Abstract
An understanding of the taxonomic status and vector distribution of anophelines is crucial to malaria control efforts. Previous phylogenetic analyses have supported the description of six species of the Neotropical malaria vector Anopheles (Nyssorhynchus) albitarsis s.l. (Diptera: Culicidae): Anopheles albitarsis, An. deaneorum, An. marajoara, An. oryzalimnetes, An. janconnae and An. albitarsis F. To evaluate the taxonomic status of An. albitarsis s.l. mosquitoes collected in various localities of the Colombian Caribbean region, specimens were analyzed using the complete mtDNA Cytochrome Oxidase I (COI) gene, the ribosomal DNA internal transcribed spacer 2 (ITS2) region and partial nuclear DNA White gene sequences. Phylogenetic analyses of the COI sequences detected a new lineage near An. janconnae in the Caribbean region of Colombia and determined its position relative to the other members of the complex. However, the ITS2 and White gene sequences lacked resolution to support a new lineage near An. janconnae or the An. janconnae clade. Nothing is known about the possible involvement in malaria transmission in Colombia of this new lineage, but its phylogenetic closeness to Anopheles janconnae, which has been incriminated in local malaria transmission in Brazil, is provocative.
Keywords: Colombia, Albitarsis Complex, a new lineage near Anopheles janconnae, malaria, COI, White gene, ITS2
INTRODUCTION
By 2006, it was estimated that 9.4% of the total Colombian population was living in moderate or high risk malaria transmission areas; the Annual Parasite Index (API), the number of confirmed malaria cases per 1,000 inhabitants, was an average of 26.2 (PAHO 2007). The National Health Institute (INS) of Colombia reported 110,480 malaria cases during 2007, caused by Plasmodium vivax Grassi & Felleti and Plasmodium falciparum Welch (Coccidia: Plasmodiidae) (INS 2007).
Because many important malaria vectors are in cryptic species complexes, it is essential to achieve precise species identification and to clarify phylogenetic status, for a better understanding of malaria transmission patterns. Several studies have defined the taxonomic status and phylogenetic relationships among members of the Albitarsis Complex using nuclear and mitochondrial markers (Lehr et al. 2005, Merritt et al. 2005, Wilkerson et al. 2005, Brochero et al. 2007, Li & Wilkerson 2007), and, have shown that it consists of at least six species: Anopheles albitarsis Lynch-Arribálzaga, An. deaneorum Rosa-Freitas, 1989, An. marajoara Galvão and Damasceno, An. albitarsis B (Wilkerson et al. 2005, Wilkerson et al. 1995a, Wilkerson et al. 1995b), An. albitarsis E (Lehr et al. 2005) and An. albitarsis F (Brochero et al. 2007). Recently, Motoki et al. (2009) presented morphological descriptions of the adult males and females, fourth-instar larvae and pupae of An. albitarsis, An. marajoara, An. deaneorum and An. albitarsis B and adult females and males of An. albitarsis E. These authors also validated and named Species B and E as An. oryzalimnetes Wilkerson and Motoki and An. janconnae Wilkerson and Sallum, respectively; however, species F has yet to be formally described. Furthermore, a new study has determined that An. deaneorum is a complex (Bourke et al. 2010), and thus the number of species in the Albitarsis Complex is no doubt more than six.
Species of the Albitarsis Complex have been reported in various South American countries (Brelsfoard et al. 2006, Brochero et al. 2007, Lehr et al. 2005, Li & Wilkerson 2005, Motoki et al. 2009). Some of these species seem to be allopatric; however, sympatric distributions have been recorded for An. marajoara and An. deaneorum, An. oryzalimnetes (as An. albitarsis B) and An. albitarsis in Brazil (Wilkerson et al. 1995, Lehr et al. 2005), and An. albitarsis F and An. marajoara in Colombia (Brochero et al. 2007). Based primarily on morphological identification, An. marajoara has been recorded in 25 of the 32 Colombian departments (González & Carrejo 2007), and in a study in Villavicencio, eastern Colombia, it was found in high abundance and had a wide distribution in urban areas, suggesting its efficient adaptation to human environments (Brochero et al. 2005). Anopheles albitarsis F is a suspected malaria vector in the Department of Vichada, eastern Colombia (Brochero et al. 2007).
