Abstract
Mammalian tooth development depends on activation of odontogenic potential in the presumptive dental mesenchyme by the Msx1 and Pax9 transcription factors. We recently reported that the zinc finger transcription factor Osr2 was expressed in a lingual-to-buccal gradient pattern surrounding the developing mouse molar tooth germs and mice lacking Osr2 developed supernumerary teeth lingual to their molars. We report here generation of a gene-targeted mouse strain that allows conditional inactivation of Pax9 and subsequent activation of expression of Osr2 in the developing tooth mesenchyme from the Pax9 locus. Expression of Osr2 from one copy of the Pax9 locus did not disrupt normal tooth development but was sufficient to suppress supernumerary tooth formation in the Osr2−/− mutant mice. We found that endogenous Osr2 mRNA expression was significantly downregulated in the developing tooth mesenchyme in Pax9del/del mice. Mice lacking both Osr2 and Pax9 exhibited early tooth developmental arrest with significantly reduced Bmp4 and Msx1 mRNA expression in the developing tooth mesenchyme, similar to that in Pax9del/del mutants but in contrast to the rescue of tooth morphogenesis in Msx1−/−Osr2−/− double mutant mice. Furthermore, we found that Osr2 formed stable protein complexes with the Msx1 protein and interacted weakly with the Pax9 protein in co-transfected cells. These data indicate that Osr2 acts downstream of Pax9 and patterns the mesenchymal odontogenic field through protein-protein interactions with Msx1 and Pax9 during early tooth development.
Keywords: Msx1, odd-skipped, odontogenic, Osr2, Pax9, tooth development
Introduction
Tooth development has long been used as a model system for studying the molecular mechanisms of organogenesis (Pispa and Thesleff, 2003; Thesleff et al., 1995). Morphologically, tooth development begins with formation of a thickened stripe of the oral epithelium, termed the dental lamina, at sites of the future dental arches in the maxilla and mandible, which occurs at around the 11th day of gestation (E11) in mice. The dental lamina cells proliferate and bud into the underlying neural crest-derived ectomesenchyme at specific sites and induce the mesenchyme to condense around the epithelial buds from E12 to E13. Subsequently, the epithelium folds and extends farther into the mesenchyme, wrapping itself around the condensing mesenchyme to form “cap” (E14) and then “bell”-shaped tooth germs (at about E16). As development proceeds, epithelial cells in contact with the dental mesenchyme differentiate into enamel-producing ameloblasts and their adjacent mesenchymal cells differentiate into dentin-producing odontoblasts. Thus, formation of each individual tooth, from initiation through morphogenesis to ultimate cytodifferentiation, involves extensive interactions between the dental epithelium and the underlying mesenchyme.
Tissue recombination experiments showed that the early oral epithelium provides the instructive signals for tooth initiation. The mouse E9-E11 rostral mandibular epithelium elicited tooth formation when combined with non-dental second branchial arch mesenchyme (Lumsden, 1988; Mina and Kollar, 1987). However, no teeth formed when the E9-E11 presumptive dental epithelium was recombined with developing limb bud mesenchyme, indicating that only the neural crest derived mesenchyme is odontogenic competent (Lumsden, 1988). As development proceeds, however, the tooth inductive potential rapidly shifts to the dental mesenchyme (Lumsden, 1988; Mina and Kollar, 1987). It was demonstrated that the dental mesenchyme from E13 mouse embryos induced tooth formation from various non-dental epithelia, including limb epithelium, causing enamel organ morphogenesis and amelogenesis (Kollar and Fisher, 1980; Lumsden, 1988; Mina and Kollar, 1987; Ruch et al., 1973; Ruch et al., 1984). Thus, whereas tooth initiation depends on a site-specific epithelium, activation of tooth inductive potential in the neural crest derived presumptive dental mesenchyme is essential for subsequent tooth morphogenesis (Lumsden, 1988).
Extensive gene expression analyses and mouse genetic studies have provided significant insight into the molecular network controlling early tooth development. Prior to tooth initiation, several signaling molecules of the bone morphogenetic protein (Bmp) and fibroblast growth factor (Fgf) families are expressed in the presumptive dental epithelium and are responsible for inducing the expression of several transcription factors, including Msx1 and Pax9, in the presumptive dental mesenchyme (Dassule and McMahon, 1998; Lyons et al., 1989; Mandler and Neubuser, 2001; Neubuser et al., 1997; Vainio et al., 1993; Wozney et al., 1988). Mice lacking either Msx1 or Pax9 had tooth developmental arrest at the early bud stage (Peters et al., 1998; Satokata and Maas, 1994). Expression of Bmp4, a critical mesenchymal odontogenic signal, was dramatically reduced in the developing tooth mesenchyme by E13.5 in either Msx1−/− or Pax9−/− mutant mice (Chen et al., 1996; Peters et al., 1998). Maintenance of Msx1 mRNA in the developing tooth mesenchyme also depends on Pax9 function (Ogawa et al., 2006; Peters et al., 1998). In vitro biochemical studies showed that Pax9 was able to activate reporter gene expression driven by either the mouse Bmp4 or Msx1 gene promoter sequences and that Msx1 and Pax9 activated the Bmp4 promoter synergistically (Ogawa et al., 2006). Moreover, addition of recombinant Bmp4 protein rescued development of Msx1−/− mutant mandibular molar tooth germs to the late bell stage in explant cultures (Bei et al., 2000; Chen et al., 1996). Furthermore, mice with tissue-specific inactivation of Bmpr1a, which encodes a type-I receptor for Bmp family ligands, in the oral epithelium exhibited tooth developmental arrest at the bud stage (Andl et al., 2004; Liu et al., 2005). These data indicate that the Msx1 and Pax9 transcription factors play essential roles during early tooth development by activating mesenchymal odontogenic signals, including Bmp4, which signal back to the epithelium to drive tooth morphogenesis beyond the bud stage.
We reported recently that mice homozygous for a targeted null mutation in the Osr2 gene had supernumerary tooth formation lingual to the molars (Zhang et al., 2009). Osr2 encodes a zinc finger protein with extensive sequence similarity to the Drosophila Odd-skipped family transcription factors that regulate multiple developmental processes during embryogenesis and tissue morphogenesis (Coulter and Wieschaus, 1988; Green et al., 2002; Hao et al., 2003; Hart et al., 1996; Lan et al., 2001; Saulier-Le Drean et al., 1998; Wang and Coulter, 1996). We found that Osr2 mRNA exhibited a lingual-to-buccal gradient, complementary to that of Bmp4, in the mesenchyme surrounding the early developing tooth buds during normal tooth development and that Bmp4 mRNA expression was upregulated and expanded into the lingual side of the developing tooth mesenchyme in the Osr2−/− mice (Zhang et al., 2009). Furthermore, whereas Msx1−/− mutant mice had tooth developmental arrest at the bud stage accompanied by loss of Bmp4 expression from the developing tooth mesenchyme, Msx1−/−Osr2−/− double mutant mice showed restoration of Bmp4 mRNA expression in the tooth mesenchyme and rescue of first molar tooth morphogenesis (Zhang et al., 2009). These data suggest that Osr2 is a negative regulator of mesenchymal odontogenic potential and patterns the tooth developmental field at least in part by antagonizing the Msx1-Bmp4 pathway.
Since mice lacking either Msx1 or Pax9 had tooth developmental arrest at the early bud stage accompanied by loss of Bmp4 expression (Chen et al., 1996; Peters et al., 1998), the rescue of molar tooth development and restoration of Bmp4 expression in the developing tooth mesenchyme in the Msx1−/−Osr2−/− double mutant mice suggests that Osr2 may also interact with Pax9 during tooth development. To address this possibility, we generated a new line of Pax9 null mutant mice and found that Pax9 functions upstream of Osr2 and is required for tooth development beyond the early bud stage even in the absence of Osr2.
