Abstract
Genetically engineered fusion proteins offer potential as multifunctional biomaterials for medical use. Fusion or chimeric proteins can be formed using recombinant DNA technology by combining nucleotide sequences encoding different peptides or proteins that are otherwise not found together in nature. In the present study, three new fusion proteins were designed, cloned and expressed and assessed for function, by combining the consensus sequence of dragline spider silk with three different antimicrobial peptides. The human antimicrobial peptides human neutrophil defensin 2 (HNP-2), human neutrophil defensins 4 (HNP-4) and hepcidin were fused to spider silk through bioengineering. The spider silk domain maintained its self-assembly features, a key aspect of these new polymeric protein biomaterials, allowing the formation of β-sheets to lock in structures via physical interactions without the need for chemical cross-linking. These new functional silk proteins were assessed for antimicrobial activity against Gram - Escherichia coli and Gram + Staphylococcus aureus and microbicidal activity was demonstrated. Dynamic light scattering was used to assess protein aggregation to clarify the antimicrobial patterns observed. Attenuated-total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) and circular dichroism (CD) were used to assess the secondary structure of the new recombinant proteins. In vitro cell studies with a human osteosarcoma cell line (SaOs-2) demonstrated the compatibility of these new proteins with mammalian cells.
Keywords: Spider silk, Antimicrobial activity, Recombinant proteins, Self-assembly, Cell viability, Bone tissue engineering
1. Introduction
Around 350 billion dollars are spent worldwide on organ replacement therapies to prolong the lives of more than 20 million patients. Most organ replacement surgeries rely on the use of organometallic devices, most of which were developed in the 1960s [1, 2]. Tissue engineering is an emerging interdisciplinary field focused on the development of tissues and organs that can be used to replace damaged and failing tissues and organs. One of the major goals of tissue engineering is to replace permanent implanted prostheses with temporary implants with reconstructive and regenerative characteristics capable of directing the full restoration of normal tissue structure and function in the body; thus they must be fully degradable over time [3]. Achieving this goal would revolutionize healthcare treatment as well as patient well being [4]. The biomaterials field contributes directly toward this goal through the development of polymers with diverse mechanical and biological characteristics [5-8] that can be used to facilitate the regeneration of damaged tissues. In the case of bone repair, meeting mechanical and functional requirements at the implant site remains a challenge for most polymeric biomaterials [9, 10].
In the present work, spider silk was selected for the core polymer due to its potential as a biomaterial to meet the requirements for both mechanical stability and biocompatibility necessary for bone tissue engineering, as well as its accessibility to bioengineering [11-13]. Furthermore, recombinant DNA technologies allow the generation of spider silk proteins fused to other protein domains not normally found in spider silks, thereby expanding the function of these protein polymers. These new chimeric or multifunctional spider silk-like molecules combine the properties of silk (e.g., self-assembly, robust material properties and physical cross-linking to stabilize materials) with characteristics of the new domains added to the silk. These types of new multifunctional polymeric biomaterials provide a new route to control biomaterial properties related to both material features and biological interactions. The addition of antimicrobial peptides to reduce or control infections at the site of implantation was the direction sought in the present study. There are more than 600 peptides known with antimicrobial activity, most with broad activity against different microorganisms, including Gram+ and Gram- bacteria, virus, protozoa and fungi [14]. The bactericidal activity of these molecules starts with direct binding to the lipid bilayer forming the bacteria membrane. After this interaction with the membrane the bactericidal peptides acquire an amphiphilic three-dimensional conformation where the positive side of the antimicrobial molecules interact directly with the negatively charged lipid head-groups. This interaction results in the formation of pores through the bacterial membrane [15, 16]. A prominent feature of defensins is the six cysteine residues which form three disulphide bonds. Cysteine residues confer cationic nature to defensins and the initial association between these peptides and bacteria occurs through electrostatic interactions between the cationic peptides and the negatively charged lipid forming the outer membrane [17]. Moreover, these peptides play an important role in inflammation [18], and chemotaxis for monocytes and T cells to activate the acquired immune response system [19]. These peptides also intervene in the wound healing [20] by stimulating the proliferation of fibroblasts and epithelial cells [21, 22], induce neovasculogenesis [23] and mobilize cytokines [24]. Human α-defensins are cationic cysteine-rich peptides containing between 29 and 35 amino acids [25, 26]. Six α-defensins have been described, human neutrophil defensins 1 to 4 (HNP-1, HNP-2, HNP-3 and HNP-4) [27], which are expressed in neutrophils, and human defensins 5 to 6 (HD-5 and HD-6) expressed by Paneth cells of the small intestine and by the epithelial cells of the female urogenital track [28]. HNP-2 manifests bactericidal activity against both Gram+ and Gram- bacteria [29]. HNP-4 was considered more effective against Gram- Escherichia coli and Enterobacter aerogenes. Moreover, HNP-4 was more effective than HNP-1, HNP-2 and HNP-3 in protecting human peripheral blood mononuclear cells from infection by both X4 and R5 HIV-1 strains [30].
Resembling the human neutrophil defensins described above, hepcidin (also named LEAP-1 for liver expressed antimicrobial peptide), is also a cysteine rich peptide. Hepcidin mRNA was found primarily in the liver and was isolated initially from human blood [31] and later detected in urine [32]. Hepcidin exhibits bactericidal and fungicidal activity with activity against Gram+ and Gram- bacteria. Hepcidin was active against the Gram- E. coli ML35 strain and Neisseria cinerea, and against the Gram+ Staphylococcus aureus, Staphylococcus epidermis, Staphylococcus carnosus, Bacillus cereus, Bacillus megaterium, Bacillus subtilis, Micrococcus luteus and group B Streptococcus [32-36]. After incubation of K562 cells (American Type Culture Collection leukemic cell line CCL-243) for 15 hours with hepcidin at a concentration of 30 μM, viability of 74% was found [32]. Thus the cytotoxicity of hepcidin was much lower than after incubation of these cells with 30 μM α-defensin HNP-1, where only 9% of the cells were viable over the same time frame [32]. Based on the antimicrobial activities of HNP-2, HNP-4 and hepcidin, these peptides were selected for fusion with the silk protein major ampullate spidroin protein (MaSpI) in the present study. The silk was selected as the main protein component as it has been shown to self-assemble into impressively strong and tough protein-based biomaterials, is biodegradable due to the activity of proteases, and can be formed into a range of material formats using water [37-39].
Due to the relatively low cytotoxicity, resistance to microorganism adaptation and broad spectra of action some antimicrobial peptides are considered strong candidates to be used in coatings of polymeric surfaces to help preventing microbial contamination. However, few studies have addressed this topic. In the past few years different antimicrobial peptides were expressed together with a carbohydrate binding module from Clostridium thermocellum, to enable attachment to different cellulosic surfaces for antimicrobial properties [40, 41]. The chemical coupling of antimicrobial peptides to different types of surfaces, such as: polyamide resins [42], polystyrene-polyethylene glycol beads [43-45] and titanium [46], has also been reported. To our knowledge this is the first study describing fusion proteins combining both silk and antimicrobial domains.
The goal of the present study was to construct three new fusion proteins combining spider silk with antimicrobial features hepcidin, HNP-2 and HNP-4 (Figure 1A). Three different antimicrobial peptides were selected due to the possible decrease in activity when the peptides were coupled with the spider silk. In this way, the activities of the three different fusion proteins could be compared to identify the most useful sequence for biomaterials formation and characterization. The silk domain in the proteins carries six repeats of the consensus repeat for the native sequence of the protein major ampullate dragline silk I (MaSpI) from the spider Nephila clavipes. Each silk repeat is formed by a hydrophilic GGX motif and a hydrophobic poly-A motif (Figure 1B). The three different silk-antimicrobial fusions were successfully expressed and characterized for structure and function. The antibacterial activity was studied by radial diffusion. Secondary structure of the recombinant proteins was assessed by attenuated-total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) and circular dichroism (CD). Dynamic light scattering (DLS) was used to assess aggregation related to antimicrobial activity in solution. Finally, cytotoxicity tests were performed to confirm the potential utility of these new multifunctional silk proteins in contact with mammalian cells.
Figure 1.
