Abstract
The factor inhibiting HIF (FIH) is one of the primary oxygen sensors in human cells, controlling gene expression by hydroxylating the α-subunit of the hypoxia inducible transcription factor (HIF). As FIH is an alpha-ketoglutarate dependent non-heme iron dioxygenase, oxygen activation is thought to precede substrate hydroxylation. The coupling between oxygen activation and substrate hydroxylation was hypothesized to be very tight, in order for FIH to fulfill its function as a regulatory enzyme. Coupling was investigated by looking for reactive oxygen species production during turnover. We used alkylsulfatase (AtsK), a metabolic bacterial enzyme with a related mechanism and similar turnover frequency, for comparison, and tested both FIH and AtsK for H2O2, O2− and OH• formation under steady and substrate-depleted conditions. Coupling ratios were determined by comparing the ratio of substrate consumed to product formed. We found that AtsK reacted with O2 on the seconds timescale in the absence of prime substrate, and uncoupled during turnover to produce H2O2; neither O2− nor OH• were detected. In contrast, FIH was unreactive toward O2 on the minutes timescale in the absence of prime substrate, and tightly coupled during steady-state turnover; we were unable to detect any reactive oxygen species produced by FIH. We also investigated the inactivation mechanisms of these enzymes and found that AtsK likely inactivated due to deoligomerizion, whereas FIH inactivated by slow autohydroxylation. Autohydroxylated FIH could not be reactivated by dithiothreitol (DTT) nor ascorbate, suggesting that autohydroxylation is likely to be irreversible under physiological conditions.
Keywords: HIF, hypoxia, non-heme iron, α-ketoglutarate, factor inhibiting HIF, reactive oxygen species
1. Introduction
Cellular oxygen-sensing in metazoans is directly controlled by enzymes which hydroxylate the alpha-subunit of the hypoxia inducible factor (HIFα or HIF-1α).[1–3] As the HIF-hydroxylases are key regulators of angiogenesis and basal metabolism, they are potential targets for treating diseases such as cancer and stroke.[4–6] The two types of HIF-hydroxylase are the factor inhibiting HIF-1 (FIH) and prolyl hydroxylase (PHD),[7–9] both of which are Fe(II), α-ketoglutarate-dependent hydroxylases. The best characterized of these enzymes is the human enzyme FIH, which hydroxylates the Asn803 residue within the C-terminal transactivation domain (CTAD) of HIFα,[10] thereby preventing transcriptional machinery from binding to HIFα.
αKG hydroxylases catalyze two half reactions resulting in the transfer of oxidizing equivalents from O2 to both αKG and the primary substrate (Chart 1).[4, 11, 12] The first half-reaction is O2-activation, in which the αKG/[Fe]2+ is oxidatively decarboxylated to form succinate/[FeO]2+ and CO2.[13, 14] The second half-reaction is transfer of the oxidant from [FeO]2+ to the prime substrate,[15, 16] which may include reactions such as desaturation, demethylations, ring-closure, and hydroxylation;[11, 12, 17] in the case of FIH, this reaction is the hydroxylation of the β-carbon of Asn803.[10] Tight coupling between these two half-reactions is challenging, and many of the αKG-dependent hydroxylases exhibit reactions with O2 that are uncoupled.[18–26] In some cases this leads to auto-hydroxylation of residues within enzyme active sites,[18, 21, 22, 25] however more commonly it leads to metal oxidation that can be rescued by ascorbate.[12, 23, 26]
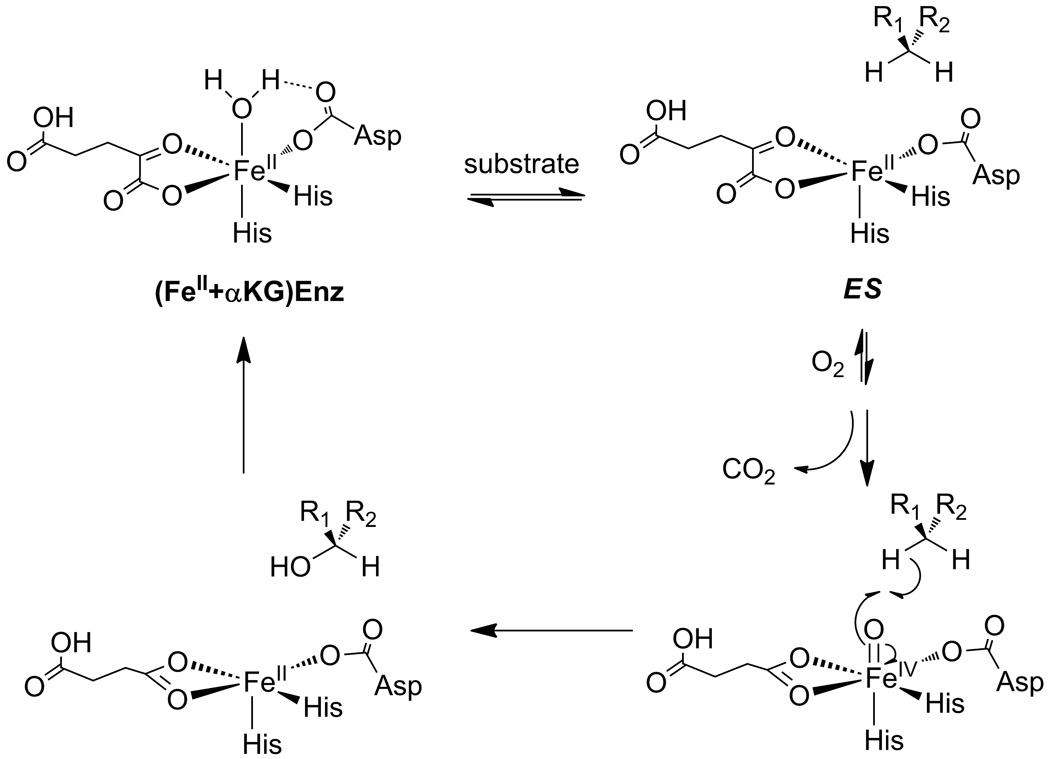
Chart 1.
Consensus chemical mechanism of αKG-dependent hydroxylases. The (Fe+αKG)Enzyme binds the primary substrate to form the ES complex.
The prevailing model for how αKG hydroxylases may achieve coupled turnover focuses on changes in the coordination number about the Fe(II) upon substrate binding. In this model, the Fe(II) is six coordinate prior to prime substrate binding, coordinated by a His2(Asp/Glu) facial triad, a bidentate αKG, and a single H2O ligand.[17] Once the prime substrate binds, the H2O ligand is released to form a five-coordinate Fe(II) center which is ready to react with O2. Crystal structures of several αKG hydroxylases support this model,[12, 27–29] as does mechanistic data indicating that poor substrates stimulate uncoupling.[20, 24, 26, 30] Perhaps the strongest evidence comes from advanced spectroscopic studies that clearly show this coordination change occurs upon substrate binding.[31–33] Tight coupling would be highly beneficial to an O2-sensing enzyme, such as FIH.
