Abstract
The Ci-Dll-B gene is an early regulator of ectodermal development in the ascidian Ciona intestinalis (Imai et al., 2006). Ci-Dll-B is located in a convergently transcribed bigene cluster with a tandem duplicate, Ci-Dll-A. This clustered genomic arrangement is the same as those of the homologous vertebrate Dlx genes, which are also arranged in convergently transcribed bigene clusters. Sequence analysis of the C. intestinalis Dll-A-B cluster reveals a 378 bp region upstream of Ci-Dll-B, termed B1, which is highly conserved with the corresponding region from the congener Ciona savignyi. The B1 element is necessary and sufficient to drive expression of a lacZ reporter gene in a pattern mimicking the endogenous expression of Ci-Dll-B at gastrula stages. This expression pattern which is specific to the entire animal hemisphere is activated preferentially in posterior, or b-lineage, cells by a central portion of B1. Expression in anterior, or a-lineage cells, can be activated by this central portion in combination with the distal part of B1. Anterior expression can also be activated by the central part of B1 plus both the proximal part of B1 and non-conserved sequence upstream of B1. Thus, cis-regulation of early Ci-Dll-B expression is activated by a required submodule in the center of B1, driving posterior expression, which works in combination with redundant submodules that respond to differentially localized anterior factors to produce the total animal hemisphere expression pattern. Interestingly, the intergenic region of the cluster, which is important for expression of the Dlx genes in vertebrates, does not have a specific activating function in the reporter genes tested, but acts as an attenuator in combination with upstream sequences.
INTRODUCTION
The Distalless or Dlx family of homeodomain transcription factors have been identified as important developmental regulatory proteins in a wide range of animal groups. First identified in Drosophila, distalless is required for proper limb and central nervous system (CNS) development. In vertebrates, the distalless orthologs, termed Dlx genes, are found in two-gene clusters. Mammals have 3 two-gene clusters, for a total of 6 Dlx genes, while teleost fishes have as many as 5 two-gene clusters and 8 Dlx genes (Sumiyama et al., 2003). Major functional roles for the vertebrate genes have been found in the brain, neural crest and branchial arch derivatives, bone and cartilage formation, and limb development (Panganiban and Rubenstein, 2002).
In protochordates, one Dlx gene has been found in the cephalochordate amphioxus, and three in ascidian tunicates, such as Ciona intestinalis. Two of the Ciona genes, Dlx-A and Dlx-B, are tightly linked in a cluster, with the same convergent transcriptional orientation seen in all vertebrate Dlx clusters, suggesting that this cluster may be homologous to those in vertebrates (Caracciolo et al., 2000; Irvine et al., 2007). The C. intestinalis Dlx genes are expressed in several tissues: Ci-Dll-A is expressed in the adhesive and sensory palps and precursors of the atrial siphon; Ci-Dll-C transcripts are found in the adhesive papillae; and Ci-Dlx-B is expressed in the early ectoderm and is soon restricted to the epidermal lineage (Caracciolo et al., 2000; Imai et al., 2004; Irvine et al., 2007). Consistent with this expression pattern, Ci-Dll-B has been identified as a pivotal gene in the regulatory network for the epidermis (Imai et al., 2006).
The mRNA expression patterns for the Dlx genes in vertebrates have been shown to be largely overlapping between the two genes from one cluster (Ellies et al., 1997; Sumiyama et al., 2002). This finding has led to the notion that shared enhancer elements may control the coordinate expression of both genes of a cluster (Ellies et al., 1997). Subsequent analysis of genomic regulatory elements in zebrafish and mouse has focused on intergenic enhancer elements that are highly conserved between orthologous clusters in all vertebrate groups (Ghanem et al., 2003). These elements have been shown to separately regulate expression of Dlx genes in the brain, limbs, branchial arches and branchial arch derivatives (Ghanem et al., 2003; Park et al., 2004; Sumiyama et al., 2002; Sumiyama and Ruddle, 2003).
In order to better understand the genomic regulation of a simple 2-gene cluster, we have examined the genomic regulation of Ci-Dll-B, using reporter transgene analysis. We have located a 400 bp enhancer upstream of the transcription start site that drives the total early endogenous pattern of expression in prospective ectoderm. We have further dissected this element to find that the anterior and posterior portions of the expression are controlled by separate portions of the enhancer. We also have evidence that the organization of the cis-regulatory elements in C. intestinalis differs considerably from the regulatory organization of the vertebrate Dlx clusters despite the fact that the convergently transcribed genomic arrangement of the genes is the same.
