Abstract
Two ceramide derivatives, bathymodiolamides A (1) and B (2), were isolated from the deep-sea hydrothermal vent invertebrate mussel Bathymodiolus thermophilus. The molecular structures of these compounds were determined using a combination of NMR spectroscopy, mass spectrometry, and chemical degradation. Biological activities were assessed in a ApopScreen cell-based screen for apoptosis induction and potential anticancer activity. To our knowledge, this is the first report of secondary metabolites from the marine hydrothermal vent mussel B. thermophilus.
The oceans are the Earth's largest ecosystem and hold great, underexplored potential for drug discovery. Within this vast ecosystem, one area remains particularly enigmatic: deep-sea hydrothermal vents, which are characterized by high concentrations of reduced sulfur compounds.1 Life is supported by the growth of chemolithoautotrophic bacteria, capable of oxidizing hydrogen sulfide, hydrogen, and other reduced inorganic compounds to provide energy that is used to fuel carbon dioxide fixation into macromolecules. The vent mussel Bathymodiolus thermophilus is characterized by a considerable flexibility in its sources of nutrition. Symbionts within the gills of this organism were found to be thiosulfate- and sulfide-oxidizing chemoautotrophs.2
In our ongoing effort to discover and develop new marine natural product biomedicinals, we have investigated the ability of the marine hydrothermal vent mussel B. thermophilus to produce novel secondary metabolites.
Thirty specimens of B. thermophilus were collected using the deep submergence vehicle DSV Alvin from an active hydrothermal vent along the Mid-Atlantic Ridge, northern region, at the Lucky Strike location with a latitude of 37°17.63′ N, longitude of 32°16.53′ W, and depth of 1733 m.3 Once aboard the research vessel, the mussels were dissected, and samples of adductor muscle, gill, and mantle tissues were frozen at −70 °C for subsequent analyses. One hundred grams wet weight of the tissues from approximately 20 gills was extracted in MeOH. One part of the MeOH-soluble extract was dissolved in DMSO and was tested for apoptosis induction as assessed by the ApopScreen protocol.4–6 Screening was expected to identify compounds that possess proapoptotic, and potentially anticancer, activity.
The extract induced apoptosis and therefore was fractionated, with subsequent purification by analytical reversed-phase HPLC. Using this strategy, two compounds were isolated. The chemical structures of these two compounds (1 and 2) were ascertained by standard spectroscopic techniques, as described below.
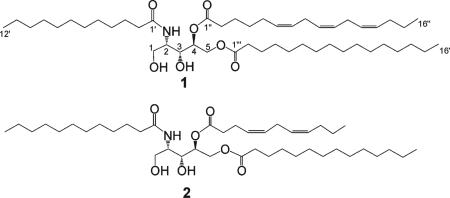
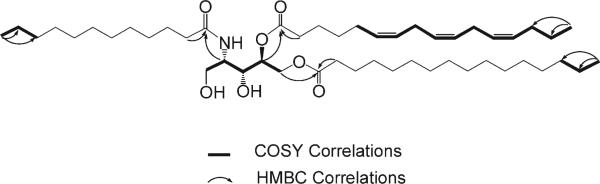
The molecular formula of 1 was established as C49H89NO7 on the basis of HRMALDITOFMS [m/z 804.6712 (M + H)+]. The 13C NMR spectrum for 1 clearly indicated resonances for three ester or amide carbonyl carbons (δC 173.2, 173.4, 173.8), as well as four oxygen-bearing carbons (δC 59.9, 62.5, 63.7, 66.1) and one nitrogen-bearing carbon (δC 53.5, δH 4.29). Analysis of the multiplicity-edited HSQC spectrum for 1 revealed that these heteroatom-substituted carbons comprise two methylenes (CH2-1, δC 63.7, δH 3.65; CH2-5, δC 59.9, δH5a 4.20, δH5b 4.45) and three methines (CH-2, δC 53.5, δH 4.29; CH-3, δC 62.5, δH 4.01; CH-4, δC 66.1, δH 5.25). The HMBC correlations between H-2 and the carbonyl at δC 173.2, H-4 and the carbonyl at δC 173.4, and H-5a and H-5b and the carbonyl at δC 173.8 suggested that 1 has an amide moiety attached to C-2 and two ester moieties attached to C-4 and C-5. Considering the molecular formula and all carbon signals for 1, the remaining oxygen-bearing carbons C-1 and C-3 are substituted with hydroxy groups, resulting in primary and secondary alcohols at C-1 and C-3, respectively. The connectivity of C-1 through C-5 of 1 was established from COSY data as shown in Figure 1, confirming the presence of the amino alcohol moiety in 1.