(A) Amino acid sequences for the chimeric proteins carrying silk block copolymer (6mer) and the HNP-2 (a), HNP-4 (b) and the hepcidin (c) peptide sequences. The 6mer block is represented by black. The HNP-2 (a), HNP-4 (b) and hepcidin (c) sequences are represented by a gray. The restriction sites for the insertion of HNP-2, HNP-4 and hepcidin sequences are underlined. (B) Amino acid sequences for the hydrophobic and hydrophilic blocks present in each of the six units forming the silk block copolymer (6mer).
2. Materials and Methods
2.1. Cloning HNP-2, HNP-4 and hepcidin into pET30L Vector Containing Silk Modules
Vector pET30a+ (Novagen, San Diego CA) was used for the construction of the vector pET30L carrying the silk block copolymer using procedures described in our previous work [47]. The silk block copolymer carries six repeats of the consensus repeat for the native sequence of the protein major ampullate dragline silk I from the specie N. clavipes. This spider silk block copolymer (6mer) was cloned with six histidine residues to facilitate protein purification, and with two restriction sites, one for SpeI and another NheI, flanking its edges to allow the addition of other DNA [47].
The HNP-2, HPN-4 and hepcidin cDNA sequences were prepared through the annealing of synthetic single strand oligonucleotide sequences (Invitrogen, CA, USA): HNP-2 top and HNP-2 bottom, HNP-4 top and HNP-4 bottom and hepcidin top and hepcidin bottom. The annealing reactions were carried out by decreasing the temperature of the oligonucleotide solution from 95°C to 20°C with a gradient of 0.1°C/s. The presence of the annealed products was verified with 5% agarose gels. At the edges of each cDNA sequence restriction sites were present for SpeI and for NheI for the insertion of the cDNA sequences into the vector pET30L. This vector already carried the DNA encoding the silk block copolymer. For the insertion of the cDNA sequences into pET30L, the vector was digested with SpeI (New England Biolabs, MA, USA, R0133S), dephosphorylated with calf intestinal phosphatase (CIP) (New England Biolabs, M0290S) and run on 0.8% agarose gels. The linearized vector was purified using the QIAquick gel extraction kit (Qiagen, CA, USA, 28706). The cDNA sequences were double digested with SpeI and NheI and the digestion products run in a 0.8% gel and the bands for the cDNA sequences were purified using a QIAquick gel extraction kit. HNP-2, HPN-4 and hepcidin cDNA were ligated individually to the silk 6mer present in the expression vector pET30L using T4 DNA ligase enzyme (New England Biolabs, M0202S).
Escherichia coli DH5α cells (Invitrogen, 18258-012) were transformed with the ligation products and successful transformants were identified by plating on agar plates containing 25 μg/mL kanamycin. The presence of the HNP-2, HPN-4 and hepcidin inserts was confirmed by DNA sequencing (Tufts Core Facility, Boston, MA) using T7 primers and the new constructs were named: 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin.
2.2. Protein Expression and Purification
The 6mer, 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin were expressed in E. coli RY-3041 strain, a mutant strain of E. coli BLR(DE3) defective in the expression of SlyD protein [47, 48]. SlyD protein was reported in previous studies as co-eluting with silk proteins during purification by Ni-NTA affinity chromatography due to the histidine content, an amino acid with known affinity for Ni-NTA columns. In E. coli RY-3041 strain the C-terminus, with the high content of histidines, of the SlyD protein was deleted to avoid co-elution with expressed recombinant proteins with his fusion tags [48]. Cells were cultivated at 37°C in LB medium, with 25 μg/mL kanamycin until an OD600 between 0.9 and 1 was reached. At this point expression was induced with isopropyl β-D-thiogalactoside (IPTG, Invitrogen, 15529019) to a final concentration of 0.5 mM. After 2 hours of expression the cells were harvested by centrifugation at 6500 rpm. The cell pellet was resuspended in denaturating buffer (100 mM NaH2PO4, 10 mM Tris HCl, 8 M urea, pH 8.0) and left overnight with stirring for complete cell lysis. Insoluble cell fragments and soluble proteins present in the cell lysate were separated through centrifugation at 11000 rpm. The supernatant was mixed with Ni-NTA resin (Qiagen, 30250) and left for 2 hours with stirring. The supernatant/Ni-NTA resin mixture was loaded onto a glass Econo-column (Biorad) and washed several times with denaturating buffer at pH 8.0 and then pH 6.0. The 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin proteins were eluted using denaturating buffer at pH 4.5. The purified proteins were dialysed first in a 20 mM sodium acetate buffer followed by extensive dialysis in MQ water using cellulose ester snake skin membranes with a 100-500 Da molecular weight cut off (Spectra/Por Biotech, 131054). The dialyzed proteins were lyophilized in a LabConco lyophilizer. Protein sequencing (Tufts Core Facility, Boston, MA), SDS-PAGE and western blot were used to confirm protein identity.
2.3. Protein identification by western blot
Proteins were mixed with NuPAGE LDS sample buffer (Invitrogen, NP0007) and heated to 80°C for 10 minutes. The samples were separated using a Bis-tris 4-12% gel (Invitrogen, NP0321BOX). For the western blot the His-Tag AP western kit provided by Novagen, EMD Biosciences, NJ, USA (70972) was used with the protocol provided. Briefly, bands were electro-transferred onto a nitrocellulose membrane at 30V for 16 hours. Membranes were blocked for one hour with 3% bovine serum albumin (BSA) in TBS buffer (blocking solution). After washing with tris buffered solution (TBS buffer) the membrane was incubated for 1 hour with mouse anti-histidine monoclonal antibody (Novagen) diluted 1:1000 in blocking solution. Following proper washing with TBSTT buffer (TBS with tween 20) the membrane was incubated for 1 hour with goat anti-mouse IgG AP conjugated antibody diluted 1:5000 in blocking solution. Colorimetric detection was performed with developing solution of 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitrobluetetrazolium (NBT).
2.4. Film Formation and ATR-FTIR Secondary Structure Analysis
Recombinant 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer proteins were dissolved in MQ water to a final concentration of 2% (w/v). Then 60 μl of each protein solution was cast onto a non adherent polystyrene surface and left to dry at room temperature. The films obtained were treated with 70% methanol for 2 hours to induce the transition of secondary structure from random coil to β-sheet. This treatment produced films with improved mechanical properties and more resistant to dissolution in water or other aqueous environments such as culture media. For cell studies the films were treated with 70% (w/w) ethanol solution for sterilization purposes. ATR-FTIR (Jasco Inc., MD, USA, model FT/IR-6200) was performed before and after methanol treatment of the films to investigate secondary structure. Spectra were collected in absorption mode at 8 cm-1 resolution and a 4000 to 400 cm-1 region was scanned. The quantification of the secondary structure was based on the analysis of the amide I and amide II regions (1700 to 1450 cm-1). The average percentage for the secondary structures, mainly β–sheet content for the 6mer (control situation), 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin proteins was calculated through the integration of the area of each deconvoluted curve followed by the normalization of the obtained value to the total area of the amide I and amide II regions [49]. OPUS deconvolution software (Bruker optics, Billerica, MA, USA) was used for spectrum deconvolution and each deconvoluted spectrum was curve-fitted with Gaussian bands using the peak pick function. After curve-fitting information concerning the percentage of amide I and II regions, bandwidth and band position, referring to β–sheet or α-helix conformations, was obtained [50].
2.5. Antibacterial Assay
The purified recombinant proteins were tested for antimicrobial activity against E. coli (American-Type Culture Collection ATCC 25922, VA, USA,) and S. aureus (ATCC 25923), purchased from American-Type Culture Collection. The radial diffusion assay [51] was used to test three concentrations of the purified proteins dissolved in phosphate buffer (pH 7.4): 10, 50 and 100 μg/mL. The 6mer protein solutions were used as controls. E. coli and S. aureus lawns were plated on LB-agar plates, using liquid E. coli and S. aureus cultures grown overnight at 37°C. A biopsy punch 8 mm in diameter was use to cut circles from filter paper (Whatman, Piscataway, NJ, USA, 09-845B) that were used as blank susceptibility disks. For sterilization, the disks were immersed in a 70% (v/v) ethanol solution for 1 hour and left to dry for two hours in a hood. After drying, the discs were immersed in the different protein solutions, placed on the bacterial lawns and incubated overnight at 37°C. The zones of growth inhibition formed around the susceptibility disks were measured using Image J software. The 6mer protein was used as a negative control in order to exclude the effect from silk itself and from possible contaminants derived from the expression and purification processes. Experiments were carried out in triplicate.