There are three criteria for effective O2-sensing by HIF hydroxylases. First, the KM(O2) should lie above the physiological pO2, so that the rate of HIF hydroxylation is proportional to the pO2. As the reported KM(O2) for FIH is much higher than the cellular pO2 under physiological conditions,[34, 35] it appears that FIH is well suited for its regulatory role by this first criterion. Second, uncoupling between the two half-reactions must be low, ideally such that no molecule of O2 reacts without HIF hydroxylation due to the potential for oxidative damage. Although αKG hydroxylases are mechanistically predisposed to some uncoupling between O2-activation and substrate hydroxylation,[12, 36] and FIH can autohydroxylate in the absence of prime substrate,[18] the extent of uncoupling during steady-state turnover by FIH is unknown. Third, release of reactive oxygen species (ROS) from the active site should be minimal in order to avoid oxidative damage to the cell. The propensity of FIH to produce ROS has never been tested.
Here we use chemical methods to test the uncoupling and ROS production from human FIH in comparison to AtsK, a metabolic αKG hydroxylase involved in bacterial sulfur metabolism.[37] As metabolic enzymes are expected to favor fast catalysis over tight coupling, we felt that AtsK would be an instructive contrast to FIH. We observed that FIH does not uncouple during turnover conditions, nor does it release ROS under any tested conditions. FIH does, however, autohydroxylate in a slow reaction in the absence of substrate, forming an inactive form FIH. In contrast, AtsK uncouples under turnover conditions and releases H2O2.
2. Experimental Procedures
2.1 Materials
The prime substrate for FIH was a 39-residue peptide corresponding to HIFα788–826 with a Cys800→Ala point mutation, which corresponds to the C-terminal transactivation domain of HIF-1α (CTAD). The CTAD sequence used (Asn803 underlined) was DESGLPQLTSYDAEVNAPIQGSRNLLQGEELLRALDQVN, which was unmodified at the peptide termini. CTAD peptide was purchased from EZBiolabs as a desalted product, and was further purified by reverse-phase HPLC. Hexylsulfate (HexSO4) and NADH were obtained from Acros Organics. The enzymes used in coupled assays were from commercial sources: alcohol dehydrogenase and Cu/Zn superoxide dismutase were from MP Biomedicals, horseradish peroxidase was from Fluka. A succinate detection kit was purchased from R-Biopharm.
2.2 Protein expression, purification, and activity
AtsK and FIH were expressed and purified as previously described.[19, 37] AtsK activity was measured continuously with both oxygen sensor using a Clark-type electrode and coupling the enzyme reaction with NADH/alcohol dehydrogenase at 25°C. In O2 consumption assay, 200 µM ascorbate, 1 mM αKG, 0–1 mM HexSO4 and 100 µM FeSO4 were premixed in 1 mL volume with 10 mM HEPES pH 7.00, and the reaction was initiated by adding 1 µM AtsK and oxygen consumption was monitored. AtsK activity was also monitored by coupling the enzyme reaction with 160 µM NADH and 5 unit/ml alcohol dehydrogenase in 100 µL volume by monitoring the absorbance change at 340 nm in UV-Visible(UV-Vis) Spectrometer.
FIH activity was monitored by a quenched time point assay and analyzied by LC-MS. In a 50 µL reaction volume, 2 mM ascorbate, 0.1 mM DTT, 5 u/mL catalase, 500 µM αKG, 25 µM FeSO4, 0–600 µM CTAD were preincubated at 37°C and 0.5–5 µM of FIH was added to initiate the reaction. At certain time points 5 µL aliquots were taken and the reaction was quenched with 45 µL 0.1% formic acid. For each reaction 5–10 samples were prepared in 3–10 min and analyzed by LC-MS using a C8 column. The +3 charge-state of parental and and hydroxylated CTAD peaks were observed at m/z of 1419.1 and 1424.4 respectively, as expected for the expected mass gain of 16 amu in product. The ratio of hydroxylated CTAD peak intensity to overall peak intensity was calculated for each sample and the rate was calculated from a linear fit of these time points.
2.3 Uncoupling
2.3.1 Absence of prime substrate
Small volumes of an AtsK stock (0.38 mM) were injected into a 1.00 mL solution containing 100 µM FeSO4, 200 µM ascorbate, 1 mM αKG and were mixed in 10 mM HEPES buffer pH 7.00, 25 °C. The FIH assay was performed with 50 µM FeSO4, 500 µM αKG, and 11.7 µM FIH in 50 mM HEPES, pH 7.50, 37°C. O2 consumption was monitored by a Clark-type electrode (YSI Incorporated).
2.3.2 Presence of prime substrate
The coupling ratio of each enzyme was determined in the presence of varied concentrations of primary substrate. For AtsK, the amount of consumed oxygen was compared to the amount of product formed by oxygen consumption and NADH coupled assays respectively. The coupling ratio of FIH was obtained by comparing the amount of succinate formed to the amount of hydroxylated peptide. Succinate was measured by UV-Vis spectroscopy using a succinate detection kit and hydroxylated CTAD was analyzed by LC-MS.
2.4 ROS production
2.4.1 H2O2/O2− assays
Hydrogen peroxide production was detected by coupling H2O2 oxidation of 50 µM 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) with 1 U/mL horseradish peroxidase (HRP). Hydrogen peroxide production was continuously monitored at 405 nm. Ascorbic acid was excluded from the reaction mixtures to prevent reduction of ABTS+. Superoxide detection was accomplished in a same manner of peroxide detection, by the use of 100 U/mL Cu/Zn superoxide dismutase (Cu/Zn SOD) to convert O2− into H2O2. The reaction conditions for H2O2 and O2− detection included the coupling reagents with the respective enzyme assays; for AtsK: 11.4 µM AtsK, 1 mM αKG, 0–100 µM HexSO4, 100 µM FeSO4; for FIH: 5 µM FIH, 500 µM αKG, 50 µM FeSO4, 0–100 µM CTAD.
2.4.2 OH• radical assay
Hydroxyl radical detection of AtsK and FIH was achieved by mixing enzyme activity assay solutions with 15 mM 2-deoxyribose.[38] Primary substrate concentrations were varied for AtsK, 0–600 µM HexSO4; and for FIH, 0–120 µM CTAD. The reactions were initiated by adding enzyme then incubating at 37 °C for 1 hour. Reactions were quenched with 100 µl 1% thiobarbituric acid in 50 mM NaOH and 100 µL 2.8 % trichloroacetic acid in water. The solution was heated at 100 °C for 20 min. After cooling down to room temperature, the absorbance at 532 nm was measured. The zero substrate sample was treated as a reference.
2.5 Inactivation and rescue of inactivated enzyme
2.5.1 Inactivation of AtsK
Inactivation of AtsK was tested by changing the concentration of AtsK in a reaction from 1 to 10 µM, in the NADH-coupled assay detected at 340 nm. 1 mM αKG, 1mM HexSO4, 100 µM Fe(II), 0–2 mM ascorbic acid, 1–10 µM AtsK, 30 µM NADH and 30 units of alcohol dehydrogenase were used in a 100 µL reaction volume.
2.5.2 Inactivation and rescue of autohydroxylated FIH
Autohydroxylated FIH was prepared to test for re-activation conditions. Under anaerobic conditions, 50 mM HEPES (pH 7.50) αKG (100 µM), FIH (49 µM) and FeSO4 (50 µM) were added to a septum-sealed UV cuvette. An initial UV-Vis absorption spectrum was collected; the septum was then removed to introduce air and initiate the reaction. The characteristic absorption peak of autohydroxylated FIH (λmax = 583 nm) grew over several hours.[18] Autohydroxylated FIH was tested for activity in an assay mixture containing αKG (500 µM), FeSO4 (25 µM), CTAD (70 µM), autohydroxylated FIH (5 µM), and a mild reductant. The mild reductants ascorbate (0 – 2 mM) and DTT (0 – 0.1 mM) were tested for their ability to re-activate auto-hydroxylated FIH. Samples were analyzed for hydroxylated CTAD by LC-ESI-MS.
