84 |
36 |
75.0 |
|
| |
rs4773144 |
110960712 |
c.99+242A>G |
|
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
21 |
84 |
36 |
75.0 |
|
| |
rs12876517 |
111009643 |
c.100–176G>A |
|
6 |
60 |
9 |
81.8 |
2 |
100 |
17 |
73.9 |
19 |
76 |
36 |
75.0 |
|
| |
rs4771678 |
111076940 |
c.181–141T>C |
|
8 |
80 |
10 |
90.9 |
2 |
100 |
20 |
87.0 |
20 |
80 |
40 |
83.3 |
|
| Ex5 |
rs4238272 |
111077197 |
c.297G>A |
Thr99Thr |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
22 |
88 |
45 |
93.8 |
|
| |
rs74967960 |
111077234 |
c.315+19T>C |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs7334986 |
111080609 |
c.361–205G>A |
|
7 |
70 |
4 |
36.4 |
1 |
50 |
12 |
52.2 |
8 |
32 |
20 |
41.7 |
|
| |
- |
111080964 |
c.477+34C>T |
|
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
0 |
0 |
8 |
16.7 |
|
| |
rs3929758 |
111082157 |
c.478–75C>A |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
22 |
88 |
43 |
89.6 |
|
| Ex9 |
rs62621885 |
111082772 |
c.574G>T |
Val192Phe |
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs60212072 |
111086650 |
c.685–98G>A |
|
0 |
0 |
3 |
27.3 |
0 |
0 |
3 |
13.0 |
6 |
24 |
9 |
18.8 |
|
| |
rs41275108 |
111088456 |
c.727–160A>T |
|
4 |
40 |
2 |
18.2 |
0 |
0 |
6 |
26.1 |
2 |
8 |
8 |
16.7 |
|
| |
rs7983487 |
111090854 |
c.862–111A>G |
|
1 |
10 |
5 |
45.5 |
0 |
0 |
6 |
26.1 |
12 |
48 |
18 |
37.5 |
|
| |
rs7984937 |
111090909 |
c.862–56T>C |
|
1 |
10 |
5 |
45.5 |
0 |
0 |
6 |
26.1 |
13 |
52 |
19 |
39.6 |
|
| |
rs7984100 |
111090924 |
c.862–41G>A |
|
1 |
10 |
5 |
45.5 |
0 |
0 |
6 |
26.1 |
13 |
52 |
19 |
39.6 |
|
| |
rs7983979 |
111091024 |
c.912+9C>T |
|
0 |
0 |
3 |
27.3 |
0 |
0 |
3 |
13.0 |
6 |
24 |
9 |
18.8 |
|
| |
rs4771680 |
111098017 |
c.958–159T>C |
|
1 |
10 |
2 |
18.2 |
0 |
0 |
3 |
13.0 |
12 |
48 |
15 |
31.3 |
|
| |
rs7489705 |
111098110 |
c.958–66C>T |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
21 |
84 |
44 |
91.7 |
|
| Ex17 |
rs4103 |
111098226 |
c.1008C>T |
Pro336Pro |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
15 |
60 |
38 |
79.2 |
|
| |
rs59905747 |
111099045 |
c.1012–100C>G |
|
5 |
50 |
5 |
45.5 |
1 |
50 |
11 |
47.8 |
5 |
20 |
16 |
33.3 |
|
| |
rs45612833 |
111099057 |
c.1012–88G>A |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
21 |
84 |
44 |
91.7 |
|
| |
rs7326449 |
111099122 |
c.1012–23G>A |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
21 |
84 |
44 |
91.7 |
|
| |
rs56676181 |
111101931 |
c.1079–95C>T |
|
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
7 |
28 |
19 |
39.6 |
|
| |
rs75082326 |
111101952 |
c.1079–74A>G |
|
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
7 |
28 |
19 |
39.6 |
|
| Ex19 |
rs76425569 |
111102042 |
c.1095G>A |
Pro365Pro |
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
7 |
28 |
19 |
39.6 |
|
| Ex19 |
rs74941798 |
111102126 |
c.1179C>T |
Ile393Ile |
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
7 |
28 |
19 |
39.6 |
|
| |
rs34734302 |
111102183 |
c.1189+47A>G |
|
9 |
90 |
9 |
81.8 |
2 |
100 |
20 |
87.0 |
13 |
52 |
33 |
68.8 |
|
| |
- |
111102853 |
c.1339+52C>G |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs72657934 |
111102865 |
c.1339+64G>A |
|
0 |
0 |
2 |
18.2 |
0 |
0 |
2 |
8.7 |
3 |
12 |
5 |
10.4 |
|
| |
rs9515218 |
111109859 |
c.1432+77A>G |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs9555703 |
111109882 |
c.1432+100A>G |
|
6 |
60 |
4 |
36.4 |
2 |
100 |
12 |
52.2 |
5 |
20 |
17 |
35.4 |
|
| |
rs9515219 |
111109960 |
c.1432+178T>C |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs9521781 |
111111023 |
c.1433–95T>C |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs9521782 |
111111043 |
c.1433–75G>A |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
15 |
60 |
36 |
75.0 |
|
| Ex22 |
rs7990214 |
111111173 |
c.1488G>A |
Pro496Pro |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| Ex22 |
rs7990383 |