Materials and methods
Generation of Pax9fneo mice
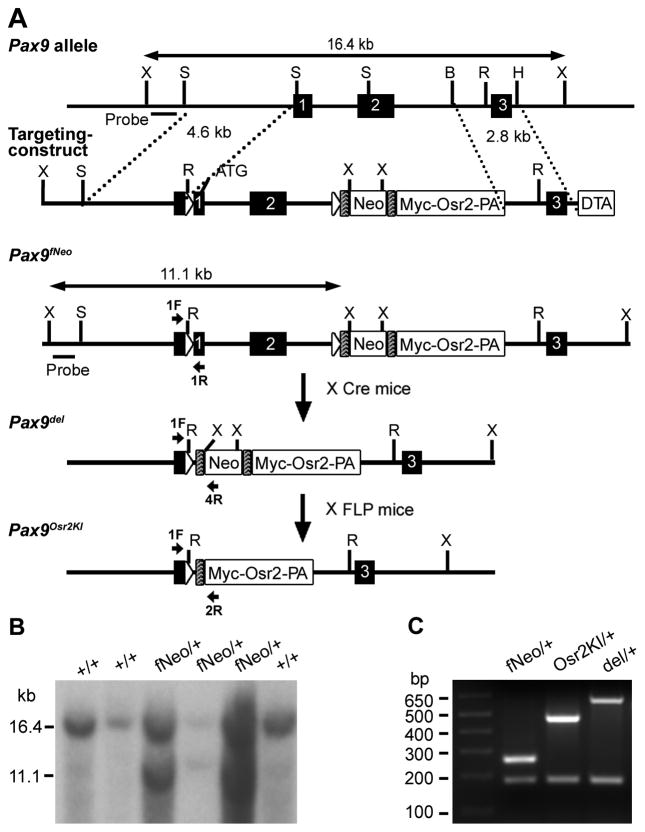
A 129/SvEv strain mouse BAC clone containing the entire Pax9 genomic region was isolated from the RPCI-22 BAC library (BACPAC Resources, Children’s Hospital of Oakland, Oakland, CA). The targeting vector used the 4.6 kb SmalI-SmalI fragment containing the beginning of the 5′ untranslated region of exon 1 as the 5′ arm and the 2.8 kb BglII-HindIII fragment containing the exon 3 region as the 3′ arm (Fig. 1A). The genomic region from the middle of the 5′ untranslated to the middle of intron 2 was subcloned in between two directly repeated loxP sites and inserted back in between the 5′ and 3′ homology arms. An FRT-flanked neo expression cassette followed by the Myc-Osr2A cDNA fusion construct (Gao et al., 2009) was then inserted in the second intron just 3′ to the loxP sequence in the targeting vector. In addition, a diphtheria toxin expression cassette (DTA) was subcloned to the 3′ end of the 3′ homology arm for selection against random integration of the targeting vector. The targeting vector was linearized and electroporated into the CJ7 mouse embryonic stem (ES) cells. ES cell culture and Southern hybridization screening of ES clones were carried out as previously described (Lan et al., 2004; Swiatek and Gridley, 1993). Two independently targeted ES cell clones were injected into blastocysts from C57BL/6J mice and the resultant chimeras bred with C57BL/6J females. F1 mice were genotyped by Southern hybridization analysis of tail DNA using the 5′ external probe (Fig. 1). Mice and embryos from subsequent generations were genotyped by PCR. PCR with Pax9 1F (5′-CCC ACG TTG CTG CTT AGA TT-3′) and Pax9 1R (5′-CGC ACT CCC AGA AAG AAA CT-3′) amplified a product of 186 bp from the wildtype Pax9 allele and a product of 260 bp from the Pax9fNeo allele. Heterozygous F1 mice were backcrossed with C57BL/6J mice and N2 heterozygous mice were intercrossed for analysis of homozygous phenotype.
Fig. 1.
Generation of mice with tissue-specific and spatiotemporally controlled expression of Osr2 in the developing tooth mesenchyme. (A) Schematic diagram of the gene-targeting and the mouse breeding strategies. At the top is the schematic representation of the region of the mouse Pax9 genomic DNA containing the first three coding exons (black boxes marked with numbers). The targeting vector contained two directly repeated loxP sequences (open triangles) inserted in 5′ to the translation start site (ATG) in exon-1 and in intron-2, respectively. Also included in the targeting vector are a frt-flanked neo expression cassette followed by a Myc-Osr2A cDNA cassette inserted in intron-2. Shaded boxes mark the frt sites. DTA, diphtheria toxin-A expression cassette. The correctly targeted allele was named Pax9fNeo. The positions for the probe used for Southern hybridization and for primers used for PCR genotyping (1F, 1R, 4R and 2R) are indicated. Pax9fNeo/+ mice were crossed with EIIa-Cre mice to generate the Pax9del/+ mice which were crossed with the FLPeR mice to generate the Pax9Osr2KI/+ mice. Restriction enzyme sites indicated are: B, BglII; H, HindIII; R, EcoRV; S, SmaI; X, XbaI. (B) Southern hybridiztion confirmation of germ line transmission of the targeted Pax9fNeo/+ allele from embryonic stem cell-derived chimeric mice. Tail DNA from F1 mice were digested with XbaI. The 16.4 kb XbaI fragment arose from the wildtype allele and the 11.1 kb fragment was specific for the Pax9fNeo allele. (C) Allele-specific PCR genotyping distinguishes Pax9fNeo/+, Pax9del/+ and Pax9Osr2KI/+ alleles by the amplified DNA fragment size.
Other mouse strains
Pax9del/+ mice were generated by crossing the Pax9fNeo/+ mice to the EIIa-Cre transgenic mice (The Jackson Laboratory, Bar Harbor, ME). Genotyping PCR with Pax9 1F (5′-CCC ACG TTG CTG CTT AGA TT-3′) and Pax9 4R (5′-GTG CCC AGT CAT AGC CGA AT-3′) amplified a product of 600 bp from the Pax9del allele. Pax9Osr2KI/+ mice were generated by crossing the Pax9del/+ mice with the FLPeR mice (The Jackson Laboratory, Bar Harbor, ME). Genotyping PCR with Pax9 1F (5′-CCC ACG TTG CTG CTT AGA TT-3′) and Pax9 2R (5′-CGC CCA AGC TCT CCA TTT CAT TCA-3′) amplified a product of 450 bp from the Pax9Osr2KI allele. Pax9del/+Osr2+/− mice were generated by crossing the Pax9del/+ mice to the Osr2+/− mice (Lan et al., 2004). The Osr2Osr2Aki mouse strain has been described previously (Gao et al., 2009).
Antibodies
The monoclonal anti-MYC epitope antibody (clone 4A6) was purchased from Millipore Corporation and anti-β-actin antibody was from Santa Cruz Biotechnology, Inc. The monoclonal anti-FLAG M2 antibody was purchased from Sigma. The Alexa Fluor® 546 goat anti-mouse IgG was purchased from Invitrogen Inc.
Western blot analysis
Mouse embryonic facial tissues were lysed in RIPA buffer (Santa Cruz). The protein concentration of supernatants was determined with Bio-Rad Assay Reagent (Bio-Rad), using BSA as the standard. Extracts were diluted in SDS-loading buffer and analyzed by standard SDS-PAGE and Western blot analysis.
Plasmid constructs, cell culture, and co-immunoprecipitation assay
The pCMV-Myc-Pax9 expression vector has been reported previously (Mensah et al., 2004) and was kindly provided by Dr. Rena D’Souza (Baylor College of Dentistry). The pCMV-3XFLAG-Osr2 expression vector was generated by subcloning a RT-PCR product containing the full-length mouse Osr2B coding sequence (Lan et al., 2001; Kawai et al., 2005) into the p3X-FLAG-CMV-7.1 vector (Sigma), which would allow production of the N-terminal 3XFLAG-tagged Osr2B protein in transfected cells. The pCMV-Myc-Msx1 and pCMV-3XFLAG-Msx1 expression vectors were constructed by subcloning the full-length mouse Msx1 coding sequence (Ogawa et al., 2006, kindly provided by Dr. Rena D’Souza) into the pCMV-Tag3B (Stratagene) and p3X-FLAG-CMV-7.1 (Sigma) vectors, respectively. All expression constructs were verified by sequencing.