Among members of the Albitarsis Complex, An. marajoara (Rubio-Palis & Zimmerman 1997, Conn et al. 2002) and An. janconnae (as An. albitarsis E, Póvoa et al. 2006) are incriminated malaria vectors, An. deaneorum is a suspected vector (Klein et al. 1991a; Klein et al. 1991b), and An. albitarsis, An. oryzalimnetes and An. albitarsis F do not have a defined role in transmission. In Colombia, a recent study on natural Plasmodium infectivity of Anopheles species from the Caribbean and the Pacific regions reported the occurrence of An. albitarsis s.l. in three of seven localities investigated, Santa Rosa de Lima, Moñitos and Tumaco (Gutiérrez et al. 2008); however, all specimens analyzed were uninfected, probably because of small sample size. To clarify the distributions of species of the Albitarsis Complex in Colombia, we used three molecular markers, the entire mitochondrial Cytochrome Oxidase I (COI) gene, the ribosomal DNA internal transcribed spacer 2 (ITS2) region and a fragment of the single copy nuclear White gene, to identify the species of the Albitarsis Complex in several localities in the Colombian Caribbean region. Phylogenetic analyses based on COI sequences identified the specimens as a new lineage near An. janconnae.
MATERIALS AND METHODS
Mosquito Collections
Adult mosquitoes were wild-caught. Collection data and species identification for Santa Rosa de Lima (SRL) and Moñitos (MON) are in Gutiérrez et al. (2008). Anopheles albitarsis s.l. specimens previously collected in Puerto Libertador, Córdoba Department (PLT) (Gutiérrez et al. 2009), were also included in this study. Species-specific ITS2 regions were amplified from mosquitoes identified morphologically as An. albitarsis s.l. following the scheme proposed by Li & Wilkerson (2005) (Table I). Location of putative and described species of the Albitarsis Complex based on COI and ITS2 sequences available in GenBank and tested in this work were depicted using DIVA-GIS software version 5.2.0.2 (Fig. 1) (Hijmans et al. 2002).
TABLE I.
Collection and molecular data for Anopheles albitarsis s.l.
| Department | Locality/ Abbreviation | n | PCR ITS2a | COIb | ITS2b | White geneb | Coordinates |
|---|---|---|---|---|---|---|---|
| Bolívar | Santa Rosa de Lima (SRL) | 73 | 73 | 6 | 5 | 1 | 10°26' N, 75°21' W |
| Córdoba | Moñitos (MON) | 2 | 2 | 1 | 2 | c | 9°15' N, 76°06' W |
| Puerto Libertador (PLT) | 10 | 10 | 7 | 5 | 1 | 7°54' N, 75°40' W |
n: Number of mosquitoes collected.
Number of specimens identified using the ITS2 PCR strategy in Li & Wilkerson (2005)
Number of specimens from each locality with DNA sequences for the respective marker
The White gene PCR amplification failed for specimens from MON.
Fig. 1.
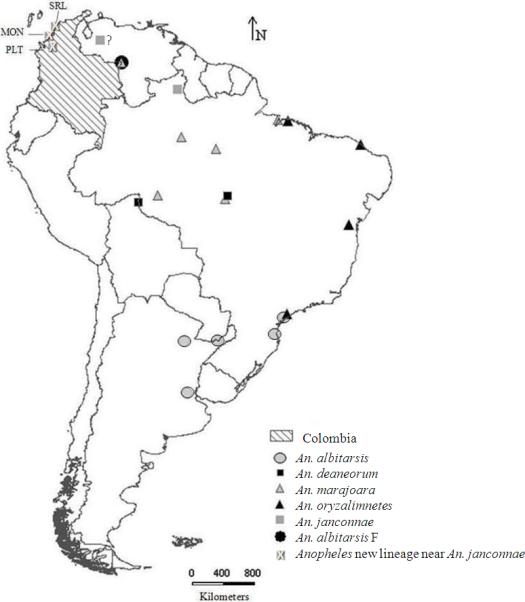
Location of putative and described species of the Albitarsis Complex based on COI and ITS2 sequences available in GenBank and obtained in this study. Based on the COI gene phylogenetic analyses the three populations from the Caribbean region of Colombia are in the same clade as the sample of An. albitarsis E from Venezuela and closely related to An. janconnae from the type locality in northern Brazil.
DNA extraction, PCR amplification and sequencing
Total DNA was extracted from individual abdomens of An. albitarsis s.l. mosquitoes using a salt precipitation protocol (Birungi & Munstermann 2002) or DNeasy® Blood & Tissue Kit (QIAGEN, Duesseldorf, Germany). Three molecular markers were amplified, sequenced and analyzed: (1) the internal transcribed spacer 2 (ITS2) of the ribosomal DNA (rDNA) using primers and conditions previously described (Li & Wilkerson et al. 2005, Zapata et al. 2007), (2) the entire COI gene amplified using two sets of conserved primer pairs described in Lunt et al. (1996), and (3) partial sequence of the single copy nuclear White gene amplified following PCR conditions recommended by Mirabello & Conn (2008). The COI and white gene PCR reactions were performed using PuReTaq™ Ready-To-Go™ PCR beads (GE Healthcare, UK) containing 0.4 μM of each primer in 25-μl volumes. The ITS2 PCR reactions were performed using BIOTAQ™ DNA Polymerase (Bioline, London, UK). The PCR products were purified using CentriSpin 40 columns (Princeton Separations, Adelphia, NJ) or the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI). Fragments were sequenced in both directions at the Wadsworth Center Applied Genomic Technologies Core Facility, New York State Department of Health Albany, NY. The nucleotide sequences were compiled and edited using Sequencher 3.0 (Gene Codes Corporation, Ann Arbor, MI).