MATERIALS AND METHODS
Animals
Adult Ciona intestinalis, sp. B (Nydam and Harrison, 2007) were collected from floating docks in the Point Judith Marina at Snug Harbor, Rhode Island. Gametes were collected by dissection and spawned in vitro. Embryos for electroporation were chemically dechorionated at spawning.
Reporter transgenes
The CiDB-A construct was made from a lambda clone (D5) obtained from a C. intestinalis, sp. B (Rhode Island, USA population) genomic library. This library was constructed in the BlueSTAR lambda vector (Novagen, Madison, WI, U.S.A.) and a 15 kilobase clone containing the entire Dll-B – Dll-A cluster and flanking sequence (Fig. 1) was subcloned in pBlueSTAR-1 by Cre-mediated excision. The reporter gene cassette from TV13 (Irvine et al., 2008), was inserted into a BglII site in the fifth exon of Ci-Dll-B using the InFusion® Dry-Down PCR Cloning Kit (Clontech) to complete the CiDB-A construct.
Fig. 1. Ciona sequence alignment, diagrams of larger reporter transgenes and scoring.
(A) mVista sequence alignment plot between C. intestinalis and C. savignyi Ci-Dll-B-A clusters. Exons shown as blue-green boxes. Curve represents levels of sequence identity in a 50 bp window. Blue-shaded peaks are in exons while pink-shaded peaks are in non-coding sequence. Peaks of conserved non-coding sequences (CNSs) tested here are denoted as B1, B2, and ig. Distances from transcription start sites of Ci-Dll-B and Ci-Dll-A shown in kilobases (kb). (B) Diagrams of transgene constructs. Non-coding sequence is represented by red bars aligned with corresponding sequence in Vista plot above. LacZ reporter gene insertion is represented by a blue bar. Extent of expression of each transgene in reporter experiments is shown at right as percentages of aggregate anterior and posterior animal quadrants with staining for ß-galactosidase. * significantly different (p<0.05) in Kruskal-Wallis test; + posterior expression significantly different (p<0.05). Refer to Supplementary Tables 1 and 2 for numbers of embryos scored and statistics.
Other reporter constructs depicted in Fig. 1b were made by amplifying the respective putative regulatory region from lambda clone D5 using primers with restriction sites designed on the 5’ ends. Upstream fragments were cloned into the AscI and NotI sites of TV13. Downstream fragments were cloned into the RsrII and PacI sites.
Reporter constructs CiDB-0.2-0.4; CiDB-0.35-0.62; CiDB-0.42-0.62; CiDB-0.38-0.62; and CiDB-0.35-0.51 were made by amplifying the non-coding sequences from lambda clone D5 by PCR with forward primers with a 5’ SalI site, and reverse primers with a 5’ BamHI site. These fragments were then cloned into the vector CiFk5'A cut with SalI and BamHI. CiFk5'A is a modification of the CiFk-1.77 reporter vector (kindly provided by A. DiGregorio and M. Levine) with a polylinker inserted between the XhoI and EcoNI sites. This produces a transcriptionally silent basal promoter lacZ reporter vector (similar to (Harafuji et al., 2002). CiFk5'A contains 349 bp upstream of the Ci-FoxA-a mRNA sequence (Genbank NM_001078564), the 5' UTR (39 bp), and the first 86 codons of the ORF (Di Gregorio et al., 2001) fused to a nuclear localization signal, lacZ, and the SV40 polyadenylation signal.
Constructs CiDB-1.0d4, d5, d7, and d10, were made by PCR deletion using the Phusion Site Directed Mutagenesis kit (New England Biolabs). Primer sequences are available upon request.
The mutated GATA site in CiDB-0.35-0.62mG was created by PCR using the primers Catgut (5'- GACGCGCTGCTCTAAGAAGTCATT) and CiDBf45 (5’-GAAAAACAATGCATTTCTCGGTAGGCT). This converts the sequence on the reverse strand from GATAA to TCTAA. All ligations, deletions, and site-directed mutagenesis were confirmed by sequencing.
Electroporation
Transgenes were delivered to single-celled embryos by electroporation, and lacZ reporter signal detected, as previously described (Irvine et al., 2008). Embryos were reared to mid to late gastrula stage and the number of animal hemisphere quadrants with detectable ß-galactosidase staining counted for each normally developing embryo. Each construct was tested in 3 or more electroporation experiments (except for CiDB-1.0d5 tested twice), and the results were pooled to derive percentages of anterior and posterior quadrants with reporter gene expression (refer to Supplementary Table 1).