111111235 |
c.1550G>A |
Arg517Lys |
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs4773186 |
111111382 |
c.1596+101G>A |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
22 |
88 |
45 |
93.8 |
|
| |
rs41275110 |
111114554 |
c.1669+21G>A |
|
4 |
40 |
6 |
54.5 |
0 |
0 |
10 |
43.5 |
6 |
24 |
16 |
33.3 |
|
| |
rs7992330 |
111114751 |
c.1776+20G>A |
|
6 |
60 |
4 |
36.4 |
2 |
100 |
12 |
52.2 |
5 |
20 |
17 |
35.4 |
|
| |
rs3803237 |
111117668 |
c.1777–84G>A |
|
4 |
40 |
6 |
54.5 |
0 |
0 |
10 |
43.5 |
6 |
24 |
16 |
33.3 |
|
| |
rs3803236 |
111117745 |
c.1777–7C>T |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs3825490 |
111117984 |
c.1978+31C>T |
|
4 |
40 |
4 |
36.4 |
1 |
50 |
9 |
39.1 |
8 |
32 |
17 |
35.4 |
|
| |
rs72657953 |
111118073 |
c.1978+120C>T |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
4 |
16 |
8 |
16.7 |
|
| |
rs1983931 |
111118102 |
c.1978+149G>A |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs1983932 |
111118221 |
c.1979–129T>C |
|
3 |
30 |
3 |
27.3 |
0 |
0 |
6 |
26.1 |
7 |
28 |
13 |
27.1 |
|
| |
rs41275112 |
111118450 |
c.2038+41C>T |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
1 |
4 |
1 |
2.1 |
|
| |
rs1927350 |
111118546 |
c.2038+137T>G |
|
3 |
30 |
3 |
27.3 |
0 |
0 |
6 |
26.1 |
7 |
28 |
13 |
27.1 |
|
| |
rs3803232 |
111119296 |
c.2039–91A>G |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs3803231 |
111119342 |
c.2039–45T>C |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
4 |
16 |
8 |
16.7 |
|
| Ex27 |
rs3803230 |
111119396 |
c.2048G>C |
Gly683Ala |
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
4 |
16 |
8 |
16.7 |
|
| |
rs9559813 |
111121444 |
c.2096–120C>A |
|
8 |
80 |
9 |
81.8 |
2 |
100 |
19 |
82.6 |
14 |
56 |
33 |
68.8 |
|
| |
rs9559814 |
111121483 |
c.2096–81A>G |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| Ex28 |
rs78829338 |
111121570 |
c.2102A>G |
Lys701Arg |
1 |
10 |
3 |
27.3 |
1 |
50 |
5 |
21.7 |
3 |
12 |
8 |
16.7 |
|
| Ex28 |
rs9583500 |
111121620 |
c.2152C>T |
Pro718Ser |
4 |
40 |
4 |
36.4 |
0 |
0 |
8 |
34.8 |
5 |
20 |
13 |
27.1 |
|
| |
rs9515229 |
111121717 |
c.2203+46A>G |
|
9 |
90 |
10 |
90.9 |
2 |
100 |
21 |
91.3 |
16 |
64 |
37 |
77.1 |
|
| |
rs9515230 |
111121847 |
c.2203+176T>C |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
4 |
16 |
8 |
16.7 |
|
| |
rs9588178 |
111125576 |
c.2425+79G>A |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
2 |
8 |
2 |
4.2 |
|
| |
rs11617206 |
111125606 |
c.2425+109A>G |
|
7 |
70 |
7 |
63.6 |
1 |
50 |
15 |
65.2 |
22 |
88 |
37 |
77.1 |
|
| |
rs9588179 |
111125747 |
c.2425+250C>A |
|
0 |
0 |
0 |
0.0 |
0 |
0 |
0 |
0.0 |
2 |
8 |
2 |
4.2 |
|
| |
rs9559818 |
111130226 |
c.2426–124G>A |
|
8 |
80 |
7 |
63.6 |
1 |
50 |
16 |
69.6 |
21 |
84 |
37 |
77.1 |
|
| |
rs2281974 |
111130519 |
c.2587+8C>T |
|
5 |
50 |
5 |
45.5 |
1 |
50 |
11 |
47.8 |
18 |
72 |
29 |
60.4 |
|
| |
rs9301457 |
111130599 |
c.2587+88G>C |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs2281973 |
111130674 |
c.2587+163C>T |
|
4 |
40 |
5 |
45.5 |
1 |
50 |
10 |
43.5 |
15 |
60 |
25 |
52.1 |
|
| |
rs72657977 |
111132413 |
c.2588–154C>T |
|
4 |
40 |
3 |
27.3 |
0 |
0 |
7 |
30.4 |
3 |
12 |
10 |
20.8 |
|
| |
rs4773194 |
111132490 |
c.2588–77A>G |
|
10 |
100 |
8 |
72.7 |
2 |
100 |
20 |
87.0 |
17 |
68 |
37 |
77.1 |
|
| |
rs9521803 |
111132556 |
c.2588–11C>T |
|
8 |
80 |
6 |
54.5 |
2 |
100 |
16 |
69.6 |
8 |
32 |
24 |
50.0 |
|
| |
rs1475438 |
111132820 |
c.2758+83G>A |
|
4 |
40 |
6 |
54.5 |
0 |
0 |
10 |
43.5 |
11 |
44 |
21 |
43.8 |
|
| |
rs58124222 |
111132947 |
c.2758+210G>A |
|
0 |
0 |
3 |
27.3 |
0 |
0 |
3 |
13.0 |
8 |
32 |
11 |
22.9 |
|
| |
rs3803229 |
111134780 |
c.2759–83G>A |
|
6 |
60 |
7 |
63.6 |
1 |
50 |
14 |
60.9 |
9 |
36 |
23 |
47.9 |
|
| |
rs3803228 |
111134858 |
c.2759–5T>C |
|
0 |
0 |
3 |
27.3 |
0 |
0 |
3 |
13.0 |
8 |
32 |
11 |
22.9 |
|
| |