2.6. Aggregation
A Zetasizer NanoZS instrument (ZEN3600, MALVERN Instruments, Worcestershire, UK) was used for DLS measurements. DLS was performed on solutions at the same concentrations used for the antibacterial assay (section 2.5): 10, 50 and 100 μg/mL. Samples were prepared with phosphate buffer (pH 7.4). The scattering light was collected at a 173° scattering angle.
2.7. Circular Dichroism Spectroscopy
Circular Dichroism (CD) spectroscopy was performed on an Aviv, Model 410 (Biomedical, Inc. NJ USA) instrument. The spectra were collected between 260 and 180 nm with a step size of 1 nm, an averaging time of 1s and five scans were collected for each sample. A baseline spectrum was subtracted from the samples. Sample cells with a 0.1 cm path length were used and measurements were performed with 1 mg/mL protein solutions in phosphate buffer (pH 7.4).
2.8. Cytotoxicity Assay
Human osteosarcoma cell line (SaOs-2) is an immortalized cell line with osteoblastic phenotype (HTB-85) and was purchased from the American-Type Culture Collection. Cells were cultured using basal medium consisting in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% penicillin-streptomycin (v/v), at 37°C with 5% CO2 in a humidified environment. After reaching confluence, cells were harvested with trypsin/EDTA and after counting cells they were seeded at 3.0×103 cells/cm2 in a 96 well plate coated with the four different types of protein films, 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin, 6mer (control) films, prepared as in section 2.3. Tissue culture plate (TCP) was used as a positive control in this assay. The Alamar blue (Invitrogen, DAL1025) assay was used to determine cell viability/proliferation after three days of culture. Alamar Blue reagent was added to the growth media in a 1:10 dilution ratio and data was collected using fluorescence at 530-560 nm excitation and 590 nm emission [52]. A standard curve with the different cell numbers (10,000, 5,000, 2,500 and 0) on the x axis and the corresponding fluorescence values on the y axis were used to determine the cell numbers as detailed by the supplier.
2.9. Statistical Analysis
SPSS 17.0 was used to perform statistical analysis. The Shapiro-Wilk test was use to test for the normality of the data. To test for significant differences between different experimental groups (6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin, 6mer) one-way ANOVA with a Dunet’s T3 post hoc comparison was used. Statistical significance was defined as p<0.05.
3. Results
3.1. Cloning and Expression of HNP-2, HNP-4 and Hepcidin
The presence of the HNP-2, HNP-4, hepcidin inserts in the cloning vector containing the silk modules was confirmed through DNA sequencing (Tufts Core Facility, Boston, MA). SDS-PAGE and Western blots (Figure 2) indicated that both expression and purification of 6mer (control), 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin proteins was successful and protein sequencing at Tufts Core Facility, Boston, MA confirmed the N-terminal amino acid sequence for the all three proteins. For 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer proteins the theoretical molecular weights were approximately 24.6, 25, 24.1 and 21.8kDa, respectively, and with SDS-PAGE and Western blot assays bands were observed at around 28kDa (Figure 2). Furthermore, with SDS-PAGE and Western blot the presence of dimers and multimers corresponding to aggregation products of the monomer forms were also observed, together with pre-terminated sequences representing proteins whose expression was incomplete and therefore present a lower molecular weight than the monomeric complete proteins. After purification, dialysis and lyophilization the yield of 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin were approximately 12 mg/L for each of the proteins, and for the 6mer control protein the yield was 25 mg/L.
Figure 2.
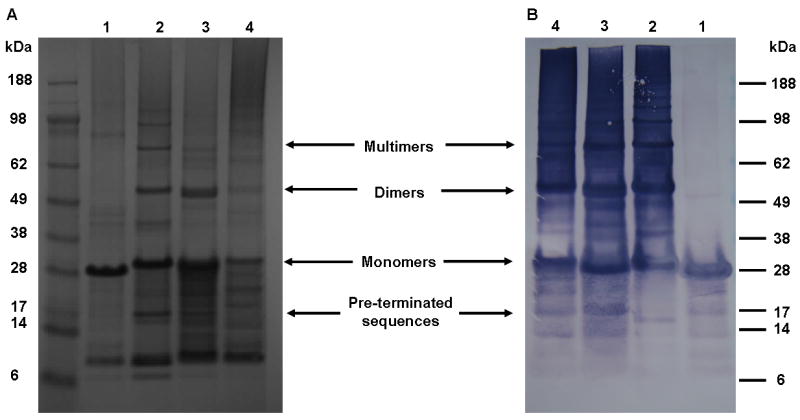
SDS-PAGE gel stained with colloidal blue (A) and western-blot using anti-histidine antibody (B). 1 - 6mer, 2 - 6mer+hepcidin, 3 - 6mer+HNP-2 and 4 - 6mer+HNP-4.
3.2. Film Formation and Secondary Structure Analysis
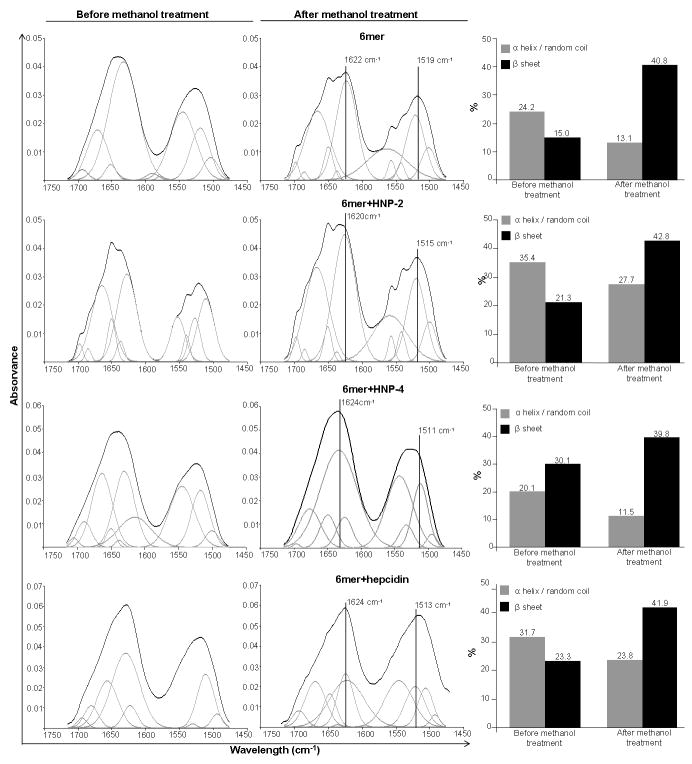
The ATR-FTIR spectra for the 6mer (control), 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin films, before and after methanol treatment, exhibited strong amide I (1700-1600 cm1) and amide II (1600-1500 cm-1) regions (Figure 3). Before methanol treatment the ATR-FTIR spectra for 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer protein films exhibited vibrational modes in the range of 1650-1647 cm-1, amide I region, indicative of helix/random coil conformations. After methanol treatment two peaks appeared, one in the range of 1620-1630 cm-1, amide I region, and another in the range of 1510-1530 cm-1, amide II region. Both peaks were indicative of antiparallel β-sheet structures [47, 53, 54]. Spectra deconvolution revealed the percentage of helix/random coil conformations and of antiparallel β-sheet structures for the 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and for 6mer films before and after methanol treatment (Figure 3). For all four films an increase of β-sheet content was observed with a corresponding decrease in the helix/random coil conformation after methanol treatment (Figure 3). The β-sheet content after methanol treatment was similar for all four constructs ranging between 39.8% for 6mer+HNP-2 and 42.8% for 6mer+HNP-4. For the 6mer and the 6mer+hepcidin films the content was 40.8% and 41.9%, respectively.
Figure 3.