Multiple sequence alignment
Twelve ITS2 and two White gene sequences from the An. albitarsis s.l. specimens obtained in this work (Table I) were aligned with available ITS2 and White gene sequences from GenBank (Table II) corresponding to An. marajoara, An. albitarsis, An. deaneorum, An. oryzalimnetes and An. albitarsis F using ClustalX1.83 (Thompson et al. 1997). All positions containing alignment gaps and missing data were eliminated for pairwise sequence comparisons (pairwise deletion option). Neighbor-Joining (NJ) analyses were conducted in MEGA4 (Tamura et al. 2007). In addition, COI DNA sequences from 14 specimens representing the three collection sites (Table I) were aligned with DNA sequences available in GenBank for An. albitarsis, An. marajoara, An. oryzalimnetes, An. deaneorum and An. janconnae (Table II). In all phylogenetic analyses conducted in this study, An. darlingi Root and An. braziliensis Chagas were included as the outgroup. COI, ITS2 and White gene sequences from the present study are deposited in GenBank under the following accession numbers, ITS2 - GQ153583 to GQ153594, White gene - GQ153595 to GQ153596, COI - GQ153597 to GQ153610.
TABLE II.
Sequences for Anopheles albitarsis s.l. from GenBank used in the alignments.
| Species | ITS2a | White geneb | COIc |
|---|---|---|---|
| An. albitarsis | DQ077807, AY828334 - AY828336 AY828320 - AY828323 AF462385-AF462387, U92332 | AF318198, AY956300, AY956299 | DQ076204-DQ076209 |
| An. oryzalimnetes | AY828324 – AY828327, AY828337, AY828338 U92333 | AY956297, AY956298 | DQ076210- DQ076215 |
| An. janconnae | d | d | DQ076231- DQ076234 |
| An. albitarsis F | DQ228315 | DQ228314 | |
| An. deaneorum | AF461751, AF461752, AY828330 - AY828333, AY828341 - AY828343, U92335 | AY956301- AY956302 | DQ076226 - DQ076230 |
| An. marajoara | DQ364141, AY028127, DQ077808, DQ848153, AY828328, AY828329, AY828339 - AY828341, AY828344 - AY828354, U92334 | DQ906919, DQ906920, DQ848154, AY956295, AY956296 | DQ076216 - DQ076221 |
| An. darlingi | AF462389 | GQ285644 | DQ076235, DQ076236 |
| An. braziliensis | AF461753 | DQ076237, DQ076238 | |
| An. albimanus | d | L76302 | d |
Authors: Li & Wilkerson (2005), Marrelli et al. (2005), Li & Wilkerson (2007), Brelsfoard et al. (2006), Brochero et al. (2007), Danoff-Burg & Conn (unpublished), Linton et al. (unpublished).
Authors: Krzywinski et al. (2001), Merritt et al. (2005), Brochero et al. (2007), Ke et al. (1997).
Authors: Lehr et al. (2005).
no data available in GenBank.
Phylogenetic analyses based on the entire mtDNA COI gene
Both Maximum Parsimony (MP) and Maximum Likelihood (ML) analyses were implemented in PAUP version 4.0b10 (Swofford 2000). Under the MP analysis, optimal trees were generated using the heuristic search option with the tree bisection-reconnection (TBR) branch-swapping algorithm. Multiple trees were saved for each run. Bootstrap support values were generated by 1,000 pseudoreplicates. For ML analysis, an appropriate model of nucleotide substitutions was determined using the program Modeltest 3.8 (Posada 2006) choosing the Akaike Information Criterion (AIC). Bootstrap support values with the heuristic search option and the tree bisection-reconnection (TBR) algorithm chosen for branch-swapping were generated by 662 pseudoreplicates. Bayesian analysis was accomplished using Mr Bayes version 3.1.2 (Ronquist & Huelsenbeck 2003). Bayesian inference was conducted under the program default values for the prior probabilities, performing two runs with four independents chains. The Markov Chain Monte Carlo (MCMC) algorithm was allowed to run 10,000,000 generations and sampled every 100 generations after a burn-in of 2,500,000 generations (25,000 trees). Convergence was assessed based on results from three different parameters: the decrease in the average standard deviations of split frequencies ranging from 1 to 0, summarizing the estimated parameters using the sump command and checking on the plot of generation versus the log probability of observing the data.