rs2296853 |
111137240 |
c.2903–12A>G |
|
3 |
30 |
3 |
27.3 |
2 |
100 |
8 |
34.8 |
6 |
24 |
14 |
29.2 |
|
| |
rs2296852 |
111137465 |
c.3025+91G>A |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
10 |
40 |
14 |
29.2 |
|
| |
rs11839527 |
111137488 |
c.3025+114G>A |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
10 |
40 |
14 |
29.2 |
|
| |
rs41315048 |
111137975 |
c.3026–27G>T |
|
1 |
10 |
3 |
27.3 |
0 |
0 |
4 |
17.4 |
2 |
8 |
6 |
12.5 |
|
| |
rs2296851 |
111138255 |
c.3207+72G>A |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
10 |
40 |
14 |
29.2 |
|
| |
rs35120918 |
111143541 |
c.3347–39G>A |
|
0 |
0 |
1 |
9.1 |
1 |
50 |
2 |
8.7 |
0 |
0 |
2 |
4.2 |
|
| |
rs413756 |
111143755 |
c.3454+68T>C |
|
8 |
80 |
11 |
100.0 |
1 |
50 |
20 |
87.0 |
22 |
88 |
42 |
87.5 |
|
| |
rs402661 |
111143851 |
c.3454+164G>C |
|
3 |
30 |
5 |
45.5 |
0 |
0 |
8 |
34.8 |
10 |
40 |
18 |
37.5 |
|
| |
rs452020 |
111144102 |
c.3455–315T>C |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs403839 |
111144321 |
c.3455–96G>A |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
7 |
28 |
11 |
22.9 |
|
| |
- |
111144382 |
c.3455–35T>C |
|
1 |
10 |
0 |
0.0 |
0 |
0 |
1 |
4.3 |
1 |
4 |
2 |
4.2 |
|
| |
rs2296849 |
111144412 |
c.3455–5C>G |
|
1 |
10 |
3 |
27.3 |
0 |
0 |
4 |
17.4 |
2 |
8 |
6 |
12.5 |
|
| |
rs421177 |
111144565 |
c.3562+41C>T |
|
1 |
10 |
3 |
27.3 |
0 |
0 |
4 |
17.4 |
2 |
8 |
6 |
12.5 |
|
| |
rs57003582 |
111145456:111145486 |
c.3563–100_3563–70del30 |
7 |
70 |
11 |
100.0 |
1 |
50 |
19 |
82.6 |
22 |
88 |
41 |
85.4 |
||
| |
rs2274544 |
111145633 |
c.3634+4C>T |
|
1 |
10 |
3 |
27.3 |
0 |
0 |
4 |
17.4 |
2 |
8 |
6 |
12.5 |
|
| |
rs2391833 |
111145676 |
c.3634+47G>C |
|
8 |
80 |
10 |
90.9 |
1 |
50 |
19 |
82.6 |
20 |
80 |
39 |
81.3 |
|
| |
rs9559826 |
111145779 |
c.3634+150C>T |
|
2 |
20 |
2 |
18.2 |
0 |
0 |
4 |
17.4 |
8 |
32 |
12 |
25.0 |
|
| |
- |
111147637 |
c.3635–52A>G |
|
3 |
30 |
4 |
36.4 |
1 |
50 |
8 |
34.8 |
0 |
0 |
8 |
16.7 |
|
| |
rs378601 |
111153934 |
c.3761–81G>A |
|
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs388222 |
111154159 |
c.3877+28C>T |
|
8 |
80 |
8 |
72.7 |
2 |
100 |
18 |
78.3 |
21 |
84 |
39 |
81.3 |
|
| |
rs2281968 |
111154160 |
c.3877+29G>A |
|
5 |
50 |
8 |
72.7 |
1 |
50 |
14 |
60.9 |
18 |
72 |
32 |
66.7 |
|
| |
rs4773198 |
111155711 |
c.4040–19C>T |
|
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
8 |
32 |
20 |
41.7 |
|
| Ex43 |
rs4773199 |
111155779 |
c.4089G>A |
Ala1363Ala |
5 |
50 |
4 |
36.4 |
1 |
50 |
10 |
43.5 |
8 |
32 |
18 |
37.5 |
|
| |
rs9301460 |
111156153 |
c.4139–41G>A |
|
5 |
50 |
6 |
54.5 |
1 |
50 |
12 |
52.2 |
8 |
32 |
20 |
41.7 |
|
| |
rs414881 |
111156411 |
c.4285+71G>A |
|
7 |
70 |
8 |
72.7 |
1 |
50 |
16 |
69.6 |
21 |
84 |
37 |
77.1 |
|
| Ex45 |
rs4771683 |
111156499 |
c.4290T>C |
Phe1430Phe |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| Ex46 |
rs445348 |
111158874 |
c.4515A>G |
Pro1505Pro |
10 |
100 |
11 |
100.0 |
2 |
100 |
23 |
100.0 |
25 |
100 |
48 |
100.0 |
|
| |
rs2479426 |
111164198 |
c.4882–83T>C |
|
7 |
70 |
6 |
54.5 |
2 |
100 |
15 |
65.2 |
20 |
80 |
35 |
72.9 |
|
| |
rs422733 |
111164614 |
c.*76T>C |
|
7 |
70 |
6 |
54.5 |
2 |
100 |
15 |
65.2 |
20 |
80 |
35 |
72.9 |
|
| |
rs3074455 |
111164639,
111164640 |
c.*101_*102del2 |
|
7 |
70 |
6 |
54.5 |
2 |
100 |
15 |
65.2 |
20 |
80 |
35 |
72.9 |
|
| |
rs10509 |
111164955 |
c.*417C>G |
|
8 |
80 |
11 |
100.0 |
2 |
100 |
21 |
91.3 |
25 |
100 |
46 |
95.8 |
|
| |
rs1049906 |
111165079 |
c.*541C>T |
|
4 |
40 |
8 |
72.7 |
1 |
50 |
13 |
56.5 |
18 |
72 |
31 |
64.6 |
|
| |
rs1049931 |
111165095 |
c.*557A>G |
|
6 |
60 |
9 |
81.8 |
1 |
50 |
16 |
69.6 |
20 |
80 |
36 |
75.0 |
|
| |
rs1049977 |
111165188 |
c.*650T>C |
|
6 |
60 |
9 |
81.8 |
1 |
50 |
16 |
69.6 |
19 |
76 |
35 |
72.9 |
|
| |
rs7711 |
111165201 |
c.*663T>C |
|
6 |
60 |
9 |
81.8 |
1 |
50 |
16 |
