Summary
Pluripotent embryonic cells become progressively lineage-restricted during development in a process that culminates in the differentiation of stable organ specific cell types that perform specialized functions. Terminally-differentiated pancreatic acinar cells do not have the innate capacity to contribute to the endocrine β-cell lineage, which is destroyed in individuals with autoimmune diabetes [1]. Some cell types can be reprogrammed using a single factor [2, 3], whereas other cell types require continuous activity of transcriptional regulators to repress alternate cell fates [4–6]. Thus, we hypothesized that a transcriptional network continuously maintains the pancreatic acinar cell fate. We found that post-embryonic antagonism of Ptf1a, a master regulator of pancreatic development [7] and acinar cell fate specification [8, 9], induced the expression of endocrine genes including insulin in the exocrine compartment. Using a genetic lineage tracing approach, we show that the induced insulin+ cells are derived from acinar cells. Cellular reprogramming occurred under homeostatic conditions, suggesting that the pancreatic micro-environment is sufficient to promote endocrine differentiation. Thus, severe experimental manipulations [10, 11] may not be required to potentiate pancreatic transdifferentiation. These data indicate that targeted post-embryonic disruption of the acinar cell fate can restore the developmental plasticity that is lost during development.
Keywords: pancreas, acinar, beta cells, endocrine, zebrafish, diabetes, lineage tracing, ptf1a
Results and Discussion
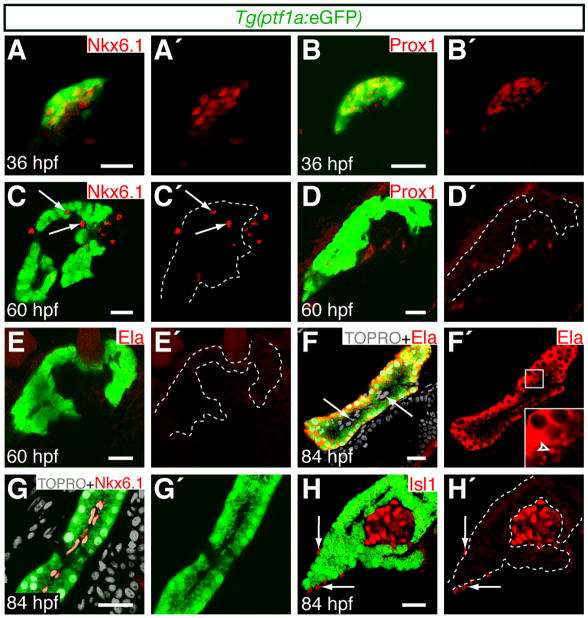
The pancreas is derived from a dorsal and a ventral bud of endodermal tissue, which together generate endocrine and exocrine cell types [12]. In zebrafish, the acinar cells are exclusively derived from the ventral pancreatic bud [13]. To analyze the temporal differentiation of acinar cells, we examined pancreatic markers at multiple time-points in Tg(ptf1a:eGFP) [14] zebrafish. The Tg(ptf1a:eGFP) BAC reporter marks the ventral pancreatic bud by 32 hpf [8, 15]. At 36 hpf, Tg(ptf1a:eGFP)-expressing cells formed a homogeneous field, and co-expressed the homeodomain transcription factors Prox1 and Nkx6.1 (Fig. 1A–B′; [8]). Co-expression of these pancreatic progenitor markers in the ptf1a expression domain suggests that the pancreatic primordium remains multi-potent at 36 hpf. By 60 hpf, Nkx6.1 and Tg(ptf1a:eGFP) were expressed in mutually exclusive domains (Fig. 1C,C′), and Prox1 was no longer detected in the pancreas (Fig. 1D,D′). However, Tg(ptf1a:eGFP) expressing cells were not fully differentiated by 60 hpf since they did not co-express the acinar cell marker Elastase (Fig. 1E,E′). Zebrafish embryonic development (0–72 hpf) is fueled by the absorption of the maternally deposited yolk. After the embryonic to larval transition at 72 hpf, a functional digestive system becomes necessary to process external nutrient sources [16]. Consistent with this requirement, Elastase was cytoplasmically localized (Fig. 1F,F′) and packaged into secretory granules on the luminal surface of Tg(ptf1a:eGFP) cells (Fig. 1F′ inset) by 84 hpf. Our results are consistent with ultrastructural studies that have shown that acinar cell cytodifferentiation after 72 hpf coincides with the expression of acinar cell markers [17]. Since Ptf1a expression becomes restricted to acinar cells in the developing mouse pancreas [18], we predicted that co-expression of Tg(ptf1a:eGFP) and Elastase would coincide with cell fate commitment of the pancreatic progenitor pool. Consistent with this prediction, Tg(ptf1a:eGFP) expression was excluded from the Nkx6.1 positive intra-pancreatic ducts (Fig. 1G,G′), and the Islet-1 positive endocrine compartment (Fig. 1H,H′) at 84 hpf. We conclude that the multi-potent pancreatic primordium gives rise to terminally differentiated exocrine and endocrine cell types by 84 hpf.