RESULTS
ITS2 and White gene analyses
The ITS2-PCR analyses showed that the An. albitarsis s.l. mosquitoes collected from the Caribbean region corresponded to the molecular pattern reported for An. marajoara (Fig. 2). However, it is important to note that the ITS2-PCR analyses (Li & Wilkerson 2005) only identify species: An. marajoara, An. deaneorum, An. albitarsis, An. oryzalimnetes, and subsequent analysis added species F to the previous scheme (Brochero et al. 2007). Therefore, further analyses using additional markers were performed to reveal the identity of the An. albitarsis s.l. specimens from the three Colombian localities.
Fig. 2.
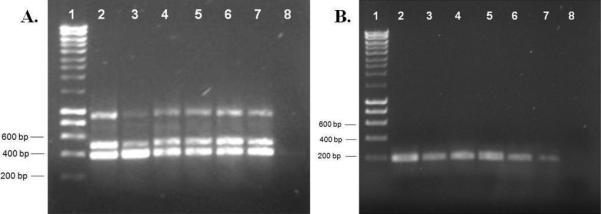
PCR products from ITS2 analyses. A: PCR amplification using specific ITS2 primers albCD, ITS2F and ITS2R, observed patterns are diagnostic for An. marajoara and An. deaneorum. B: PCR amplification using specific ITS2 primers albC, ITS2F and ITS2R, observed patterns are diagnostic for An. marajoara (~194 bp). Lanes: 1: DNA ladder, 2-7: Amplicons corresponding to An. albitarsis s.l. specimens from Santa Rosa de Lima (SRL): 2: SRL55, 3: SRL56, 4: SRL57, 5: SRL58, 6: SRL59, 7: SRL60, 8: PCR negative control.
The ITS2 sequences obtained from specimens collected in SRL, MON and PLT showed 100% identity among them and White gene sequences from SRL and PLT were also identical. A BLASTn search (GenBank) based on ITS2 and White gene sequences of samples from the Caribbean region detected 99% identity with an ITS2 sequence corresponding to An. marajoara from Chocó, Colombia (accession number AY028127) and 98% identity with White gene sequence reported as An. marajoara from Puerto Carreño, Colombia (accession number DQ906920). In this study, NJ analyses based on ITS2 sequences basically clustered all sequences into two groups, one including all species of the Albitarsis Complex together and another including both sequences from An. darlingi and An. braziliensis. The NJ tree based on White gene sequences showed individual clustering for An. albitarsis, An. deaneorum and An. albitarsis F, one sequence of An. oryzalimnetes clustering with An. albitarsis and samples from this study collected in the Caribbean region clustering with sequences of An. marajoara (see additional file 1 and 2).
COI gene analyses
COI sequences from An. albitarsis s.l. collected in the Caribbean localities produced an alignment without indels, with 31 variable and four parsimony informative sites, all at the third nucleotide position. Alignment of all COI sequences from the Albitarsis Complex specimens (this work and GenBank) detected 209 (14.2%) variable and 155 (10.5%) parsimony informative sites distributed at the first and third nucleotide positions in the alignment. The strict consensus of 14 most parsimonious trees was estimated (see additional file 3). A maximum likelihood tree (see additional file 4) was estimated considering the best-fit DNA substitution model selected by AIC criterion (General Time Reversible model: GTR+I+G) for 1,470 sites of the COI dataset (Tavaré 1986), with invariable sites (I=0.7261) and Gamma distribution shape parameter (G=1.1297)). The Bayesian analysis was visualized using the majority rule consensus tree employing the GTR+I+G model and six categories of rates (Fig. 3). Results of the maximum likelihood (662 replicates), Maximum parsimony (1000 replicates) and Bayesian Inference trees based on COI sequence analyses including An. darlingi and An. braziliensis as outgroups illustrated similar topological conformation. Three sequences of An. janconnae from the type locality of Roraima, Brazil, consistently grouped together with high bootstrap support (96 for each of MP and ML, 1.00 for BI) whereas the specimens from the Colombian Caribbean localities consistently grouped with one sequence of purported An. janconnae, from Portuguesa, Venezuela (Fig. 1). In summary, monophyly of the An. albitarsis, An. oryzalimnetes and An. deaneorum clades was observed under MP, ML and Bayesian analyses. Anopheles marajoara was consistently more closely related to An. deaneorum, and An. oryzalimnetes to An. albitarsis. Anopheles janconnae (as An. albitarsis E in Fig. 3) is polyphyletic. The mean genetic distance (GTR and Kimura two-parameter models) computed by pairwise comparisons for the different specimens of An. janconnae, from Venezuela and Brazil, were comparable with specimens from the Caribbean region of Colombia, was 0.028 (range 0.024 to 0.031).
Fig. 3.