69.6 |
19 |
76 |
35 |
72.9 |
|
| rs15457 | 111165265 | c.*727G>C | 4 | 40 | 8 | 72.7 | 1 | 50 | 13 | 56.5 | 17 | 68 | 30 | 62.5 | |||
dbSNP ref ID: identity numbers of observed sequence variants; chromosome position (NCBI build 37.1).
The sequencing of the genomic region containing the common promoter of COL4A1 and COL4A2 revealed no sequence changes.
Statistical analysis and in silico predictions
PolyPhen analyses of non-synonymous changes in COL4A1 and COL4A2 predicted that only the Gln1334His variant in COL4A1 was possibly damaging for protein function and structure (Table 3). The multiple sequence alignment of COL4A1 orthologs shows that the amino acid glutamine at position 1,334 is conserved throughout the analyzed species (Figure 1). Gln1334His substitution was observed more frequently in patients than in healthy individuals in family KTCN-014 (p=0.056). There was no difference in the c.4002A>C allele distribution between the analyzed affected individuals from the remaining KTCN families and the Ecuadorian control subjects (p=0.17).
Table 3. Prediction of effect of amino acid substitutions found in COL4A1 and COL4A2.
|
|
|
PolyPhen |
SIFT |
PMUT |
PANTHER |
SNAP |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Sequence variant | PSIC score | Prediction | Score | Prediction | NN | Prediction | subPSEC | Pdeleterious | Expected Accuracy | Prediction |
| COL4A1 |
Val7Leu |
N/A |
benign |
1 |
tolerated |
0.2367 |
neutral |
- |
- |
92% |
neutral |
|
|
Thr555Pro |
N/A |
benign |
0.65 |
tolerated |
0.0250 |
neutral |
−0.52603 |
0.0777 |
94% |
neutral |
|
|
Gln1334His |
1.66 |
possibly damaging |
0.12 |
tolerated |
0.1039 |
neutral |
−1.0433 |
0.12382 |
69% |
neutral |
| COL4A2 |
Val192Phe |
1.13 |
benign |
64 |
tolerated |
0.1921 |
neutral |
- |
- |
78% |
neutral |
|
|
Arg517Lys |
0.1 |
benign |
0.96 |
tolerated |
0.0861 |
neutral |
- |
- |
92% |
neutral |
|
|
Gly683Ala |
N/A |
benign |
0.96 |
tolerated |
0.4841 |
neutral |
- |
- |
85% |
neutral |
|
|
Lys701Arg |
N/A |
benign |
0.97 |
tolerated |
0.0166 |
neutral |
- |
- |
89% |
neutral |
| Pro718Ser | N/A | benign | 0.98 | tolerated | 0.2039 | neutral | - | - | 89% | neutral | |
The PolyPhen tool predicts which missense substitution affects the structure and function of protein, and uses Position-Specific Independent Counts software to assign profile scores. The SIFT tool evaluates conserved positions, and calculates a score for the amino acid change at a particular position. A score of <0.05 is considered as pathogenic for the protein structure. The PMUT calculates the pathological significance of non-synonymous amino acid substitution using neural networks (NN). NN output >0.5 is considered to be deleterious. PANTHER generates the substitution Position-Specific Evolutionary Conservation score. The value −3 is cutoff point for functional significance and corresponds to a Pdeleterious of 0.5. If the substitution occurs at a position not appearing in the multiple sequence alignment, a subPSEC score cannot be calculated and change is not likely to be pathogenic. The SNAP output shows prediction neutral or non-neutral, and the expected accuracy.
Figure 1.
Multiple sequence alignment of the amino acid sequences of COL4A1 orthologs in different species. Conservation of glutamine (Q) at the 1334 position is shown in gray.
The SIFT, PMUT, PANTHER, and SNAP analyses defined all missense amino acid substitutions in COL4A1 and COL4A2 as neutral/tolerated and lacking any effect on protein function. All prediction results are summarized in Table 3.
Haplotype reconstruction
Haplotypes of sequence variants observed in family KTCN-014 are shown in Figure 2. The coding sequence variants in COL4A1 are surrounded by markers rs13260 and col4a1_snp2. Exons of COL4A2 are localized between rs35466678 and rs422733.