Figure 1. pt f1a expression becomes restricted to differentiated acinar cells during early larval development.
Embryos express Tg(ptf1a:eGFP) in the pancreatic domain. Expression of pancreatic markers was analyzed by immuno-fluorescence. Scale bars = 20μm. (A–B′) Confocal projections of pancreatic tissue at 36 hpf. (C–H′) Confocal sections of pancreatic tissue at 60 (C–E′) and 84 hpf (F–H′). Dotted lines delineate Tg(ptf1a:eGFP) expressing tissue. (A–B′) Tg(ptf1a:eGFP) expressing cells initially co-express Nkx6.1 and Prox1. (C–D′) By 60 hpf, N kx6.1 (C,C′, arrow s) and Prox1 (D,D′) expression becomes excluded from the Tg(ptf1a:eGFP) expression domain. (E–F′) Co-expression of Tg(ptf1a:eGFP) with the acinar cell marker Elastase (Ela) is detected by 84 hpf. Elastase positive granules are detected on the apical surface of acinar cells (F′, arrow head ), adjacent to Tg(ptf1a:eGFP) negative intra-pancreatic cells (F, arrow s). (G–H′) The duct marker Nkx6.1 (G,G′) and the endocrine marker Isl1 (H,H′) appear to be restricted to Tg(ptf1a:eGFP) negative ductal and islet cells respectively by 84 hpf. Isl1 is also expressed in the pancreatic mesenchyme (H,H′, arrows).
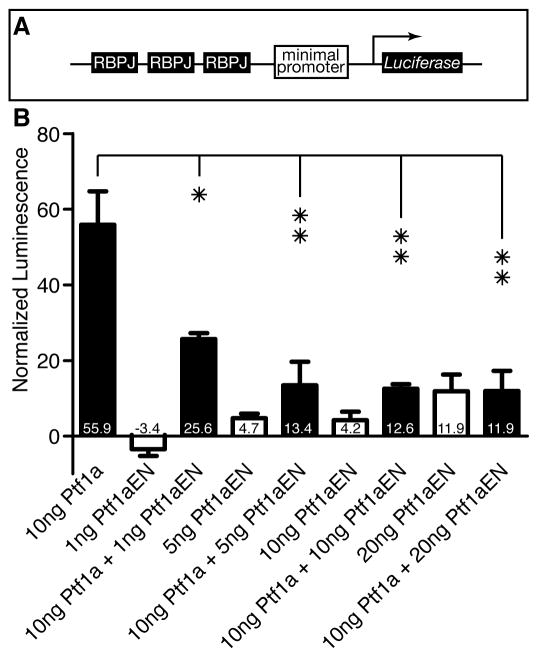
During embryogenesis (0–72 hpf), the level of Ptf1a activity influences the fate of pancreatic cells; high levels of Ptf1a activity promote exocrine cell fates, while low levels of Ptf1a activity are compatible with endocrine differentiation [8]. Ptf1a activity requires assembly of the tripartite PTF1 complex, which consists of Ptf1a, an E protein and RBPJ(L), and which directly activates exocrine target gene expression [19]. In addition, the PTF1 complex autoregulates the Ptf1a promoter resulting in sustained expression of high levels of Ptf1a in acinar cells [20]. To further address the function of PTF1, we generated an Engrailed-Ptf1a fusion protein, in which we fused the Engrailed transcriptional repressor domain to the amino-terminus of the full length Ptf1a protein. The Engrailed repressor domain recruits chromatin-modifying complexes to repress target gene transcription [21]. To explore the regulation of PTF1 targets, we generated a stable cell line that expresses Luciferase under the control of a tandem array of PTF1 binding sites in human embryonic kidney 293 cells, which require exogenous Ptf1a to form a PTF1 complex [22] (Fig. 2A). We found that Engrailed -Ptf1a repressed the transcriptional activity of wild-type Ptf1a in a dose dependent manner (Fig. 2B). Our results suggest that overexpression of Engrailed-Ptf1a antagonizes transcription initiation at Ptf1a binding sites, and perm its only low level transcription of PTF1 target genes.