3. Results
Uncoupling for FIH and AtsK was evaluated by comparing the steady-state kinetic signatures of the two half reactions. As O2 reacts with αKG to form succinate and CO2 in the first half-reaction, the stoichiometry of O2-activation was measured by O2 consumption or succinate formation. This assumes that O2 is only activated by oxidative decarboxylation of αKG. The rate of prime substrate hydroxylation was monitored by mass spectrometry for FIH or by a UV-Vis assay for AtsK. A fully coupled reaction would exhibit an [O2]:[hydroxylated product] ratio of unity; deviations from this ratio would indicate that O2 was being activated in the absence of substrate hydroxylation.
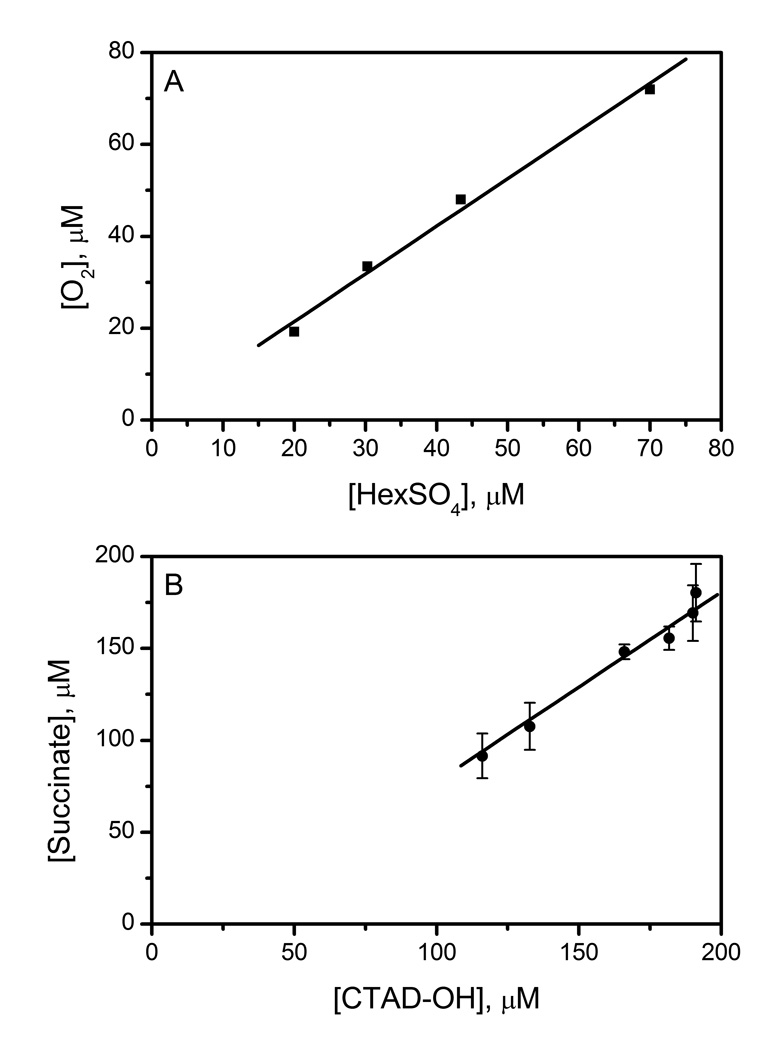
Uncoupling for AtsK and FIH was measured with variable concentrations of substrate. The solution conditions for AtsK included 0 – 1000 µM HexSO4, with up to 3.0 µM AtsK. AtsK was tightly coupled under steady-state conditions, as the number of enzyme turnovers was equivalent by both Clark electrode and by the UV-Vis assay. These assays indicated a coupling ratio of 1.04 ± 0.07 in the steady-state for AtsK (Fig 1). As the KM(HexSO4) is 15 µM for AtsK, the measured steady-state coupling ratio for AtsK reflected that of an ES complex, indicating that once AtsK bound HexSO4 and entered its catalytic cycle it completed the hydroxylation with relatively high fidelity (ca. 96%). This indicated that AtsK turnover was much faster than uncoupling from the ES complex.
Fig 1.
Coupling of O2 to prime substrate for AtsK and FIH. A) O2 consumption vs. HexSO4 hydroxylation for AtsK. AtsK (0.38 – 3.0 µM), ascorbate (200 µM), αKG (1 mM), FeSO4 (100 µM), HexSO4 (1 mM), NADH (160 µM), alcohol dehydrogenase (5 U/mL) in 10 mM HEPES (10 mM, pH 7.00). B) Succinate production vs. CTAD hydroxylation for FIH. FIH (2.0 µM), ascorbate (2000 µM), DTT (100 µM), αKG (500 µM), FeSO4 (50 µM), CTAD (240 µM) in HEPES (50 mM, pH 7.50).
Similarly, FIH was tightly coupled as shown by the stoichiometry of succinate to hydroxylated peptide present in quenched reactions. FIH (2 µM) was incubated with CTAD (200 µM) and the reaction quenched at several time points for analysis. These assays indicated a coupling ratio of 1.03 ± 0.17 in the steady-state for FIH (Fig 1). FIH distributed between predominantly an (Fe2+αKG)FIH form at low [CTAD], and an ES complex at high [CTAD], as we measured the KM(CTAD) as 77 µM.[39] Nevertheless, FIH exhibited a tightly coupled reaction under steady-state conditions, indicating that O2-activation by FIH is tightly controlled. It should be noted that the succinate assay used to measure O2-activation by FIH was less precise than the simpler UV-Vis assay used for AtsK, due to the many reaction components and manipulations needed.