Figure 1.
Key HMBC and selected COSY correlations for bathymodiolamide A (1).
The remaining partial structures for 1 could be assigned as three acyl moieties. Given the six degrees of unsaturation of 1 based on its molecular formula, three double bonds must be present in one or more of the three acyl moieties. The HMBC correlations from δH 5.35 to δC 26.5 and from δH 2.81 to δC 127.5, taken together with the 4H integral of the signal at δH 2.81 in the 1H NMR spectrum of 1, indicated the occurrence of four bis-allylic protons and two bis-allylic methylene carbons, suggesting that all three double bonds are located in one branch of the aliphatic side chain. Since bis-allylic carbon signals for Z and E isomers are observed at ca. δC 27 and 32, respectively,7,8 the 26.5 ppm shift suggested that all double bonds have a cis geometry (Z).
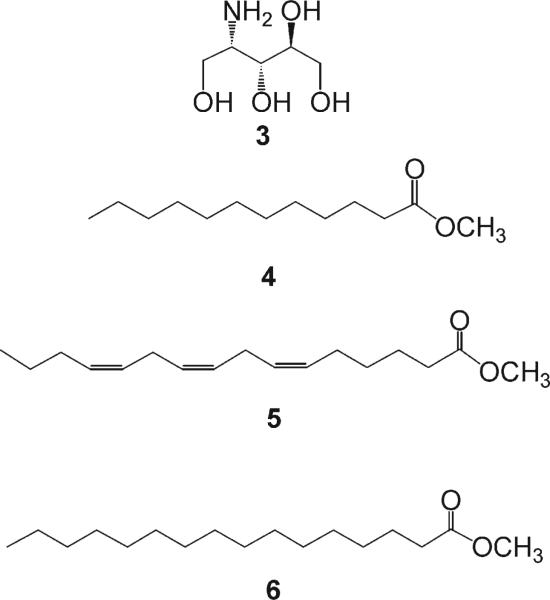
To deduce the identity of the fatty acids, 1 was subjected to a transesterification reaction in MeOH/NaOMe (2 h). After routine workup of the reaction, the nonpolar organic extract was analyzed by GC-MS and ESIMS, and three ion peaks [M + H]+ at m/z 215.2010, 265.2166, and 271.2636 were observed, corresponding to dodecanoic acid methyl ester (4), hexadecatrienoic acid methyl ester (5), and hexadecanoic acid methyl ester (6), respectively (Figure 2).
Figure 2.
Compounds 3-6 resulting from chemical degradation of 1.
The regiochemical distribution of these three acyl chains was established using different ionization mass spectrometry analyses. The amide bond was most readily recognized21 by MALDITOFMS/MS [M + H]+ at m/z 184.1, corresponding to the cleavage of the dodecanoyl moiety. ESIMS9 was used to differentiate the acyl attached at C-4 and C-5. Fragmentation by positive-mode ESIMS generated a more stable fragment ion from cleavage of the acyl chain attached at C-5 than cleavage of the acyl chain attached to C-4. In contrast, the negative-mode ESIMS fragment ion resulting from cleavage of the acyl chain attached at C-4 is more stable. Successive losses of dodecanoyl and hexadecatrienoyl moieties were observed in the negative-mode ESIMS (m/z 584.2 and 367.1, respectively). From these analyses it was concluded that the dodecanoyl moiety is attached to C-2, while the hexadecatrienoyl and hexadecanoyl moieties are attached to C-4 and C-5, respectively. To verify this conclusion, a selective enzyme reaction was performed. Upon regioselective enzymic hydrolysis of 1 with lipase enzyme type III in dioxane/H2O (1:1) at 37 °C for 4 h,10 only hexadecanoic acid was obtained, as evidenced by ESIMS and comparison with an authentic sample. Thus, it was concluded that the hexadecanoyl residue is attached to C-5 of 1.
