Abstract
Based on SAR in the alkyne class of mGlu5 receptor negative allosteric modulators and a set of amide-based positive allosteric modulators, optimized substitution of the aryl ‘b’ ring was used to create substituted N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamides. Results from an mGlu5 receptor functional assay, using calcium fluorescence, revealed varying efficacies and potencies that provide evidence that subtle changes in compounds within a close structural class can have marked effects on functional activity including switches in modes of efficacy (i.e. negative to positive allosteric modulation).
Glutamate (L-Glutamic acid) is the major excitatory neurotransmitter within the mammalian central nervous system and regulates a variety of neuronal activities through ionotropic glutamate receptors and metabotropic glutamate receptors (mGlu receptors)1. mGlu receptors are G protein-coupled receptors (GPCRs) that have been cloned, sequenced, and classified into Group I (mGlu1 and mGlu5 receptors), Group II (mGlu2 and mGlu3 receptors), or Group III (mGlu4, 6, 7, 8 receptors) based on sequence homology, pharmacology and 2nd messenger coupling.1 The mGlu5 receptor is primarily located postsynaptically and is coupled with phospholipase C. Activation of mGlu5 receptor stimulates phospholipase C, which results in the hydrolysis of phosphoinositide and increases intracellular calcium concentrations.1,2 The mGlu5 receptors have been targeted in the development of drugs to treat a variety of neurological and psychiatric illnesses, including anxiety, depression, pain, Parkinson’s disease, schizophrenia and Fragile X syndrome.2-8 Preclinical studies suggest that mGlu5 receptor may also play a role in drug abuse and addiction.9 A large number of potent noncompetitive antagonists for mGlu5 receptor have been developed based on the structure of the leading compounds 6-(methyl-2(phenylethynyl)pyridine (MPEP, 1, Fig. 1) and 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]pyridine (MTEP). However, cross-target activity and in vivo metabolism may limit further development of these alkynes as medications.10 In our attempts to design nonalkynyl mGlu5 receptor antagonists (negative allosteric modulators), several moderately active diarylamides were discovered (e.g. 2b, in Fig. 1).11 Based on the binding and functional data for amide-linked derivatives, we previously considered that the marked differences in functional potency as compared to binding affinities might be due to different conformational states of the mGlu5 receptor.11a The discovery of amide-linked positive allosteric modulators of mGlu5 receptor, such as 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide12 (CDPPB; 3, Fig. 1), provided further support of this idea. Thus, we hypothesized that the ‘b’ ring in our amide series primarily influences affinity at the mGlu5 receptor, whereas the ‘a’ ring determines efficacy that leads to potentiation rather than inhibition of glutamate stimulation.
Figure 1.
Structures of mGlu5 receptor allosteric modulators
In this study, we explore i) the substituent effects at the 6-position of the pyridyl ‘a’ ring of the 3-CN, 5-F phenyl ‘b’ ring amide (Scheme 1, Table 1), and ii) structural modifications of the ‘b’ ring of CDPPB by incorporating previously described11b,c and optimized substitutions of the ‘b’ ring (Schemes 2 and 3, Table 2). 1-(2-Chlorophenyl)-3-phenyl-1H-pyrazol-5yl was used to replace the ‘a’ ring of CDPPB since a chloro substitution at that position was reported to increase mGlu5 receptor binding affinity of CDPPB13.
Scheme 1.
Synthesis of (6-substituted pyridine-2-yl)benzamides.a
aReagents and conditions: (a) SOCl2, reflux, 2h; (b) 5- or 6-substituted 2-aminopyridines (6a-e), Et3N, CH2Cl2, r.t., 1h.
Table 1.
