Abstract
One of the major limitations to effective siRNA delivery is the lack of a siRNA-specific delivery system. Currently, the same delivery systems that are used for plasmid DNA (pDNA) delivery to the cell nucleus are used for siRNA delivery to the cytoplasm. To fill this gap, the objective of this study was to design a biopolymer that can be programmed via its amino acid sequence to deliver siRNA specifically to cytoplasm. For pDNA delivery, a nuclear localization signal (NLS) was added to the biopolymer structure to facilitate active translocation of the genetic material towards nucleus. The biopolymers were complexed with pEGFP and GFP-siRNA and used to transfect SKOV-3 (HER2+) cells. The intracellular trafficking of the nanoparticles was also monitored in real-time and live cells. The results demonstrated that the biopolymer with NLS is a suitable carrier for pDNA delivery but not siRNA delivery. Conversely, the biopolymer without NLS was suitable for siRNA delivery to the cytoplasm but not pDNA to the cell nucleus. The potential use of the designed biopolymer for combination therapy of cancer cells with gene (thymidine kinase) and siRNA (BCL2) was also examined in SKOV-3 cancer cells.
Keywords: siRNA, Gene Delivery, Cancer Therapy, vector development, targeted therapy
1. INTRODUCTION
One of the major limiting factors in successful targeted siRNA and plasmid DNA (pDNA) delivery is the lack of suitable vectors that by design have the ability to target cells and specifically deliver siRNA to the cytoplasm or pDNA to the cell nucleus. Unlike gene therapy where the gene must be delivered to the nucleus of the cell to initiate transcription, the site of action for siRNA is the cytoplasm. Therefore, for maximum efficacy, a delivery system must be designed that is capable of differentiating between delivery to the cytoplasm and the cell nucleus. So far, pDNA delivery systems such as cationic lipids and polymers are also utilized to form complexes with siRNA in order to carry them to the cytoplasm for mRNA knockdown[1–5]. While relatively effective, liposomes and synthetic polymers allow the siRNA to enter any cellular compartment non-discriminatingly and do not necessarily localize the cargo (i.e., siRNA) to the cytoplasm (site of action). This is due to the fact that such delivery systems (e.g., liposomes) were originally designed for pDNA delivery to the cell nucleus [6–8].
We have recently introduced a new class of genetically engineered multifunctional vectors for pDNA delivery [9–13]. In one design, we have genetically engineered a prototype multi-functional polymer for gene delivery to the cell nucleus. More details about the characterization of the biopolymer including biodegradation, efficiency, non-toxicity and cell targeting can be found in reference [14]. The described biopolymer, features a DNA condensing and endosomolytic (DCE) domain consisted of repeating units of arginine-histidine (RH) with the general structure of (RRXRRXHHXHHX)n, where X is any amino acid except D and E, and n is the number of repeating units. Other domains include a pH-dependent fusogenic peptide for destabilization of endosomal membrane, a high affinity HER2 targeting affibody for targeted gene delivery, and a M9 nuclear localization signal (NLS) to enhance active translocation of genetic material toward the cell nucleus. In addition, the RRXRR in the DCE domain is engineered to not only condense pDNA but be a substrate for the intracellular furin enzyme, which will facilitate biodegradation of the cationic domain intracellularly, resulting in less biopolymer related toxicity[14]. This biopolymer which is composed of a Fusogenic peptide, a DNA condensing and endosomolytic motif, a Nuclear localization signal and a Targeting motif is named FDNT.
The objective of this study was to design a biopolymeric platform that can selectively deliver siRNA to cytoplasm and pDNA to the cell nucleus. It was hypothesized that by removing the nuclear localization signal sequence from the FDNT biopolymer structure, the function of the biopolymer could be tailor-made for siRNA delivery. Because we have previously demonstrated that FDNT can target SKOV-3 (HER2+) ovarian cancer cells and mediate gene expression [14], we used this cell line as a model to demonstrate the potential use of the designed biopolymers for combination therapy of cancer cells with gene and siRNA. Schematics of the two biopolymers are shown in Figure 1. While biopolymer FDNT (contains NLS) is designed for pDNA delivery, biopolymer FDT (lacks NLS) is expected to be suitable for siRNA delivery.
Figure 1.