Figure 2. Engrailed-Ptf1a exhibits dose-dependent antagonism of wild-type Ptf1a activity.
(A) A stable cell line (293-3xRBPJ-Luc) was generated in which Luciferase was placed under the control of RBPJ binding sites. (B) Normalized luminescence of 293-3xRBPJ-Luc cells that were transfected with Ptf1a and / or Ptf1aEN (mean +/− SD). *p<0.05; **p<0.01.
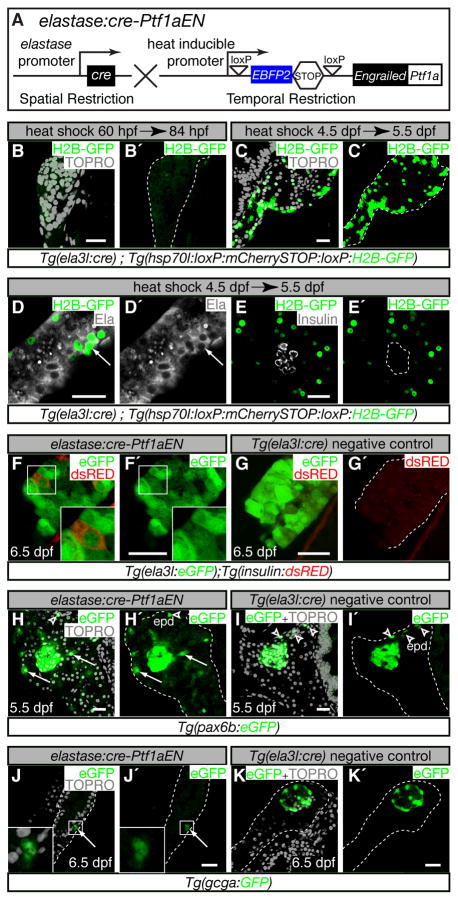
To test whether PTF1 activity is required to maintain acinar cell fate in vivo, we employed the Heat-Inducible overexpression of transgenes in a cre restricted domain (HOTcre) system [15] to express the Engrailed-Ptf1a fusion protein specifically in acinar tissue (Fig. 3A). We placed the Cre recombinase under the control of the elastase3l promoter to provide spatial control over gene expression. We used the hsp70l promoter to control the temporal expression of the Engrailed-Ptf1a fusion protein in cells that had undergone a Cre mediated excision event. For simplicity, we will refer to this double transgenic line as elastase:cre-Ptf1aEN (Fig. 3A).
Figure 3. Inhibition of Ptf1a activity induces ectopic endocrine gene expression.