In vitro data for amide-linked mGlu5 receptor antagonists

| Compd | X | R1 | R2 | R3 | R4 | R5 | cLogPf | mGluR5 binding (Ki, nM)a |
mGluR5 function IC50 (nM) (Ca+2 flux) |
|---|---|---|---|---|---|---|---|---|---|
| 2ad,e | C | CN | H | H | Me | H | 2.1 | 330±20c,d | 490±94 |
| 2b e | C | CN | Ph | H | Me | H | 4.0 | 9.8±2.1 b | 13.7±2.54 |
| 2c e | C | CN | 3′FPh | H | Me | H | 4.1 | 22±5.3 b | 25.3±1.90 |
| 2d e | C | CN | 4′FPh | H | Me | H | 4.1 | 134±31c | 4.57±0.38 |
| 2e e | C | CN | 1-naphth | H | Me | H | 5.1 | 72±12c | 640±32.2 |
| 2f e | N | H | 3,5-diFPh | H | Me | H | 3.8 | 43±10 | 98.1±18.9 |
| 7a | C | CN | H | F | Me | H | 2.2 | 65.5±20c | 21.7±5.41 |
| 7b | C | CN | H | F | Et | H | 2.8 | 762±177c | 169±21.7 |
| 7c | C | CN | H | F | n-Pr | H | 3.3 | 325±62c | 165±21.7 |
| 7d | C | CN | H | F | H | Me | 2.2 | 2831±567c | 4170±528 |
| 7e | C | CN | H | F | n-Bu | H | 3.8 | 1054±262c | 1550±146 |
|
1,
MPEP c |
- | - | - | - | - | - | 3.8 | 13±1 | 3.54±1.39 |
Data provided by NIMH-PDSP
cloned15
rat brain (http://pdsp.med.unc.edu)
Compound previously reported11a
Compound and data reported previously11b,c
determined using Sybyl 7.2.3, Tripos Inc.
Scheme 2.
Synthesis of substituted N-(1,3-diphenyl-1H-pyrazol-5yl]benzamides.a
a Reagents and conditions: (a) KOBut, CH3CN, C6H6; (b) p-ClC6H4NHNH2, HCl; (c) BnBr, K2CO3, acetone; (d) Zn(CN)2, Pd(PPh3)4, DMF; (e) NaOH(aq), H2O/EtOH; (f) i. SOCl2, DMF(cat.), CH2Cl2; ii. EtOH, Et3N, CH2Cl2; (g) i. H2, Pd/C, EtOH; ii Tf2O, pyridine, CH2Cl2; (h) R2B(OH)2, Pd(PPh3)4, KF·2H2O, DME/H2O; (i) NaOH(aq),H2O/EtOH; (j) i. SOCl2, DMF(cat.), CH2Cl2; ii. 10, Et3N, CH2Cl2.
Scheme 3.
Synthesis of substituted N-(1,3-diphenyl-1H-pyrazol-5yl]pyridylamide 22.a
aReagents and conditions: (a) i. SOCl2, DMF(cat.), CH2Cl2; ii. 10, Et3N, CH2Cl2, 60%; (b) 3,4-difluorophenylboronic acid, Pd(PPh3)4, Na2CO3, DME/H2O, 80 °C, 78%.
Table 2.
In vitro data for amide-linked mGlu5 receptor potentiators

| Compd | X | R1 | R2 | cLogPb | mGluR5 binding (Ki, nM)a |
mGluR5 function EC50 (nM) (Ca+2 flux) |
% Glu Max |
|---|---|---|---|---|---|---|---|
| 19a | C | CN | Ph | 7.5 | 37±9 | 4140±807 | 61.84±3.48 |
| 19b | C | CN | 3′FPh | 7.7 | 2060±520 | >30,000 | |
| 19c | C | CN | 4′FPh | 7.7 | 23±5 | 2430±481 | 67.98±1.9 |
| 19d | C | CN | 1-naphth | 8.7 | >10,000 | 2170±1000 | 50.23±12.62 |
| 22 | N | H | 3,5- diFPh |
7.5 | 344±63 | 646±242c | 67.6±7.28 |
| 3, CDPPB d | 4.8 | 3670±430 | 77±15 |
Data provided by NIMH-PDSP using rat brain (http://pdsp.med.unc.edu)
determined using Sybyl 7.2.3, Tripos Inc.
Agonist-potentiator
Compound and data reported previously.13
Synthesis of the N-(5- or 6-substituted pyridin-2-yl)-3-cyano-5-fluorobenzamides 7 started from 3-cyano-5-fluorobenzoic acid 4 (Scheme 1). The acid 4 was first converted to the corresponding acid chloride 5 followed by reaction with 2-aminopyridines 6 to give the benzamides 7(a-e) in good yields.