For statistical testing of differences between reporter experiments, percentages of anterior or posterior embryonic animal quadrants with reporter expression for individual electroporation experiments were taken as data points. The mean anterior or posterior percentages for sets of electroporation experiments for different reporter constructs were compared using a nonparametric test, the Kruskal-Wallis test, implemented in SPSS, since it is appropriate for the small sample sizes available, which may not be normally distributed. p-values less than or equal to 0.05 were considered significant.
Sequence analysis
Similarity between putatively homologous or sequences were assessed using mVista (http://genome.lbl.gov/vista/index.shtml)(Mayor et al., 2000). For alignment the LAGAN option was used (Brudno et al., 2003). For LAGAN, all default parameters were used. Prediction of protein binding sequences in the conserved B1 element was done using CONSITE (Sandelin et al., 2004b) which uses transcription factor binding profiles from the JASPAR database (Sandelin et al., 2004a).
RESULTS
A 15 kilobase DNA fragment encompassing the whole Ci-Dll-A – Dll-B cluster with a reporter gene inserted into Dll-B recapitulates the endogenous early Dll-B expression pattern
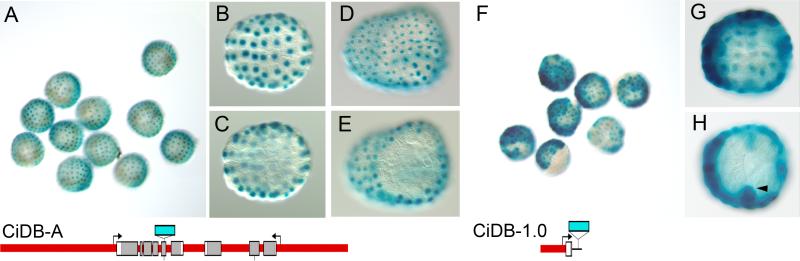
A 15 kilobase lambda clone (CiDB-A) was isolated from a C. intestinalis genomic DNA library which encompassed the Ci-Dll-B and Ci-Dll-A genes. This clone contains a number of conserved non-coding sequences revealed by a sequence alignment comparing the corresponding regions of the C. intestinalis Dll cluster with that of the congeneric ascidian C. savignyi (Fig. 1A). A lacZ reporter gene inserted into the fifth exon of Ci-Dll-B was able to recapitulate the early expression pattern of Ci-Dll-B from the 64-cell to late gastrula stages, which includes, and is specific to, all animal hemisphere cells (Fig. 2A-E). Endogenous transcript expression of Ci-Dll-B is downregulated at the neurula stage to a few cells in the anterior dorsal part of the embryo (Irvine et al., 2007). The CiDB-A construct does not show this downregulation, either in β-galactosidase protein expression or in Ci-Dll-B transcript expression, assayed by whole-mount in situ hybridization (data not shown). This paper will therefore concentrate on the control of the early expression pattern at the early and late gastrula stages.
Fig. 2. Reporter transgene expression – whole cluster and upstream 1 kb.
(A-E) CiDB-A whole cluster construct β-galactosidase staining pattern. (B, C) Animal and vegetal views, respectively, at early gastrula stage. (D, E) Animal and vegetal views, respectively, at late gastrula stage. (F-H) Ci-DB-1.0 transgenic embryos. (G) Animal and (H) vegetal pole views at early gastrula stage. Cells in H which appear on the vegetal side (arrowheads) are b8.17 and b8.19 cells derived from the animal hemisphere (Nishida, 1986).