The aqueous phase from the hydrolysis product of 1 yielded an aminotetraol (3), which was identified as (2S,3S,4R)-2-amino-1,3,4,5-pentane tetraol. The relative configuration of 3 was elucidated using the 3JH,H coupling constant analysis from an NMR database for a given stereocluster.11 Eight stereoisomeres are possible for aminotetraol (3), depending on the relative position of H-2 and H-3 (S = syn or A = anti) and H-3 and H-4 (S = syn or A = anti), and the eight isomers are classified as follows: one pair of SS, one pair AA, one pair of SA, and one pair of AS. According to the value recorded for 3 of the 3JH,H coupling constant between H-2 and H-3 (J = 2.5 Hz) and H-3 and H-4 (J =7 Hz), H-2 and H-3 are on the same side (S = syn) and H-3 and H-4 are on the opposite side (A = anti); thus the remaining probable stereoisomers is the pair of SA: (2S,3S,4R)-2-amino-1,3,4,5-pentane tetraol and (2R,3R,4S)-2-amino-1,3,4,5pentane tetraol. Theoretically these two isomers should have opposite optical rotation sign. The positive optical rotation of 3 ([α]24D +8 (c 0.05, H2O)) favors the (2S,3S,4R)-2-amino-1,3,4,5-pentane tetraol configuration as that of seen in l-arabino-phytosphingosine ([α]25D +5 (c 0.51, pyridine)),22 which is also the opposite of that observed in d-arabino-phytosphingosine ([α]25D −4.3 (c 0.50, pyridine)).22 The identical coupling constants between H-2 and H-3 (J = 3 Hz) and H-3 and H-4 (J = 6 Hz) along with the optical rotation reported ([α]27D +9.2 (c 0.05, MeOH)) for the sphingosine derivative12–14 and bathymodiolamides A (1) further suggested a (2S,3S,4R)-configuration of the sphingosine-like moiety in 1. From the above data, the structure of 1 was established as shown.
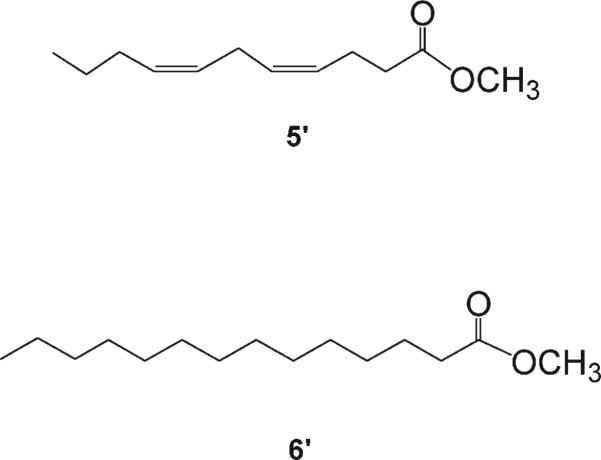
The molecular formula of 2 was established as C42H77NO7 on the basis of HRMALDITOFMS (m/z 730.5594 [M + Na]+). The strong similarity of its 1H NMR spectrum to that+of bathymodiolamide A (1) revealed that 1 and 2 share the same general structural features. Upon regioselective enzymic hydrolysis of 2 with lipase enzyme type III in dioxane/H2O (1:1) at 37 °C for 4 h,10 only tetradecanoic acid was obtained, as established by ESIMS analysis and comparison with an authentic sample. Thus, it was concluded that the tetradecanoyl residue is attached to the C-5 position of 2. The hydrolysis of 2 gave similar results to those observed for 1, with the exception of the presence of undecadienoic acid methyl ester (5′) and tetradecanoic acid methyl ester (6′) (Figure 3). The structure of 2 is similar to 1 with undecadienoyl and tetradecanoyl residues at C-4 and C-5, respectively. Thus, the structure of 2 was established as shown.
Figure 3.
Compounds 5′ and 6′ resulting from chemical degradation of 2.