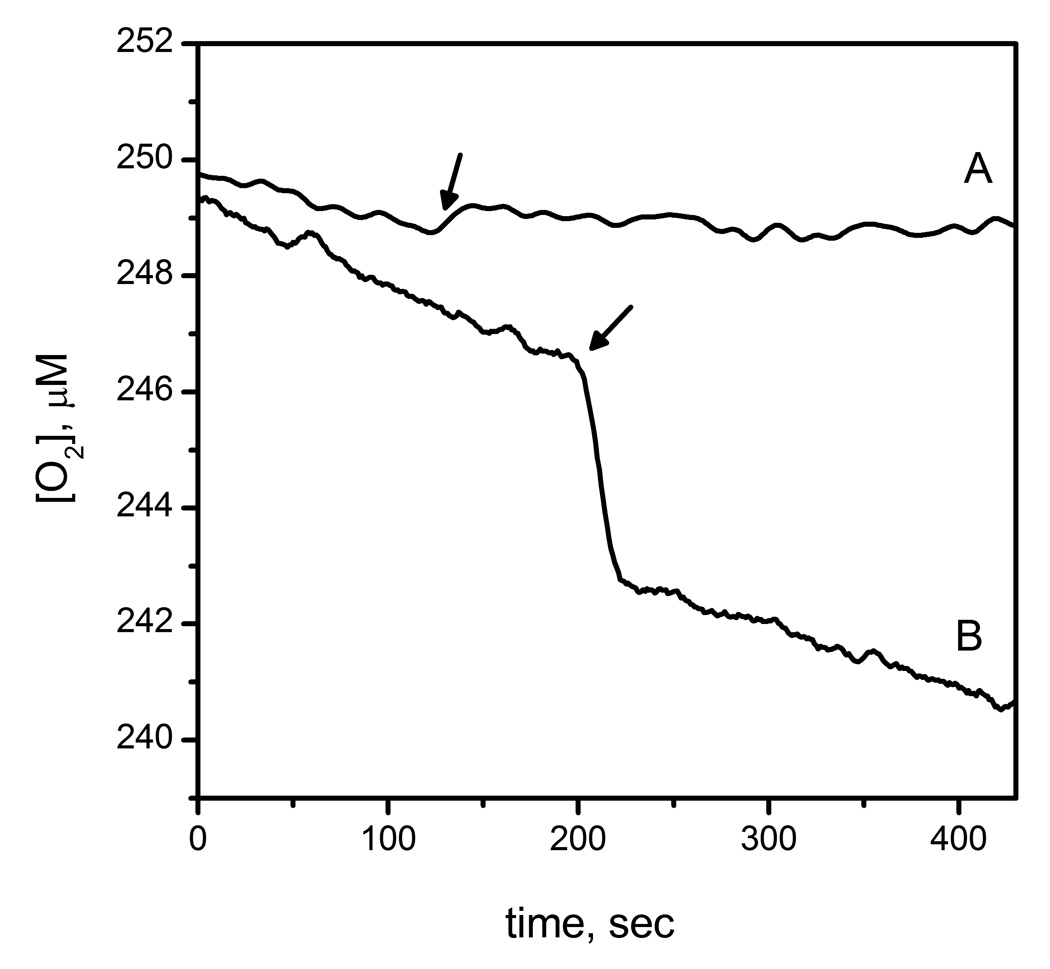
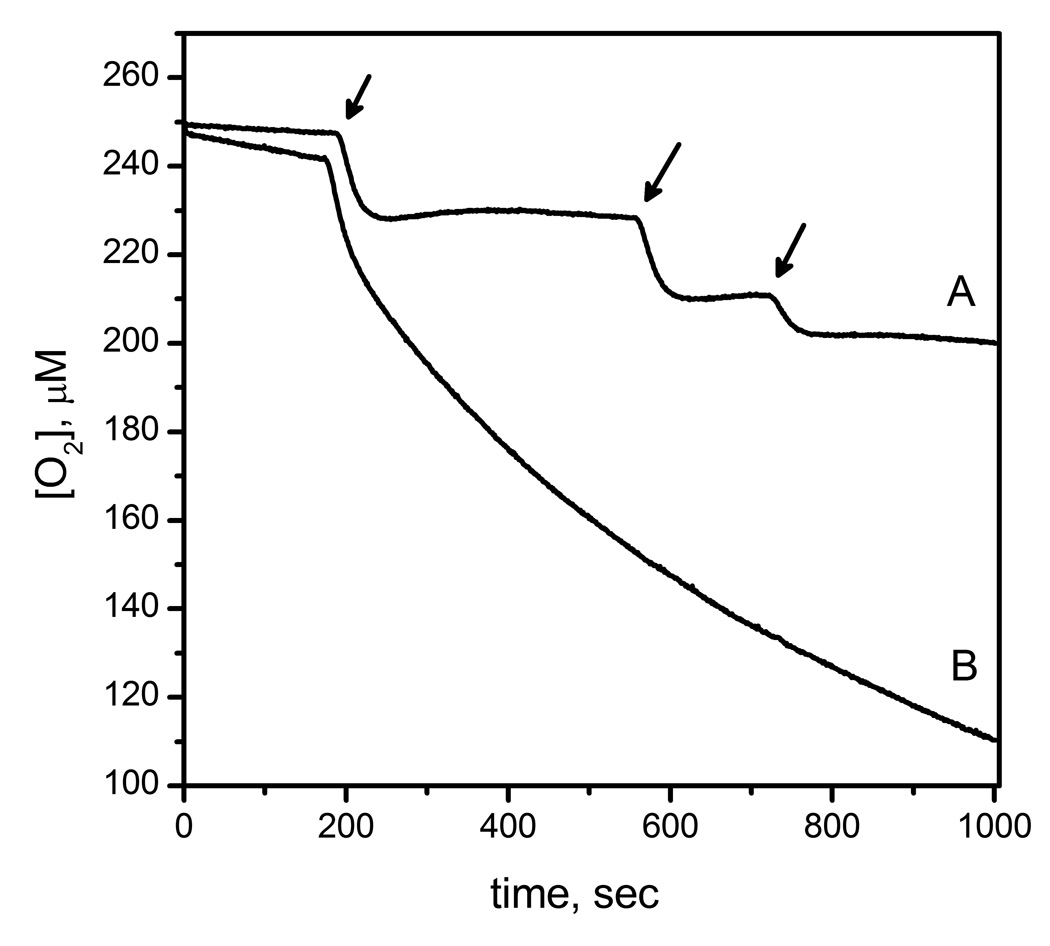
The consumption of O2 by AtsK and FIH in the absence of prime substrate was directly monitored by use of a Clark electrode (Fig 2). Small volumes of enzyme (1–3 µL) were injected into thermostated 1.00 mL reaction buffer containing FeSO4 (100 µM), ascorbate (200 µM), and αKG (1mM). Despite the tight coupling ratios observed during steady-state turnover, O2 consumption was not absolutely tied to substrate hydroxylation for either enzyme. In the case of AtsK, O2 was consumed within 30 seconds at a molar stoichiometry of 2.5 (+/− 0.2) O2 per AtsK active site (Figs 1B and 2). Such a rapid reaction with O2 is reminiscent of the fast inactivation observed for several other αKG hydroxylases, such as TauD and TfdA.[25, 26] In contrast, injections of FIH at a final concentration of 11.7 µM consumed no measurable O2 on the minutes timescale (Fig 1B). Although we were unable to measure any uncoupled O2-consumption for FIH on the minutes timescale, we did reproduce the autohydroxylation reaction.[18] Autohydroxylation required O2, and formed a hydroxylated Trp296 residue on FIH,[18] which is formed via hydroxylase activity in the absence of CTAD.
Fig 2.
Oxygen consumption of FIH and AtsK measured with O2 sensor; A) FIH (11.7 µM) mixed with FeSO4 (50 µM), αKG (500 µM) in 50 mM HEPES pH 7.5 B) AtsK (1.14 µM) mixed with ascorbate (200 µM), FeSO4 (100µM), αKG (1 mM) in 10 mM HEPES pH 7.00.
Although the coupling ratios for both FIH and AtsK were near unity during the steady-state, the consumption of O2 by AtsK in the absence of prime substrate suggested that ROS may be produced by the resting form of AtsK, (Fe+ αKG)AtsK. As an enzyme will partition amongst various forms depending on substrate concentrations, ROS formation was measured while varying prime substrate concentrations for both FIH and AtsK. The tested ROS species were superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (OH•), which may form depending on the number of electrons transferred to O2.