COS7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were transfected with the expression vectors using FuGene-6 transfection reagents (Roche), cultured for 48 hours and harvested in RIPA lysis buffer (Santa Cruz). Co-immunoprecipitation was carried out as previously described (Xie et al., 2006). Briefly, cell lysates containing equal amounts of total proteins were added to 3 μl of anti-Myc antibody (clone 4A6, agarose conjugate, Millipore) and rotated overnight at 4°C. The agarose beads were collected and washed four times with RIPA buffer and the precipitated proteins analyzed by standard SDS-PAGE and Western blotting.
Skeletal analysis, histology, immunofluorescent staining and in situ hybridization
Skeletal preparations of newborn mice were carried out as previously described (Martin et al., 1995). For histology, embryos were dissected from timed pregnant heterozygous female mice and fixed for 1–2 days at room temperature in Bouin’s fixative. Fixed embryos were dehydrated through a graded series of ethanol and embedded in paraffin wax. Serial sections of 7 m thickness were stained with hematoxylin and eosin for histology analyses. For immunofluorescent staining and in situ hybridization, embryos were fixed in 4% paraformaldehyde overnight at 4ºC and processed for paraffin sections as described above. Indirect immunofluorescent staining was performed as described (Xie et al., 2006). In situ hybridization of sections was performed as described previously (Zhang et al., 1999).
Results
Generation of a gene-targeted mouse strain that allows conditional inactivation of Pax9 and spatiotemporally regulated expression of Osr2 in the developing tooth mesenchyme using the Cre/loxP and FLP/frt in vivo DNA recombination systems
The complementary patterns of expression of Osr2 and Bmp4 mRNAs along the buccolingual axis of the developing tooth mesenchyme and supernumerary tooth formation in the Osr2−/− mutant mice suggested that Osr2 functions to restrict odontogenic potential in the developing tooth mesenchyme (Zhang et al., 2009). To investigate further the role of Osr2 in patterning the tooth developmental field, we decided to test the effect of misexpression of Osr2 in the early tooth mesenchyme on early tooth development. Previous studies showed that Pax9 mRNA expression was activated in the presumptive tooth mesenchyme at the onset of tooth initiation (Neubuser et al., 1995; Peters et al., 1998). Thus, the regulatory sequences driving endogenous Pax9 gene expression during tooth development would be ideal for driving transgenic Osr2 expression in mice. However, since the cis-regulatory elements controlling Pax9 gene expression in the developing tooth mesenchyme have not been identified, we decided to generate mice expressing Osr2 from the endogenous Pax9 locus using a targeted insertion approach. Because the Pax9 gene is also expressed in many other tissues, including the developing pharyngeal endoderm, the developing somites and limb tissues (Peters et al., 1998), mis-expression of Osr2 in all Pax9-expressing cells might impair the survival of the gene-targeted mice. Thus, we designed a gene targeting strategy to generate mice containing two loxP sites flanking the first two coding exons of the Pax9 gene and an insertion of the frt-flanked neo expression cassette followed by a MYC-Osr2A cDNA construct (Gao et al., 2009) in the second intron (Fig. 1A). This new mouse strain would enable conditional inactivation of the Pax9 gene function by Cre/loxP-mediated deletion of the first two coding exons and subsequently expression of the MYC-Osr2A transgene can be activated from the endogenous Pax9 promoter following FLP-mediated removal of the frt-flanked neo cassette (Fig. 1A).
Two independently targeted mouse embryonic stem cell clones were used to generate chimeric mice and germ line transmission was obtained from one of these lines. Mice carrying the targeted allele, named Pax9fNeo, were confirmed by Southern hybridization analysis of tail DNA samples (Fig. 1B). The Pax9fNeo/+ heterozygous mice appeared normal and fertile, but the Pax9fNeo/fNeo homozygous mice exhibited malformed mandibular incisors, similar to mice homozygous for a neo cassette insertion in the second intron of the Pax9 gene reported previously (Kist et al., 2005). We crossed the Pax9fNeo/+ heterozygous mice to the FLPeR mice that express ubiquitously the FLP recombinase (Farley et al., 2000) to remove the neo cassette from this allele. The resultant Pax9fOsr2/+ heterozygous mice were intercrossed. The Pax9fOsr2/fOsr2 homozygous mice, in which both Pax9 alleles contained the MYC-Osr2A cassette in the second intron, were born at expected Mendelian ratio and exhibited normal life span and fertility, indicating that the neo cassette, not the MYC-Osr2A cassette, inserted in the second intron interfered with Pax9 gene function.
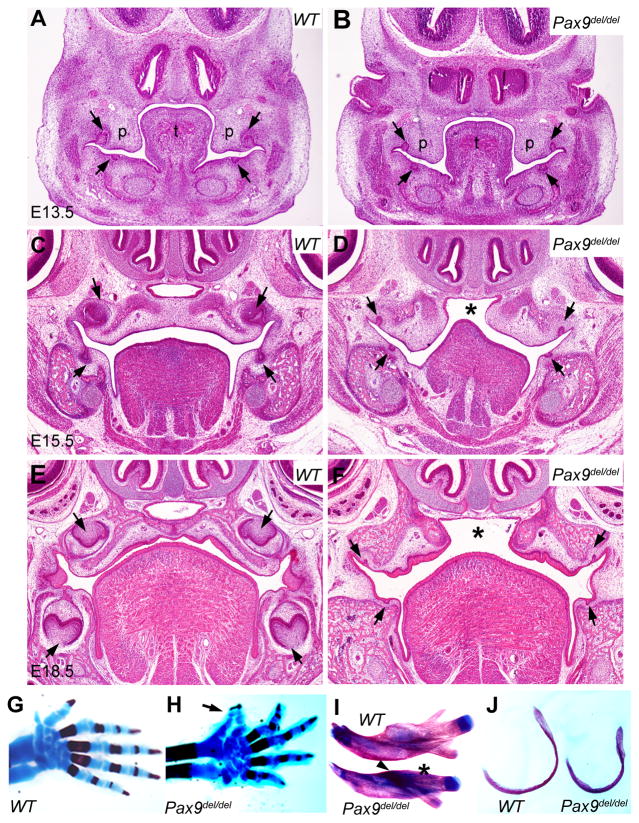
The Pax9fNeo/+ mice were crossed with EIIa-Cre transgenic mice (Lakso et al., 1996) to generate the Pax9del/+ mice carrying deletion of the first two coding exons. The Pax9del/+ mice lived and bred normally. We intercrossed Pax9del/+ mice and analyzed the phenotypes of Pax9del/del homozygous mice. The Pax9del/del homozygous mice were born alive but died shortly after birth. We carefully examined the development of the palate, tooth and skeleton structures in the Pax9del/del homozygous mutant mice by histological and skeletal preparations at different developmental stages. We found that the Pax9del/del homozygous mutant mice displayed all of the developmental defects reported previously in Pax9-deficient mice (Peters et al., 1998), including cleft secondary palate (Fig. 2A–F), tooth developmental arrest at the bud stage (Fig. 2A–F), preaxial polydactyly (Fig. 2G, H), and craniofacial skeletal defects (Fig. 2I, J). These data confirm that the Pax9del allele is a new Pax9 null allele.
Fig. 2.
The Pax9del/del mutant mice have cleft palate, tooth developmental arrest and skeletal defects. (A – F) Frontal sections E13.5 (A, B), E15.5 (C, D), and E18.5 (E, F) wildtype control (A, C, E) and Pax9del/del mutant (B, D, F) embryos. At E13.5, the Pax9del/del mutant embryos exhibited broadened palatal shelves (B) in comparison with the control littermates (A). By E15.5, the palatal shelves had elevated and fused with at the midline to separate the oral and nasal cavities in the control embryos (C), but the palatal shelves in the Pax9del/del mutant embryos (D) were malformed and failed to contact each other at the midline (asteroid in D). In addition, the mutants exhibited tooth developmental arrest at the early bud stage (arrows in D) while the tooth germs had developed to the cap stage in control embryos at E15.5 (C). At E18.5, the tooth germs had developed to the bell stage in control embryos (E) while the tooth germs still arrested at the rudimentary bud stage in the mutant embryos (arrows in F). Asteroid in F marks the gap between the bilateral palatal shelves. (G, H) In comparison to the wildtype controls (G), the mutant mice showed preaxial polydactyly (arrow in H). (I, J) The Pax9del/del mutant neonates showed smaller mandible (I) and tympanic ring (J) in comparison with wildtype littermates. Asteroid in I points to the lack of coronoid process and arrowhead points to the defect in alveolar bone in the mutant mandible.