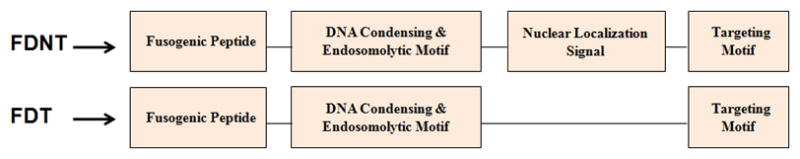
Schematics of the multi-domain biopolymers designed for siRNA delivery (FDT) and pDNA delivery (FDNT).
2. EXPERIMENTAL METHODS
2.1. Genetic engineering of the biopolymers
The details of the methods used to genetically engineer FDNT have been reported previously[14]. FDT biopolymer was genetically engineered using the same methods as for FDNT. In brief, the genes encoding the FDT and FDNT biopolymers were designed in our lab and synthesized by Integrated DNA Technologies (San Diego, CA) with C-terminal NdeI and N-terminal XhoI restriction sites. Synthesized genes were double digested using NdeI and XhoI restriction enzymes (New England Biolabs, Ipswich, MA) and subcloned into a pET21b expression vector (EMD Biosciences, Gibbstown, NJ). The biopolymer genes were expressed in E. coli BL21(DE3) pLysS (Novagen, San Diego, CA) expression system and purified using Ni-NTA affinity chromatography [14]. The expression of the biopolymers as well as purity was determined by Western Blot analysis and SDS-PAGE.
2.2. Particle size analysis
pEGFP or GFP-siRNA was complexed with FDT or FDNT in Bis-Tris buffer at pH 6.0. A Malvern Nano ZS90 (Malvern Instruments, UK) was used to determine the particle sizes of the particles. For example, for plasmid DNA delivery, 21 μg FDNT or 18 μg FDT was used to complex with 1μg of pEGFP for an NP (N-atoms in biopolymer to P-atoms in plasmid DNA) ratio of 14. This ratio was selected based on our previously published data [14]. For siRNA delivery, 5 μg of biopolymer (FDNT or FDT) was used to complex with 1 μg of siRNA to prepare complexes at a weight ratio of 5:1. This optimum 5:1 ratio was chosen based on the findings from our preliminary cell transfection studies. After 15 minutes of incubation, the size of the complexes were measured and reported as mean ± SEM, (n=3). Each mean is the average of 15 measurements and n represents the number of separate batches prepared for the measurements.
2.3. Preparation of stable SKOV-3 cell line that overexpress GFP
SKOV-3 cells (human ovarian cancer) were seeded in T-75 flasks (BD-Biosciences, San Jose, CA) and grown to 80% confluence in McCoy’s 5A media (Hyclone) supplemented with 10% fetal bovine serum (Hyclone). Using lipofectamine (Invitrogen), 2 × 106 cells were co-transfected with 1 μg pUB-GFP (under the control of ubiquitin promoter) and 0.2 μg pBABE (plasmid containing a pyromycin selectable marker) in serum free McCoy’s 5A media containing transferrin, selenium, ovalbumin, dexamethasone, and fibronectin (McCoy’s-SFM). Transfected SKOV3 cells were treated with pyromycin to kill the non-transfected ones and select for GFP expressing SKOV-3 cells (SKOV3/GFP). Stocks of SKOV3/GFP cells were frozen and stored in liquid nitrogen for future use. The level of GFP expression was determined using flowcytometry (FACSCalibur, BD Biosciences, San Jose, CA) and visualized by epifluoresent microscopy (AxioObserver, Carl Zeiss). Flowcytometry data are reported as mean ± s.d. and statistical significance was evaluated using t-test.
2.4. Preparation of stable SKOV-3 cell line that overexpress BCL2
1×105 SKOV-3 cells were seeded in 6 well plates. After 24 hours media was removed and replaced with McCoy’s-SFM. Using lipofectamine, cells were transfected with 1 μg pCMV:BCL2 (Addgene) under the control of a CMV promotor. Media was removed after 4 hours and replaced with McCoy’s 5A media containing 10% FBS and neomycin to select for SKOV-3 cells overexpressing BCL2 (SKOV3/BCL2).
To verify overexpression of BCL2 in SKOV3/BCL2 cells, from each well cells were collected and lysed on ice using extraction buffer composed of 25 mM HEPES, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 0.1 mM PMSF, and protease inhibitor cocktail (Calbiochem). Cell lysate was centrifuged and supernatant was removed. Equivalent of 50 μg protein was loaded onto SDS PAGE gel and electrophoresed. The expression of BCL2 in SKOV3/BCL2 cells was then verified by Western Blot analysis using anti-BCL2 antibody (Calbiochem).