Figure 2.
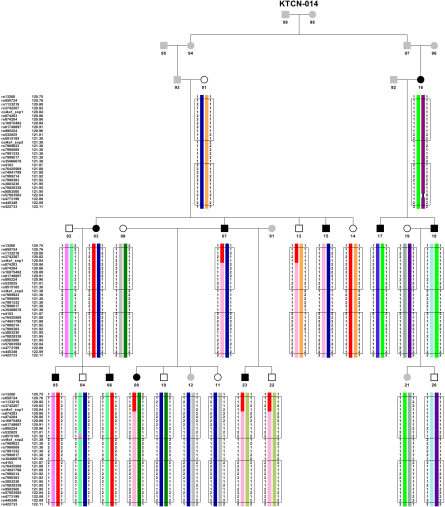
Pedigree of the family KTCN-014. Black-filled symbols: individuals with KTCN; open symbols: individuals without KTCN; gray-filled symbols: individuals with unknown KTCN status. Below each symbol the haplotypes are shown for the coding sequence in genes COL4A1, COL4A2 and UTRs between them. In COL4A1, the coding regions are surrounded by the markers rs13260 and col4a2_snp, and by rs35466678 and rs422733 in COL4A2, which were marked by a black frame. Haplotype regions in different colors indicate patterns of inheritance in the two branches in the pedigree.
KTCN-014 consists of two family branches. Distinct haplotypes in the branches were identified (Figure 2). In the first one, initiated by parents KTCN-93 and KTCN-01, six subjects with KTCN had the same haplotype in the COL4A1 region, extending from rs13260 to col4a1_snp1. Three unaffected individuals, KTCN-13, KTCN-14, and KTCN-22, share that part of the haplotype with their affected relatives. One of four variants in this region, rs3742207, causes a change in the protein sequence, replacing Gln in position 1334 with His (Gln1334His). That haplotype region, from rs13260 to col4a1_snp1, represents a short fragment of the haplotype which covers the whole COL4A1 and COL4A2 sequence in KTCN-03, KTCN-05, KTCN-06, and KTCN-14. In addition, individuals KTCN-07, KTCN-09, KTCN-13, KTCN-22, and KTCN-23 share the rs874203-rs422733 region (Figure 2 – pink bars). For markers rs13260-col4a1_snp1, a different haplotype was observed in the second family branch, initiated by parents KTCN-92 and KTCN-16. This haplotype covered the entire length of the analyzed region, and was identified in all affected individuals and KTCN-21, whose phenotype was unknown. Subject KTCN-17 had the same allele pattern for markers s13260-col4a1_snp1, as individuals from the first branch of the family. However, in this case, analysis indicated that these markers are inherited from KTCN-92, who is unrelated to KTCN-93 and KTCN-01.
Discussion
To our knowledge, this is the first report describing complete sequence analysis of the coding regions and the exon-intron boundaries of COL4A1 and COL4A2 in families with KTCN. Previous studies have revealed a correlation between KTCN development and histopathological alterations in the structure of the corneal stroma and basement membrane, including a loss of collagen concentration [42] and rearrangement of collagen fibers [26]. Moreover, several types of collagen, including collagen type IV have been identified in the cornea [24], and COL4A1 and COL4A2 expression has been detected in the human cornea [29]. Finally, we had mapped a locus for KTCN to 13q32, in close proximity of which COL4A1 and COL4A2 are localized [21]. Given that information, we hypothesized that COL4A1 and COL4A2 genes are good candidates for causing KTCN in families with linkage to that locus.
Different studies have revealed several loci and a few candidate genes for familial KTCN. The first gene proposed as playing a significant role in KTCN pathogenesis was the VSX1 (visual system homeobox 1, OMIM 605020) gene. It was suggested that a few disease-causing mutations were present in this gene [43,44], but recent studies have not confirmed these findings [21,45-47]. Next, heterozygous genomic 7-bp deletion in intron 2 of SOD1 (superoxide dismutase 1; OMIM 147450) was identified in two families with KTCN [48,49]. In contrast, other studies have shown that mutations in this gene are not associated with KTCN pathogenesis [21,47]. Genetic analyses of COL4A3, COL4A4, COL8A1, and COL8A2 genes have revealed no pathogenic mutations in patients with KTCN, indicating that other genetic factors cause the disease [50-52].
We identified several single base pair substitutions in the coding regions of COL4A1 and COL4A2, including one novel heterozygous change, c.3693G>A in exon 42 of COL4A1. None of the detected alterations segregated fully with the affected phenotype in the analyzed members of the Ecuadorian KTCN families. Among the identified missense substitutions in COL4A1, one change, c.4002A>C (p. Gln1334His), was observed more frequently in KTCN patients than in healthy individuals in family KTCN-014. However, no significant statistical association of this change with familial disease could be proven (p=0.056), and no difference in the c.4002A>C allele distribution between the analyzed affected individuals from the remaining KTCN families and the Ecuadorian control subjects was discovered (p=0.17). To predict the impact of the substitutions on the structure and function of the protein, we used different tools. All identified missense substitutions in COL4A1 and COL4A2 were predicted by the SIFT, PMUT, PANTHER, and SNAP tools to have no effect, but PolyPhen defined the Gln1334His change in COL4A1 as possibly damaging. Glutamine at this position is highly conserved in different species. Moreover, this change is present in the collagenous domain of the α1(IV) chain with Gly-X-Y repeats, which plays a role in the assembly into a triple-helical structure of the protein [22]. Replacement of the neutral residue (Gln) with the polar amino acid (His) at the Y position is likely to affect the protein structure. Nevertheless, further studies should be performed to determine the functional significance of this substitution.