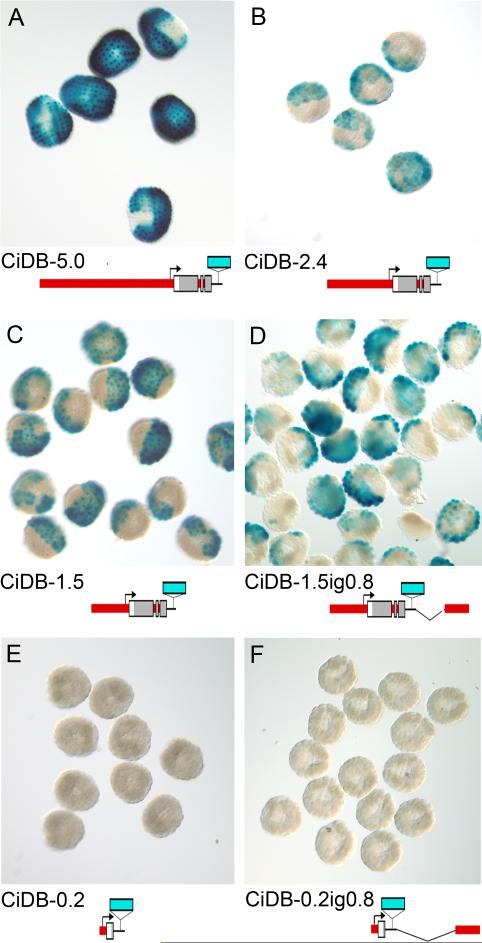
To locate the regulatory elements driving the expression pattern we tested a series of reporter gene constructs with fragments both upstream of Ci-Dll-B and from the intergenic region between Ci-Dll-B and Ci-Dll-A. Fragments that included the conserved upstream sequence denoted B1 were able to drive the animal hemisphere expression in much the same way as the whole cluster CiDB-A construct (Figs. 1B, 2F-H). However, the reporter with 5 kb of upstream sequence (CiDB-5.0; Fig. 3A) was able to drive expression in a higher proportion of animal hemisphere cells, like the whole-cluster construct, than a shorter construct lacking 5 short segments of conserved sequence (CiDB-2.4; Fig. 3B). Interestingly, eliminating the next conserved segment, B2 in Fig. 1A (CiDB-1.5; Fig. 3C), increases the extent of expression, suggesting that B2 is an attenuator. Cutting another 500 bp of sequence reduces the extent of expression to the level of the 2.4 kb fragment, indicating that there may be enhancer binding sites in the non-conserved region from 1.0 to 1.5 kb upstream (Fig. 1B). This reporter, CiDB-1.0, which includes only the B1 conserved element was still able to drive expression in the endogenous gastrula stage pattern (Fig. 2H-J). A reporter gene with only 200 bp of upstream sequence (CiDB-0.2; Fig. 3E) showed no lacZ signal. However, when linked with a fragment known to drive expression with the strong heterologous Ci-FoxA-a promoter, CiDBp-0.35-0.62, expression was seen in the expected region, although at a lower level than when the Ci-FoxA-a promoter is used (Figs. 4, 5G). This result indicates that a functional basal promoter region is included in CiDB-0.2.
Fig. 3. Reporter transgene expression.
(A-F) Representative animal pole views of transgenic embryos. Name and a diagram of the reporter transgenes electroporated in each case are below the corresponding photos.
Fig. 4. Diagrams of reporter transgenes associated with the B1 conserved sequence element.
Representation of the position of the B1 CNS within the CiDB-1.0 construct (top). Predicted transcription factor binding sites are shown by colored ovals. Below are diagrams of reporter transgene constructs. The Ci-Dll-B promoter is incorporated, except where noted “fkh” which indicates the non-coding sequence linked to a silent basal Ci-FoxA-a promoter (Harafuji et al., 2002) (Constructs CiDB-0.2-0.44, CiDB-0.35-0.62 and CiDB-0.35-0.62mG. CiFk5'A is the empty Ci-FoxA-a promoter vector.) Chart at right shows the extent of expression of each transgene as percentages of aggregate number of anterior and posterior animal quadrants with staining for ß-galactosidase. Refer to Supplementary Tables 1 and 2 for numbers of embryos scored and statistics.
Fig. 5. Reporter transgene expression – B1 region analysis.
(A-H) Representative animal pole views of transgenic embryos. Name and a diagram of the reporter transgenes electroporated in each case are below the corresponding photos. (A) Deletion of the distal 170 bp of B1 has little effect on expression compared with CiDB-1.0., while deletion of the proximal 140 bp (B) causes some loss of expression. (C) 440 bp upstream of Ci-Dll-B drives expression only in the posterior animal hemisphere, similar to that seen when the proximal 211 bp of B1 is tested upstream of a heterologous Ci-FoxA-a promoter (D). On the other hand the more distal 276 bp of B1 is able to drive expression in all quadrants of the animal hemisphere, upstream of both the native Ci-Dll-B promoter (G), and the stronger Ci-FoxA-a promoter (E) Mutation of a canonical GATA site in this fragment (F) reduces, but does not eliminate expression in the same domain. Truncation of the same fragment to 163 bp near the middle of B1 still drive expression, but mostly in the posterior quadrants.