We next generated the Pax9Osr2KI/+ mice by crossing the Pax9del/+ mice to the FLPeR mice. Allele-specific PCR primers were designed to genotype and clearly distinguish mice carrying the Pax9fNeo, Pax9del and Pax9Osr2KI alleles (Fig. 1C). As expected, the Pax9Osr2KI/+ embryos, but not the Pax9del/+ embryos, expressed MYC-Osr2A protein in the craniofacial tissues (Fig. 3). Immunofluorescent staining of frontal sections of E12.5 and E13.5 Pax9Osr2KI/+embryos confirmed spatially localized expression of MYC-Osr2A protein in the developing tooth mesenchyme (Fig. 4, A and B). These data demonstrate that the gene-targeted Pax9fNeo mice are useful for both the genetic analysis of the developmental and physiological roles of Pax9 by Cre/loxP-mediated inactivation and generation of mice expressing Osr2 from the endogenous Pax9 locus.
Fig. 3.
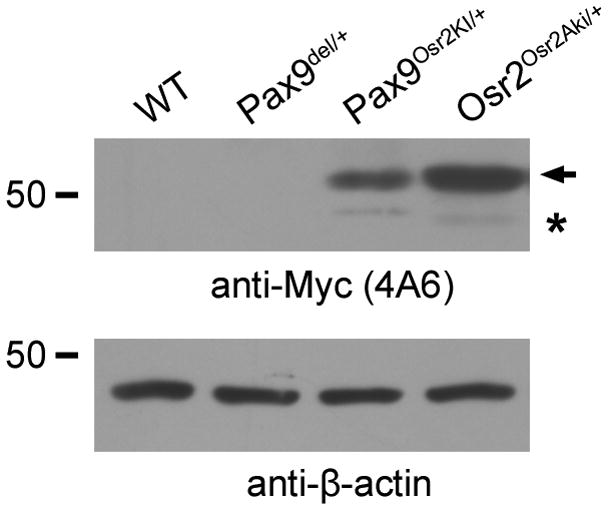
Myc-Osr2A protein is produced in the embryonic facial tissues in Pax9Osr2KI/+ embryos but not in the Pax9del/+ embryos. Protein extracts from E13.5 wildtype (WT), Pax9del/+, Pax9Osr2KI/+, Osr2Osr2Aki/+ embryonic facial tissues were compared by western blot analyses. The position of the 50 kilodalton molecular weight marker was indicated with the number 50 marked on the left. The top panel shows protein bands detected by the anti-MYC antibody, while the bottom panel shows -actin protein detected using the anit- -actin antibody. Arrow points to the predicted MYC-Osr2A full-length protein. Asteroid indicates a faint band in the Pax9Osr2KI/+and Osr2Osr2Aki/+ with a molecular weight corresponding to the product of a cryptically-spliced transcript from the Myc-Osr2A cDNA.
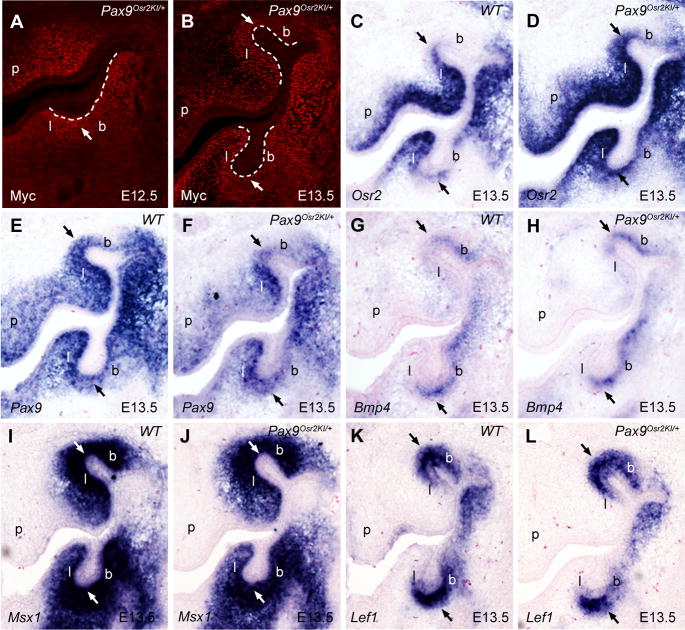
Fig. 4.
Comparison of Osr2, Pax9, Bmp4, Msx1 and Lef1 expression in the developing tooth mesenchyme in the wildtype and Pax9Osr2KI/+ embryos. In situ hybridization signals are shown in blue, while anti-MYC antibody staining shown in red. The buccal and lingual sides of the tooth mesenchyme are marked with b and l, respectively. (A, B) Detection of MYC-Osr2 protein expression in the developing tooth mesenchyme in E12.5 (A) and E13.5 (B) Pax9Osr2KI/+ embryos. Arrow points to the developing tooth mesenchyme underlying the distal region of the tooth bud. White dashed line marks the boundary between the dental epithelium and mesenchyme. (C, D) Osr2 mRNA expression in the E13.5 wildtype (C) and Pax9Osr2KI/+ (D) embryos. (E) Pax9 mRNA expression exhibited a graded pattern along the buccolingual axis of the developing tooth mesenchyme, with higher levels on the lingual side, in wildtype embryos. (F) Pax9 mRNA expression was decreased in Pax9Osr2KI/+ embryos in comparison with the wildtype littermate (E). (G - L) Expression of Bmp4 (G and H), Msx1 (I and J), and Lef1 (K and L) mRNAs in the developing tooth mesenchyme was not significantly altered in the E13.5 Pax9Osr2KI/+ embryos (H, J, L) in comparison with the wildtype (G, I, K) littermates. p, palatal shelf.
Expression of Osr2 from the Pax9 locus did not disrupt primary tooth morphogenesis but suppressed supernumerary tooth formation in the Osr2−/− mutant mice
Whereas Myc-Osr2A protein expression was clearly detected in the developing tooth mesenchyme in Pax9Osr2KI/+ embryos (Fig. 4, A and B), the Pax9Osr2KI/+ mice were born at expected Mendelian ratios and did not show any obvious developmental defects. In particular, all molar and incisor teeth developed normally in these mice (data not shown). Interestingly, we found, while MYC-Osr2A protein was detected in the developing tooth mesenchyme as early as E12.5 in the Pax9Osr2KI/+ embryos (Fig. 4A), that its expression exhibited a clear buccolingual gradient pattern by E13.5, with high levels of nuclearly localized MYC-Osr2A proteins in the mesenchyme lingual to the tooth bud and much lower levels buccal to the tooth bud (Fig. 4B). To investigate whether the gradient pattern of MYC-Osr2A proteins in the developing tooth mesenchyme resulted from differential mRNA expression, we performed in situ hybridization with a cRNA probe complementary to the Osr2-coding sequence so that it would detect both endogenous Osr2 mRNA as well as the MYC-Osr2A mRNA. In comparison to the preferential expression of endogenous Osr2 mRNA in the lingual side of the developing tooth mesenchyme in wildtype embryos (Fig. 4C), the E13.5 Pax9Osr2KI/+ embryos showed increased total Osr2 mRNA expression, but the overall Osr2 mRNA expression pattern still exhibited a strong preference to the mesenchyme lingual to the tooth buds and only very low levels of Osr2 mRNA was detected in the mesenchyme buccal to the tooth buds (Fig. 4D). We analyzed endogenous Pax9 mRNA expression and found that it also exhibited preferential expression in the mesenchyme lingual to the tooth buds than that on the buccal side at E13.5 (Fig. 4E), suggesting that the gradient pattern of MYC-Osr2A protein distribution in the developing tooth mesenchyme in Pax9Osr2KI/+ embryos was due to differential activity of the Pax9 gene promoter. Whereas the overall level of Pax9 mRNA expression was significantly reduced in the Pax9Osr2KI/+ embryos compared with the wildtype littermate, most likely due to the replacement of one of the two Pax9 alleles by the MYC-Osr2A cDNA in the mutant mice, the Pax9 mRNA expression in the developing tooth mesenchyme still exhibited the lingual-buccal gradient with lower levels on the buccal side (Fig. 4F). These changes in Osr2 and Pax9 expression in the Pax9Osr2KI/+ embryos, with the added MYC-Osr2 protein in the Pax9 heterozygous state, however, did not have an obvious impact on expression of downstream tooth developmental genes, including Bmp4, Msx1 and Lef1 (Fig. 4, G–L).