To the best of our knowledge, no mutations in COL4A1 were associated with corneal disease. The spectrum of COL4A1-related disorders included porencephaly (OMIM 175780) [53-55], Hereditary Angiopathy with Nephropathy, Aneurysm and Muscle Cramps (HANAC; OMIM 611773) [56], and brain small vessel disease with hemorrhage (OMIM 607595) [57]. Recent studies have also revealed an association between mutations in exon 29 of COL4A1 and Axenfeld-Rieger anomaly with leukoencephalopathy and stroke [58]. In our study, none of the previously reported COL4A1 mutations were identified. The absence of these changes in patients with KTCN suggests that they are specific to the above-mentioned disorders only, and are not associated with KTCN in the tested families. To date, no mutations responsible for COL4A2-related human diseases have been reported.
Besides changes identified in the coding regions of COL4A1 and COL4A2, our study revealed numerous alterations in introns and UTRs of both genes, including single base pair substitutions, deletions, and insertions. Fourteen of these were novel and their clinical significance is not known. Each of the changes was observed in affected and healthy individuals in the tested families. Because important functional elements are located in non-coding regions of genes [59] and intronic alterations can result in a deleterious effect on pre-mRNA splicing [60], identification of these sequence variants could be non-accidental. Further research is needed to delineate the role of these sequence variants.
Recent studies have shown that a mouse with a mutation in a splice acceptor site of Col4a1 has ocular dysgenesis. The mutation results in a lack of exon 40 from mice’s transcripts and leads to the accumulation of mis-folded protein in the lens epithelial cells. Col4a1∆ex40 mice show optic nerve hypoplasia and anterior segment dysgenesis (ASD) including pigment dispersion, cataracts, and corneal opacifications [61]. Splice acceptor sites are highly conserved regions in different species [56]. We detected no alterations in the splice acceptor site in intron 39 of human COL4A1.
Extended genetic studies executed in families with KTCN have shown a high level of genetic heterogeneity [62]. The presence of many putative loci supports the hypothesis that KTCN is an oligogeneic disease in which accumulation of sequence variants at several loci cause a specific KTCN haplotype and may trigger the phenotypic effect. The absence of mutations in COL4A1 and COL4A2 genes indicates that other genes are involved in KTCN pathogenesis in Ecuadorian families.
Acknowledgments
Supported by the Polish Ministry of Science and Higher Education, Grant NN 402097837. The authors thank Genomed Company (Warsaw, Poland) for support in sequencing service.
References
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Yang H, Rabinowitz YS. Longitudinal study of keratoconus progression. Exp Eye Res. 2007;85:502–7. doi: 10.1016/j.exer.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen JF, Butler HG. Mongolism (Down’s syndrome) and keratoconus. Br J Ophthalmol. 1963;47:321–30. doi: 10.1136/bjo.47.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuming BS, Joffe L. Ehlers-Danlos syndrome associated with keratoconus. A case report. S Afr Med J. 1977;52:403–5. [PubMed] [Google Scholar]
- 5.Elder MJ. Leber congenital amaurosis and its association with keratoconus and keratoglobus. J Pediatr Ophthalmol Strabismus. 1994;31:38–40. doi: 10.3928/0191-3913-19940101-08. [DOI] [PubMed] [Google Scholar]
- 6.McMonnies CW. Mechanisms of rubbing-related corneal trauma in keratoconus. Cornea. 2009;28:607–15. doi: 10.1097/ICO.0b013e318198384f. [DOI] [PubMed] [Google Scholar]
- 7.Steahly LP. Keratoconus following a contact lens wear. Ann Ophthalmol. 1978;10:1177–9. [PubMed] [Google Scholar]
- 8.Rahi A, Davies P, Ruben M, Lobascher D, Menon J. Keratoconus and coexisting atopic disease. Br J Ophthalmol. 1977;61:761–4. doi: 10.1136/bjo.61.12.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards M, McGhee CN, Dean S. The genetics of keratoconus. Clin Experiment Ophthalmol. 2001;29:345–51. doi: 10.1046/j.1442-9071.2001.d01-16.x. [DOI] [PubMed] [Google Scholar]
- 10.Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G. Familial keratoconus with cataract: linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest Ophthalmol Vis Sci. 2003;44:5063–6. doi: 10.1167/iovs.03-0399. [DOI] [PubMed] [Google Scholar]
- 11.Tyynismaa H, Sistonen P, Tuupanen S, Tervo T, Dammert A, Latvala T, Alitalo T. A locus for autosomal dominant keratoconus: linkage to 16q22.3-q23.1 in Finnish families. Invest Ophthalmol Vis Sci. 2002;43:3160–4. [PubMed] [Google Scholar]