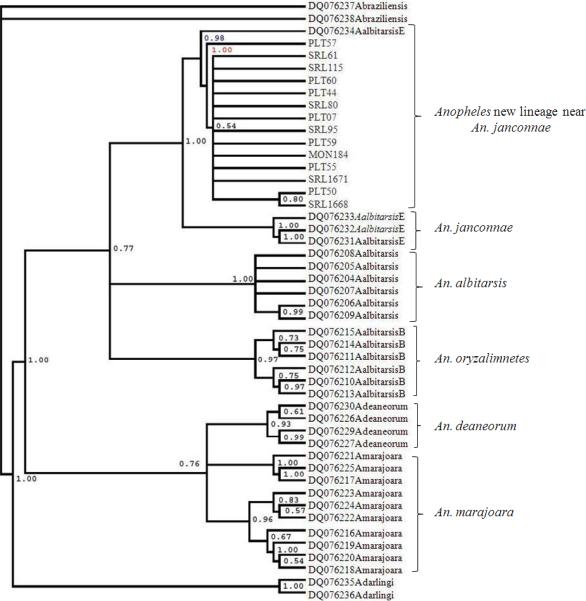
Bayesian topology of the COI sequences. Numbers above branches indicate posterior probabilities. DQ076234, An. albitarsis E (?) specimen from Portuguesa, Venezuela; DQ076231 - DQ076233, An. albitarsis E (An. janconnae) from the type locality in Roraima, Brazil. Outgroup comprises An. darlingi and An. braziliensis.
DISCUSSION
In this study, after applying the scheme to identify specimens of the Albitarsis Complex from Caribbean localities to species based on ITS2-PCR (Li & Wilkerson 2005), it was not possible to determine if they were An. marajoara, An. janconnae or a new lineage near An. janconnae. However, phylogenetic analyses of mitochondrial COI gene sequences of An. albitarsis s.l. specimens from the Caribbean localities of Colombia (Fig. 3) strongly supported monophyly with specimens previously identified as An. albitarsis E from Venezuela (Lehr et al. 2005), but not with An. janconnae from the type locality in Roraima, Brazil.
The DNA sequences of ribosomal and nuclear genes placed sequences of a new lineage near An. janconnae and An. janconnae in close relationship to An. marajoara. Results of the NJ analyses of the ITS2 is unresolved with most sequences clustered as a polytomy, and the White gene topology showed the new An. janconnae E lineage within a strongly supported clade (99% bootstrap support) formed by An. marajoara. The NJ analyses showed that genetic differentiation for species of the Albitarsis Complex based on ITS2 and White gene sequences lacked resolution to support a new lineage near An. janconnae or the An. janconnae clade, probably due to (1) homoplasy; (2) linage divergence within the Albitarsis Complex, in particular between An. marajoara and a new lineage near An. janconnae, and An. janconnae is still undetectable using these molecular markers, and/or (3) the possibility that the mtDNA lineages are the result of allopatric fragmentation rather than speciation. This result suggests that the ITS2 and single copy nuclear White gene markers have limitations for discriminating between incipient or very recently separated species/lineages proposed for the Albitarsis Complex and that the mtDNA genome is a more sensitive indicator of that divergence (Zink & Barrowclough 2008).
As with other species complexes, females of the Albitarsis Complex are difficult to distinguish using the available morphological keys: they may exhibit high intraspecific morphological variability (Rubio-Palis et al. 2003) and also interspecific similarity; hence, multivariate analysis of measurements of morphological features (Motoki et al. 2009) and molecular analyses based on mtDNA sequences have been shown to be effective approaches for distinguishing morphologically similar species in the complex (Lehr et al. 2005).
A recent population genetics study using microsatellite markers detected population structure for An. marajoara in Colombia (Brochero & Ruiz-García 2006; Brochero et al. 2010). These authors found that populations at different locations in Colombia showed departures from HW equilibrium associated with heterozygote deficits because of the Wahlund effect. However, a different study (Posso et al. 2006), using RAPD-PCR, reported the occurrence of gene flow among An. marajoara populations from eastern and western Colombia that was higher among the eastern populations. Results of the present study based on the phylogenetic analyses of COI sequences suggest the presence of more than one species of the Albitarsis Complex in Colombia; therefore, results of the previous studies may have been biased, since only Brochero et al. (2010) included molecular analyses to support or confirm species identification. The differentiation detected in An. marajoara populations from Colombia may have two possible explanations: (1) The genetic variation observed among An. marajoara populations from Colombia may be influenced by allopatric distribution (potential isolation by distance), in addition to possible reproductive isolation, or (2) different lineages/species of An. albitarsis s.l., in particular a new lineage near An. janconnae, may be present in these areas of Colombia.