(A) elastase:cre-Ptf1aEN, heat-inducible overexpression of Engrailed-Ptf1a in an elastase3l:Cre restricted domain. Expression of Cre in acinar cells excises the EBFP2-STOP cassette and perm its heat-inducible expression of Engrailed -Ptf1a. (B–K′) Confocal sections through pancreata marked with immuno-fluorescence (D–E′) or fluorescent transgenes (F–K′). Scale bars = 20μm. (B–E′) Tg(ela3l:cre); (hsp70l:loxP:mCherrySTOP:loxP:H2B-GFP) embryos were heat-shocked at 60 hpf (B,B′) or 4.5 dpf (C–E′) and analyzed 24 h post heat-shock. (F–I′) elastase:cre-Ptf1aEN (F,F′,H,H′,J,J′) and Tg(ela3l:cre) negative control embryos (G,G′,I,I′,K,K′) were heat-shocked at 4.5 dpf and analyzed at 5.5 dpf (H–I′) or 6.5 dpf (F–G′, J–K′). (B,B′) Tg(ela3l:cre) did not mark pancreatic cells by 60 hpf (dotted outline). (C,C′) By 4.5 dpf, Tg(ela3l:cre) marked a large fraction of pancreatic cells (dotted outline). Cells that were marked by Tg(ela3l:cre) at 4.5 dpf co-expressed the acinar marker Elastase (D,D′, arrow ) but never co-expressed the endocrine marker Insulin (E,E′) at 5.5 d pf. (F,F′) elastase:cre-Ptf1aEN induction caused down-regulation of Tg(elastase:eGFP) expression in a subset of acinar cells (F′, inset). Some acinar cells that expressed low levels of Tg(elastase:eGFP) co-expressed Tg(insulin:d sRED) (F, inset). (G,G′) Tg(insulin:d sRED) expression was neverdetected in Tg(ela3l:cre) negative controls. (H–I′) elastase:cre-Ptf1aEN expression induced expression of the endocrine progenitor marker Tg(pax6b:eGFP) in cells outside of the principal islet (arrow s, H,H′). In Tg(ela3l:cre) negative controls at this stage, Tg(pax6b:eGFP) was expressed in the principal islet and in endocrine cells that differentiated in the extra-pancreatic duct (arrow head s, I,I′). (J–K′) elastase:cre-Ptf1aEN also induced Tg(gcga:GFP) expression in the exocrine compartment (J,J′), whereas Tg(gcga:GFP) expression was restricted to the principal islet in Tg(ela3l:cre) negative controls (K,K′).
To determine whether Tg(ela3l:cre) specifically marked differentiated acinar cells, we heat-shocked Tg(ela3l:cre); Tg(hsp70l:loxP:mCherrySTOP:loxP:H2B-GFP) double transgenic embryos, which induced H2B-GFP expression in cells that had undergone Cre mediated recombination. When the double transgenic embryos were heat-shocked at 60 hpf, a time-point before the onset of elastase expression (Fig. 1E′), H2B-GFP expression was not detected in the pancreas at 84 hpf (Fig. 3B,B′). However, when the double transgenics were heat-shocked during larval development at 4.5 dpf, at least 24 h after the onset of Elastase expression (Fig. 1F′), broad expression of H2B-GFP was observed specifically in the pancreatic domain at 5.5 dpf (Fig. 3C,C′). Heat-shock induced expression of Tg(hsp70l:loxP:m CherrySTOP:loxP:H2B-GFP) was observed throughout the exocrine compartment in Tg(ela3l:cre) negative controls (Fig. S1). The cells that expressed H2B-GFP after heat-induction at 4.5 dpf co-expressed Elastase (Fig. 3D,D′, arrow, n=10 animals) but did not co-express the β-cell marker Insulin (Fig. 3E,E′, n=10 animals) at 5.5 dpf. We conclude that Tg(ela3l:cre) specifically marks differentiated acinar cells.