2.5. Cell transfections
SKOV-3 cells were seeded in 96 well plates at 2.0 × 104 cells per well. Cells were incubated overnight in 37°, 5% CO2 environment until 80–90% confluent. Plasmid DNA encoding green fluorescent protein (pEGFP) was complexed with FDNT or FDT at N:P ratio of 14 to form nanoparticles. This optimum N:P ratio for cell transfection was based on our previous published data [14]. At this N:P ratio, 18 μg of FDT or 21 μg of FDNT was complexed with 1ug of pEGFP for cell transfection. SKOV-3 cells were then transfected in McCoy’s-SFM with the biopolymer/pEGFP complexes. Four hours post transfection, media was replaced with McCoy’s 5A supplemented with 10% FBS and antibiotics. Visualization of the green fluorescence was conducted using a Zeiss epifluoresence microscope. Total green fluoresence intensity was measured 48 hours post transfection using a FACS Calibur flow cytomter. Flow cytometry data are reported as mean ± s.d, n=3.
For GFP knockdown studies, SKOV3/GFP cells were seeded in 96 well plates at 2.0 × 104 cells per well. After 24 hours, unmodified GFP-siRNA (Ambion, Cat#AM4626) or scrambled-siRNA (Ambion, Cat#AM4613) was complexed with the biopolymers (i.e., FDT or FDNT) at biopolymer/siRNA weight ratios of 1:1, 5:1 and 10:1 for gene knockdown. Four hours post transfection the media was replaced with McCoy’s 5A supplemented with 10% FBS and antibiotics. The GFP expression level was evaluated 48 hours post transfection using flow cytometry.
For BCL2 knockdown studies, SKOV3/BCL2 cells were used as described above. Various amounts of unmodified BCL2-siRNA (Ambion, Cat#AM16706, GGAUUGUGGCCUUCUUUGAtt) or scrambled siRNA (Ambion, Cat# AM4613, UGUACUGCUUACGAUUCGGtt) was complexed with FDT and used to transfect cells. After 2 days cells were harvested, lysed and equivalent of 50 μg proteins was loaded onto SDS PAGE gel. The proteins were then transferred onto PVDF membrane and blotted with anti-BCL2 (1:500) or anti GADPH (1:1000) (Calbiochem) antibodies.
2.6. Cancer cell killing by combination gene and siRNA delivery
The general protocol used for combination gene and siRNA therapy is shown in Figure 2.
Figure 2.
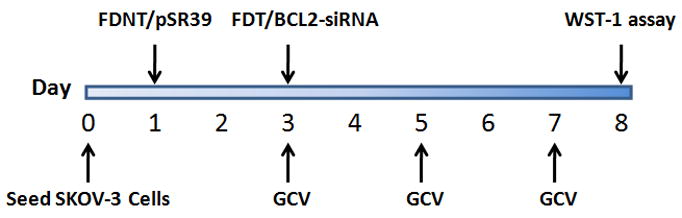
Schematics of the treatment protocol used to evaluate the potential use of the biopolymeric platform for combination gene (pSR39) and siRNA (BCL2) therapy.
First, 2×104 SKOV-3 cells were seeded in 96 well plates. At the time of transfection, media was removed and replaced with McCoy’s–SFM. 1 μg Plasmid DNA encoding mutant Thymidine Kinase gene, namely pSR39, was complexed with FDNT at the NP ratio of 14. pSR39 was generously provided by Dr. Margaret Black in the Department of Pharmaceutical Sciences at Washington State University. FDNT/pSR39 complexes were added to each well. After 4 hours, media was removed and replaced with McCoy’s 5A containing 10% FBS.
Subsequently (48 hours post transfection with pSR39), media was changed to McCoy’s-SFM to prepare cells for siRNA delivery. FDT in complex with BCL2-siRNA or scrambled-siRNA were added to transfect cells. 4 hours post siRNA transfection, media was removed and replaced with McCoy’s 5A containing 10% FBS and 50 μM Gancyclovir. On alternating days media containing 50 μM GCV was added to cells. Eight days following initial transfection with FDNT/pSR39 complexes, cell survival was assessed using WST-1 reagent (Roche, Applied Science, Indianapolis, IN). 10 μL of reagent was added per 100 μL media and plates were incubated for 4 hr followed by measuring absorbance at 420 nm. The measured absorbance for test groups is expressed as a percent of the control where the control is defined as 100% viable. The data are reported as mean ± SEM, n=6 and statistical significance was evaluated using t-test.