ATR-FTIR spectra of 6mer, 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin films before and after treatment with 70% methanol and respective percentage of β-sheet and random coil/helix conformations after ATR-FTIR spectra deconvolution.
3.3. Antimicrobial Activity of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin Proteins
The antimicrobial activity of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin against the Gram- E. coli and the Gram+ S. aureus was studied using the radial diffusion assay. The three proteins displayed antimicrobial activity against E. coli with the formation of inhibition zones at all three concentrations tested (Figures 4,5). In the case of E. coli, 6mer+HNP-4 and 6mer+hepcidin registered higher bactericidal values when compared with the 6mer+HNP-2 peptide. For all three proteins the highest antimicrobial activity was at a concentration of 10 μg/mL (p<0.05). Against S. aureus there was a decrease in bactericidal activity for all the three peptides when compared to E. coli, based on the smaller diameter inhibition zone, especially in the case of the 6mer+hepcidin and 6mer+HNP-2 (Figure 5). For the proteins tested there was a decrease in the bactericidal activity from 10 to 100 μg/mL (p<0.05). The 6mer protein was used as a negative control and no inhibition zones were detected for the three concentrations tested (Figure 5).
Figure 4.
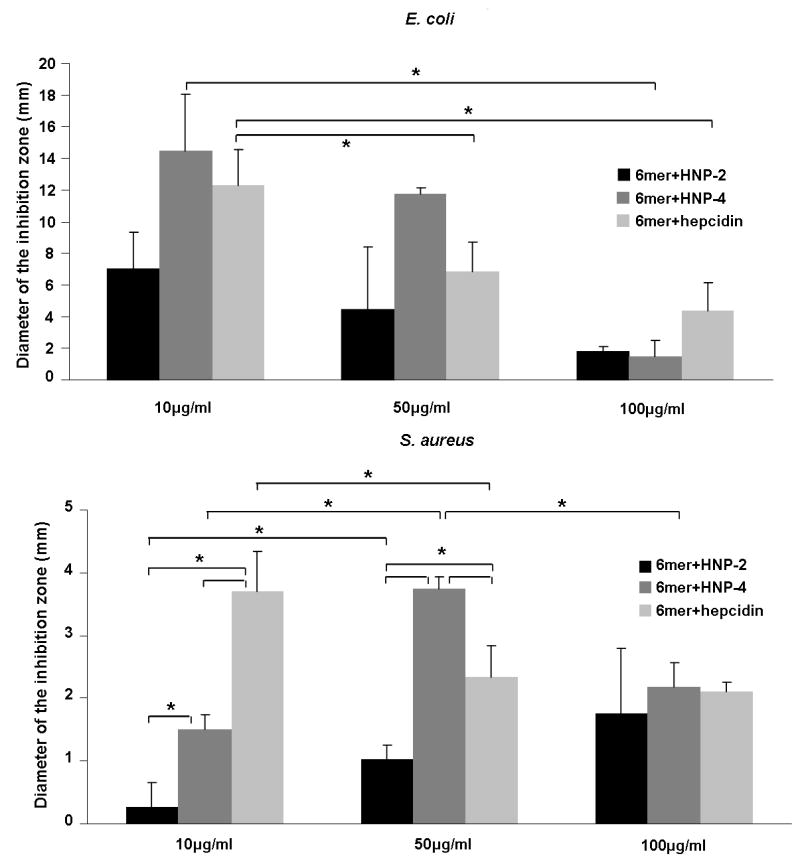
Graphics representing the antimicrobial activity of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin using a radial diffusion assay. Recombinant 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin proteins were used in concentrations of 10 μg/ml, 50 μg/ml and 100 μg/ml against E. coli and S. aureus. 6mer was used as control. As mentioned in section 3.4. No inhibitory effect was detected for the 6mer protein and for that reason it was not included in the graphic representation. * stands for p<0.05.
Figure 5.
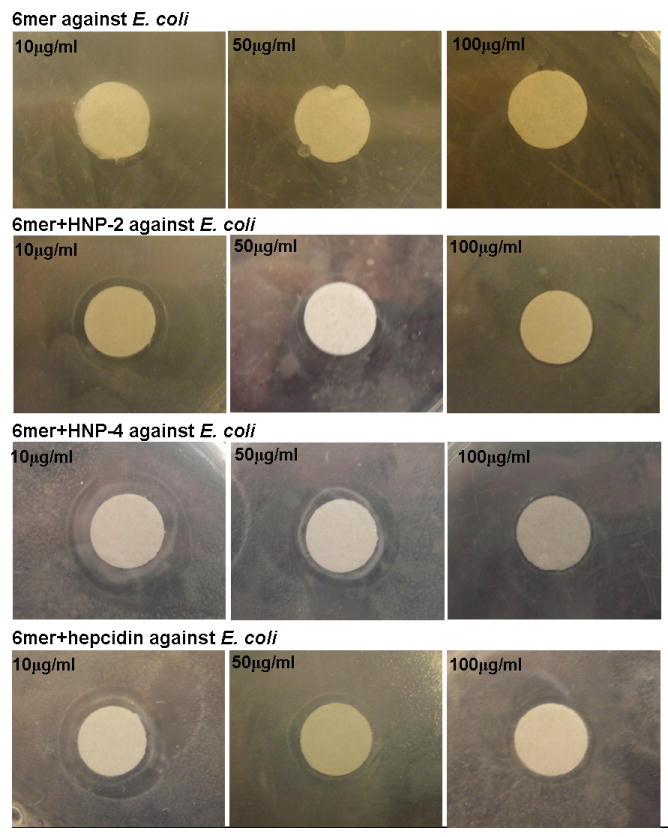

Images showing the antimicrobial activity of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin using radial diffusion assay. Recombinant 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin proteins were used at concentrations of 10 μg/ml, 50 μg/ml and 100 μg/ml against E. coli and S. aureus. 6mer was used as control. Discs were 8 mm in diameter.
3.4. Aggregation
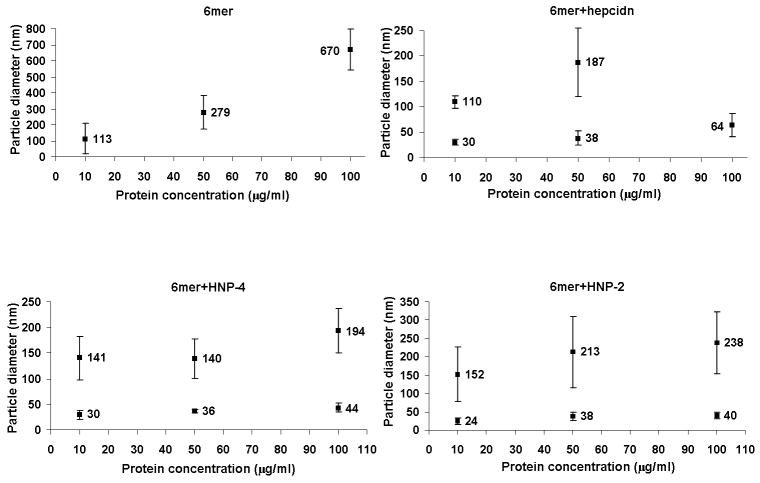
For the 6mer, 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin, DLS measurements detected an increase in particle diameter with increasing protein concentration, indicating that aggregation was dependent on protein concentration (Figure 6). In the case of the 6mer, only one peak was detected for each of the three concentrations measured. In contrast, for 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin two peaks were always detected, except for 6mer+hepcidin at concentration of 100 μg/mL. These data indicated the coexistence of smaller (between 24 and 44 nm) and larger (between 110 and 238 nm) aggregates, the size of which decreased with decreasing protein concentration.
Figure 6.
DLS measures of particle diameter (nm) as a function of protein concentration.
3.5. Circular Dichroism Spectroscopy
CD spectroscopy was carried out for the four recombinant proteins, 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer, in phosphate buffer (pH 7.4). The spectra of the 6mer indicated a dominant β-hairpin conformation characterized by a negative ellipticity with a minimum at approximately 203 nm (Figure 7). In the case of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin the spectra suggested the presence of both β and α conformations with two minima at 204-208 and 220nm [55-58].
Figure 7.