Next, we asked whether over-expression of Engrailed-Ptf1a impacted the gene expression profile of differentiated acinar cells. We crossed elastase:cre-Ptf1aEN to reporter fish for both the acinar cell marker Tg(ela3l:eGFP) [23] and the β-cell marker Tg(insulin:d sRED) [24]. Heat-shock induction of elastase:cre-Ptf1aEN at 4.5 dpf resulted in mosaic expression of Tg(ela3l:eGFP) at 6.5 dpf (Fig. 3F′ inset). Because the half-life of eGFP in cells is ~24h [25], Tg(ela3l:eGFP) expression was likely down-regulated shortly after elastase:cre-Ptf1aEN induction. This observation is consistent with the dominant-negative activity exhibited by Engrailed-Ptf1a in vitro (Fig. 2B). In contrast, Tg(ela3l:eGFP) expression was uniform in Tg(ela3l:cre) negative controls (Fig. 3G). Strikingly, a subset of cells (3–10 per animal, n=20) that exhibited low levels of Tg(ela3l:eGFP) expression also expressed Tg(insulin:dsRED) (Fig. 3F, inset); the origin of these insulin+ cells will be discussed below. Tg(insulin:d sRED) expression was not observed in cells that expressed high levels of Tg(ela3l:eGFP) (Fig. 3F) or in Tg(ela3l:cre) negative controls (Fig. 3G′). To determine whether acinar cells might have acquired a pluripotent endocrine progenitor fate prior to activating Tg(insulin:dsRED), we analyzed the expression of the Tg(pax6b:eGFP) pan-endocrine progenitor reporter [26] one day after elastase:cre-Ptf1aEN heat-shock induction (Fig. 3H–I′). elastase:cre-Ptf1aEN larvae exhibited broad low-level expression of Tg(pax6b:eGFP) throughout the pancreas (com pare Fig. 3H′, I′). In addition, scattered cells outside of the islet expressed high levels of Tg(pax6b:eGFP) (Fig, 3H,H′ arrow s). In contrast, Tg(pax6b:eGFP) expression was only observed in the extra-pancreatic duct and principal islet in Tg(ela3l:cre) negative controls (Fig. 3I–I′, arrow heads). To further investigate the fate of cells that expressed Tg(pax6b:eGFP), we analyzed glucagon expression using Tg(gcga:GFP) [27], a marker of α-cell fate, after elastase:cre-Ptf1aEN induction. As with the expression of Tg(insulin:d sRED) (Fig. 3F), we observed a small number of glucagon+ cells in the exocrine compartment of animals that expressed elastase:cre-Ptf1aEN (Fig. 3J,J′; 4–12 cells per animal, n=20), and we did not observe glucagon+ cells outside of the principal islet in Tg(ela3l:cre) negative controls (Fig. 3K,K′). Thus, elastase:cre-Ptf1aEN expression altered the gene expression profile of many exocrine cells but only a subset of these cells initiated an endocrine gene expression program.
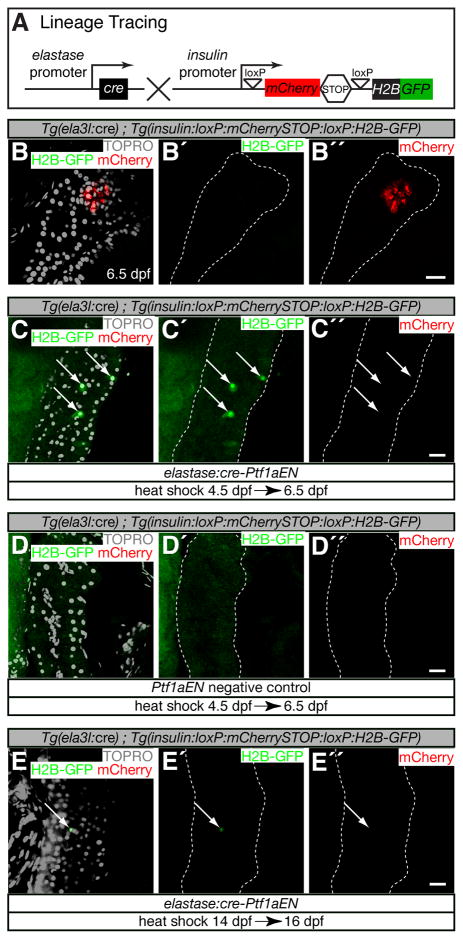
There are at least two possible origins for the ectopic insulin+ cells (Fig. 3F). A form al possibility is that elastase:cre-Ptf1aEN acted cell non-autonomously to induce partial differentiation towards both acinar and endocrine cell fates in a small population of pancreatic progenitors. A second possibility is that acinar cells down-regulated Tg(ela3l:eGFP) and up-regulated Tg(insulin:d sRED) expression. To distinguish between these possibilities, we developed a genetic lineage tracing approach to mark acinar cells that activated insulin expression. We generated animals that expressed Tg(insulin:loxP:mCherrySTOP:loxP:H2B-GFP) to specifically label with H2B-GFP insulin+ cells that derived from a lineage that expressed Tg(ela3l:cre) (Fig. 4A). insulin+ cells derived from lineages that did not express Tg(ela3l:cre) will exhibit mCherry expression. In Tg(ela3l:cre); Tg(insulin:loxP:mCherrySTOP:loxP:H2B-GFP) animals, β-cells in the principal islet exhibited mCherry expression at 6.5 dpf (Fig. 4B,B″), whereas H2B-GFP expression was not observed (Fig. 4B,B′, n>20 animals). Therefore, in wild-type animals, β-cells did not derive from acinar cells. However, after elastase:cre-Ptf1aEN induction at 4.5 dpf, H2B-GFP expression was detected in cells scattered throughout the exocrine compartment of the pancreas (4–10 cells per animal, n=20, Fig. 4C–C″, arrow s). H2B-GFP expression was not observed in Ptf1aEN negative controls (Fig. 4D–D″, n>20 animals). We conclude that elastase:cre-Ptf1aEN can cell-autonomously induce the conversion of acinar cells into an insulin+ cell type.