- 12.Brancati F, Valente EM, Sarkozy A, Feher J, Castori M, Del Duca P, Mingarelli R, Pizzuti A, Dallapiccola B. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet. 2004;41:188–92. doi: 10.1136/jmg.2003.012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchings H, Ginisty H, Le Gallo M, Levy D, Stoesser F, Rouland JF, Arne JL, Lalaux MH, Calvas P, Roth MP, Hovnanian A, Malecaze F. Identification of a new locus for isolated familial keratoconus at 2p24. J Med Genet. 2005;42:88–94. doi: 10.1136/jmg.2004.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burdon KP, Coster DJ, Charlesworth JC, Mills RA, Laurie KJ, Giunta C, Hewitt AW, Latimer P, Craig JE. Apparent autosomal dominant keratoconus in a large Australian pedigree accounted for by digenic inheritance of two novel loci. Hum Genet. 2008;124:379–86. doi: 10.1007/s00439-008-0555-z. [DOI] [PubMed] [Google Scholar]
- 15.Tang YG, Rabinowitz YS, Taylor KD, Li X, Hu M, Picornell Y, Yang H. Genomewide linkage scan in a multigeneration Caucasian pedigree identifies a novel locus for keratoconus on chromosome 5q14.3-q21.1. Genet Med. 2005;7:397–405. doi: 10.1097/01.gim.0000170772.41860.54. [DOI] [PubMed] [Google Scholar]
- 16.Bisceglia L, De Bonis P, Pizzicoli C, Fischetti L, Laborante A, Di Perna M, Giuliani F, Delle Noci N, Buzzonetti L, Zelante L. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive Loci. Invest Ophthalmol Vis Sci. 2009;50:1081–6. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Rabinowitz YS, Tang YG, Picornell Y, Taylor KD, Hu M, Yang H. Two-stage genome-wide linkage scan in keratoconus sib pair families. Invest Ophthalmol Vis Sci. 2006;47:3791–5. doi: 10.1167/iovs.06-0214. [DOI] [PubMed] [Google Scholar]
- 18.Liskova P, Hysi PG, Waseem N, Ebenezer ND, Bhattacharya SS, Tuft SJ. Evidence for keratoconus susceptibility locus on chromosome 14: a genome-wide linkage screen using single-nucleotide polymorphism markers. Arch Ophthalmol. 2010;128:1191–5. doi: 10.1001/archophthalmol.2010.200. [DOI] [PubMed] [Google Scholar]
- 19.Hameed A, Khaliq S, Ismail M, Anwar K, Ebenezer ND, Jordan T, Mehdi SQ, Payne AM, Bhattacharya SS. A novel locus for Leber congenital amaurosis (LCA4) with anterior keratoconus mapping to chromosome 17p13. Invest Ophthalmol Vis Sci. 2000;41:629–33. [PubMed] [Google Scholar]
- 20.Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S. Identity-by-descent approach to gene localisation in eight individuals affected by keratoconus from north-west Tasmania, Australia. Hum Genet. 2002;110:462–70. doi: 10.1007/s00439-002-0705-7. [DOI] [PubMed] [Google Scholar]
- 21.Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, Ratnamala U, Ewing K, Molinari A, Pitarque JA, Lee K, Leal SM, Bejjani BA. Localization of a gene for Keratoconus to a 5.6 Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–9. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech. 2008;71:357–70. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer G, Schmidt C, Opitz J, Cully Z, Kuhn K, Poschl E. Identification of a novel sequence element in the common promoter region of human collagen type IV genes, involved in the regulation of divergent transcription. Biochem J. 1993;292:687–95. doi: 10.1042/bj2920687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abalain JH, Dossou H, Colin J, Floch HH. Levels of collagen degradation products (telopeptides) in the tear film of patients with keratoconus. Cornea. 2000;19:474–6. doi: 10.1097/00003226-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 25.Patey A, Savoldelli M, Pouliquen Y. Keratoconus and normal cornea: a comparative study of the collagenous fibers of the corneal stroma by image analysis. Cornea. 1984;3:119–24. [PubMed] [Google Scholar]
- 26.Meek KM, Tuft SJ, Huang Y, Gill PS, Hayes S, Newton RH, Bron AJ. Changes in collagen orientation in distribution in keratoconus corneas. Invest Ophthalmol Vis Sci. 2005;46:1948–56. doi: 10.1167/iovs.04-1253. [DOI] [PubMed] [Google Scholar]
- 27.Kenney MC, Nesburn AB, Burgeson RE, Butkowski RJ, Ljubimov AV. Abnormalities of the extracellular matrix in keratoconus corneas. Cornea. 1997;16:345–51. [PubMed] [Google Scholar]
- 28.Tuori AJ, Virtanen I, Aine E, Kalluri R, Miner JH, Uusitalo HM. The immunohistochemical composition of corneal basement membrane in keratoconus. Curr Eye Res. 1997;16:792–801. doi: 10.1076/ceyr.16.8.792.8989. [DOI] [PubMed] [Google Scholar]
- 29.Jun AS, Liu SH, Koo EH, Do DV, Stark WJ, Gottsch JD. Microarray analysis of gene expression in human donor corneas. Arch Ophthalmol. 2001;119:1629–34. doi: 10.1001/archopht.119.11.1629. [DOI] [PubMed] [Google Scholar]
- 30.Stachs O, Bochert A, Gerber T, Koczan D, Thiessen HJ, Guthoff RF. The extracellular matrix structure in keratoconus. Ophthalmologe. 2004;101:384–9. doi: 10.1007/s00347-003-0902-3. [DOI] [PubMed] [Google Scholar]
- 31.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–7. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 32.Weeks DE, Sobel E, O’Connell JR, Lange K. Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet. 1995;56:1506–7. [PMC free article] [PubMed] [Google Scholar]