2.7. Real-time live cell particle tracking
2×105 SKOV-3 cells were seeded in 35 mM glass bottom MaTek plates (MaTek Corp, Ashland, MA) in the presence of McCoy’s 5A plus 10% FBS. After 24 hours media was replaced with McCoy’s 5A media without FBS to starve the cells. 24 hours later, biopolymers were used to transfect SKOV-3 cells with pDNA which was labeled with Cy3 (Mirus LabelIT kit, Mirus Bio, Madison, WI) according to manufacturer’s instructions. Images were captured sequentially every 5 minutes for 80 minutes using an epifluorescene microscope (Carl Zeiss Axiobserver Z1 with 40X objective). For z-stack images, depths of 7 μM were captured with images taken every 300 nM. Image compilation was performed using AxioObserver software (Zeiss) and National Institutes of Health Image J (Bethesda, MD) software.
3. RESULTS
3.1. Genetic engineering of FDT and FDNT biopolymers
Both FDNT and FDT biopolymers were genetically engineered, expressed in E. coli and purified using nickel column affinity chromatography. The expression and purification protocol for FDNT is reported previously [14]. The FDT was also purified and its expression verified by Western Blot analysis (Supplementary Figure 1).
3.2. Biopolymer mediated pDNA delivery
To examine the ability of the biopolymers to deliver pDNA, where the site of action is the cell nucleus, each biopolymer was complexed with pEGFP and used to transfect SKOV-3 cells. The sizes of the FDT/pEGFP nanoparticles were 78±4 nm, whereas the sizes of FDNT/pEGFP nanoparticles were 66±5nm. SKOV-3 cells that were transfected with FDNT biopolymer (contains NLS) showed significantly more GFP expression in comparison to FDT biopolymer which lacks the NLS domain (Figure 3).
Figure 3.
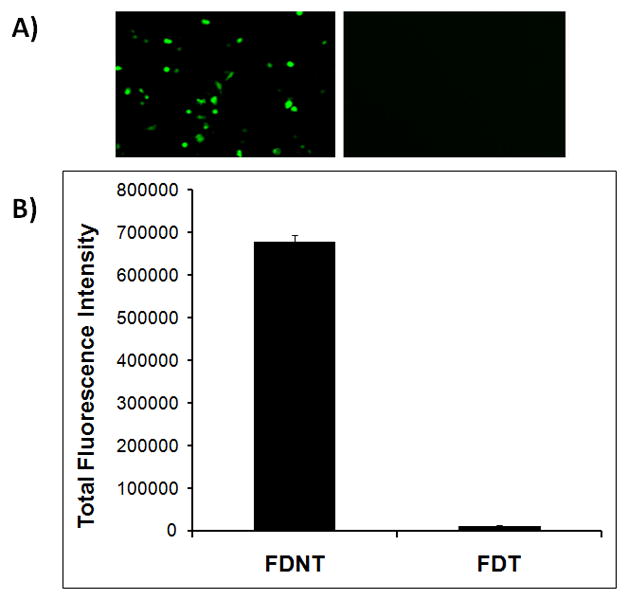
Evaluation of the delivery of pEGFP to SKOV-3 cells using FDNT and FDT biopolymers. A) Epifluorescent images of transfected SKOV-3 cells by FDNT/pEGFP (left panel) and FDT/pEGFP (right panel). B) Quantitative analysis of the gene expression in cells transfected with FDNT/pEGFP and FDT/pEGFP. The gene expression was measured using flow cytometry.
3.3. Dynamic imaging of the nanoparticles in live cells
To better understand the reason behind the observed differences in transfection efficiencies of the biopolymers, the intracellular trafficking of the biopolymer/pDNA complexes was studied using dynamic imaging (real-time in live cells). Labeled pDNA was complexed with FDNT or FDT and used to transfect cells. It was observed that the FDNT/pEGFP complexes are trafficked towards the nuclear membrane actively and in less than 80 minutes (Figure 4A). In contrast, FDT/pEGFP complexes remain stagnant, are more localized in their movements, and rarely reach the nuclear membrane area (Figure 4B).