Figure 4. Genetic lineage tracing of acinar to insulin+ cell fate conversion.
(A) Tg(ela3l:cre); Tg(insulin:loxP:mCherrySTOP:loxP:H2B-GFP) lineage tracing. Expression of Cre in acinar cells excises the mCherry -STOP cassette and perm its expression of H2B-GFP in cells that activate the insulin promoter. (B–E″) Confocal sections through pancreata marked with fluorescent transgenes and TOPRO to mark DNA. Scale bars = 20μm. (B–B″) Tg(ela3l:cre); Tg(insulin:loxP:mCherrySTOP:loxP:H2B-GFP) did not mark any pancreatic cells in wild-type animals. (C–C″) elastase:cre-Ptf1aEN expression induced scattered H2B-GFP positive cells throughout the pancreatic domain (arrows), which were not induced in Ptf1aEN negative controls (D–D″). (E–E″) elastase:cre-Ptf1aEN expression induced isolated H2B-GFP positive cells at 14 dpf (arrow).
To determine whether acinar cell maturation affects lineage plasticity, we repeated the lineage tracing experiments in older animals. A less intense heat-shock induction protocol was used because older animals were more heat - sensitive (see Experimental Procedures). Following elastase:cre-Ptf1aEN induction at 14 dpf, H2B-GFP was detected in the exocrine compartment in most animals two days later (0–2 cells per animal, n=10, Fig. 4E–E″, arrow ), whereas H2B-GFP expression was not observed in Ptf1aEN negative controls (data nor show n). By 21 dpf, acinar cells appeared to be resistant to Ptf1aEN mediated cell type conversion (data not shown). Thus, higher levels of Ptf1aEN might be required after 14 dpf to disrupt acinar cell fate or maturing acinar cells might activate additional maintenance mechanisms.
It has been shown that inflammation in response to adenoviral packaging proteins is essential for cellular reprogramming of liver cells to pancreatic endocrine fates [28]. It is unclear whether inflammation is broadly required for in vivo reprogramming of other cell types. By the onset of larval development (72 hpf), the zebrafish innate immune system can infiltrate tissues in response to injury and infection [29]. We predicted that genetically encoded elastase:cre-Ptf1aEN expression did not initiate an immune response. However, it is possible that antagonism of normal acinar cell function or the endogenous heat-shock response could result in pancreatic injury and inflammation due to the inappropriate release of digestive enzymes. To address this possibility, we induced elastase:cre-Ptf1aEN expression in a Tg(mpx:GFP) background, which marks granulocytes during larval development [30]. In response to physical injury, zebrafish granulocytes immediately exhibit chemotaxis towards the wound and accumulate within the target tissue by 2 h post injury [31]. We analyzed pancreatic granulocytes at 2 h (Fig. S2A,B) and 24 h post heat-shock (data not shown) to determine whether heat-shock treatment or elastase:cre-Ptf1aEN induction triggered an inflammatory response. The number of pancreatic granulocytes was unchanged at both time-points in elastase:cre-Ptf1aEN animals compared to Tg(ela3l:cre) negative controls (Fig. S2C) suggesting that Engrailed-Ptf1a induced cell fate conversion does not involve an inflammatory response.