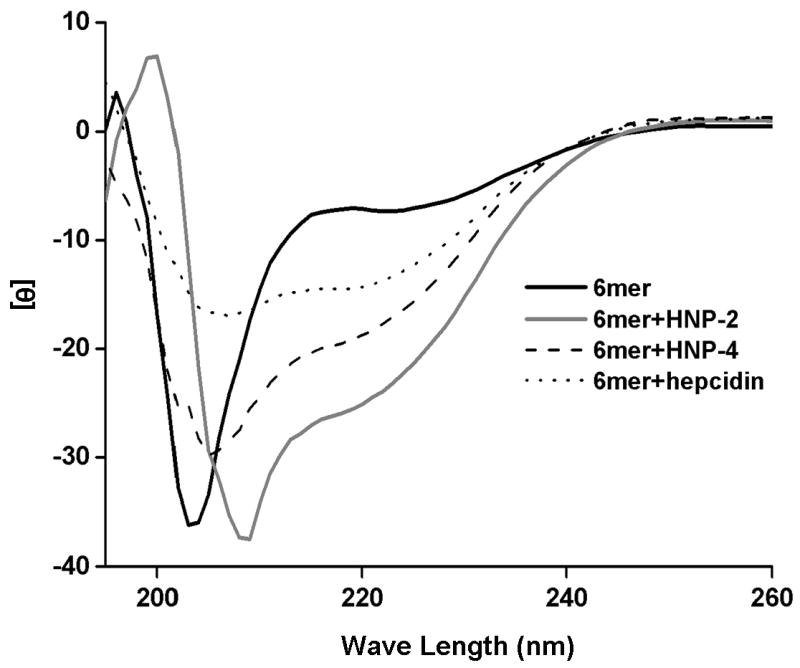
CD spectra for the recombinant 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer.
3.6. Cytotoxicity
Cell viability/proliferation with the SaOs-2 cells was measured after 3 days of culture using the Alamar Blue assay. The results indicated that there were no significant differences (p<0.05) in cell viability/proliferation between cells cultured on 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer films. Furthermore, when cell viability/proliferation results (expressed in cell number) for 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin films were compared with the results for the positive control TCP, only the 6mer+HNP-4 films had a cell number significantly lower than TCP, p<0.05 (Figure 8).
Figure 8.
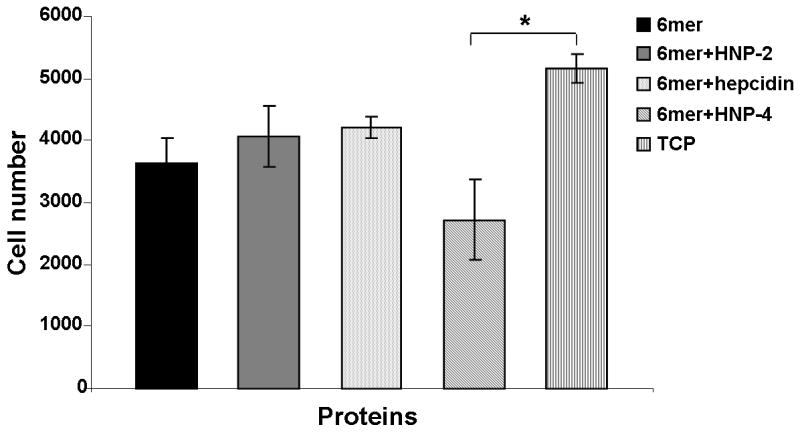
Alamar blue for cells cultured on 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer films after three days. TCP stands for tissue culture plastic, used as a positive control, and * stands for p<0.05.
4. Discussion
The design and successful expression of three new chimeric proteins is presented through the fusion of the consensus repeat for spider silk protein MaSp1 (6mer) with hepcidin, HNP-2 and HNP-4 (Figure 1). SDS-PAGE and Western blots showed the presence of bands around 28 kDa (Figure 2). The difference between the expected and the observed molecular weights for 6mer+HNP-2 (24.6 kDa), 6mer+HNP-4 (25 kDa) and 6mer+hepcidin (24.1 kDa) proteins is probably due to the formation of cysteine bridges which interfere with the migration of the proteins during electrophoresis and also lead to the formation of multimers. This behaviour was also detected for other proteins such as bone morphogenetic proteins with six cysteine residues involved in the formation of intra and inter-molecular disulphide bonds [59, 60]. Furthermore, additional DLS measurements with silk proteins in SDS solution showed the formation of aggregates (data not shown) which can also interfere with the migration of these proteins during electrophoresis and lead to the formation of higher molecular weight structures as well as multimers.
ATR-FTIR was used to confirm the integrity of the 6mer sequence in the three proteins (Figure 3). The spectra collected for the three new proteins films, 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin, demonstrated the presence of β-sheet, with peaks in 1620-1630 cm-1 for amide I region and 1510-1530 cm-1 for amide II region [47, 53, 54], after methanol treatment. Furthermore, spectral deconvolution indicated an increase of β-sheet content for 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin, after methanol treatment, reaching values similar to those obtained for the 6mer control (Figure 3); with 39.8% for 6mer+HNP-2, 40.8% for the 6mer, 42.8% for 6mer+HNP-4 and 41.9% in the case of 6mer+hepcidin. These data demonstrate that the core self-assembling spider silk domain retained function related to the generation of beta sheet crystals in these new fusion systems. This is important for the material performance of these new proteins to attain self-standing material formats, such as the films studied here.
The three new fusion proteins, 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin, also retained antimicrobial activity attributed to the inserted motifs HNP-2, HNP-4 and hepcidin, respectively (Figure 4 and 5). Previous studies have demonstrated the efficacy of HNP-2, HNP-4 and hepcidin against different strains of E. coli [32]. 6mer+HNP-4 presented the highest antibacterial activity, except for the concentration 100 μg/mL, whereas 6mer+HNP-2 manifested the lowest bactericidal activity. These data support the functional attributes of the antimicrobial component of the fusion proteins, the second critical component of the protein polymer designs, to add to the silk material properties above. Additionally, besides testing the concentrations of 10, 50 and 100 μg/mL other concentrations were also evaluated (data not shown for, 1 and 5 μg/mL), however the antimicrobial effect was either very reduced or nonexistent. For this reason the concentration of 10 μg/mL was selected in the present study as the lower concentration with an antimicrobial effect.
The three proteins had a decrease in their antimicrobial activity against S. aureus when compared to E. coli. This reduction in activity was also detected previously in the case of hepcidin. Park and co-workers isolated two different hepcidin peptides, hepcidin 20 and hepcidin 25, from human urine and tested their bactericidal activity against different bacterial strains, E. coli ML35p and S. aureus. Both peptides were antimicrobial against E. coli ML35p but showed a reduction in activity against S. aureus [32]. Ericksen and colleagues [61] studied the activity of different α-defensins (HNP-1, HNP-2, HNP-3, HNP-4, HD-5 and HD-6) against different bacterial strains and demonstrated that HNP-4 was the most effective against Gram- bacterial strains, one of which was E. coli ATCC 25922, the same strain as used in this present study.
Furthermore, in the present work for a concentration of 10 μg/mL the 6mer+hepcidin was more efficient against S. aureus than the other two proteins. However, with an increase in the protein concentration from 10 to 50 μg/mL the 6mer+HNP-4 became more effective. For both E. coli and S. aureus 6mer+HNP-2 proved to be the less bactericidal. Wilde and colleagues [62] compared the antimicrobial activities of HNP-4 and a mixture of HNP-1, HNP-2 and HNP-3 and concluded that HNP-4 was approximately 100-fold more potent against Gram- E. coli and about 4-fold more potent against Gram+ Streptococcus faecalis. This difference in efficacy may be one of the reasons why human neutrophils produce HNP-1, HNP-2 and HNP-3 at a 60-fold greater abundance when compared with the production of HNP-4 [61, 63]. These results are similar to those obtained in the present work where 6mer+HNP-4 had higher antimicrobial activity when compared with 6mer+HNP-2.