The isolated compounds were evaluated in the apoptosis induction assay. The results showed that 1 and 2 inhibit the growth of two cancer cell lines [HeLa (cervical cancer) (IC50 0.4 μM for 1 and IC50 0.5 μM for 2) and MCF7 (breast cancer) (IC50 0.1 μM for 1 and IC50 0.2 μM for 2)]. The compounds described herein represent new chemical structures and may have potential in future drug discovery efforts. Another interesting observation is that although the deep-sea hydrothermal vents are characterized by high concentrations of reduced sulfur compounds, the presence of sulfur-containing secondary metabolites from the deep-sea hydrothermal vent invertebrate mussel Bathymodiolus thermophilu is not detected.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured on a JASCO P 1010 polarimeter. UV and FT-IR spectra were obtained using Hewlett-Packard 8452A and Nicolet 510 instruments, respectively. CD spectra were acquired on a JASCO J-720 spectropolarimeter. All NMR spectra were recorded on a Bruker Avance DRX400 spectrometer, a Varian-400 instrument (400 MHz), and a Varian-500 instrument (500 MHz). Spectra were referenced to residual solvent resonances at δH/C 3.31/49.15 (CD3OD). ESIMS data were acquired on a Waters Micromass LCT Classic mass spectrometer and a Varian 500-MS LC ion trap instrument. MALDITOFMS data were acquired on an ABI-MDS SCIEX 4800 instrument capable of performing true MS/MS for peptide analysis. HPLC separations were performed using Waters 510 HPLC pumps, a Waters 717 plus autosampler, and a Waters 996 photodiode array detector. All solvents were purchased as HPLC grade.
Animal Material
Bathymodiolus thermophilus was collected using the deep submergence vehicle DSV Alvin from an active hydrothermal vent along the Mid-Atlantic Ridge (region: north, location: Lucky Strike (LS), dive number: 3120, date: July 10, 1997, latitude: 37°17.63′ N, longitude: 32°16.53′ W, depth: 1733 m) and identified by Dr. R. C. Vrijenhoek (Department of Genetics, Rutgers, The State University of New Jersey) and Dr. C. Vetriani (IMCS, Rutgers, The State University of New Jersey). A voucher specimen is available at the Center for Marine Biotechnology, IMCS, Rutgers, The State University of New Jersey, New Brunswick, with collection number AD-MUS-LS-7/10/97.
Extraction and Isolation
B. thermophilus tissue (100 g wet mass) was extracted three times with MeOH to give a polar organic extract (850 mg). A portion of this extract (20 mg) was tested for induction of apoptosis and was found to be active. Therefore, the remaining organic extract was subjected to fractionation using solid-phase extraction on normal-phase silica to give four fractions, F1 to F4, eluted with hexanes, hexanes/EtOH, EtOH, and MeOH, in a series of increasingly hydrophilic solvent systems. The MeOH fraction (F4) had apoptosis induction activity. This fraction was further chromatographed by analytical RP HPLC (Phenomenex Luna C18, 250 × 4.60 mm, isocratic elution 4:1 MeOH/H2O, flow rate 1 mL/min) to yield 11.4 mg of 1 (tR = 11.5 min) and 8.7 mg of 2 (tR = 9.5 min).