In this study, MP, ML and Bayesian analyses of COI sequences of An. albitarsis s.l. from Colombian Caribbean specimens grouped into a strongly supported clade together with the single sequence reported as An. albitarsis E from Venezuela, and separately from the monophyletic lineage of An. janconnae (Fig. 3). However, a low to moderate level of genetic variability was detected among the specimens forming the An. janconnae clade and samples from Colombia, as could be expected for mosquitoes from different populations. In summary, the topology of all optimal trees under MP, ML, and Bayesian methods, in addition to the strong nodal support and the values of genetic distance calculated for An. albitarsis s.l. specimens from the Caribbean localities, showed that a new lineage near An. janconnae is distributed in this region. The occurrence of a new lineage near An. janconnae in sympatry with other known malaria vectors in the Caribbean region (Gutiérrez et al. 2008, 2009) adds additional complexity to the understanding of malaria transmission dynamics in Colombia, since An. janconnae is known as a locally important malaria vector in Brazil. Further studies will be essential to verify if this putative species plays an important role as a secondary or local vector in the Caribbean region of Colombia. Also, these data suggest that more intensive studies of morphological and molecular characterization of the species of the Albitarsis Complex, not only from Colombia but throughout their distribution in the Neotropical biogeographic region, should be undertaken to complete the descriptions of the unknown life stages for all members of this complex (for instance, fourth-instar larvae and pupae of An. janconnae and species F remain undescribed), including the putative new lineage suggested here.
Supplementary Material
LEGENDS FOR SUPPLEMENTARY DATA
Additional file 1: Neighbor-Joining analyses based on 64 ITS2 sequences from individuals of the different species of Albitarsis Complex. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches, retaining only groups with frequency ≥60%. Outgroups are An. darlingi and An. braziliensis.
Additional file 2: Neighbor-Joining analyses based on 17 nuclear protein-coding white gene sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches, retaining only groups with frequency ≥60%. Outgroups are An. darlingi and An. braziliensis.
Additional file 3: Maximum parsimony tree based on COI sequences. Numbers above branches indicate the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates). DQ076234, An. albitarsis E (?) specimen from Portuguesa, Venezuela; DQ076231 - DQ076233, An. albitarsis E (An. janconnae) from the type locality in Roraima, Brazil. Bootstrap values >70% are shown. Outgroup comprises An. darlingi and An. braziliensis.
Additional file 4: Maximum likelihood tree based on COI sequences. Numbers above branches indicate ML bootstrap proportions (662 replicates). DQ076234, An. albitarsis E (?) specimen from Portuguesa, Venezuela; DQ076231 - DQ076233, An. albitarsis E (An. janconnae) from the type locality in Roraima, Brazil. Bootstrap values >70% are shown. Outgroup comprises An. darlingi and An. braziliensis.
ACKNOWLEDGMENTS
We are grateful to R.C. Wilkerson, Walter Reed Systematics Unit, Smithsonian Institution, Washington, D.C for a thorough review of an earlier draft of this manuscript.
Sponsorships: This research was supported by the Comité para el Desarrollo de la Investigación - CODI, Universidad de Antioquia, Grant number 8700-039 to MMC and by the United States National Institutes of Health, Grants R03AI076710 to MMC and 2R01AI054139 to JEC. LAG received financial support for her doctoral training from the Instituto Colombiano para el Desarrollo de la Ciencia y la Tecnología Francisco José de Caldas, COLCIENCIAS.
REFERENCES
- Birungi J, Munstermann L. Genetic structure of Aedes albopictus (Diptera: Culicidae) populations based on mitochondrial ND5 sequences: evidence for an independent invasion into Brazil and United States. Ann Entomol Soc Am. 2002;95:125–132. [Google Scholar]
- Bourke BP, Foster PG, Bergo ES, Calado DC, Sallum MA. Phylogenetic relationships among species of Anopheles (Nyssorhynchus) (Diptera, Culicidae) based on nuclear and mitochondrial gene sequences. Acta Trop. 2010;114:88–96. doi: 10.1016/j.actatropica.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Brelsfoard CL, Fritz GN, Rodriguez R. Identification of Anopheles (Nyssorhynchus) marajoara (Diptera: Culicidae) in Bolivia using Polymerase Chain Reaction and a Restriction Endonuclease. Ann Entomol Soc Am. 2006;99:707–713. [Google Scholar]
- Brochero HH, Li C, Wilkerson RC. A newly recognized species in the Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) from Puerto Carreno, Colombia. Am J Trop Med Hyg. 2007;76:1113–1117. [PubMed] [Google Scholar]