One possibility as to why the Pax9Osr2KI/+ mice did not show any obvious developmental defects might be that the MYC-Osr2A protein expressed in these mice did not have the biochemical activities of the endogenous Osr2 protein. To investigate this possibility, we introduced the Pax9Osr2KI allele into the Osr2−/− mutant background to examine whether the Myc-Osr2A protein expressed from the Pax9Osr2KI allele could suppress supernumerary tooth formation in the Osr2−/− mutants. We found that none of the Pax9Osr2KI/+Osr2−/− mutant mice examined at birth had any supernumerary tooth germs in either the maxilla or mandible (n = 10) while all Osr2−/− mutant littermates (n = 8) exhibited the supernumerary tooth phenotype as previously reported [(Zhang et al., 2009) Fig. 5, A–C and E–G]. In addition, 60% of the Pax9Osr2KI/+Osr2−/− mutant mice exhibited fused secondary palate (Fig. 5C) while 100% of the Osr2−/− mutant mice exhibited cleft secondary palate (Fig. 5B). Because the Pax9Osr2KI allele is also a Pax9-null allele, we crossed the Pax9del/+ mice to Osr2+/− mice and examined palatal and tooth developmental defects in the Pax9del/+Osr2−/− newborn pups. All Pax9del/+Osr2−/− newborn pups examined exhibited cleft palate at birth (n = 6), indicating that expression of MYC-Osr2A from the Pax9Osr2KI allele rather than Pax9 heterozygosity was responsible for rescuing the cleft palate in the Pax9Osr2KI/+Osr2−/− mutant mice. Interestingly, the Pax9del/+Osr2−/− pups had supernumerary tooth germs lingual to the maxillary molars but did not have supernumerary teeth in the mandible (Fig. 5, D and H).
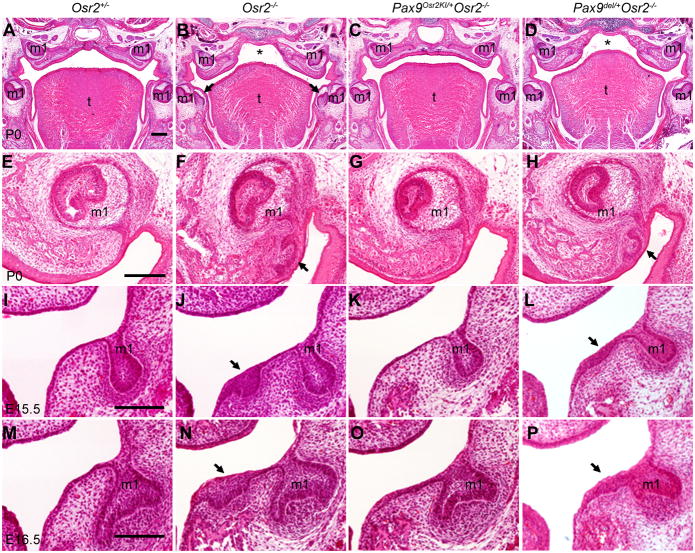
Fig. 5.
Expression of Osr2 from the Pax9 locus rescued palate and tooth developmental defects in the Osr2−/− mutant mice. (A – D) Hematoxylin-eosin (HE)-stained frontal sections through the first molar region of the newborn Osr2+/− (A), Osr2−/− (B), Pax9Osr2KI/+Osr2−/− (C), and Pax9del/+Osr2−/− (D) pups. Compared with the Osr2+/− littermate (A), Osr2−/− mutant mice (B) had cleft palate (marked by asteroid) and supernumerary teeth (arrows) lingual to the first molars. The Pax9Osr2KI/+Osr2−/− exhibited normal secondary palate and teeth (C). The Pax9del/+Osr2−/− mouse (D) had cleft palate (marked by asteroid) but did not have supernumerary teeth in the mandible. (E H) Frontal sections through the posterior region of the upper first molar tooth germs of newborn Osr2+/− (E), Osr2−/− (F), Pax9Osr2KI/+Osr2−/− (G), and Pax9del/+Osr2−/− (H) pups. (I L) Frontal sections through the posterior region of the mandibular first molar tooth germs of E15.5 Osr2+/− (I), Osr2−/− (J), Pax9Osr2KI/+Osr2−/− (K), and Pax9del/+Osr2−/− (L) embryos. (M P) Frontal sections through the posterior region of the mandibular first molar tooth germs of E16.5 Osr2+/− (M), Osr2−/− (N), Pax9Osr2KI/+Osr2−/− (O), and Pax9del/+Osr2−/− (P) embryos. Panel A - D are shown at the same magnification with scale bar representing 200 m. Panels E H are shown at the same magnification with scale bar representing 100 m. Panels I P are shown at the same magnification with the scale bar representing 100 m. m1, first molar tooth germ; t, tongue.
To investigate further the phenotypic differences in the Pax9Osr2KI/+Osr2−/− and Pax9del/+Osr2−/− mutant mice, we carried out detailed histological studies of the mutant embryos at E15.5 to E16.5 and compared with the Osr2+/− control and Osr2−/− mutant littermates. At E15.5, the Osr2−/− mutant embryos exhibited an obvious supernumerary tooth bud with condensed mesenchyme underneath, lingual to each of the mandibular first molar tooth germs (Fig. 5J). By E16.5, the mandibular supernumerary tooth germs in the Osr2−/− mutant embryos had progressed to the early cap stage (Fig. 5N), as previously reported (Zhang et al., 2009). In contrast, no supernumerary tooth germs were detected in the Pax9Osr2KI/+Osr2−/− mutant embryos at either E15.5 or E16.5 (Fig. 5, K and O). In the E15.5 Pax9del/+Osr2−/− mutant embryos, the oral epithelium lingual to the mandibular first molar tooth germs was much thickened (Fig. 5L), in comparison with that in the Osr2+/− control (Fig. 5I) and Pax9Osr2KI/+Osr2−/− mutant (Fig. 5K) embryos, but did not form an obvious supernumerary tooth bud as in the Osr2−/− mutant embryos at this stage (compare Fig. 5L with Fig. 5J). At E16.5, the thickened oral epithelium lingual to the mandibular first molar tooth germs in the Pax9del/+Osr2−/− mutant embryos still did not progress to form distinct tooth buds (Fig. 5P), in contrast to the Osr2−/− mutant embryos (Fig. 5, J and N). These data indicate, while expression of MYC-Osr2A from the Pax9 locus suppressed supernumerary tooth formation and partially rescued the cleft palate defect in the Pax9Osr2KI/+Osr2−/− mutant mice, that Pax9 was haploinsufficient to support complete morphogenesis of the mandibular supernumerary teeth in the Pax9del/+Osr2−/− mice.