Synthesis of substituted N-[1-(2-chlorophenyl)-3-phenyl-1H-pyrazol-5-yl]benzamides 19 is shown in Scheme 2. The preparation of 1-(2-chlorophenyl)-3-phenyl-5-amino-1H-pyrazol 10 was achieved according to a literature procedure13. Benzylic protection of both the OH and COOH groups in compound 11 gave intermediate 12. Cyanation of 12 with Zn(CN)2 in the presence of Pd(PPh3)4 catalyst gave intermediate 13. Hydrolysis of 13 under basic conditions resulted in selective deprotection to give the carboxylic acid 14. The free acid 14 was protected as the ethyl ester 15 via its acid chloride. Pd/C catalyzed hydrogenation successfully deprotected 15, followed by treatment with trifluoromethanesulfonic anhydride to give triflate 16, which was reacted with various arylboronic acids under Suzuki-coupling conditions to give the desired ethyl (3-cyano-4-aryl)benzoates 17(a-d). Hydrolysis of the esters 17(a-d) under basic conditions gave the free acids 18(a-d). Finally, 18(a-d) were converted to the amides 19(a-d) as shown.
Synthesis of substituted N-[1-(2-chlorophenyl)-3-phenyl-1H-pyrazol-5yl]pyridylamides 22 is depicted in Scheme 3. 4-Bromopicolinic acid 20 was converted to amide 21 by refluxing with SOCl2 in CH2Cl2 to form the acid chloride, followed by treatment of the acid chloride with pyrazolamine 10. Suzuki-coupling of 21 with 3,5-difluorophenyl-boronic acid in the presence of Pd(PPh3)4 catalyst gave the product 22.
All the compounds synthesized were assessed in a radioligand displacement binding assay for mGlu5 receptor, using [3H]MPEP as the radioligand in rat brain membranes or HEK293-T cells transfected with cloned rat mGlu5 receptor cDNA. An assay utilizing calcium fluorescence was employed to test functional activity of compounds by measuring receptor-induced intracellular release of calcium utilizing a kinetic imaging plate reader that makes simultaneous measurements of calcium levels in each well of a 384-well plate. Briefly, either vehicle or a test compound was added to cells expressing rat mGluR5 that were loaded with calcium-sensitive fluorescent dye. After a 2.5 minute incubation period, an EC20 concentration of glutamate was added followed by an EC80 concentration added 1 minute later. The methods for this triple add protocol, which allows simultaneous testing of antagonists and potentiators, are described in detail.14 The results of these in vitro tests for the amides are in Tables 1 and 2.
We previously optimized the substitution of the phenyl ‘b’ ring of a series of N-(6-methylpyridin-2-yl)benzamides and found that a CN at the 3-position and an aromatic substituent at the 4-position resulted in compounds with high binding affinity at the mGlu5 receptor.11 In addition, the 3-CN, 5-F substitution in the amide ‘b’ ring demonstrated high affinity at mGlu5 receptor, (e.g. 7a) and was chosen for exploration of other substitutions in the pyridyl ‘a’ ring. If the substitutions were well-tolerated, the 3-CN, 5-F substituent would also be utilized to synthesize CDPPB analogues, with reduced lipophilicity.
Several amides were synthesized with substitutions larger than methyl (7b, 7c, 7e) or at another position of the pyridyl ring (e.g. 7d). mGlu5 receptor binding results showed a 5-43-fold decrease in affinities compared to the parent compound 7a, and functional test results demonstrated a 8-192-fold decrease. This trend was especially noted with compound 7d, which has a methyl group at the 5-position of the pyridyl ‘a’ ring and showed the lowest potencies in both mGlu5 receptor binding affinity and functional assays. The results clearly showed that pyridyl ‘a’ ring substitutions in the amide series, other than the 6-methyl group, were not well tolerated at mGlu5 receptor and hence the 3-CN, 5-F substituent was not chosen for the CDPPB series of compounds.
In the CDPPB analogue series, 19a (R2=Ph) and 19c (R2=4-FPh) showed high binding affinities, while 19d (R2=naphth) did not (Ki > 10,000). Moreover, all the CDPPB analogues showed lower mGlu5 receptor binding affinities than their corresponding N-(6-methylpyridin-2-yl)benzamides except the 4-fluorophenyl substituted compound 19c, which showed a 5.8-fold increase in binding affinity. Several N-(6-methylpyridin-2-yl)benzamide analogs were potent antagonists in the functional assay. However, all the CDPPB analogs showed lower potency in the functional assay, especially 19b, which was completely inactive. In addition, their intrinsic activities differed from the benzamide series and included mGluR5 pure potentiators (e.g. 19a, 19c, 19d) and agonist-potentiator (22), which induced a small response when added alone (EC50 > 10 μM) and potentiated the response to an EC20 concentration of glutamate.