The Ci-Dll-B – Ci-Dll-A intergenic region does not activate cell-specific Ci-Dll-B expression in a reporter gene
We also tested the effects of the intergenic region on expression of the reporter genes, since important regulatory elements are located in the intergenic region in zebrafish and mouse Dlx clusters (Ghanem et al., 2003; Park et al., 2004; Sumiyama et al., 2002; Sumiyama and Ruddle, 2003; Zerucha et al., 2000). In our constructs the intergenic region does not activate a specific aspect of the Ci-Dll-B expression pattern by itself. This finding is demonstrated by construct CiDB-0.2ig0.8 (Fig. 3F), which links the intergenic region to the non-expressing basal promoter region, and exhibits no detectable β-galactosidase staining. However, the intergenic region does appear to have an attenuation effect on Ci-Dll-B expression, as shown by the comparison of CiDB-1.5 (Fig. 3C) to CiDB-1.5ig0.8 (Fig. 3D). The latter construct is expressed in a lower percentage of animal hemisphere quadrants, similar to the level seen with CiDB-2.4, which incorporates the B2 element, but lacks the intergenic region (Fig. 1 & Suppl. Tables 1 & 2).
The major animal hemisphere enhancer is localized to a 400 bp region and has separable anterior and posterior control elements
Deletion of the entire 378 bp conserved B1 sequence (construct CiDB-1.0d10; Fig. 4) completely obliterates reporter gene expression. However, deletions of small portions of the B1 element within CiDB-1.0 fail to alter expression (data not shown). Even with relatively large deletions from proximal or distal parts of B1, e. g. CiDB-1.0d4 and CiDB-1.0d5 (Figs. 4, 5A&B), expression in all four animal quadrants is maintained. These results suggest that there is redundancy within the B1 element, which is able to compensate for small deletions. However, these experiments do not exclude the possibility that elements distal to B1 within the 1.0 kb upstream DNA are compensating for the deletions within B1. This is apparently the case in comparison of CiDB-0.44 and CiDB-1.0d4 (Figs. 4, 5A&C). CiDB-1.0d4 is the same as CiDB-0.44, except that 316 bp of sequence upstream of the B1 element is added. While CiDB-0.44 only drives expression in a small proportion of posterior animal quadrants, CiDB-1.0d4 can activate expression equivalent to the whole -1.0 kb upstream region.
To further analyze the animal hemisphere enhancer we made a series of reporter constructs with either smaller fragments of the upstream region or other deletions from the CiDB-1.0 reporter within the B1 element. Fig. 4 (top) diagrams the B1 conserved element with the locations of putative transcription factor binding sites predicted by an analysis using the program CONSITE (Sandelin et al., 2004b) (Sequence included as Suppl. Fig. 1). Only sites for transcription factors that are expressed in the animal hemisphere at the stages examined here were included. Transcription factor expression patterns were obtained from examination of the ANISEED database, (Tassy et al.), including data from the Ghost database (Satou et al., 2005). The reliability of these predictions is hampered by the lack of Ciona-specific transcription factor binding profiles. The freely accessible JASPAR database was used for profiles to support CONSITE, and these profiles come mostly from organisms distantly related to C. intestinalis, and certainly do not give a complete prediction of the complement of transcription factors binding the B1 element. However, this binding site prediction does give a starting point to identify candidate trans factors influencing Ci-Dll-B expression.