- Brochero HL, Rey G, Buitrago LS, Olano VA. Biting activity and breeding sites of Anopheles species in the municipality Villavicencio, Meta, Colombia. J Am Mosq Control Assoc. 2005;21:182–186. doi: 10.2987/8756-971X(2005)21[182:BAABSO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Brochero HL, Ruiz-García M. Estructura genética poblacional de Anopheles (Nyssorhynchus) marajoara Galvão & Damasceno 1942 (Diptera: Culicidae) en Colombia. Universitas Scientiarum. 2006;11:87–93. [Google Scholar]
- Brochero H, Li C, Wilkerson R, Conn JE, Ruiz-García M. Genetic structure of Anopheles (Nyssorhynchus) marajoara (Diptera: Culicidae) in Colombia. Am J Trop Med Hyg. 2010;83:585–595. doi: 10.4269/ajtmh.2010.09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn JE, Mirabello L. The biogeography and population genetics of neotropical vector species. Heredity. 2007;99:245–256. doi: 10.1038/sj.hdy.6801002. [DOI] [PubMed] [Google Scholar]
- Conn JE, Wilkerson RC, Segura MN, de Souza RT, Schlichting CD, Wirtz RA, Póvoa MM. Emergence of a new neotropical malaria vector facilitated by human migration and changes in land use. Am J Trop Med Hyg. 2002;66:18–22. doi: 10.4269/ajtmh.2002.66.18. [DOI] [PubMed] [Google Scholar]
- González R, Carrejo N. Introducción al estudio taxonómico de Anopheles de Colombia: claves y notas de distribución. Universidad del Valle; Cali: 2007. p. 237. [Google Scholar]
- Gutiérrez LA, Naranjo N, Jaramillo LM, Muskus C, Luckhart S, Conn JE, Correa MM. Natural infectivity of Anopheles species from the Pacific and Atlantic Regions of Colombia. Acta Trop. 2008;107:99–105. doi: 10.1016/j.actatropica.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Gutiérrez LA, González JJ, Gómez GF, Castro MI, Rosero DA, Luckhart S, Conn JE, Correa MM. Species composition and natural infectivity of anthropophilic Anopheles (Diptera: Culicidae) in the states of Córdoba and Antioquia, Northwestern Colombia. Mem Inst Oswaldo Cruz. 2009;104:1117–1124. doi: 10.1590/s0074-02762009000800008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans R, Guarino L, Cruz M, Rojas E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA GIS. Plant Genet Resour Newsl. 2002;127:15–19. [Google Scholar]
- INS, Instituto Nacional de Salud Boletín Epidemiológico Semanal. Estadísticas del Sistema de Vigilancia en Salud Pública – SIVIGILA, Casos Totales en la Semana Epidemiológica 52 y Acumulados del Año, Subdirección de Vigilancia y Control en Salud Pública. 2007.
- Klein TA, Lima JB, Tada MS. Comparative susceptibility of anopheline mosquitoes to in Rondonia Brazil. Am J Trop Med Hyg. 1991a;44:598–603. doi: 10.4269/ajtmh.1991.44.598. [DOI] [PubMed] [Google Scholar]
- Klein TA, Lima JB, Tada MS, Miller R. Comparative susceptibility of anopheline mosquitoes in Rondonia Brazil to infection by Plasmodium vivax. Am J Trop Med Hyg. 1991b;45:463–470. doi: 10.4269/ajtmh.1991.45.463. [DOI] [PubMed] [Google Scholar]
- Lehr MA, Kilpatrick CW, Wilkerson RC, Conn JE. Cryptic species in the Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) Complex: incongruence between Random Amplified Polymorphic DNA-Polymerase Chain Reaction identification and analysis of mitochondrial DNA COI gene sequences. Ann Entomol Soc Am. 2005;98:908–917. doi: 10.1603/0013-8746(2005)098[0908:CSITAN]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wilkerson RC. Identification of Anopheles (Nyssorhynchus) albitarsis complex species (Diptera: Culicidae) using rDNA internal transcribed spacer 2-based polymerase chain reaction primes. Mem Inst Oswaldo Cruz. 2005;100:495–500. doi: 10.1590/s0074-02762005000500009. [DOI] [PubMed] [Google Scholar]
- Li C, Wilkerson RC. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). J Hered. 2007;98:51–59. doi: 10.1093/jhered/esl037. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Zhang DX, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Merritt TJ, Young CR, Vogt RG, Wilkerson RC, Quattro JM. Intron retention identifies a malaria vector within the Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). Mol Phylogenet Evol. 2005;35:719–724. doi: 10.1016/j.ympev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Conn JE. Population analysis using the nuclear white gene detects Pliocene/Pleistocene lineage divergence within Anopheles nuneztovari in South America. Med Vet Entomol. 2008;22:109–119. doi: 10.1111/j.1365-2915.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Motoki MT, Wilkerson RC, Sallum MA. The Anopheles albitarsis complex with the recognition of Anopheles oryzalimnetes Wilkerson and Motoki, n. sp. and Anopheles janconnae Wilkerson and Sallum, n. sp. (Diptera: Culicidae). Mem Inst Oswaldo Cruz. 2009;104:823–850. doi: 10.1590/s0074-02762009000600004. [DOI] [PubMed] [Google Scholar]