Pax9 acts upstream of Osr2 and is required for activation of mesenchymal odontogenic potential even in the absence of Osr2
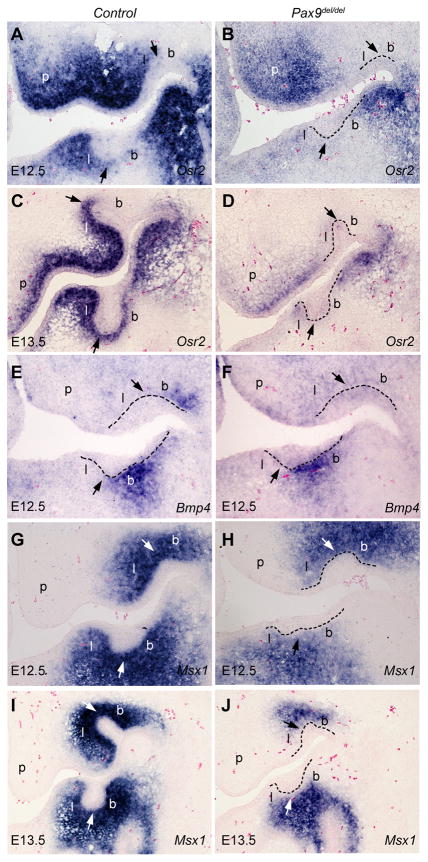
The disruption of mandibular supernumerary tooth development in the Pax9del/+Osr2−/− mutant embryos suggested genetic interactions between Osr2 and Pax9 in the control of tooth development. To investigate further the relationship between Osr2 and Pax9 in tooth development, we analyzed expression of Osr2 in the Pax9del/del null mutants. We found that Osr2 mRNA expression in the developing tooth mesenchyme was dramatically reduced in the Pax9del/del embryos by E12.5, compared with wildtype littermates (Fig. 6A, B). At E13.5, very little Osr2 mRNA was detected in the developing tooth mesenchyme in the Pax9del/del embryos, compared with the robust gradient pattern of Osr2 mRNA expression in the developing tooth mesenchyme in the control littermates (Fig. 6C, D). The Pax9del/del embryos also showed significantly reduced Osr2 mRNA expression in the palatal mesenchyme and the lateral mandibular mesenchyme (Fig. 6A–D), tissues that normally co-express Osr2 and Pax9, but Osr2 mRNA expression in the developing eyelid, a tissue that does not normally express Pax9, was not significantly altered in the Pax9del/del embryos in comparison with wildtype littermates (data not shown).
Fig. 6.
Expression of Osr2, Bmp4, and Msx1 was significantly downregulated at E12.5 and E13.5 in the developing tooth mesenchyme in Pax9del/del embryos in comparison with wildtype littermates. mRNA signals are shown in blue color. Arrows point to the first molar tooth buds. Black dashed line marks the boundary between dental epithelium and mesenchyme. The buccal and lingual sides of the tooth mesenchyme are marked with b and l, respectively. (A–D) Frontal sections of E12.5 (A, B) and E13.5 (C, D) showing expression of Osr2 mRNA in the developing palatal shelf and first molar tooth germs. (E, F) Frontal sections of E12.5 embryos showing Bmp4 mRNA expression in the developing first molar tooth region. (G–J) Frontal sections of E12.5 (G, H) and E13.5 (I, J) embryos showing Msx1 mRNA expression in the developing first molar tooth region. p, palatal shelf.
Previously, it was reported that expression of Bmp4, Msx1, and Lef1 was indistinguishable in Pax9−/− and wildtype littermates at E12.0 and each was substantially downregulated in the Pax9−/− embryos in comparison with wildtype littermates at E13.5 (Peters et al., 1998). Since we found that Osr2 mRNA expression was dramatically reduced in the E12.5 Pax9del/del embryos, we investigated whether expression of Bmp4 and Msx1 was also affected at this early stage of tooth development. We found that expression of both Bmp4 and Msx1 was indeed significantly downregulated in the developing tooth mesenchyme in the Pax9del/del embryos at E12.5 (Fig. 6, E–H). Interestingly, Msx1 mRNA expression persisted at relatively high levels in the distal tooth mesenchyme adjacent to the developing osteogenic regions at E13.5 (Fig. 6I, J), suggesting that the osteogenic cells can induce Msx1 mRNA expression through a Pax9-independent mechanism.
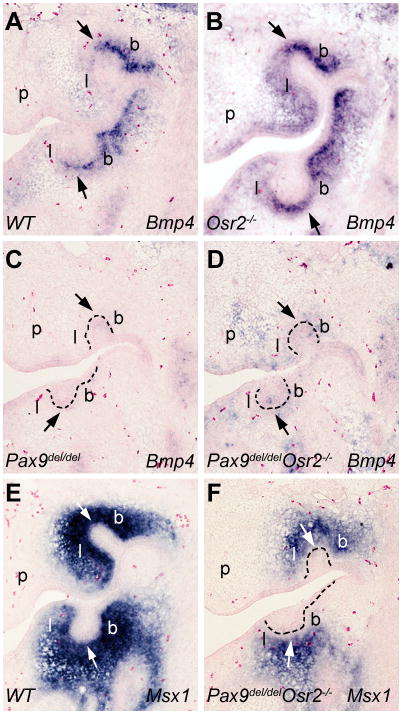
We previously showed that deleting Osr2 rescued first molar tooth development in the Msx1−/− mutant mice (Zhang et al., 2009). To investigate whether inactivation of Osr2 could rescue tooth development in the Pax9del/del mutant mice, we analyzed tooth development in embryos from intercrosses of Pax9del/+Osr2+/− double heterozygous mice. Histological analyses revealed that tooth development was arrested at the early bud stage in the Pax9del/delOsr2−/− double mutant mice, similar to that in the Pax9del/del mutant mice (Fig. 7). We previously showed that expression of Bmp4, a critical odontogenic signal, was restored in the developing tooth mesenchyme in E13.5 Msx1−/−Osr2−/− double mutant embryos in comparison with the Msx1−/− littermates (Zhang et al., 2009). Examination of Bmp4 expression showed that the E13.5 Pax9del/delOsr2−/− double mutant embryos lacked Bmp4 expression in the developing tooth mesenchyme, similar to the Pax9del/del mutant littermate (Fig. 8, A-D). Msx1 mRNA expression was also similarly downregulated in the developing tooth mesenchyme in Pax9del/delOsr2−/− double mutant and Pax9del/del mutant embryos in comparison with control littermates (Fig. 6I, J, and Fig. 8E, F). Taken together, these data indicate that Pax9 acts upstream of Osr2 and is required for the activation of odontogenic potential of the developing tooth mesenchyme even in the absence of Osr2.
Fig. 7.
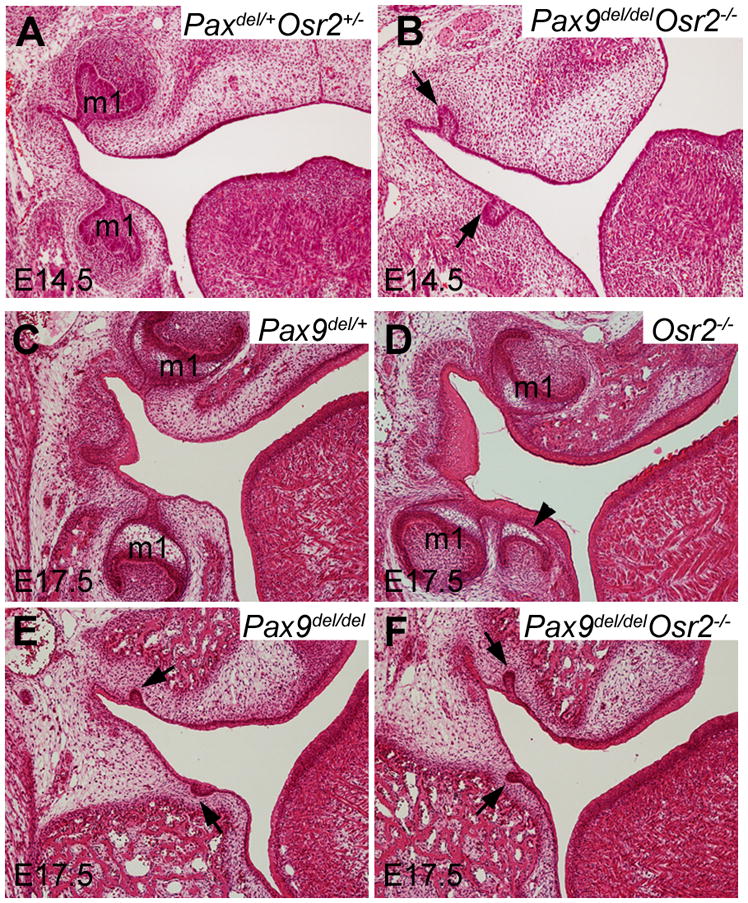
Pax9del/delOsr2−/− double homozygous mutant mice exhibited tooth developmental arrest at the early bud stage, similar to Pax9del/del mutant mice. Frontal sections through the first molar tooth germ (m1) region of E14.5 (A, B) and E17.5 (C–F) Pax9del/+Osr2+/− control (A), Pax9del/+ control (C), Osr2−/− (D), Pax9del/del (E), and Pax9del/delOsr2−/− mutant (B and F) embryos. Compared with the tooth development in the control littermates at E14.5 (cap stage) (A) and E17.5 (bell stage) (C), Pax9del/delOsr2−/− mutant embryos showed tooth development arrested at early bud stage (B, F), similar to the Pax9del/del mutant embryos (E). Arrowhead in D points to the supernumerary tooth germ lingual to the first molar tooth germs in the Osr2−/− mutant embryo. Arrows in B, E and F point to the arrested first molar tooth germs at the early bud stage in these mutant embryos.