The entire 400 bp conserved region is not required for reporter gene expression in all animal hemisphere blastomeres. A 270 bp fragment at the distal side of the B1 element, CiDB-0.35-0.62, is sufficient to drive expression in all four animal quadrants (Figs. 4 & 5E) when cloned into a Ci-FoxA-a basal promoter-lacZ reporter vector (similar to that used by Harafuji et al. (2002) refer to methods section). As mentioned above, the same fragment when cloned upstream of the 200 bp putative promoter region, CiDBp-0.35-0.62, also activates expression in all animal quadrants, albeit in a lower proportion of total quadrants. (Figs. 4 & 5G). Overlapping proximal fragments, CiDB-0.2-0.4 or CiDB-0.44, confer expression in only the posterior half of the animal hemisphere, derived from the b-line blastomeres (Figs. 4, 5C&D). Another short fragment, CiDBp-0.35-0.5 (Fig. 5H) also drives posterior expression, in this case with only a low level of anterior (a-line) quadrants staining. While the sequence from 200 bp to 350 bp upstream is dispensable for expression in all quadrants, as shown by CiDB-1.0d5, a further deletion from 350 bp to 450 bp upstream eliminates all transgene activation. Taken together these data suggest that a center portion of B1, from -350 to -440 bp, is necessary and sufficient to activate expression in posterior (b-line) cells. Anterior (a-line) activation requires the addition of either the distal part of B1, such as constructs CiDB-0.35-0.62, CiDBp-0.35-0.62, and CiDB-1.0d5, or the proximal part of B1 and sequence upstream of B1, as shown by CiDB-1.0d4 (Figs. 4 & 5). Therefore, central, distal and proximal portions of the B1 element, and likely additional upstream sequence, respond to factors differentially localized in a-line (anterior) vs. b-line (posterior) animal hemisphere cells.
A GATA transcription factor canonical binding sequence is the only putative binding site we identified within the region included in all expressing constructs. (See Suppl. Fig. 1) To test whether this site is required for expression we inserted a site-directed mutation to change the sequence from GATAA to TCTAA in the construct CiDB-0.35-0.62mG (Figs. 4 & 5F). The mutation significantly reduced (p=0.05, Suppl. Table 2) but did not obliterate expression. This result suggests that a GATA DNA binding protein may be important, but not essential at this one binding site, for activation of Ci-Dll-B transcription.
DISCUSSION
Overall organization of Ci-Dll-B cis-regulation
We have dissected the Dll-B-A bigene cluster and located a 378 bp non-coding region about 200 bp 5’ of the transcription start site, which we call B1, highly conserved with the congener C. savignyi that is sufficient to drive expression in all animal hemisphere cells at gastrula stages. This animal hemisphere reporter gene expression matches that of the endogenous transcript in the gastrula (Irvine et al., 2007; Satou et al., 2005). This small element exhibits reporter gene expression in the same cells as a large reporter construct comprising the entire Dll-B-A cluster (Fig. 2).
We also tested the effects of other parts of the non-coding DNA associated with the cluster. These other parts outside the B1 element have no effect on the cellular localization of reporter gene expression. However, sequence between 2.4 kb and 5 kb upstream of Ci-Dll-B amplifies expression, while sequence between 1.5 and 2.4 kb, containing the B2 conserved sequence attenuates expression. Likewise, the intergenic region containing the AB1 element attenuates expression in combination with 1.5 kb of upstream sequence (Figs. 1 - 3).
From these deletion experiments it is apparent that the enhancers required for the basic animal hemisphere expression in the gastrula reside within 1 kb of the transcription start site, as shown by construct CiDB-1.0 (Fig. 2F-H). Further deletions within this element show that the central region of the B1 element between -440 and -350 bp are required for any expression, and that this region primarily drives expression in the posterior, b-line, animal blastomeres (cf. CiDBp-0.35-0.5, CiDB-0.2-0.44, CiDB-1.0d5, Fig. 4, and Fig. 5B, D, & H).
None of our reporter construct experiments showed any involvement of repressor elements that might be required to exclude expression from the vegetal hemisphere. We never saw expression in vegetal cells, regardless of which non-coding DNA was excluded from the reporter transgene. This negative finding indicates that spatial specificity of the expression domain is due to upstream regulation of the activators of Ci-Dll-B, which must be confined to the animal hemisphere, unless any repressor binding sites are small, and within the required sequence between -440 and -350 bp.
The only transcription factor binding site that our analysis predicted to fall within this critical -440 to -350 bp region, is a canonical GATA site on the minus strand (5'-GATAA). We mutated this site in the CiDB-0.35-0.62 construct (CiDB-0.35-0.62mG) to assay its effect on transcription. There was a statistically significant drop in expression without the GATAA sequence, indicating that a protein, probably a GATA factor binds the DNA at this site and helps to activate transcription (Figs. 4, 5E&F, Suppl. Table 2). This involvement of a GATA protein in activation of Ci-Dll-B is consistent with the finding that Ci-fog transcription, which commences before Ci-Dll-B and occupies the same domain, depends on activation by CiGATAa (Rothbacher et al., 2007). However, Ci-Dll-B expression does not appear to be as dependent on GATAa as Ci-fog expression, unless there are undetected GATA sites in our constructs that are required along with the one that we have mutated.