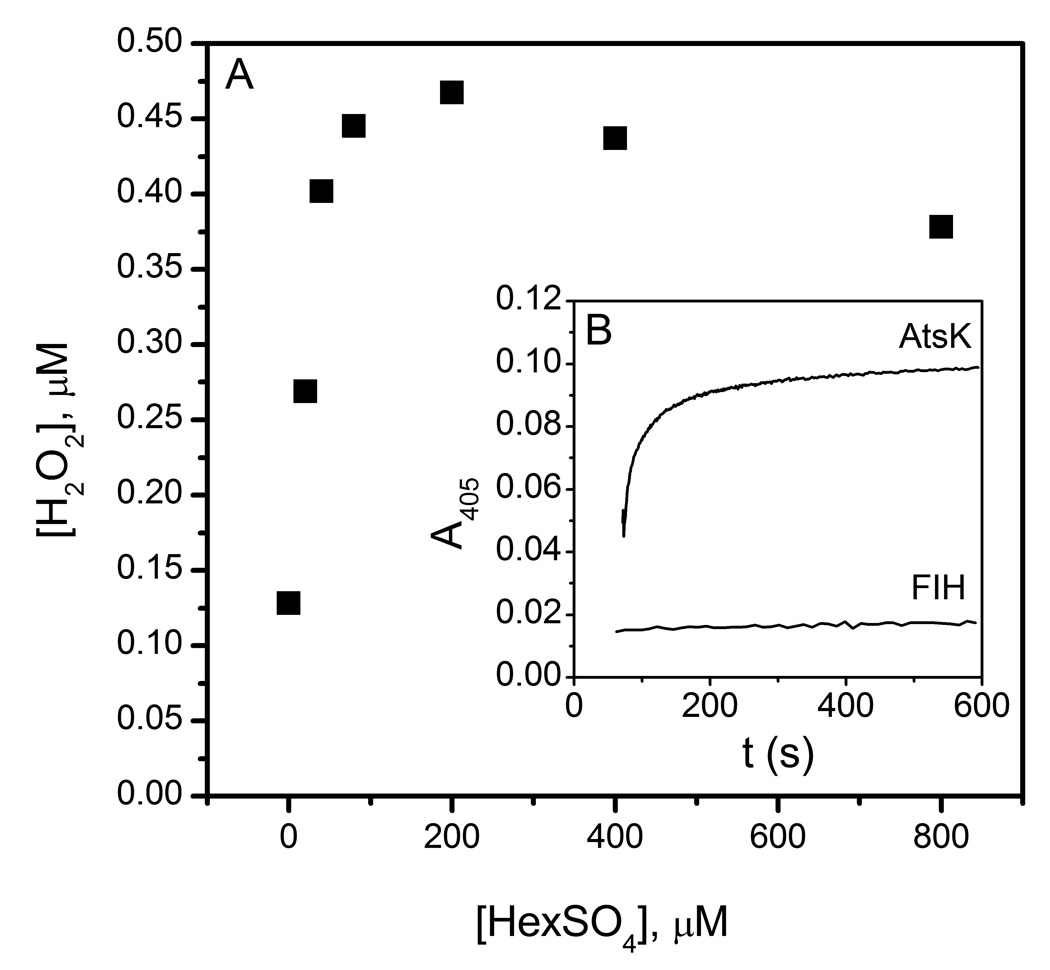
Both H2O2 and O2− were assayed by a peroxidase/ABTS assay in which H2O2 was indicated by a characteristic absorption at 405 nm due to the formation of ABTS+. In the absence of HexSO4, AtsK (11.4 µM) produced 0.12 µM H2O2; this was much less than the anticipated 2.5 equivalents of O2 consumed by AtsK. Under steady-state conditions, AtsK produced up to 0.47 µM H2O2, with [HexSO4] ranging from 0 – 800 µM. Notably, H2O2 production was a saturable function of [HexSO4], with an apparent half-maximal value of 10 µM HexSO4 (Fig 4). As this is similar to the reported KM(HexSO4),[37] it suggested that H2O2 was also released from the ES complex of AtsK, (Fe2+αKG+HexSO4)AtsK.
Fig 4.
A) H2O2 produced by AtsK (11.4 µM) during steady state turnover. B) Timecourse for H2O2 production by AtsK (upper line) and FIH (lower line). AtsK (11.4 µM) was added into a reaction mixture containing FeSO4 (100 µM), αKG (1 mM), HexSO4 (100 µM), ABTS (50 µM) and HRP (1 u/mL) in 10 mM HEPES pH 7.00. FIH (5 µM) was added into a solution wich has FeSO4 (25 µM), αKG (500 µM), CTAD (50 µM), ABTS (50 µM) and HRP (1 u/mL) in 50 mM HEPES pH 7.50.
FIH (5 µM) failed to produce measurable levels of H2O2 under any tested condition. FIH concentrations were varied between 5 and 50 µM in the absence of CTAD to test for H2O2 production by (Fe2++αKG)FIH, however H2O2 was not detected even at 50 µM FIH. FIH was also tested under steady-state conditions with 50 µM CTAD; however, H2O2 was not detected suggesting that the ES complex, (Fe2+αKG+CTAD)FIH, did not produce H2O2. This was consistent with the near unity coupling ratio for FIH.
Superoxide was measured by the use of Cu/Zn SOD to convert two equivalents of O2− into one equivalent of H2O2 for the peroxidase/ABTS assay. Neither AtsK nor FIH produced detectable O2− by this coupled assay in the absence of prime substrate; experiments in the presence of added prime substrate similarly yielded no detectable O2−.
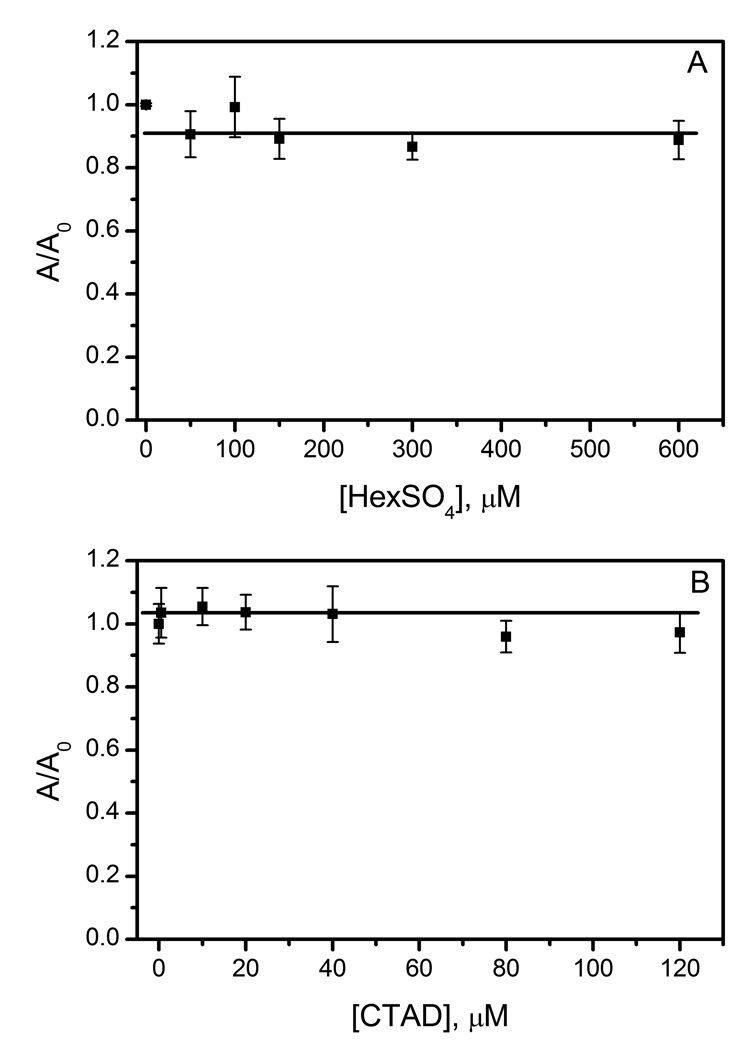
Hydroxyl radicals were tested by the deoxyribose method, a qualitative colorimetric assay that compares OH• production to a baseline condition by normalized absorptivities (A/A0); an increase in A/A0 would indicate that more OH• radical were produced. AtsK (1.14 µM) was tested at varied concentrations of HexSO4 (0–600 µM). This allowed a comparison of different enzyme forms, with free enzyme prevalent at sub-saturating concentrations of HexSO4, and an ES complex prevalent at saturation. A plot of the normalized data indicated a slight decrease in OH• production in the presence of HexSO4, however this was independent of [HexSO4] (A/A0 = 0.91 +/− 0.05) (Fig 5A). This showed that OH• radical production was not a function of enzyme form, suggesting that neither the free enzyme nor the ES complex produced diffusible OH•.