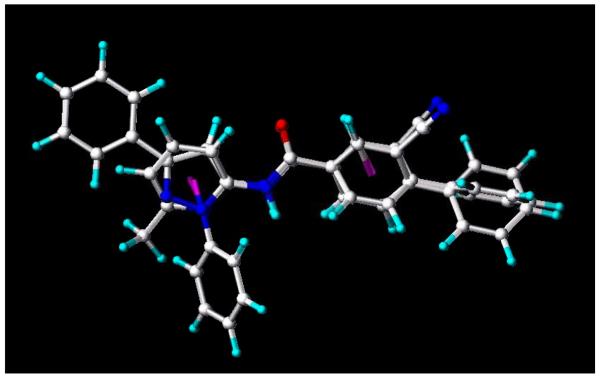
In superimposing 2b and 19a (Fig. 2), the two molecules fit quite well on the ‘b’ ring side, and showed similar mGlu5 receptor binding affinities. On the other hand, there were big differences on the ‘a’ ring side of the two molecules. These differences may contribute to their different intrinsic activities corresponding to our initial hypothesis that the ‘b’ ring in the amide series primarily influences affinity at mGlu5 receptor, whereas the ‘a’ ring determines efficacy and may be stabilizing a protein conformation that leads to potentiation rather than inhibition of glutamate stimulation.
Fig. 2.
3D superimposition of 2b and 19a.
All the CDPPB analogues, except 19d, showed relatively high mGlu5 receptor binding affinities, especially 19a and 19c that were 99- and 169-fold higher than CDPPB, respectively. However, all the analogues showed much lower potency in the functional assay than CDPPB and their relatively high cLogP values render these compounds poorly water soluble with predicted poor bioavailability.
This affinity-potency disconnect has been previously observed and may be explained by differing degrees of positive cooperativity between the potentiator and glutamate.12a It is also possible these potentiators do not act entirely through the MPEP binding site and instead bind to an overlapping or distinct allosteric binding site (in the case of 19d) as is the case for mGlu5 receptor potentiator CPPHA.12b
In summary, a series of N-(6-methylpyridin-2-yl)benzamide analogues (7a-e) were synthesized to explore substitution on the pyridyl ‘a’ ring of the N-(6-methylpyridin-2-yl)benzamides. In vitro testing showed that a 6-methyl substituent in the pyridyl ‘a’ ring is optimal. Incorporating the previously discovered optimal Ar-substitution into the phenyl ‘b’ ring of CDPPB (19a-d, 22) revealed varying efficacies and potencies in the calcium fluorescence mGlu5 receptor functional assay suggesting a difference in intrinsic activity that may reflect distinct binding modes at the protein, as reported recently for a series of mGlu5 receptor alkynyl analogues.16
Supplementary Material
Acknowledgements
This work was funded by the NIDA-Intramural Research Program and an NRSA Fellowship F32 NS049865 awarded by NINDS to ALR. mGlu5 receptor binding data were provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program, Contract no. NO1MH32004 (NIMH PDSP). We thank Dr. Tom Keck for helpful comments on an earlier version of this manuscript.