Additionally, in order to facilitate the comparison between the present study and previous studies, using the same antimicrobial domains, the peptide concentrations studied here were kept within the same range of concentrations as those used previously [35, 36, 61, 62]. However, in the present study and in contrast to what was observed in previous reports, the antimicrobial activity of the new chimeric proteins was not always concentration dependent. The presence of the 6mer silk domain explains this behaviour, since silk proteins are known to aggregate in solution with increased concentration. The 6mer protein, similar to the silk fibroin collected from the silk worm Bombyx mori, is rich in hydrophobic amino acids having a high percentage of the hydrophobic amino acids alanine and glycine. The presence of these hydrophobic cores induce the assembly of 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin proteins toward the more thermodynamically stable assembly resulting in a structural reorganization with reduction of exposure of hydrophobic residues [64]. In the case of the silk domains, this stability is achieved by self-assembly or by aggregation. In the present study a relationship between protein concentration and size of the aggregates was found as measured by DLS (Figure 6). The 6mer registered the highest values of aggregate size for the 50 and 100 μg/mL concentrations with an average of 279 and 670 nm, respectively. The other three proteins, 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin, presented bi-modal distributions of sizes, except for 6mer+hepcidin at a concentration of 100μg/mL. In most cases the aggregate size generated by these proteins was smaller than in the case of the 6mer alone. Moreover, the dependence between protein concentration and aggregatesd size for 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin was not as evident as in the case of the 6mer alone. This fact can be explained by the presence of the antimicrobial domain which increases the number of charged amino acid residues resulting in a reduction of the tendency of these proteins to aggregate by enhancing interactions with the solvent. A previous study where DLS was applied to the dynamic behaviour and the conformational characteristics of B. mori silk fibroin demonstrated that this molecule forms large and stable aggregates even in very dilute solutions, supporting the tendency of silk molecules to form aggregates even in dilute conditions [65].
In the present work the decrease in antimicrobial activity of the new chimeric proteins as their concentration in solution increased can be explained by the occurrence of aggregation resulting in larger and more stable aggregates as protein concentration was increased. This interaction leads to a depletion of protein monomers in solution leaving fewer molecules free in solution to act against the bacteria. A possible reason for the decrease in antimicrobial activity is the absence of post-translational modifications leading to the formation of disulphide bonds between cysteine residues. The results obtained by Varkey and co-workers with HNP-1 peptides indicate that the presence of all three disulphide bonds and the order of connectivity are not essential for maintaining the activity of HNP-1. Also, linear HNP-1 peptides maintained antibacterial activity, however there was a 10 to 20-fold reduction in activity [55].
CD analyses indicated that in aqueous medium 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin displayed both β-hairpin and α conformations. In the case of the 6mer β-hairpin or β-sheet are the predominant conformations (Figure 7) [55-58]. Previous CD conformation studies with synthetic HNP-1 in aqueous medium showed a spectrum with a minimum of 205 nm indicating the presence of β conformation [66]. β structure was also observed for HNP-1 peptides with two or three disulphide bonds [66]. These spectra have a minimum at 215 nm and differ from the ideal β-sheet spectra due to distortions and structural flexibility [66]. Additionally, a later study also with HNP-1 peptides dissolved in water showed spectra characteristic of an unordered structure. However, when these peptides are dissolved in sodium dodecyl sulphate or trifluoroethanol their spectra showed the presence of both β and α conformations with minimums at 205 and 223 nm [55], very similar to the spectra obtained for 6mer+HNP-2 and 6mer+HNP-4 proteins in the present study. In the case of hepcidin a previous study showed a CD spectrum for this peptide in phosphate buffer (pH 7.4) with β-turns, loops and β-sheet [32]. In the present study the spectrum for 6mer+hepcidin showed both β and α conformations.
For quantitative evaluation of the cytotoxic response to the chimeric proteins, the osteosarcoma cell line SaOs-2 was cultured on 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin films. The 6mer+HNP-2 and 6mer+hepcidin 2% films showed the same ability as TCP to support viable cells (p>0.05), while the 6mer+HNP-4 films showed a significant difference (p<0.05) from TCP (Figure 8). No significant difference was observed between TCP and 6mer+HNP-2, 6mer+HNP-4, 6mer+hepcidin and 6mer films. These results indicated that these chimeric proteins, when processed into films, are capable of supporting cell proliferation, and the bactericidal activity does not impact mammalian cell function.
Many drug delivery systems for antibiotics have been described. Hydroxyapatite microspheres can be loaded with an antibiotic and used as a local drug delivery system for the treatment of periodontitis [67], chronic osteomyelitis [68], as just two examples. Nonetheless, hydroxyapatite-carrying systems tend to have a initial burst releasing the entire antibiotic in the first hours [67] and in many cases the antibiotics elute only for a short period of time [69, 70]. This can be problematic if the final purpose is to obtain a more controlled release of the drug through time. In recent studies, different biopolymers have also been tested for potential applications as drug delivery systems. Silk fibroin [71, 72], collagen [73, 74] and hyaluronic acid [75] are examples of natural polymers that have been used as a carrier systems for growth factors, such as bone morphogenetic proteins (BMPs), and antibiotics, in the form of micro-particles, membranes and scaffolds.
Antibiotic-doped biomaterials can be used to address growing concerns with the selection and spreading of multiresistant pathogens [76]. Antimicrobial peptides, as new chemotherapeutic molecules due to their activity against different types of microorganisms, are also important options, including antimicrobial functions as well as immune response [77] and wound repair [20] impacts. However, due to their peptidic nature these biomolecules are more susceptible to proteolytic digestion [78]. Therefore, their combination with a suitable delivery system, such as the one presented in this work, may represent an advantage by decreasing proteolitic digestion [79] and by allowing for more controlled release of these peptides to prolong effectiveness over time. This more controlled release is important in maintaining regional or localized effects, more precisely at the interface between the implant/host tissues, where bacterial colonization and posterior inflammation development first take place [76].
Conclusions
The present study outlines the formation of new silk-based chimeric proteins with antimicrobial potential. Secondary structure analysis, performed by ATR-FTIR, indicated that silk maintains β-sheet formation capability even after adding the antimicrobial domains, HNP-2, HNP-4 and hepcidin. The maintenance of the capacity to form β-sheet is important, since these physical cross-links are responsible for the exceptional mechanical properties, stability and slow degradability of silk. Moreover, activity tests against Gram- and Gram+ organisms showed that the antimicrobial domains present in 6mer+HNP-2, 6mer+HNP-4 and 6mer+hepcidin proteins maintained bactericidal activity. Cytotoxicity/proliferation studies demonstrated that the new proteins were capable of sustaining the proliferation of mammalian cells. These new chimeric proteins suggest a new multifunctional approach to generate biomaterials with useful properties, in this case, control of infections due to the addition of the antimicrobial peptides.