Comparison with cis-regulatory organization of vertebrate Dlx cluster regulation
A major difference between the regulation of Ci-Dll-B and that of vertebrate Dlx genes is the greater relative importance of intergenic enhancers in the vertebrate clusters. All the Dlx bigene clusters in zebrafish and mouse have pairs of highly conserved sequences in the intergenic region. These two sequence elements have been verified by reporter gene experiments to be enhancers that together explain the major expression patterns in the brain, branchial arches, and limbs (Ghanem et al., 2003; Ghanem et al., 2007; Ghanem et al., 2008; Park et al., 2004; Sumiyama et al., 2002; Sumiyama and Ruddle, 2003). Upstream sequences from the clusters have not been as well characterized, although elements have been found that confer expression in brain (Ghanem et al., 2007) and teeth (Jackman and Stock, 2006). Taken together, a major part of the expression pattern for both genes of each of the vertebrate clusters appears to be controlled by the intergenic enhancers. This arrangement is in stark contrast to our finding for the C. intestinalis Ci-Dll-B/A cluster. Here the intergenic region is much smaller than in the vertebrate clusters, being only about 700 bp in Ciona, as opposed to several kilobases in zebrafish (Ghanem et al., 2003). There is one stretch of sequence highly conserved with the congener C. savignyi, indicating possible cis-regulatory activity. However, in our reporter transgene experiments, the Ciona intergenic region is not responsible for any tissue-specific expression on its own, as read out through the Ci-Dll-B promoter. Rather, it appears to function to attenuate expression activated by the B1 element (Figs. 1, 3C&D).
On the other hand, it is possible that the intergenic region controls Ci-Dll-A expression rather than Ci-Dll-B. We have not tested this possibility directly. Even if Ci-Dll-A is regulated by the intergenic region, this would still differ from the vertebrate pattern, where each of the conserved intergenic elements controls a portion of the expression pattern of both clustered genes.
Model for activation of the Ci-Dll-B pan-ectodermal module
As a working model, the Ci-Dll-B proximal cis-regulatory region can be divided into four regions (Fig. 6). Region “a” in the center of the B1 conserved element is required for any expression, and alone can drive transcription in posterior animal, or b-line, cells at gastrula stages. Anterior quadrant expression can be gained by the addition of element “b”. Alternatively, anterior expression can be gained by the addition of elements “c” & “d” to “a”. Thus, at least the anterior domain of expression is due to redundant enhancer elements, possibly serving to dampen developmental noise from variation in the levels or timing of expression of upstream activating elements. Interestingly, we found no evidence for repressor elements that might be required for silencing expression in the vegetal hemisphere, and no reporter assay ever showed any expression in vegetal cells. This observation suggests that spatial regulation of Ci-Dll-B relies on DNA binding proteins that are limited to the animal hemisphere prior to the initiation of transcription. Further mutational and biochemical analysis of the B1 module will be necessary to verify this working model.
Fig. 6. Model for Ci-Dll-B regulation.
Diagram of working model of Ci-Dll-B B1 element organization. In this scheme, consistent with our results, a central part of B1 (labeled “a”) is required for any expression, which is confined to the posterior (b-line) cells of the animal hemisphere. Anterior expression is gained when either the distal part of B1, “b” is added, or the combination of proximal B1, “d” and the region upstream of B1, “c”. Elements “b”,“c”, or “d” on their own or together, without “a”, do not drive expression.
Supplementary Material
Suppl. Fig. S1. Sequence of Ci-Dll-B upstream “B1 element” aligned with the homologous region in C. savignyi. Predicted putative transcription factor binding sites shown in gray boxes. Sequences included in reporter gene constructs are denoted by colored lines under the corresponding sequence. See legend for color code.
ACKNOWLEDGEMENTS
This work benefited from discussion and technical advice from Brad Davidson and Thomas H. Meedel, and insightful comments from two anonymous reviewers. The Point Judith Marina generously provided access to their docks for animal collection. The work was supported by National Institutes of Health grant 1R15GM07373701 from the National Institute for General Medical Sciences, and grant P20RR016457 from the RI-BRIN/RI-Inbre program of the National Center for Research Resources.