In conclusion, we have identified a novel mechanism to induce conversion of acinar cells to endocrine fates under homeostatic conditions. Our data support a model in which cues from the pancreatic micro-environment can reprogram acinar cells with low levels of Ptf1a activity into hormone expressing cells. Thus, if targeted knockdown of Ptf1a phenocopies Ptf1a antagonism, emerging methods for tissue-specific delivery of siRNAs [32, 33] could potentiate β-cell regeneration therapy.
Experimental Procedures
Zebrafish strains and lines
Zebrafish were raised under standard laboratory conditions at 28°C. Heat-shocks before 5 dpf were performed at 39.5°C for 25 minutes. Heat-shocks after 5 dpf were performed at 38.5°C for 25 minutes. We used the following lines: Tg(ptf1a:eGFP)jh1 [14], Tg(ela3l:Cre; cryaa:Venus)s932; Tg(hsp70l:loxP-EBFP2-STOP-loxP-Ptf1aEN ; cryaa:Cerulean)s933 (defined here as “elastase:cre-Ptf1aEN ”), Tg(hsp70l:loxP-mCherry-STOP-loxP-H2B-GFP; cryaa:Cerulean)s923 [15], Tg(ela3l:GFP)gz2 [23], Tg(ins:dsRed)m1018 [24], Tg(P0-pax6b:GFP)ulg515 [26], Tg(gcga:GFP)ia1 [27], Tg(insulin:loxP:mCherrySTOP:loxP:H2B-GFP; cryaa:Cerulean)s934, and Tg(mpx:GFP)i114 [30].
DNA Constructs and Transgenic lines
ela3l:Cre; cryaa:Venus was generated by placing the Cre coding sequence downstream of 2.9kb of the proxim al ela3l promoter. The cryaa:Venus cassette was inserted downstream of Cre in the reverse orientation using 0.54kb of the cryaa promoter [34]. hsp70l:loxP-EBFP2-STOP-loxP-Ptf1aEN ; cryaa:Cerulean was constructed by sequentially replacing mCherry and H2B-GFP fragments in hsp70l:loxP-mCherry-STOP-loxP-H2B-GFP; cryaa:Cerulean [15] with EBFP2-STOP and Ptf1aEN respectively. insulin:loxP:mCherrySTOP:loxP:H2B-GFP; cryaa:Cerulean was constructed by replacing the hsp70l promoter in hsp70l:loxP-mCherry-STOP-loxP-H2B-GFP; cryaa:Cerulean with 1.5kb of the proxim al insulin promoter. All constructs were generated in a pBluescript backbone that contains I-SceI meganuclease sites. Multiple transgenic lines were established for each construct using the I-SceI meganuclease method as described [35]. A single representative transgenic line for each construct was used for all experiments.
Immunofluorescence and confocal microscopy
Antibody staining was performed as described [36] using the following antibodies: Islet-1 (1:100, DSH B clone 39.4D5), Nkx6.1 (1:100, DSH B clone F55A10), Prox1 (1:1000, Millipore AB5474), Elastase (1:200, Millipore AB1216), and Alexa secondary antibodies (1:500, Invitrogen). Cell nuclei were visualized with TOPRO3 (1:2000, Invitrogen T3605). All confocal sections and z-stacks were acquired on a Zeiss LSM5 Pascal microscope and processed in ImageJ.
Generation of 293-3xRBPJ-Luc cell line
A fragment containing three 32-bp PTF1 binding sites from the mouse Rbpjl promoter upstream of a minim al Ela1 promoter (−92 to +8) [22] was subcloned into the luciferase reporter vector pGL4.17 (Promega). Following transfection, 293 cells that contained stable integrations were selected using 800 μg/m L G418. A single representative line that exhibited low levels of basal luciferase expression was used for all experiments.