Figure 4.
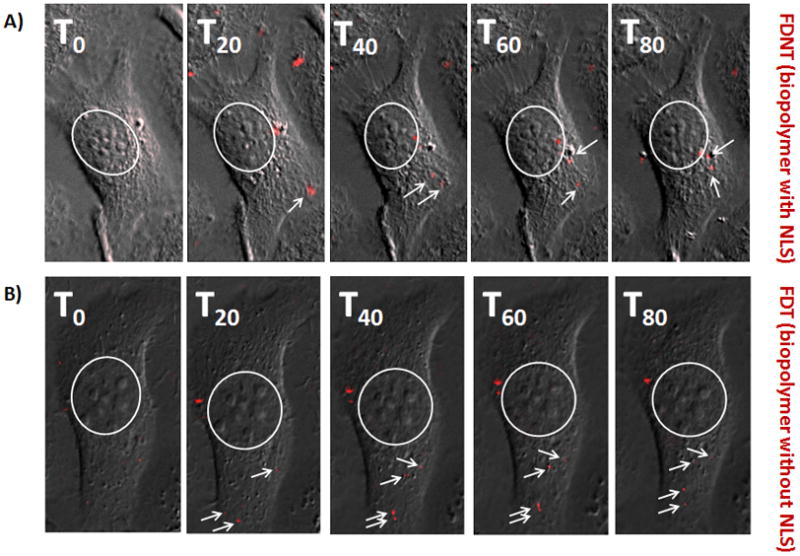
Real time imaging of the FDNT/pDNA and FDT/pDNA nanoparticles in live SKOV-3 cells by using an epifluorescent microscope. A) Transfection of SKOV-3 cells with FDNT/pDNA complexes and their intracellular trafficking. B) Transfection of SKOV-3 cells with FDT/pDNA complexes and their movements inside the cell. The nuclear membrane is shown by a white circle whereas white arrows point at the red fluorescent nanoparticles.
To verify that the nanoparticles were indeed internalized and not immobilized on the cell surface, z-stack images were taken each 300 nm apart. Z-stack images of the SKOV-3 cells that were transfected with FDNT/pDNA and FDT/pDNA complexes are shown in Figure 5A and B. Highest levels of red fluorescence were observed in midsection slices (z-stacks) highlighting the internalization of the nanoparticles.
Figure 5.
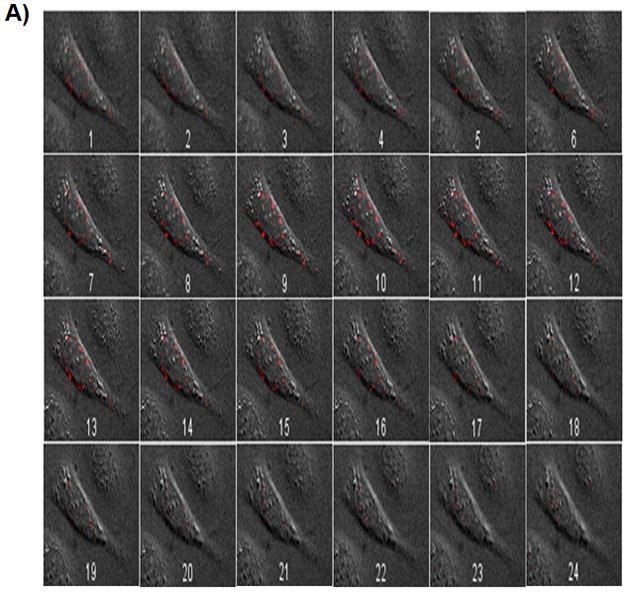
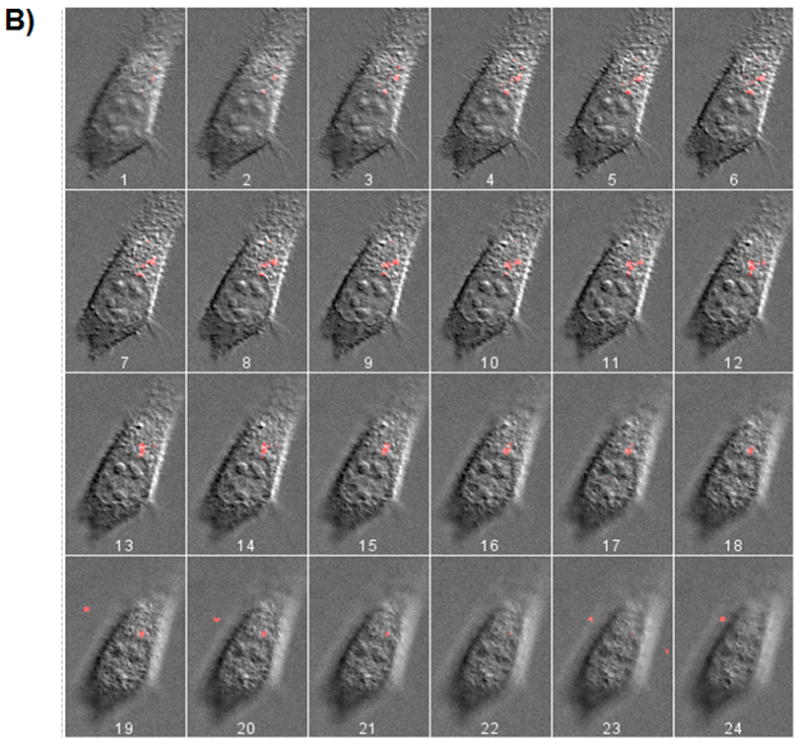
Z-stack images taken from SKOV-3 cells transfected with FDNT/pDNA (A), and FDT/pDNA (B) nanoparticles. pDNA (pEGFP) was labeled with Cy3 dye and used to complex with biopolymers. Z-stack images 7 to 12 (inside the cell) show the highest levels of nanoparticle fluorescence intensity.
3.4. Biopolymer mediated siRNA delivery