Fig 5.
Hydroxyl radical assays of AtsK and FIH. A) AtsK (1.14 µM), ascorbate (200 µM), αKG (1 mM), FeSO4 (100 µM), HexSO4 (0–600 µM), 2-deoxyribose (15 mM) in 10 mM HEPES pH 7.0 B) FIH (0.5 µM), ascorbate (2 mM), αKG (500 µM), FeSO4 (25 µM), CTAD (0–120 µM), 2-deoxyribose (15 mM) in 50 mM HEPES pH 7.50.
Similarly, FIH (0.5 µM) was assayed for OH• production at varied CTAD concentrations. The normalized data was a line of zero slope with respect to [CTAD] (A/A0 = 1.03 +/− 0.03) (Fig 5B). This indicated that OH• radical production was not a function of enzyme form. As with AtsK, this suggested that FIH did not produce diffusible OH• radicals.
During steady-state kinetics assays, we noted a biphasic timecourse for AtsK, indicating enzyme inactivation. Inactivation could result from the oxidation of the iron pool into Fe(III), or from specific changes to AtsK. Oxidation of iron was excluded as the reason for inactivation by serial injections of AtsK (0.5–1 µM, 1.25–2.5 µL) into a 1.0 mL reaction solution containing HexSO4 (1 mM) and FeSO4 (100 µM); ascorbate was omitted to simplify data interpretation. Each injection of AtsK consumed O2 for approximately 60 seconds, followed by a return to baseline; subsequent AtsK injections were similarly active (Fig 6A). This showed that the Fe(II) and HexSO4 in solution were sufficient for turnover, and that diluted AtsK (1 µM) itself became inactive within one min.
Fig 6.
Inactivation of AtsK measured by O2-electrode. A) Sequential injection of AtsK (1 µM, 1 µM, 0.5 µM) into a mixture of FeSO4 (100 µM), ascorbate (200 µM), αKG (1 mM), HexSO4 (1 mM) in 10 mM HEPES pH 7.00. B) Injection of high concentration of AtsK (7 µM) into same reaction conditions as in A.
As AtsK is a tetramer,[29] we tested de-oligomerization as the cause of the rapid inactivation. Timecourses for varied concentrations of AtsK (0.5 – 10 µM) were monitored by either the UV-Vis assay or the O2-consumption assay. High concentrations of AtsK (5 – 10 µM) were active for hundreds of seconds, whereas low concentrations (0.5 – 1 µM) inactivated within 50 seconds (Fig 6A, 6B), suggesting that de-oligomerization was the root cause of the rapid inactivation.
FIH exhibited linear progress curves for more than 4 min in steady-state assays that included ascorbate, indicating that enzyme inactivation was not a rapid process. However, we previously observed that FIH would autohydroxylate over several hours to form an Fe(III) form of the enzyme called ‘purple FIH’ in which Fe(III) was coordinated by the hydroxy-group of hydroxylated Trp296.[18] As Saari and Hausinger noted that inactivated TfdA was partially rescued by ascorbate,[26] we tested two common reductants for their ability to rescue auto-hydroxylated FIH. First, we tested auto-hydroxylated FIH for activity; purple FIH (5 µM) exhibited no activity following incubation with CTAD (70 µM), FeSO4 (25 µM) and αKG (500 µM), indicating that this enzyme form was inactive. Next, we tested ascorbate (2 mM) and/or DTT (100 µM) for their ability to re-activate purple FIH by including these reductants in purple FIH assay mixtures, however no activity was observed with these mild reductants (Table 1).
Table 1.
Effect of reducing agents on auto-hydroxylated FIH.
| FIH (µM) |
Ascorbate (mM) |
DTT (mM) |
υ (µM/min) |
|---|---|---|---|
| 5 (purple) | 0 | 0.1 | 0.05±0.04 |
| 5 (purple) | 2 | 0.1 | 0.06±0.05 |
| 5 (purple) | 2 | 0 | 0.06±0.08 |
| 5 (fresh) | 0 | 0 | 4.4±0.4 |
| 5 (fresh) | 2 | 0.1 | 7.3±0.5 |
4. Discussion
Although the αKG hydroxylases are mechanistically predisposed to uncouple the oxidative and reductive parts of their catalytic cycle, the rate and products of uncoupling depend greatly on the identity of the enzyme and on reaction conditions. The reported coupling ratio during turnover was within error of unity for all reported examples,[20, 26, 40, 41] indicating that uncoupling occurs on less than ~ 5% of turnovers in the presence of prime substrate. However, reactivity toward O2 in the absence of prime substrate is variable, as some αKG oxygenases release ROS whereas some autohydroxylate, suggesting that structural differences may control both the rates and products of uncoupling. Neither FIH nor AtsK were significantly uncoupled during turnover, however they differed in both their reactivity toward O2 in the absence of prime substrate, as well as in their uncoupling products.