Fig. 8.
Pax9 is required for maintenance of Bmp4 and Msx1 mRNA expression in the developing tooth mesenchyme even in the absence of Osr2. (A–D) Frontal sections through the first molar tooth region of E13.5 wildtype (A), Osr2−/− (B), Pax9del/del (C), and Pax9del/delOsr2−/− mutant (D) embryos hybridized with Bmp4 cRNA probe. Arrows point to the distal end of the first molar tooth buds. The buccal and lingual sides of the tooth mesenchyme are marked as b and l, respectively. Compared with the wildtype embryo (A), Osr2−/− mutant embryo (B) showed increased Bmp4 mRNA expression that expanded to the lingual side of the tooth buds. In contrast, little Bmp4 mRNA was detected in the developing tooth region in either the Pax9del/del (C) or Pax9del/delOsr2−/− mutant (D) embryos. (E, F) Frontal sections through the first molar tooth region of E13.5 wildtype (E) and Pax9del/delOsr2−/− mutant (F) embryos showing Msx1 mRNA expression. p, palatal shelf.
Physical interactions of Osr2, Msx1, Pax9 proteins
Previous studies showed that Msx1 and Pax9 proteins physically interact in co-transfected COS7 cells (Ogawa et al., 2006). To investigate whether Osr2 interacts with either Msx1 or Pax9 protein in vivo, we carried out immunoprecipitation and western blot analyses of epitope-tagged proteins expressed in co-transfected COS7 cells. We found that a large amount of Osr2 protein was co-precipitated with Msx1 (Fig. 9), indicating that Osr2 and Msx1 form stable protein complexes in vivo. A small amount of Osr2 protein was co-precipitated with Pax9, in contrast to the strong co-precipitation of Osr2 with Msx1 and of Msx1 with Pax9 (Fig. 9), suggesting that Osr2 may interact transiently or weakly with Pax9 in vivo.
Fig. 9.
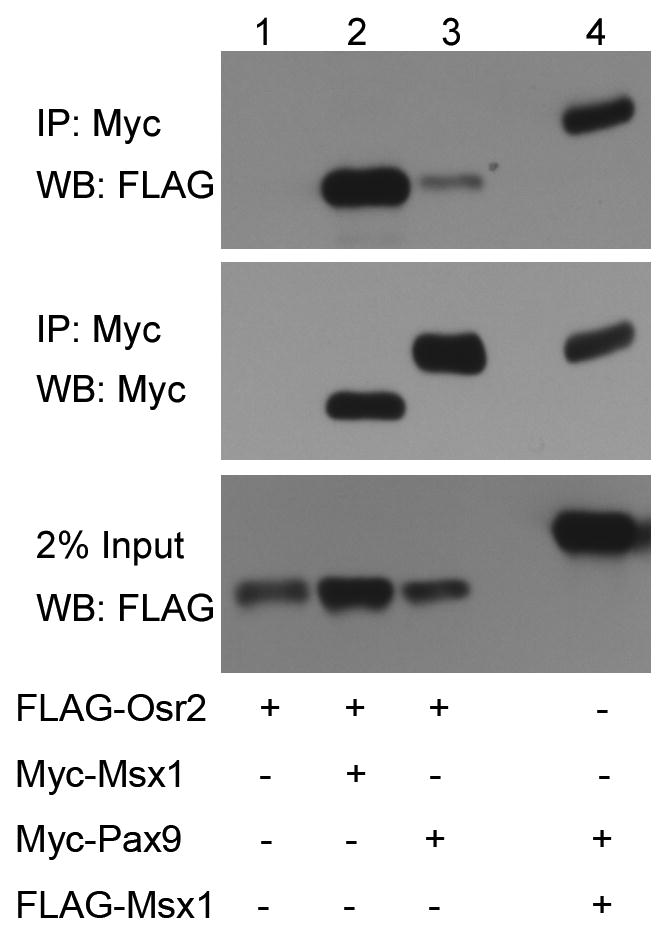
Interactions of Osr2 with Msx1 and Pax9 proteins. Lanes 1 – 3 are from COS7 cells transfected with pCMV-3XFLAG-Osr2 expression vector and co-transfected with, respectively, a control pCMV-MYC-tag vector (Lane 1), pCMV-Myc-Msx1 expression vector (Lane 2), or pCMV-Myc-Pax9 (Lane 3). Lane 4 is from COS7 cells transfected with pCMV-3XFLAG-Msx1 and pCMV-Myc-Pax9 expression vectors as positive control. Cell lysates containing equal amounts of total proteins were immunoprecipitated with anti-Myc monoclonal antibody followed by western blot analysis with the anti-FLAG antibody (top panel). The western blot membrane was re-probed with the anti-Myc antibody (middle panel). The bottom panel shows western blot detection of the 3XFLAG-Osr2 (Lanes 1–3) and 3XFLAG-Msx1 (Lane 4) proteins in the cell lysates.
Discussion
Previous studies showed that the transcription factors Msx1 and Pax9 are both required and act synergistically during early tooth development. Mice lacking either Msx1 or Pax9 had tooth developmental arrest at the bud stage with loss of Bmp4 expression in the developing tooth mesenchyme (Satokata and Maas, 1994; Chen et al., 1996; Peters et al., 1998). Biochemical studies showed that Msx1 and Pax9 proteins physically interact and synergistically activate the Bmp4 gene promoter in co-transfected COS7 cells (Ogawa et al., 2006). Moreover, while mice heterozygous for either Msx1 or Pax9 were normal, mice doubly heterozygous for Msx1 and Pax9 mutations had defects in development of the lower incisors, with loss of expression of Fgf3 and Fgf10 in the developing tooth mesenchyme (Nakatomi et al., 2010). These data firmly establish Msx1 and Pax9 as essential activators of mesenchymal odontogenic activity. In this report, we show that Pax9 is also required for maintenance of expression of Osr2, a suppressor of mesenchymal odontogenic activity (Zhang et al., 2009). Our data reveal complex interplay between Osr2, Pax9, and Msx1 during early tooth development.