- 33.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- 34.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, Hyland FC, Kennedy GC, Kong X, Murray SS, Ziegle JS, Stewart WC, Buyske S. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–6. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–2. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 36.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrer-Costa C, Gelpí JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–8. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 39.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherwin T, Brookes NH. Morphological changes in keratoconus: pathology or pathogenesis. Clin Experiment Ophthalmol. 2004;32:211–7. doi: 10.1111/j.1442-9071.2004.00805.x. [DOI] [PubMed] [Google Scholar]
- 43.Héon E, Greenberg A, Kopp KK, Rootman D, Vincent AL, Billingsley G, Priston M, Dorval KM, Chow RL, McInnes RR, Heathcote G, Westall C, Sutphin JE, Semina E, Bremner R, Stone EM. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11:1029–36. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 44.Bisceglia L, Ciaschetti M, De Bonis P, Campo PA, Pizzicoli C, Scala C, Grifa M, Ciavarella P, Delle Noci N, Vaira F, Macaluso C, Zelante L. VSX1 mutational analysis in a series of Italian patients affected by keratoconus: detection of a novel mutation. Invest Ophthalmol Vis Sci. 2005;46:39–45. doi: 10.1167/iovs.04-0533. [DOI] [PubMed] [Google Scholar]
- 45.Aldave AJ, Yellore VS, Salem AK, Yoo GL, Rayner SA, Yang H, Tang GY, Piconell Y, Rabinowitz YS. No VSX1 gene mutations associated with keratoconus. Invest Ophthalmol Vis Sci. 2006;47:2820–2. doi: 10.1167/iovs.05-1530. [DOI] [PubMed] [Google Scholar]
- 46.Tang YG, Picornell Y, Su X, Li X, Yang H, Rabinowitz YS. Three VSX1 gene mutations, L159M, R166W, and H244R, are not associated with keratoconus. Cornea. 2008;27:189–92. doi: 10.1097/ICO.0b013e31815a50e7. [DOI] [PubMed] [Google Scholar]
- 47.Stabuc-Silih M, Strazisar M, Hawlina M, Glavac D. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29:172–6. doi: 10.1097/ICO.0b013e3181aebf7a. [DOI] [PubMed] [Google Scholar]
- 48.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, Kenney MC. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–51. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 49.Udar N, Atilano SR, Small K, Nesburn AB, Kenney MC. SOD1 haplotypes in familial keratoconus. Cornea. 2009;28:902–7. doi: 10.1097/ICO.0b013e3181983a0c. [DOI] [PubMed] [Google Scholar]
- 50.Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15:2848–60. [PMC free article] [PubMed] [Google Scholar]
- 51.Aldave AJ, Bourla N, Yellore VS, Rayner SA, Khan MA, Salem AK, Sonmez B. Keratoconus is not associated with mutations in COL8A1 and COL8A2. Cornea. 2007;26:963–5. doi: 10.1097/ICO.0b013e31811dfaf7. [DOI] [PubMed] [Google Scholar]
- 52.Stabuc-Silih M, Strazisar M, Ravnik-Glavac M, Hawlina M, Glavac D. Genetics and clinical characteristics of keratoconus. Acta Dermatovenerol Alp Panonica Adriat. 2010;19:3–10. [PubMed] [Google Scholar]
- 53.Breedveld G, de Coo IF, Lequin MH, Arts WF, Heutink P, Gould DB, John SW, Oostra B, Mancini GM. Novel mutations in three families confirm a major role of COL4A1 in hereditary porencephaly. J Med Genet. 2006;43:490–5. doi: 10.1136/jmg.2005.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, Schimenti JC, Aguglia U, van der Knaap MS, Heutink P, John SW. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308:1167–71. doi: 10.1126/science.1109418. [DOI] [PubMed] [Google Scholar]
- 55.de Vries LS, Koopman C, Groenendaal F, Van Schooneveld M, Verheijen FW, Verbeek E, Witkamp TD, van der Worp HB, Mancini G. COL4A1 mutation in two preterm siblings with antenatal onset of parenchymal hemorrhage. Ann Neurol. 2009;65:12–8. doi: 10.1002/ana.21525. [DOI] [PubMed] [Google Scholar]
- 56.Plaisier E, Gribouval O, Alamowitch S, Mougenot B, Prost C, Verpont MC, Marro B, Desmettre T, Cohen SY, Roullet E, Dracon M, Fardeau M, Van Agtmael T, Kerjaschki D, Antignac C, Ronco P. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N Engl J Med. 2007;357:2687–95. doi: 10.1056/NEJMoa071906. [DOI] [PubMed] [Google Scholar]
- 57.Gould DB, Phalan FC, van Mil SE, Sundberg JP, Vahedi K, Massin P, Bousser MG, Heutink P, Miner JH, Tournier-Lasserve E, John SW. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med. 2006;354:1489–96. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 58.Sibon I, Coupry I, Menegon P, Bouchet JP, Gorry P, Burgelin I, Calvas P, Orignac I, Dousset V, Lacombe D, Orgogozo JM, Arveiler B, Goizet C. COL4A1 mutation in Axenfeld-Rieger anomaly with leukoencephalopathy and stroke. Ann Neurol. 2007;62:177–84. doi: 10.1002/ana.21191. [DOI] [PubMed] [Google Scholar]
- 59.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010;20:311–9. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–48. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gould DB, Marchant JK, Savinova OV, Smith RS, John SW. Col4a1 mutation causes endoplasmic reticulum stress and genetically modifiable ocular dysgenesis. Hum Mol Genet. 2007;16:798–807. doi: 10.1093/hmg/ddm024. [DOI] [PubMed] [Google Scholar]
- 62.Rabinowitz YS. The genetics of keratoconus. Ophthalmol Clin North Am. 2003;16:607–20. doi: 10.1016/s0896-1549(03)00099-3. vii.14741001. [DOI] [PubMed] [Google Scholar]