Grant support: National Institutes of Health Grant 1R15GM07373701 from the National Center for General Medical Sciences, and Grant P20 RR016457 from the BRIN/INBRE Program of the National Center for Research Resources
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Research Highlights
> 400 bp element 200-600 bp upstream of Ci-Dll-B activates transcription > 100 bp submodule essential and drives posterior animal expression > 3 other submodules drive anterior expression in combination with posterior module > The region between Ci-Dll-A and Ci-Dll-B is not an activator for Ci-Dll-B
LITERATURE CITED
- Brudno M, Do CD, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S, Program NCS. LAGAN and Multi-LAGAN: Efficient Tools for Large-Scale Multiple Alignment of Genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracciolo A, Gregorio AD, Aniello F, Lauro RD, Branno M. Identification and developmental expression of three Distal-less homeobox containing genes in the ascidian Ciona intestinalis. Mech. Dev. 2000;99:173–176. doi: 10.1016/s0925-4773(00)00474-3. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Corbo J, Levine M. The regulation of forkhead/HNf-3 beta expression in the Ciona embryo. Dev. Biol. 2001;229:31–43. doi: 10.1006/dbio.2000.9964. [DOI] [PubMed] [Google Scholar]
- Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, Ekker M. Relationship between the genomic organization and the overlapping embryonic expression patterns of the zebrafish dlx genes. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- Ghanem N, Jarinova O, Amores A, Long Q, Hatch G, Park BK, Rubenstein JLR, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Research. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long QM, Hatch G, Rubenstein JLR, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J. Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Poitras L, Rubenstein JLR, Ekker M. Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia thorough the activity of the I56ii enhancer. Dev. Biol. 2008;322:415–424. doi: 10.1016/j.ydbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Harafuji N, Keys DN, Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. USA. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Irvine SQ, Cangiano MC, Millette BJ, Gutter ES. Non-overlapping expression patterns of the clustered DllA/B genes in the ascidian Ciona intestinalis. J. Exp. Zool. (Mol. Dev. Evol.) 2007;308B:428–441. doi: 10.1002/jez.b.21169. [DOI] [PubMed] [Google Scholar]
- Irvine SQ, Fonseca VC, Zompa MA, Antony R. Cis-regulatory organization of the Pax6 gene in the ascidian Ciona intestinalis. Developmental Biology. 2008;317:649–659. doi: 10.1016/j.ydbio.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Stock DW. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc. Natl. Acad. Sci. USA. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics. 2000:1046. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell division pattern during gastrulation of the ascidian, Halocynthia roretzi. Dev. Growth Diff. 1986;28:191–201. doi: 10.1111/j.1440-169X.1986.00191.x. [DOI] [PubMed] [Google Scholar]
- Nydam ML, Harrison RG. Geneological relationships within and among shallow-water Ciona species (Ascidiacea). Mar. Biol. 2007;151:1839–1847. [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–86. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Park BK, Sperber SM, Choudhury A, Ghanem N, Hatch GT, Sharpe PT, Thomas BL, Ekker M. Intergenic enhancers with distinct activities regulate Dlx gene expression in the mesenchyme of the branchial arches. Dev. Biol. 2004;268:532–545. doi: 10.1016/j.ydbio.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Rothbacher U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134:4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman W, Lenhard B. JASPAR: an open access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004a;32 doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004b;32:W249–52. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zoological Science. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Ruddle FH. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. J. Struc. Func. Genomics. 2003;3:151–159. [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Stock DW, Weiss KM, Kawasaki K, Shimizu N, Shashikant CS, Miller W, Ruddle FH. Genomic structure and functional control of the Dlx3-7 bigene cluster. Proc. Natl. Acad. Sci. USA. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, Ruddle F. Regulation of Dlx3 gene expression in visceral arches by evolutionarily conserved enhancer elements. Proc. Natl. Acad. Sci. USA. 2003;100:4030–4034. doi: 10.1073/pnas.0530119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, Laporte B, Rios A, Luxardi G, Kusakabe T, Joly JS, Darras S, Christiaen L, Contensin M, Auger H, Lamy C, Hudson C, Rothbacher U, Gilchrist MJ, Makabe KW, Hotta K, Fujiwara S, Satoh N, Satou Y, Lemaire P. The ANISEED database: Digital representation, formalization, and elucidation of a chordate developmental program. Genome Research. 20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JLR, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. S1. Sequence of Ci-Dll-B upstream “B1 element” aligned with the homologous region in C. savignyi. Predicted putative transcription factor binding sites shown in gray boxes. Sequences included in reporter gene constructs are denoted by colored lines under the corresponding sequence. See legend for color code.