Bathymodiolamide A (1)
[α]24D +10.8 (c 0.08, MeOH); IR νmax (neat) 3350, 3250, 2900, 2850, 1640, 1450, 1050 cm−1; 1H NMR and 13C NMR, see Table 1; HRMALDITOFMS m/z 804.6712 [M + H]+ (calcd for C49H90NO7, 804.6717).
Table 1.
NMR Spectroscopic Data for Bathymodiolamide A (1) (500 MHz, CD3OD)
| position | δC, mult. | δH (J in Hz) | HMBCa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-a | 1-b | 63.7, CH2 | 3.65, d (4.5), 3.65, d (4.5) | 2, 3 | |||||||
| 2 | 53.5, CH | 4.29, dd (4.5, 2) | 1, 3, 1′ | ||||||||
| 3 | 62.5, CH | 4.01, dd (2, 6.5) | 2, 4 | ||||||||
| 4 | 66.1, CH | 5.25, dt (6.5, 3) | 3, 5, 1″ | ||||||||
| 5-a | 5-b | 59.9, CH2 | 4.20, dd (12, 3), 4.45, dd (12, 3) | 4, 1‴ | |||||||
| 1′ | 1″ | 1‴ | 173.2, C | 173.4, C | 173.8, C | ||||||
| 2′ | 2″ | 2‴ | 30.0, CH2 | 30.1, CH2 | 30.2, CH2 | 2.31, t (7.5) | 2.35, t (7.2) | 2.36, t (7.5) | 1′, 4′ | 1″, 4″ | 1‴, 4‴ |
| 3′ | 3″ | 3‴ | 29.9, CH2 | 22.5, CH2 | 29.9, CH2 | 1.29, brs | 1.68, m | 1.29, brs | 1′, 5′ | 1″, 5″ | 1‴, 5‴ |
| 4′ | 4″ | 4‴ | 29.8, CH2 | 27.5, CH2 | 29.8, CH2 | 1.29, brs | 1.33, m | 1.29, brs | 2′, 6′ | 2″, 6″ | 2‴, 6‴ |
| 5′ | 5″ | 5‴ | 29.8, CH2 | 24.5, CH2 | 29.8, CH2 | 1.29, brs | 2.08, m | 1.29, brs | 3′, 7′ | 3″, 7″ | 3‴, 7‴ |
| 6′ | 6″ | 6‴ | 29.8, CH2 | 129.5, CH | 29.8, CH2 | 1.29, brs | 5.38, m | 1.29, brs | 4′, 8′ | 4″, 8″ | 4‴, 8‴ |
| 7′ | 7″ | 7‴ | 29.7, CH2 | 127.8, CH | 29.7, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 5′, 9′ | 5″, 9″ | 5‴, 9‴ |
| 8′ | 8″ | 8‴ | 29.7, CH2 | 26.5, CH2 | 29.7, CH2 | 1.29, brs | 2.81, m | 1.29, brs | 6′, 10′ | 6″,10″ | 6‴, 10‴ |
| 9′ | 9″ | 9‴ | 29.5, CH2 | 127.9, CH | 29.6, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 7′, 11′ | 7″, 11″ | 7‴, 11‴ |
| 10′ | 10″ | 10‴ | 29.5, CH2 | 128.1, CH | 29.6, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 8′, 12′ | 8″, 12″ | 8‴, 12‴ |
| 11′ | 11″ | 11‴ | 28.9, CH2 | 26.5, CH2 | 29.5, CH2 | 1.29, brs | 2.81, m | 1.29, brs | 9′, 12′ | 9″, 13″ | 9‴, 13‴ |
| 12′ | 12″ | 12‴ | 13.2, CH3 | 128.0, CH | 29.5, CH2 | 0.89, t (7.0) | 5.36, m | 1.29, brs | 10′, 11′ | 10″, 14″ | 10‴, 14‴ |
| 13″ | 13‴ | 128.4, CH | 29.5, CH2 | 5.36, m | 1.29, brs | 11″, 15″ | 11‴, 15‴ | ||||
| 14″ | 14‴ | 25.5, CH2 | 29.4, CH2 | 2.01, m | 1.29, brs | 12″, 16″ | 12‴, 16‴ | ||||
| 15″ | 15‴ | 22.5, CH2 | 28.9, CH3 | 1.40, m | 1.29, brs | 13″, 16″ | 13‴, 16‴ | ||||
| 16″ | 16″ | 13.1, CH3 | 13.0, CH3 | 0.91, t (7.5) | 0.90, t (7.5) | 13″, 15″ | 14‴, 15‴ | ||||
HMBC correlations, optimized for 8 Hz, are from proton(s) stated to the indicated carbon.
Bathymodiolamide B (2)
[α]24D +10.9 (c 0.08, MeOH); IR νmax (neat) 3350, 3250, 2900, 2850, 1640, 1450, 1050 cm−1; 1H NMR and 13C NMR, see Table 2; HRMALDITOFMS m/z 730.5594 [M + Na]+ (calcd for C42H77NNaO7, 730.5598).
Table 2.