Footnotes
This manuscript is dedicated to Professor James M. Cook on the occasion of his 65th birthday.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conn PJ, Pin JP. Ann. Rev. Pharmacol. Toxicol. 1997;37:205. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- 2.Pin J-P, Acher F. Curr.Drug Targets: CNS Neurol. Disord. 2002;1:297. doi: 10.2174/1568007023339328. [DOI] [PubMed] [Google Scholar]
- 3.Brodkin J, Busse C, Sukoff SJ, Varney MA. Pharmacol., Biochem. Behav. 2002;73:359. doi: 10.1016/s0091-3057(02)00828-6. [DOI] [PubMed] [Google Scholar]
- 4.Cosford NDP, Tehrani L, Roppe J, Schweiger E, Smith ND, Anderson JJ, Bristow L, Brodkin J, Jiang XH, McDonald I, Rao S, Washburn M, Varney MA. J. Med. Chem. 2003;46:204. doi: 10.1021/jm025570j. [DOI] [PubMed] [Google Scholar]
- 5.(a) Walker K, Reeve A, Bowes M, Winter J, Wotherspoon G, Davis A, Schmid P, Gasparini F, Kuhn R, Urban L. Neuropharmacology. 2001;40:10. doi: 10.1016/s0028-3908(00)00114-3. [DOI] [PubMed] [Google Scholar]; (b) Walker K, Bowes M, Panesar M, Davis A, Gentry C, Varney MA, Urban L, Kuhn R. Neuropharmacology. 2001;40:1. doi: 10.1016/s0028-3908(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 6.(a) Marino MJ, William DL, Jr., O’Brien JA, Valenti O, McDonald TP, Clements MK, Wang R, DiLella AG, Hess JF, Kinney GG, Conn PJ. PNAS. 2003;100:13668. doi: 10.1073/pnas.1835724100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Battaglia G, Busceti CL, Molinaro G, Biagioni F, Traficante A, Nicoletti F, Bruno V. J. Neurosci. 2006;26:7222. doi: 10.1523/JNEUROSCI.1595-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietraszek M, Nagel J, Gravius A, Schafer D, Danysz W. Amino Acids. 2006;32:173. doi: 10.1007/s00726-006-0319-9. [DOI] [PubMed] [Google Scholar]
- 8.Dölen G, Carpenter RL, Ocain TD, Bear MF. Pharmacol Ther. 2010:78. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Kenny PJ, Markou A. Trends Pharmacol. Sci. 2004;25:265. doi: 10.1016/j.tips.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 10.(a) Pagano A, Ruegg D, Litschig S, Stoehr N, Stierlin C, Heinrich M, Floersheim P, Prezeau L, Carroll F, Pin JP, Cambria A, Vranesic I, Flor PJ, Gasparini F, Kuhn R. J. Biol. Chem. 2000;275:33750. doi: 10.1074/jbc.M006230200. [DOI] [PubMed] [Google Scholar]; (b) Malherbe P, Kratochwil N, Zenner MT, Piussi J, Diener C, Kratzeisen C, Fischer C, Porter RH. Mol. Pharmacol. 2003;64:823. doi: 10.1124/mol.64.4.823. [DOI] [PubMed] [Google Scholar]
- 11.(a) Kulkarni SS, Nightingale B, Dersch CM, Rothman RB, Newman AH. Bioorg Med Chem Lett. 2006;16:3371. doi: 10.1016/j.bmcl.2006.04.032. [DOI] [PubMed] [Google Scholar]; (b) Kulkarni SS, Newman AH. Bioorg Med Chem Lett. 2007;17:2074. doi: 10.1016/j.bmcl.2006.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kulkarni SS, Zou MF, Cao J, Deschamps JR, Rodriguez AL, Conn PJ, Newman AH. J Med Chem. 2009;52:3563. doi: 10.1021/jm900172f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Chen Y, Nong Y, Goudet C, Hemstapat K, de Paulis T, Pin J-P, Conn PJ. Mol Pharmcol. 2007;71:1389. doi: 10.1124/mol.106.032425. [DOI] [PubMed] [Google Scholar]; (b) Chen Y, Goudet C, Pin JP, Conn PJ. Mol. Pharmacol. 2008;73:909. doi: 10.1124/mol.107.040097. [DOI] [PubMed] [Google Scholar]
- 13.De Paulis T, Hemstapat K, Chen Y, Zhang Y, Saleh S, Alagille D, Baldwin RM, Tamagnan GD, Conn PJ. J. Med. Chem. 2006;49:3332. doi: 10.1021/jm051252j. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez AL, Grier MD, Jones CK, Herman EJ, Kane AS, Smith RL, Williams R, Zhou Y, Marlo JE, Days EL, Blatt TN, Jadhav S, Menon UN, Vinson PN, Rook JM, Stauffer SR, Niswender CM, Lindsley CW, Weaver CD, Conn PJ. Mol. Pharmacol. 2010:1105. doi: 10.1124/mol.110.067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iso Y, Grajkowska E, Wroblewski JT, Davis J, Goeders NE, Johnson KM, Sanker S, Roth BL, Tueckmantel W, Kozikowski AP. J. Med. Chem. 2006;49:1080. doi: 10.1021/jm050570f. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Kedrowski J, Rook JM, Smith RL, Jones CK, Rodriguez AL, Conn PJ, Lindsley CW. J. Med. Chem. 2009;52:4106. doi: 10.1021/jm900654c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.