Acknowledgments
The authors acknowledge Olena Rabotyagova for advice in protein sequence design. Sílvia Gomes thanks the Portuguese Foundation for Science and Technology (FCT) for providing her a PhD grant (SFRH/BD/28603/2006). This work was carried out under the scope of the European NoE EXPERTISSUES (NMP3-CT-2004-500283), the FIND & BIND project funded by the agency EU-EC (FP7 program), the FCT R&D project ProteoLight (PTDC/FIS/68517/2006) funded by the FCT agency, the Chimera project (PTDC/EBB-EBI/109093/2008) funded by the FCT agency, the NIH (P41 EB002520) Tissue Engineering Resource Center and the NIH (EB003210 and DE017207).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hodgkinson R. Internal fixation of fractures of long bones with metallic devices. Med J Aust. 1961;48(1):691–693. doi: 10.5694/j.1326-5377.1961.tb69062.x. [DOI] [PubMed] [Google Scholar]
- 2.Osbon O, Lilly G, Thompson C, Jost T. Bone grafts with surface decalcified allogeneic and particulate autologous bone: report of cases. J Oral Surg. 1977;35(4):276–284. [PubMed] [Google Scholar]
- 3.Kohna J, Welshb WJ, Knight D. A new approach to the rationale discovery of polymeric biomaterials. Biomaterials. 2007;28(29):4171–4177. doi: 10.1016/j.biomaterials.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams DJ, Sebastine IM. Tissue engineering and regenerative medicine: manufacturing challenges. IEE Proc Nanobiotechnol. 2005;156(6):207–210. doi: 10.1049/ip-nbt:20050001. [DOI] [PubMed] [Google Scholar]
- 5.Santos MI, Pashkuleva I, Alves CM, Gomes ME, Fuchs S, Unger RE, et al. Surface-modified 3D starch-based scaffold for improved endothelialization for bone tissue engineering. J Mater Chem. 2009;19(24):4091–4101. [Google Scholar]
- 6.Santo VE, Frias AM, Carida M, Cancedda R, Gomes ME, Mano JF, et al. Carrageenan-based hydrogels for the controlled delivery of PDGF-BB in bone tissue engineering applications. Biomacromolecules. 2009;16(6):1392–1401. doi: 10.1021/bm8014973. [DOI] [PubMed] [Google Scholar]
- 7.Reis RL, Neves NM, Mano JF, Gomes ME, Marques AP, Azevedo HS. Natural-based polymers for biomedical applications: Woodhead Publishing. 2008 [Google Scholar]
- 8.Mano JF, Silva GA, Azevedo HS, Malafaya PB, Sousa RA, Silva SS, et al. Natural origin biodegradable systems in tissue engineering and regenerative medicine: present status and some moving trends. J R Soc Interface. 2007;4(17):999–1030. doi: 10.1098/rsif.2007.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris LD, Kim B-S, Mooney DJ. Open pore biodegradable matrices formed with gas foaming. J Biomed Mater Res B Appl Biomater. 1998;42(3):396–402. doi: 10.1002/(sici)1097-4636(19981205)42:3<396::aid-jbm7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 10.Suh H. Recent advances in biomaterials. Yonsei Med J. 1998;39(2):87–96. doi: 10.3349/ymj.1998.39.2.87. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Wong C, George A, Kaplan DL. The effect of genetically engineered spider silk-dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation. Biomaterials. 2007;28(14):2358–2367. doi: 10.1016/j.biomaterials.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Hedhammar M, Blom T, Leifer K, Johansson J, Habibovic P, et al. Biomimetic calcium phosphate coatings on recombinant spider silk fibres. Biomed Mater. 2010;5(4):1–10. doi: 10.1088/1748-6041/5/4/045002. [DOI] [PubMed] [Google Scholar]
- 13.Foo CWP, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, et al. Novel nanocomposites from spider silk–silica fusion (chimeric) proteins. Proc Natl Acad Sci USA. 2006;103(25):9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marr AK, Gooderham WJ, Hancock REW. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6(5):468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Huang HW. Action of antimicrobial peptides: two-state model. Biochemistry. 2000;39(29):8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 16.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462(1-2):55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 17.Hancock REW, Chapple DS. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Befus AD, Mowat C, Gilchrist M, Hu J, Solomon S, Bateman A. Neutrophil defensins induce histamine secretion from mast cells: mechanisms of action. J Immunol. 1999;163(2):947–953. [PubMed] [Google Scholar]
- 19.Grigat J, Soruri A, Forssmann U, Riggert J, Zwirner J. Chemoattraction of macrophages, T lymphocytes, and mast cells is evolutionarily conserved within the human α-defensin family. J Immunol. 2007;179(6):3958–3965. doi: 10.4049/jimmunol.179.6.3958. [DOI] [PubMed] [Google Scholar]
- 20.Aarbiou J, Verhoosel RM, Wetering Sv, Boer WId, Krieken JHJMv, Litvinov SV, et al. Neutrophil defensins enhance lung epithelial wound closure and mucin gene expression In vitro. Am J Respir Cell Mol Biol. 2004;30(2):193–201. doi: 10.1165/rcmb.2002-0267OC. [DOI] [PubMed] [Google Scholar]
- 21.Aarbiou J, Ertmann M, Wetering S, Noort P, Rook D, Rabe KF, et al. Human neutrophil defensins induce lung epithelial cell proliferation in vitro. J Leukoc Biol. 2002;72(1):167–174. [PubMed] [Google Scholar]
- 22.Murphy CJ, Foster BA, Mannis MJ, Selsted ME, Reid TW. Defensins are mitogenic for epithelial cells and fibroblasts. J Cell Physiol. 1993;155(2):408–413. doi: 10.1002/jcp.1041550223. [DOI] [PubMed] [Google Scholar]
- 23.Chavakis T, Cines DB, Rhee J-S, Liang OD, Schubert U, Hammes H-P, et al. Regulation of neovascularization by human neutrophil peptides (α-defensins): a link between inflammation and angiogenesis. FASEB J. 2004;18(11):1306–1308. doi: 10.1096/fj.03-1009fje. [DOI] [PubMed] [Google Scholar]
- 24.Bals R. Epithelial antimicrobial peptides in host defense against infection. Respir Res. 2000;1(3):141–150. doi: 10.1186/rr25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider JJ, Unholzer A, Schaller M, Schäfer-Korting M, Korting HC. Human defensins. J Mol Med. 2005;83(8):587–595. doi: 10.1007/s00109-005-0657-1. [DOI] [PubMed] [Google Scholar]
- 26.Smet KD, Contreras R. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol Lett. 2005;27(18):1337–1347. doi: 10.1007/s10529-005-0936-5. [DOI] [PubMed] [Google Scholar]
- 27.Selsted ME, Harwig SSL, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest. 1985;76(4):1436–1439. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267(15):23216–23225. [PubMed] [Google Scholar]
- 29.Pazgier M, Lubkowski J. Expression and purifcation of recombinant human α-defensins in Escherichia coli. Protein Expr Purif. 2006;49(1):1–8. doi: 10.1016/j.pep.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Cocchia F, Gentlesa D, Ericksena B, Lubkowskib J, DeVicoa A, et al. Human neutrophil a-defensin 4 inhibits HIV-1 infection in vit. FEBS Lett. 2005;579(1):162–166. doi: 10.1016/j.febslet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 31.Krause A, Neitz S, Mägert H, Schulz A, Forssmann W, Schulz-Knappe P, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Letters. 2000;480:147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 32.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276(11):7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 33.Koliaraki V, Marinou M, Samiotaki M, Panayotou G, Pantopoulos K, Mamalaki A. Iron regulatory and bactericidal properties of human recombinant hepcidin expressed in Pichia pastori. Biochimie. 2008;90(5):1–10. doi: 10.1016/j.biochi.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Krause A, Neitz S, Mägert H, Schulz A, Forssmann W, Schulz-Knappe P, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480(2-3):147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 35.Wallace DF, Jones MD, Pedersen P, Rivas L, Sly LI, Subramaniam VN. Purification and partial characterisation of recombinant human hepcidin. Biochimie. 2006;88(1):31–37. doi: 10.1016/j.biochi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Yuan Q, Zhu Y, Ma R. Expression and preparation of recombinant hepcidin in Escherichia coli. Protein Expr Purif. 2005;41(2):409–416. doi: 10.1016/j.pep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Xu M, Lewis RV. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci USA. 1990;87(18):7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheibel T. Spider silks: recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. Microb Cell Fact. 2004;3(1):14–24. doi: 10.1186/1475-2859-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, Wang B. Biodegradation of silk biomaterials. Int J Mol Sci. 2009;10(4):1514–1524. doi: 10.3390/ijms10041514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerreiro CIPD, Fontes CMGA, Gama M, Domingues L. Escherichia coli expression and purification of four antimicrobial peptides fused to a family 3 carbohydrate-binding module (CBM) from Clostridium thermocellum. Protein Expr Purif. 2008;59:161–168. doi: 10.1016/j.pep.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Ramos R, Domingues L, Gama M. Escherichia coli expression and purification of LL37 fused to a family III carbohydrate-binding module from Clostridium thermocellum. Protein Expr Purif. 2010;71(1):1–7. doi: 10.1016/j.pep.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Haynie SL, Crum GA, Doele BA. Antimicrobial activities of amphiphilic peptides covalently bonded to a water-insoluble resin. Antimicrob Agents Chemother. 1995;39(2):301–307. doi: 10.1128/aac.39.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagheri M, Beyermann M, Dathe M. Immobilization reduces the activity of surface-bound cationic antimicrobial peptides with no influence upon the activity spectrum. Antimicrob Agents Chemother. 2009;53(3):1132–1141. doi: 10.1128/AAC.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appendini P, Hotchkiss JH. Surface modification of poly(styrene) by the attachment of an antimicrobial peptide. J Appl Polymer Sci. 2001;81(3):609–616. [Google Scholar]