NMR Spectroscopic Data of Bathymodiolamide B (2) (400 MHz, CD3OD)
| position | δC, mult. | δH (J in Hz) | HMBCa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-a | 1-b | 63.7, CH2 | 3.65, d (4.5), 3.65, d (4.5) | 2, 3 | |||||||
| 2 | 53.5, CH | 4.29, dd (4.5, 2) | 1, 3, 1′ | ||||||||
| 3 | 62.5, CH | 4.01, dd (2, 6.5) | 2, 4 | ||||||||
| 4 | 66.1, CH | 5.25, dt (6.5, 3) | 3, 5, 1″ | ||||||||
| 5-a | 5-b | 59.9, CH2 | 4.20, dd (12, 3), 4.45, dd (12, 3) | 4, 1‴ | |||||||
| 1′ | 1″ | 1‴ | 173.2, C | 173.4, C | 173.8, C | ||||||
| 2′ | 2″ | 2‴ | 30.0, CH2 | 30.1, CH2 | 30.2, CH2 | 2.31, t (7.5) | 2.35, t (7.2) | 2.36, t (7.5) | 1′, 4′ | 1″, 4″ | 1‴, 4‴ |
| 3′ | 3″ | 3‴ | 29.9, CH2 | 24.5, CH2 | 29.9, CH2 | 1.29, brs | 2.08, m | 1.29, brs | 1′, 5′ | 1″, 5″ | 1‴, 5‴ |
| 4′ | 4″ | 4‴ | 29.8, CH2 | 127.5, CH | 29.8, CH2 | 1.29, brs | 5.38, m | 1.29, brs | 2′, 6′ | 2″, 6″ | 2‴, 6‴ |
| 5′ | 5″ | 5‴ | 29.8, CH2 | 128.1, CH | 29.8, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 3′, 7′ | 3″, 7″ | 3‴, 7‴ |
| 6′ | 6″ | 6‴ | 29.8, CH2 | 26.5, CH2 | 29.8, CH2 | 1.29, brs | 2.81, m | 1.29, brs | 4′, 8′ | 4″, 8″ | 4‴, 8‴ |
| 7′ | 7″ | 7‴ | 29.7, CH2 | 128.0, CH | 29.7, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 5′, 9′ | 5″, 9″ | 5‴, 9‴ |
| 8′ | 8″ | 8‴ | 29.7, CH2 | 26.5, CH2 | 29.7, CH2 | 1.29, brs | 5.36, m | 1.29, brs | 6′, 10′ | 6″, 10″ | 6‴, 10‴ |
| 9′ | 9″ | 9‴ | 29.5, CH2 | 127.9, CH | 29.6, CH2 | 1.29, brs | 2.01, m | 1.29, brs | 7′, 11′ | 7″, 11″ | 7‴, 11‴ |
| 10′ | 10″ | 10‴ | 29.5, CH2 | 128.1, CH | 29.6, CH2 | 1.29, brs | 1.40, m | 1.29, brs | 8′, 12′ | 8″, 12″ | 8‴, 12‴ |
| 11′ | 11″ | 11‴ | 28.9, CH2 | 26.5, CH2 | 29.5, CH2 | 1.29, brs | 0.91, t (7.5) | 1.29, brs | 9′, 12′ | 9″, 13″ | 9‴, 13‴ |
| 12′ | 12‴ | 13.2, CH3 | 128.0, CH | 29.5, CH2 | 0.89, t (7.0) | 1.29, brs | 10′, 11′ | 10‴, 14‴ | |||
| 13‴ | 28.4, CH2 | 1.29, brs | 11‴, 14‴ | ||||||||
| 14‴ | 13.0, CH2 | 0.90, t (7.5) | 12‴, 13‴ | ||||||||
HMBC correlations, optimized for 8 Hz, are from proton(s) stated to the indicated carbon.
Preparation of Methyl Esters
Compound 1 (7.5 mg) was dissolved in MeOH (1.5 mL), and NaOMe (2.5 mg) was added; the reaction mixture was stirred at room temperature for 3 h. The reaction mixture was quenched with the acidic ion-exchange resin Amberlite IRC-50 (Rohm and Hass, H+ form), and the resin was removed by filtration. The filtrate was dried under reduced pressure and partitioned between CHCl3 and H2O. The organic layer was analyzed by GC-MS. The aqueous layer was also purified and analyzed.
Enzymatic Hydrolysis
Compounds 1 and 2 (2 mg) were dissolved separately in 4 mL of dioxane/H2O (1:1) and treated with lipase enzyme type III (2 mg, 50 unit, from a Pseudomonas species, Sigma-Aldrich) at 37 °C, with shaking for 4 h. Each reaction mixture was quenched with 5% AcOH (1 mL), and the product was dried under reduced pressure. The resulting crude residues were each dissolved in H2O and extracted with EtOAc, concentrated under reduced pressure, and analyzed by ESIMS.
Biological Evaluation (Apoptosis Induction)
Testing for induction of apoptosis in the presence of compounds 1 and 2 was carried out as described in Andrianasolo et al. using the ApoScreen assay.5,18–20 In this assay, the viability of treated W2 (apoptosis competent) and D3 (apoptosis defective)15 cells is measured using a modification of the MTT assay.16 For the current study, viability was measured at 0 and 48 h, and differential growth was calculated in the presence and absence of the compounds. Staurosporine (an apoptosis inducer) was used as a positive control, and DMSO as a negative control.