Coupling between O2-activation and substrate hydroxylation likely relies on three structural factors. The first being local structures which link the binding of prime substrate to coordination changes at Fe, such as proposed for hydrogen bonding between the facial triad and the Fe-bound H2O.[31] In a highly organized active site, the (Fe2++αKG) form of enzyme would be unable to react with O2 until prime substrate bound, and would tightly couple O2 activation to substrate oxidation. The second factor is a closed active site to prevent solvent access, such as by loop closure or the binding of a large substrate. The (Fe2++αKG+Substrate) form of enzyme, the ES complex, would be tightly coupled if solvent were unable to reach reactive intermediates, such as (FeO)2+. The third factor is the presence of oxidizable residues near the active site which may react with (FeO)2+ or scavenge any generated ROS. Any ROS formed by O2-activation in the absence of prime substrate could then damage the protein rather than diffusing away, and has been proposed as a general protective strategy for αKG-dependent hydroxylases.[36]
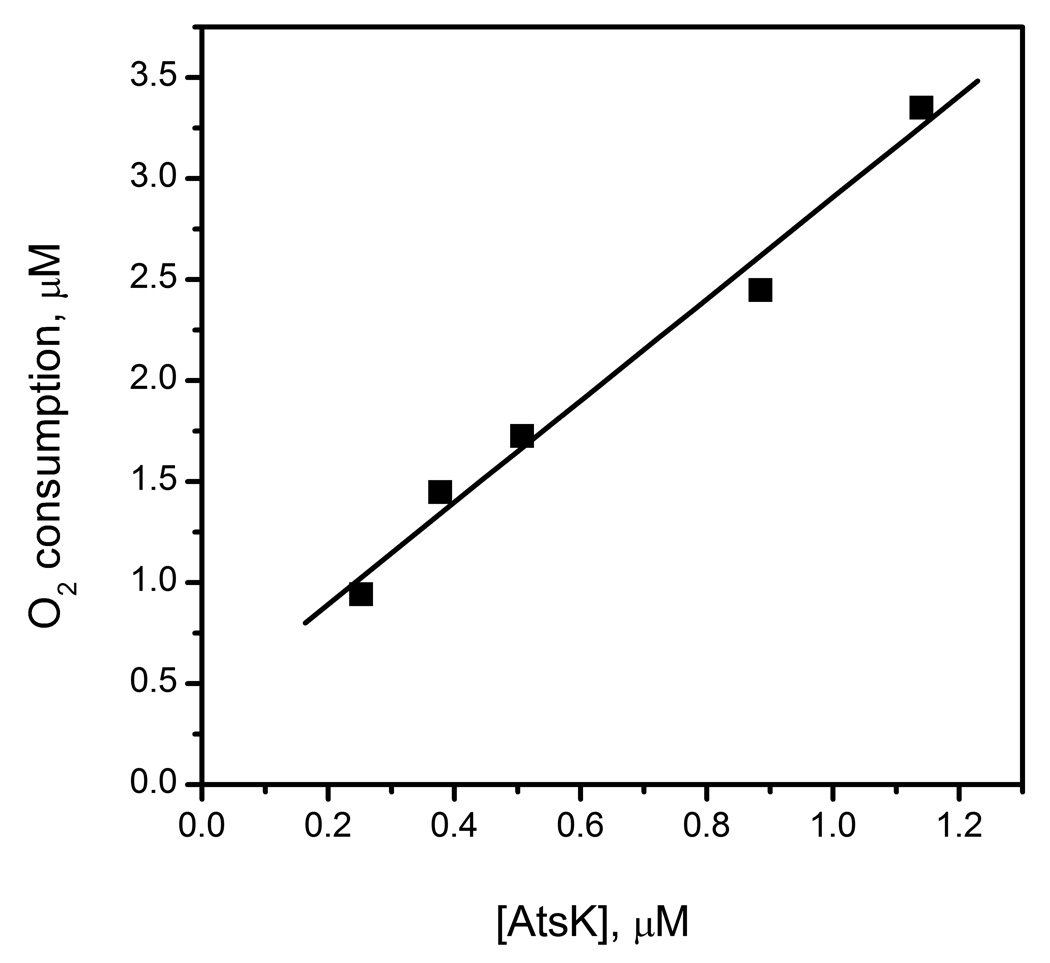
The free enzyme forms of AtsK and FIH were quite distinct in their propensity to react with O2, suggesting differences in their structures. As described below, the primary factor is likely to be hydrogen bonding between the facial triad and the coordinated H2O. In the context of the inner-sphere O2-activation model, the Fe(II) in the free enzyme (eg: (Fe+αKG)FIH) should be 6-coordinate due to a H2O ligand which is hydrogen bonded to the Asp/Glu ligand. In order for free enzyme to react with O2, this H2O ligand must be released in the absence of prime substrate. As (Fe+αKG)FIH was unreactive toward O2 on the minutes timescale (Fig 2), the H2O-ligand is likely to be tightly bound in FIH. In contrast, (Fe2+αKG)AtsK consumed 2.5 equivalents of O2 (Fig 3), suggesting weaker hydrogen bonding from the facial triad in AtsK.
Fig 3.
Oxygen consumption of AtsK in the absence of HexSO4. AtsK (0.2–1.2 µM) mixed with ascorbate (200 µM), FeSO4 (100 µM) and αKG (1 mM) in 10 mM HEPES pH 7.00.
While the reactivity data presented herein do not directly probe the strength of this hydrogen bond, the structural data for FIH and AtsK are consistent with weakened hydrogen bonding in AtsK. In particular, while Asp201 of the facial triad in FIH forms a hydrogen bond to the Fe-bound H2O,[27] the orientation of AtsK Asp110 precludes any hydrogen-bond from the facial triad of AtsK.[29, 31] The confluence of structural data with our reactivity data suggest that hydrogen bonding from the facial triad is a significant factor in deactivating FIH towards uncoupled O2-activation.
Differential solvent access to the active sites of FIH and AtsK is unlikely to be the origin of the very different uncoupling behavior in the absence of prime substrate, as both active sites are highly solvent exposed. The crystal structures of AtsK suggested that the active site would be very solvent accessible, as a ‘lid’ formed by residues 80 – 102 was so disordered in the crystal structure that it could not be refined.[29] This is also consistent with the broad substrate tolerance of AtsK, and suggests that solvent access to the active site should be facile. In a similar vein, FIH has no structural obstruction to solvent access to the active site in the (Fe+ αKG)FIH enzyme form, as the active site only becomes closed upon binding of the prime substrate, CTAD.[27] As a consequence, any uncoupled O2-activation in the absence of prime substrate should lead to a highly solvent exposed (FeO)2+ center in both AtsK and FIH. That AtsK released ROS, but FIH did not, indicated that some combination of slow initial reactivity and internal scavenging (see below) is most likely operative in FIH.
The only ROS produced by (Fe+αKG)AtsK was H2O2, suggesting that simple hydrolysis of the (FeO)2+ intermediate formed H2O2 and regenerated the Fe2+ cofactor. ROS have been implicated during uncoupling of αKG oxygenases, but seldom observed. Two negative examples are enzymes structurally related to AtsK, which also inactivated during turnover: TFdA and TauD. The irreversible inactivation of TfdA was suggested to involve oxidative damage from hydroxyl radicals, however diffusible OH• was not observed;[26] neither H2O2 nor O2− were observed during uncoupling by TauD.[41] The rare positive example is for CS2, which releases H2O2 in the absence of substrate as does AtsK.[40] A crucial difference between AtsK and CS2 is that unlike CS2, AtsK also released H2O2 during normal turnover. This may relate to the size of the active site pocket, as the AtsK active site is highly disordered[29] and may therefore be more prone to hydrolytic attack at the FeO2+ intermediate than CS2.
The presence of a sacrificial site in FIH may explain the absence of diffusible ROS from this enzyme. FIH autohydroxylation led to irreversibly inactivated enzyme on the timescale of hours. The autohydroxylated form of FIH, in which the Fe(III) is coordinated by the hydroxylated ring of Trp296,[18, 19] could not be rescued by the addition of common reducing agents nor by excess αKG (Table 1). Irreversible inactivation by ‘sacrificial’ reactivity near the active site has precedent in the autohydroxylation reactions of TauD, AlkB and TfdA, where new chromophores arise from Fe(III) coordination of the newly formed hydroxylated residue, and for which no reactivation pathway has been identified.[21, 22, 25] We propose that FIH is deactivated from aberrant O2 reactivity by strong hydrogen bonding from the facial triad, and further insured against ROS release by the internal reaction with Trp296.