- PAHO, Pan American Health Organization . Technical notes & sources: Health situation in the Americas: Basic Indicators. Regional Core Health Data and Country Profiles Initiative; Washington, DC. USA: 2007. [Google Scholar]
- Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006;34:700–703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posso C, González R, Cárdenas H, Tascón Y. Estructura genética de Anopheles darlingi Root, An. nuneztovari Gabaldon y An. marajoara Galvao & Damasceno de Colombia mediante RAPD-PCR. Revista Colombiana de Entomologia. 2006;32:49–56. [Google Scholar]
- Póvoa MM, de Souza RT, Lacerda RN, Rosa ES, Galiza D, de Souza JR, Wirtz RA, Schlichting CD, Conn JE. The importance of Anopheles albitarsis E and An. darlingi in human malaria transmission in Boa Vista, state of Roraima, Brazil. Mem Inst Oswaldo Cruz. 2006;101:163–168. doi: 10.1590/s0074-02762006000200008. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosa-Freitas MG. Anopheles (Nyssorhynchus) deaneorum: a new species in the albitarsis complex (Diptera: Culicidae). Mem Inst Oswaldo Cruz. 1989;84:535–543. [Google Scholar]
- Rubio-Palis Y, Wilkerson R, Guzman H. Morphological characters of adult Anopheles (Nyssorhynchus) marajoara in Venezuela. J Am Mosq Control Assoc. 2003;19:107–114. [PubMed] [Google Scholar]
- Rubio-Palis Y, Zimmerman RH. Ecoregional classification of malaria vectors in the neotropics. J Med Entomol. 1997;34:499–510. doi: 10.1093/jmedent/34.5.499. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (and Other Methods) Sinauer Associates, Inc.; Sunderland, Massachusetts: 2000. PAUP* version 4.0. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tavaré S. Some mathematical questions in biology - DNA sequence analysis. Amer. Math. Soc.; Providence, RI: 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson RC, Foster PG, Li C, Sallum MA. Molecular phylogeny of neotropical Anopheles (Nyssorhynchus) Albitarsis Species Complex (Diptera: Culicidae). Ann Entomol Soc Am. 2005;98:918–925. doi: 10.1603/0013-8746(2005)098[0918:mponan]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson RC, Gaffigan TV, Bento Lima J. Identification of species related to Anopheles (Nyssorhynchus) albitarsis by random amplified polymorphic DNA-polymerase chain reaction (Diptera: Culicidae). Mem Inst Oswaldo Cruz. 1995a;90:721–732. doi: 10.1590/s0074-02761995000600013. [DOI] [PubMed] [Google Scholar]
- Wilkerson RC, Parsons TJ, Klein TA, Gaffigan TV, Bergo E, Consolim J. Diagnosis by random amplified polymorphic DNA polymerase chain reaction of four cryptic species related to Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) from Paraguay, Argentina, and Brazil. J Med Entomol. 1995b;32:697–704. doi: 10.1093/jmedent/32.5.697. [DOI] [PubMed] [Google Scholar]
- Zapata MA, Cienfuegos AV, Quiros OI, Quinones ML, Luckhart S, Correa MM. Discrimination of seven Anopheles species from San Pedro de Uraba, Antioquia, Colombia, by polymerase chain reaction-restriction fragment length polymorphism analysis of its sequences. Am J Trop Med Hyg. 2007;77:67–72. [PubMed] [Google Scholar]
- Zink RM, Barrowclough GF. Mitochondrial DNA under siege in avian phylogeography. Mol Ecol. 2008;17:2107–2121. doi: 10.1111/j.1365-294X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LEGENDS FOR SUPPLEMENTARY DATA
Additional file 1: Neighbor-Joining analyses based on 64 ITS2 sequences from individuals of the different species of Albitarsis Complex. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches, retaining only groups with frequency ≥60%. Outgroups are An. darlingi and An. braziliensis.
Additional file 2: Neighbor-Joining analyses based on 17 nuclear protein-coding white gene sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches, retaining only groups with frequency ≥60%. Outgroups are An. darlingi and An. braziliensis.
Additional file 3: Maximum parsimony tree based on COI sequences. Numbers above branches indicate the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates). DQ076234, An. albitarsis E (?) specimen from Portuguesa, Venezuela; DQ076231 - DQ076233, An. albitarsis E (An. janconnae) from the type locality in Roraima, Brazil. Bootstrap values >70% are shown. Outgroup comprises An. darlingi and An. braziliensis.
Additional file 4: Maximum likelihood tree based on COI sequences. Numbers above branches indicate ML bootstrap proportions (662 replicates). DQ076234, An. albitarsis E (?) specimen from Portuguesa, Venezuela; DQ076231 - DQ076233, An. albitarsis E (An. janconnae) from the type locality in Roraima, Brazil. Bootstrap values >70% are shown. Outgroup comprises An. darlingi and An. braziliensis.