Supplementary Material
ACKNOWLEDGMENT
We thank S. Kim for NMR data from NMR facilities in the Department of Chemistry at Rutgers University. We also thank H. Zheng for mass spectrometry analysis at the Center for Advanced Biotechnology and Medicine, Rutgers University. This research was funded by Rutgers University through an Academic Excellence award, by NSF grants OCE 03-27373 (R.A.L and C.V.) and MCB 04-56676 (C.V.), by the New Jersey Agricultural and Experiment Station (C.V.), and by NIH grant R37 CA53370 (E.W.).
Footnotes
Supporting Information. NMR and MS data for 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Gärtner A, Wiese J, Imhoff JF. Int. J. Syst. Evol. Microbiol. 2008;58:34–39. doi: 10.1099/ijs.0.65234-0. [DOI] [PubMed] [Google Scholar]
- 2.Nelson DC, Hagen KD, Edwards DB. Mar. Biol. 1995;121:487–495. [Google Scholar]
- 3.Nees HA, Moore TS, Mullaugh KM, Holyoke RR, Janzen CP, Ma S, Metzer E, Waite TJ, Yücel M, Lutz RA, Shank TM, Vetriani C, Nuzzio DB, Luther GW., III J. Shellfish Res. 2008;27:169–175. [Google Scholar]
- 4.Andrianasolo EH, Haramaty L, Degenhardt K, Mathew R, White E, Lutz R, Falkowski P. J. Nat. Prod. 2007;70:1551–1557. doi: 10.1021/np070088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathew R, Degenhart K, Haramaty L, Karp CM, White E. Methods Enzymol. 2008;453:53–81. [Google Scholar]
- 6.Karantza-Wadsworth V, White E. In: Programmed Cell Death. Cancer: Principles and Practice of Oncology. DeVita VT, Lawrence TS, Rosenberg SA, editors. Lippincott, Williams, and Wilkins; Philadelphia, PA: 2008. pp. 93–101. [Google Scholar]
- 7.Ishii T, Okino T, Mino Y. J. Nat. Prod. 2006;69:1080–1082. doi: 10.1021/np050530e. [DOI] [PubMed] [Google Scholar]
- 8.Scribe P, Guezennec J, Dagaut J, Pepe C, Saliot A. Anal. Chem. 1988;60:928–931. [Google Scholar]
- 9.Guella G, Frassanito R, Mancini I. Rapid Commun. Mass Spectrom. 2003;17:1982–1994. doi: 10.1002/rcm.1142. [DOI] [PubMed] [Google Scholar]
- 10.Kitagawa I, Hayashi K, Kobayashi M. Chem. Pharm. Bull. 1989;37:849–851. doi: 10.1248/cpb.37.1676. [DOI] [PubMed] [Google Scholar]
- 11.Seike H, Ghosh I, Kishi Y. Org. Lett. 2006;8:3861–3864. doi: 10.1021/ol061580t. [DOI] [PubMed] [Google Scholar]
- 12.Garg HS, Agrawal S. J. Nat. Prod. 1995;58:442–445. [Google Scholar]
- 13.Natori T, Morita M, Akimoto K, Koezuka Y. Tetrahedron. 1994;50:2771–2774. [Google Scholar]
- 14.Mulzer J, Brand C. Tetrahedron. 1986;42:5961–5964. [Google Scholar]
- 15.Degenhardt KSR, Chen G, Lindsten T, Thomson C, White E. J. Biol. Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- 16.Denyer SP, Maillard J-Y. J. Immunol. Methods. 1983;65:55–63. [Google Scholar]
- 17.Danial N, Korsmeyer S. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 18.Gelinas C, White E. Genes Dev. 2005;19:1263–1268. doi: 10.1101/gad.1326205. [DOI] [PubMed] [Google Scholar]
- 19.Degenhardt K, White E. Clin. Cancer Res. 2006;12:5274–5276. doi: 10.1158/1078-0432.CCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 20.Fesik SW. Nat. Rev. Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 21.Koster S, Duursma MC, Boon JJ, Heeren RMA, Ingemaan S, Van Benthem RATM, Kostert CG. J. Am. Soc. Mass Spectrom. 2003;14:332–341. doi: 10.1016/S1044-0305(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 22.Park J-J, Lee JH, Li Q, Diaz K, Chang Y-T, Chung S-K. Bioorg. Chem. 2008;36:220–228. doi: 10.1016/j.bioorg.2007.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.