5. Conclusions
FIH is a proximate O2-sensor for human cells, controlling vital processes such as basal metabolism and angiogenesis. Understanding the link between O2-activation and substrate hydroxylation is crucial to developing therapies targeting FIH function. Our findings show that FIH minimizes uncoupled O2 reactivity at two levels: through an inherently low reactivity in the absence of prime substrate; and through presentation of a sacrificial acceptor (Trp296) near the active site. The tight control over O2-reactivty for FIH is in contrast to the facile production of H2O2 by the bacterial enzyme AtsK, both in the presences and absence of the prime substrate for AtsK. We propose that this low reactivity for FIH may be due to strong hydrogen bonding between the coordinated water and the facial triad. Such tight control over O2-activation by FIH is compatible with the significance of O2 and O2-derived species to both gene expression and cellular toxicity.
Supplementary Material
Acknowledgements
We thank the NIH for funding (R01-GM077413); Dr. Michael Kertesz (U. Manchester) for providing the expression plasmid for AtsK.
Abbreviations
- ABTS
2,2'-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)
- αKG
alpha-ketoglutarate
- AtsK
oxygenative alkylsulfatase
- CS2
clavaminate synthase-2
- CTAD
C-terminal transactivation domain of HIFα
- ESI-MS
electrospray ionization mass spectrometry
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- FIH-1
the factor inhibiting HIF
- HexSO4
hexylsulfate
- HIF
Hypoxia Inducible Factor
- HRP
horseradish peroxidase
- SOD
superoxide dismutase
- TauD
taurine dioxygenase
- TfdA
(2, 4-dichlorophenoxy)acetate dioxygenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Materials: Steady state kinetics data and fits for AtsK.
References
- 1.Bruick RK. Genes Dev. 2003;17:2614–2623. doi: 10.1101/gad.1145503. [DOI] [PubMed] [Google Scholar]
- 2.Ozer A, Bruick RK. Nat. Chem. Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL. Physiology. 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 4.Hanauske-Abel HM, Gunzler V. J. Theor. Biol. 1982;94:421–455. doi: 10.1016/0022-5193(82)90320-4. [DOI] [PubMed] [Google Scholar]
- 5.Hewitson KS, Schofield CJ. Drug Discovery Today. 2004;9:704–711. doi: 10.1016/S1359-6446(04)03202-7. [DOI] [PubMed] [Google Scholar]
- 6.Nagel S, Talbot NP, Mecinovic J, Smith TG, Buchan AM, Schofield CJ. Antioxid. Redox Signal. 2010;12:481–501. doi: 10.1089/ars.2009.2711. [DOI] [PubMed] [Google Scholar]
- 7.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, Ratcliffe PJ, Pugh CW, Schofield CJ. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 8.Bruick RK, McKnight SL. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Biochem. J. 2002;367:571–575. doi: 10.1042/BJ20021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costas M, Mehn MP, Jensen MP, Que L., Jr Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 12.Hausinger RP. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 13.Price JC, Barr EW, Tirupati B, Bollinger JM, Jr, Krebs C. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 14.Proshlyakov DA, Henshaw TF, Monterosso GR, Ryle MJ, Hausinger RP. J. Am. Chem. Soc. 2004;126:1022–1023. doi: 10.1021/ja039113j. [DOI] [PubMed] [Google Scholar]
- 15.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Proc. Natl. Acad. Sci. USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price JC, Barr EW, Glass TE, Krebs C, Bollinger JM., Jr J. Am. Chem. Soc. 2003;125:13008–13009. doi: 10.1021/ja037400h. [DOI] [PubMed] [Google Scholar]
- 17.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS. J. Zhou, Chem. Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 18.Chen YH, Comeaux LM, Eyles SJ, Knapp MJ. Chem. Commun. 2008:4768–4770. doi: 10.1039/b809099h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YH, Comeaux LM, Herbst RW, Saban E, Kennedy DC, Maroney MJ, Knapp MJ. J. Inorg. Biochem. 2008;102:2120–2129. doi: 10.1016/j.jinorgbio.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Counts DF, Cardinale GJ, Udenfriend S. Proc. Natl. Acad. Sci. USA. 1978;75:2145–2149. doi: 10.1073/pnas.75.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henshaw TF, Feig M, Hausinger RP. J. Inorg. Biochem. 2004;98:856–861. doi: 10.1016/j.jinorgbio.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Liu A, Ho RY, Que L, Jr, Ryle MJ, Phinney BS, Hausinger RP. J. Am. Chem. Soc. 2001;123:5126–5127. doi: 10.1021/ja005879x. [DOI] [PubMed] [Google Scholar]
- 23.Myllyla R, Majamaa K, Gunzler V, Hanauske-Abel HM, Kivirikko KI. J. Biol. Chem. 1984;259:5403–5405. [PubMed] [Google Scholar]
- 24.Rao NV, Adams E. J. Biol. Chem. 1978;253:6327–6330. [PubMed] [Google Scholar]
- 25.Ryle MJ, Liu A, Muthukumaran RB, Ho RY, Koehntop KD, McCracken J, Que L, Jr, Hausinger RP. Biochemistry. 2003;42:1854–1862. doi: 10.1021/bi026832m. [DOI] [PubMed] [Google Scholar]
- 26.Saari RE, Hausinger RP. Biochemistry. 1998;37:3035–3042. doi: 10.1021/bi972388p. [DOI] [PubMed] [Google Scholar]
- 27.Dann CE, Bruick RK, Deisenhofer J. Proc. Natl. Acad. Sci. USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elkins JM, Ryle MJ, Clifton IJ, Dunning Hotopp JC, Lloyd JS, Burzlaff NI, Baldwin JE, Hausinger RP, Roach PL. Biochemistry. 2002;41:5185–5192. doi: 10.1021/bi016014e. [DOI] [PubMed] [Google Scholar]
- 29.Muller I, Kahnert A, Pape T, Sheldrick GM, Meyer-Klaucke W, Dierks T, Kertesz M, Uson I. Biochemistry. 2004;43:3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- 30.Welford RW, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ. J. Biol. Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]
- 31.Neidig ML, Brown CD, Light KM, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM, Krebs C, Walsh CT, Solomon EI. J. Am. Chem. Soc. 2007;129:14224–14231. doi: 10.1021/ja074557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Kelly WL, Bachmann BO, Gunsior M, Townsend CA, Solomon EI. J. Am. Chem. Soc. 2001;123:7388–7398. doi: 10.1021/ja004025+. [DOI] [PubMed] [Google Scholar]
- 33.Pavel EG, Zhou J, Busby RW, Gunsior M, Townsend CA, Solomon EI. J. Am. Chem. Soc. 1998;120:743–753. [Google Scholar]
- 34.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. J. Biol. Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 35.Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ. Biochem. J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryle MJ, Hausinger RP. Curr. Opin. Chem. Biol. 2002;6:193–201. doi: 10.1016/s1367-5931(02)00302-2. [DOI] [PubMed] [Google Scholar]
- 37.Kahnert A, Kertesz MA. J. Biol. Chem. 2000;275:31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
- 38.Halliwell B, Gutteridge JMC, Aruoma OI. Anal. Biochem. 1987;165:215–219. doi: 10.1016/0003-2697(87)90222-3. [DOI] [PubMed] [Google Scholar]
- 39.Saban E, Chen YH, Holmes B, Knapp MJ. 2010 in prep. [Google Scholar]
- 40.Salowe SP, Marsh EN, Townsend CA. Biochemistry. 1990;29:6499–6508. doi: 10.1021/bi00479a023. [DOI] [PubMed] [Google Scholar]
- 41.McCusker KP, Klinman JP. Proc. Natl. Acad. Sci. USA. 2009;106:19791–19795. doi: 10.1073/pnas.0910660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.