Luciferase Assay
293-3xRBPJ-Luc cells were plated at 80% confluence in 96-well plates and transfected with 40 ng total DNA/well and 1 μL Lipofectamine transfection reagent (Invitrogen) in 75 μL serum free DMEM. After 4 h the media was replaced with DMEM containing 10% FBS. Each transfection contained 10 ng renilla (pRL-TK, Promega) and variable amounts of mouse Ptf1a (pcDNA3.1/Ptf1a) and / or mouse Ptf1a-Engrailed (pcDNA3.1/Ptf1aEN) expression plasmids. An empty expression vector (pcDNA3.1, Invitrogen) was used to normalize the total amount of DNA/well. Two days post-transfection, cells were analyzed for luciferase and renilla activity using the Dual-Glo reporter assay system (Promega) per manufacturer’s protocol.
Supplementary Material
Acknowledgments
We thank Ana Ayala and Milagritos Alva for expert help with the fish, Philipp Gut, Nikolay Ninov, and Olov Andersson for critical reading of the manuscript, and members of the Stainier and German laboratories for helpful discussions. This work was supported by LLH F and JDRF postdoctoral fellowships to D.H., a JDRF postdoctoral fellow ship to R.M.A and grants from the National Institutes of Health (DK075032), JDRF and Packard Foundation to D.Y.R.S.
References
- 1.Desai BM, Oliver-Krasinski J, De Leon DD, Farzad C, Hong N, Leach SD, Stoffers DA. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet beta cell, regeneration. J Clin Invest. 2007;117:971–977. doi: 10.1172/JCI29988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 4.Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 5.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 6.Thayer MJ, Weintraub H. Activation and repression of myogenesis in somatic cell hybrids: evidence for trans-negative regulation of MyoD in primary fibroblasts. Cell. 1990;63:23–32. doi: 10.1016/0092-8674(90)90285-m. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 8.Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18:1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wessells NK, Cohen JH. Early Pancreas Organogenesis: Morphogenesis, Tissue Interactions, and Mass Effects. Developmental Biology. 1967;15:237–270. doi: 10.1016/0012-1606(67)90042-5. [DOI] [PubMed] [Google Scholar]
- 13.Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 14.Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- 15.Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci USA. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 17.Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 18.Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, Macdonald RJ. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28:5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolkunova EN, Fujioka M, Kobayashi M, Deka D, Jaynes JB. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan H, Korzh S, Li Z, Mudumana SP, Korzh V, Jiang YJ, Lin S, Gong Z. Analyses of pancreas development by generation of gfp transgenic zebrafish using an exocrine pancreas-specific elastaseA gene promoter. Exp Cell Res. 2006;312:1526–1539. doi: 10.1016/j.yexcr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Shin CH, Chung WS, Hong SK, Ober EA, Verkade H, Field HA, Huisken J, Stainier DY. Multiple roles for Med12 in vertebrate endoderm development. Dev Biol. 2008;317:467–479. doi: 10.1016/j.ydbio.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 26.Delporte FM, Pasque V, Devos N, Manfroid I, Voz ML, Motte P, Biemar F, Martial JA, Peers B. Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX and PREP factors. BMC Dev Biol. 2008;8:53. doi: 10.1186/1471-213X-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zecchin E, Filippi A, Biemar F, Tiso N, Pauls S, Ellertsdottir E, Gnugge L, Bortolussi M, Driever W, Argenton F. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev Biol. 2007;301:192–204. doi: 10.1016/j.ydbio.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 28.Wang AY, Ehrhardt A, Xu H, Kay MA. Adenovirus transduction is required for the correction of diabetes using Pdx-1 or Neurogenin-3 in the liver. Mol Ther. 2007;15:255–263. doi: 10.1038/sj.mt.6300032. [DOI] [PubMed] [Google Scholar]
- 29.Meeker ND, Trede NS. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 31.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 34.Kurita R, Sagara H, Aoki Y, Link BA, Arai K, Watanabe S. Suppression of lens growth by alphaA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev Biol. 2003;255:113–127. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- 35.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 36.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.