- 45.Choa W-M, Joshia BP, Choa H, Lee K-H. Design and synthesis of novel antibacterial peptide-resin conjugates. Bioorg Med Chem Lett. 2007;17(21):5772–5776. doi: 10.1016/j.bmcl.2007.08.056. [DOI] [PubMed] [Google Scholar]
- 46.Gabriel M, Nazmi K, Veerman EC, Amerongen AVN, Zentner A. Preparation of LL-37-grafted titanium surfaces with bactericidal activity. Bioconjug Chem. 2006;17(2):548–550. doi: 10.1021/bc050091v. [DOI] [PubMed] [Google Scholar]
- 47.Rabotyagova O, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10(2):229–236. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 48.Yan S-Z, Beeler JA, Chen Y, Shelton RK, Tang W-J. The regulation of type 7 adenylyl cyclase by Its C1b region and Escherichia coli peptidylprolyl isomerase, SlyD. J Biol Chem. 2001;276(11):8500–8506. doi: 10.1074/jbc.M010361200. [DOI] [PubMed] [Google Scholar]
- 49.Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39(18):6161–6170. [Google Scholar]
- 50.Arrondo J, Goñi F. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog Biophys Mol Biol. 1999;72(4):367–405. doi: 10.1016/s0079-6107(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 51.Lehrer RI, Rosenman M, Harwig SSSL, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137(2):167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 52.Lancaster MV, Fields RD. inventors. Antibiotic and cytotoxic drug susceptibility assays using resazurin and poising agents. 5501959. USA Patent No. 1996
- 53.Foo CWP, Bini E, Huang J, Lee SY, Kaplan DL. Solution behavior of synthetic silk peptides and modified recombinant silk proteins. Appl Phys A Mater Sci Process. 2006;82:193–203. [Google Scholar]
- 54.Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in β-sheet formation in spider silk block copolymers. Macromol Biosci. 2010;10(1):49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- 55.Varkey J, Nagaraj R. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob Agents Chemother. 2005;49(11):4561–4566. doi: 10.1128/AAC.49.11.4561-4566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751(2):119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Cerpa R, Cohen FE, Kuntz ID. Conformational switching in designed peptides: the helix/sheet transition. Fold Des. 1996;1(2):91–101. doi: 10.1016/S1359-0278(96)00018-1. [DOI] [PubMed] [Google Scholar]
- 58.Mamdal M, Nagaraj R. Antibacterial activities and conformations of synthetic α-defensin HNP-1 and analogs with one, two and three disulfite bridges. J Pept Res. 2002;59:95–104. doi: 10.1034/j.1399-3011.2002.01945.x. [DOI] [PubMed] [Google Scholar]
- 59.Bessa PC, Pedro AJ, Klosch B, Nobre A, Griensven Mv, Reis RL, et al. Osteoinduction in human fat-derived stem cells by recombinant human bone morphogenetic protein-2 produced in Escherichia coli. Biotechnol Lett. 2008;30(1):15–21. doi: 10.1007/s10529-007-9501-8. [DOI] [PubMed] [Google Scholar]
- 60.Long S, Truong L, Bennett K, Phillips A, Wong-Staal F, Ma H. Expression, purification, and renaturation of bone morphogenetic protein-2 from Escherichia coli. Protein Expr Purif. 2006;46(2):374–378. doi: 10.1016/j.pep.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 61.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother. 2005;49(1):269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilde CG, Griffith JE, Marra MN, Snable JL, Scott RW. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J Biol Chem. 1989;264(5):11200–11120. [PubMed] [Google Scholar]
- 63.Harwig SS, Park AS, Lehrer RI. Characterization of defensin precursors in mature human neutrophils. Blood. 1992;79:1532–1537. [PubMed] [Google Scholar]
- 64.Calamai M, Canale C, Relini A, Stefani M, Chiti F, Dobson CM. Reversal of protein aggregation provides evidence for multiple aggregated states. J Mol Biol. 2005;346(2):603–616. doi: 10.1016/j.jmb.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 65.Hossain KS, Nemoto N. Dynamic and static light scattering of dilute aqueous solutions of silk fibroin collected from Bombyx mori silkworms. Langmuir. 1999;15:4114–4119. [Google Scholar]
- 66.Mandal M, Nagaraj R. Antibacterial activities and conformations of synthetic α-defensin HNP-1 and analogs with one, two and three disulfite bridges. J Pept Res. 2002;59(3):95–104. doi: 10.1034/j.1399-3011.2002.01945.x. [DOI] [PubMed] [Google Scholar]
- 67.Ferraz MP, Mateus AY, Sousa JC, Monteiro FJ. Nanohydroxyapatite microspheres as delivery system for antibiotics: Release kinetics, antimicrobial activity, and interaction with osteoblasts. J Biomed Mater Res A. 2007;81(4):994–1004. doi: 10.1002/jbm.a.31151. [DOI] [PubMed] [Google Scholar]
- 68.Krisanapiboon A, Buranapanitkit B, Oungbho K. Biocompatability of hydroxyapatite composite as a local drug delivery system. J Orthop Surg. 2006;14(3):315–318. doi: 10.1177/230949900601400315. [DOI] [PubMed] [Google Scholar]
- 69.Kanellakopoulou K, Giamarellos-Bourboulis EJ. Carrier systems for the local delivery of antibiotics in bone infections. Drugs. 2000;59(6):1223–1232. doi: 10.2165/00003495-200059060-00003. [DOI] [PubMed] [Google Scholar]
- 70.Shinto Y, Uchida A, Korkusuz F, Araki N, Ono K. Calcium hydroxyapatite ceramic used as a delivery system for antibiotics. J Bone Joint Surg. 1992;74(4):600–604. doi: 10.1302/0301-620X.74B4.1320622. [DOI] [PubMed] [Google Scholar]
- 71.Bessa PC, Balmayor ER, Azevedo HS, Nürnberger S, Casal M, Griensven Mv, et al. Silk fibroin microparticles as carriers for delivery of human recombinant BMPs. Physical characterization and drug release. J Tissue Eng Regen Med. 2010;4(5):349–355. doi: 10.1002/term.245. [DOI] [PubMed] [Google Scholar]
- 72.Lia C, Veparia C, Jin H-J, Kim HJ, Kaplan DL. Electrospun silk-BMP-2 scaffolds for bone tissue engineering. Biomaterials. 2006;27(16):3115–3124. doi: 10.1016/j.biomaterials.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 73.Ruszczak Z, Friess W. Collagen as a carrier for on-site delivery of antibacterial drugs. Adv Drug Deliv Rev. 2003;55(12):1679–1698. doi: 10.1016/j.addr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 74.Schlapp M, Friess W. Collagen/PLGA microparticle composites for local controlled delivery of gentamicin. J Pharm Sci. 2003;92(11):2145–2151. doi: 10.1002/jps.10460. [DOI] [PubMed] [Google Scholar]
- 75.Luo Y, Kirker KR, Prestwich GD. Cross-linked hyaluronic acid hydrogel films: new biomaterials for drug delivery. J Control Release. 2000;69(1):169–184. doi: 10.1016/s0168-3659(00)00300-x. [DOI] [PubMed] [Google Scholar]
- 76.Campoccia D, Montanaro L, Speziale P, Arciola CR. Antibiotic-loaded biomaterials and the risks for the spread of antibiotic resistance following their prophylactic and therapeutic clinical use. Biomaterials. 2010;31(25):6363–6377. doi: 10.1016/j.biomaterials.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 77.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nature. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 78.Bruschi M, Pirri G, Giuliani A, Nicoletto SF, Baster I, Scorciapino MA, et al. Synthesis, characterization, antimicrobial activity and LPS-interaction properties of SB041, a novel dendrimeric peptide with antimicrobial properties. Peptides. 2010;31(8):1459–1467. doi: 10.1016/j.peptides.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 79.Numata K, Subramanian B, Currie HA, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2010;30(29):5775–5784. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]