At the initiation stage of tooth development, expression of Msx1 and Pax9 in the presumptive tooth mesenchyme is each induced by signals from the presumptive dental epithelium (Dassule and McMahon, 1998; Mandler and Neubuser, 2001; Neubuser et al., 1997; Vainio et al., 1993). As the tooth buds form, Bmp4 mRNA expression was preferentially activated in the buccal side and Osr2 mRNA expression activated specifically in the lingual side of the developing tooth mesenchyme (Zhang et al., 2009). By the early tooth bud stage (E12.5), the levels of expression of Bmp4, Msx1, and Osr2 were all significantly reduced in the Pax9del/del mutant embryos in comparison with wildtype littermates (Fig. 6). Expression of Bmp4 but not that of Osr2 mRNA was downregulated in the developing tooth mesenchyme in Msx1−/− mutant embryos by E13.5 (Chen et al., 1996; Zhang et al., 2009). Bmp4 mRNA expression was partially restored in the developing tooth mesenchyme in the Msx1−/−Osr2−/− embryos (Zhang et al., 2009) but not in the Pax9del/delOsr2−/− embryos (Fig. 8). These data indicate that Pax9 plays dual roles in patterning the developing tooth mesenchyme, by activating mesenchymal odontogenic factors to drive tooth morphogenesis from the early bud stage and by maintaining Osr2 expression to restrict the Msx1-mediated propagation of mesenchymal odontogenic activity along the buccolingual axis of the tooth developmental field.
Whereas our previous mutant mouse studies showed antagonistic genetic interactions between Osr2 and Msx1 during tooth development (Zhang et al., 2009), our co-immunoprecipitation results suggest that Osr2 forms a stable protein complex with the Msx1 protein in vivo. Since Msx1 has been shown to physically interact and synergize with Pax9 in the activation of the Bmp4 gene promoter (Ogawa et al., 2006), it is possible that Osr2 competes with Pax9 in the interactions with Msx1 to pattern odontogenic gene expression in the developing tooth mesenchyme. Alternatively, since previous biochemical studies showed that Msx1 is a potent transcriptional repressor and physically interacts with the Groucho-related gene family of co-repressors (Catron et al., 1995; Rave-Harel et al., 2005; Zhang et al., 1996), it is possible that interaction of Osr2 and Msx1 may stabilize the repressor complex and thus prevent activation of important odontogenic genes such as Bmp4. The development of the first molar teeth in the Msx1−/−Osr2−/− mutant mice, compared with the tooth developmental arrest in the Msx1−/− mutant mice, however, indicates that Osr2 also suppressed mesenchymal odontogenic gene expression in the absence of Msx1. We found that a small amount of Osr2 protein was co-precipitated with Pax9 (Fig. 9). Together with the restoration of Bmp4 mRNA expression in the developing tooth mesenchyme in the Msx1−/−Osr2−/− mutant but not in the Osr2−/−Pax9−/− mutant embryos, compared with the Msx1−/− and Pax9−/− mutant embryos, these data suggest that Osr2-Pax9 interactions may underlie the tooth developmental arrest in the Msx1−/− mutant mice and that deletion of Osr2 in the Msx1−/− mutant mice restored Pax9-mediated activation of Bmp4 mRNA expression in the developing tooth mesenchyme.
In summary, while previous genetic and tissue recombination studies established that both Pax9 and Msx1 are essential for the activation of the mesenchymal odontogenic potential and that Osr2 suppressed the mesenchymal odontogenic program lingual to the developing tooth buds during tooth development in mice (Bei et al., 2000; Chen et al., 1996; Peters et al., 1998; Zhang et al., 2009), our data demonstrate that Pax9 has critical Msx1-independent functions and acts genetically upstream of both Msx1 and Osr2 to pattern the mesenchymal odontogenic field. Our biochemical analyses suggest that Osr2 suppresses the mesenchymal odontogenic program through physical interactions with Msx1 and Pax9 proteins.
Acknowledgments
We thank Dr. Thomas Gridley for the CJ7 mouse ES cells. We thank Dr. Rena D’Souza for the mouse Msx1 and Pax9 expression plasmids. This work was supported by NIH grants R01DE013681 and R01DE018401 to RJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, Lyons KM, Mishina Y, Seykora JT, Crenshaw EB, 3rd, Millar SE. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Wieschaus E. Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes Dev. 1988;2:1812–1823. doi: 10.1101/gad.2.12b.1812. [DOI] [PubMed] [Google Scholar]
- Dassule HR, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Gao Y, Lan Y, Ovitt CE, Jiang R. Functional equivalence of the zinc finger transcription factors Osr1 and Osr2 in mouse development. Dev Biol. 2009;328:200–209. doi: 10.1016/j.ydbio.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RB, Hatini V, Johansen KA, Liu XJ, Lengyel JA. Drumstick is a zinc finger protein that antagonizes Lines to control patterning and morphogenesis of the Drosophila hindgut. Development. 2002;129:3645–3656. doi: 10.1242/dev.129.15.3645. [DOI] [PubMed] [Google Scholar]
- Hao I, Green RB, Dunaevsky O, Lengyel JA, Rauskolb C. The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev Biol. 2003;263:282–295. doi: 10.1016/j.ydbio.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Hart MC, Wang L, Coulter DE. Comparison of the structure and expression of odd-skipped and two related genes that encode a new family of zinc finger proteins in Drosophila. Genetics. 1996;144:171–182. doi: 10.1093/genetics/144.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kist R, Watson M, Wang X, Cairns P, Miles C, Reid DJ, Peters H. Reduction of Pax9 gene dosage in an allelic series of mouse mutants causes hypodontia and oligodontia. Hum Mol Genet. 2005;14:3605–3617. doi: 10.1093/hmg/ddi388. [DOI] [PubMed] [Google Scholar]
- Kollar EJ, Fisher C. Tooth induction in chick epithelium: expression of quiescent genes for enamel synthesis. Science. 1980;207:993–995. doi: 10.1126/science.7352302. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho ES, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- Lyons KM, Pelton RW, Hogan BL. Patterns of expression of murine Vgr-1 and BMP-2a RNA suggest that transforming growth factor-beta-like genes coordinately regulate aspects of embryonic development. Genes Dev. 1989;3:1657–1668. doi: 10.1101/gad.3.11.1657. [DOI] [PubMed] [Google Scholar]
- Mandler M, Neubuser A. FGF signaling is necessary for the specification of the odontogenic mesenchyme. Dev Biol. 2001;240:548–559. doi: 10.1006/dbio.2001.0490. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Mensah JK, Ogawa T, Kapadia H, Cavender AC, D’Souza RN. Functional analysis of a mutation in PAX9 associated with familial tooth agenesis in humans. J Biol Chem. 2004;279:5924–5933. doi: 10.1074/jbc.M305648200. [DOI] [PubMed] [Google Scholar]
- Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen Y, Maas RL, Peters H. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Koseki H, Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kapadia H, Feng JQ, Raghow R, Peters H, D’Souza RN. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J Biol Chem. 2006;281:18363–18369. doi: 10.1074/jbc.M601543200. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Rave-Harel N, Miller NLG, Givens ML, Mellon PL. The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J Biol Chem. 2005;280:30975–30983. doi: 10.1074/jbc.M502315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch JV, Karcher-Djuricic V, Gerber R. Determinants of morphogenesis and cytodifferentiations of dental anloges in mice. J Biol Buccale. 1973;1:45–56. [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Karcher-Djuricic V, Meyer JM. Extracellular matrix-mediated interactions during odontogenesis. Prog Clin Biol Res. 1984;151:103–114. [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Saulier-Le Drean B, Nasiadka A, Dong J, Krause HM. Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development. 1998;125:4851–4861. doi: 10.1242/dev.125.23.4851. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Vaahtokari A, Kettunen P, Aberg T. Epithelial-mesenchymal signaling during tooth development. Connect Tissue Res. 1995;32:9–15. doi: 10.3109/03008209509013700. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- Wang L, Coulter DE. bowel, an odd-skipped homolog, functions in the terminal pathway during Drosophila embryogenesis. EMBO J. 1996;15:3182–3196. [PMC free article] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Xie ZH, Huang YN, Chen ZX, Riggs AD, Ding JP, Gowher H, Jeltsch A, Sasaki H, Hata K, Xu GL. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum Mol Genet. 2006;15:1375–1385. doi: 10.1093/hmg/ddl059. [DOI] [PubMed] [Google Scholar]
- Zhang H, Catron KM, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YD, Zhao X, Hu Y, St Amand TR, Ramamurthy R, Qiu MS, Chen YP. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]