To examine the ability of the biopolymers to deliver siRNA, where the site of action is the cytoplasm, FDT and FDNT biopolymers were complexed with GFP-siRNA and used to transfect SKOV3/GFP cells. The sizes of the nanoparticles formed with FDNT/GFP-siRNA and FDT/GFP-siRNA (weight/weight 5:1) were 121 ± 7 and 140 ± 5nm, respectively. As shown in Figures 6A and B, the SKOV3/GFP cells (GFP+) have significantly higher green fluorescent intensity above the background level (GFP̄). SKOV3/GFP cells treated with GFP-siRNA alone showed no decrease in GFP expression. While SKOV3/GFP cells that were transfected with FDT/GFP-siRNA showed significant GFP knockdown (*t-test, p<0.05), no significant GFP knockdown with FDNT/GFP-siRNA was observed. Scrambled siRNA sequences also had no effect on measured GFP levels. In addition, no significant gene knockdown was observed with biopolymer/GFP-siRNA complexes formed at weight/weight ratios of 1:1 and 10:1. Therefore, in subsequent studies 5:1 weight ratio was used.
Figure 6.
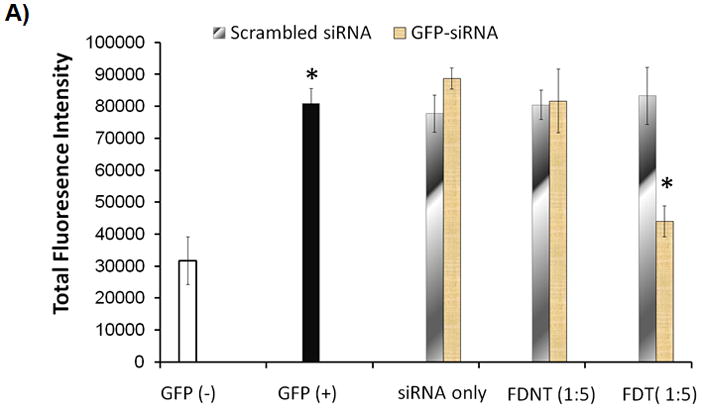
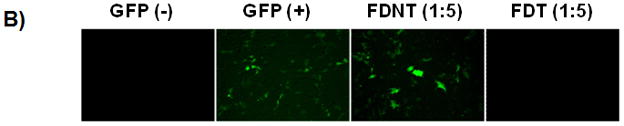

Evaluation of the gene silencing by the biopolymers. A) SKOV3/GFP cells were transfected with GFP-siRNA alone, FDNT/GFP-siRNA and FDT/GFP-siRNA to knock down GFP expression. 1:5 ratio refers to 1 ug of GFP-siRNA to 5 ug of biopolymer. Scrambled siRNA was used as a control. The level of GFP expression was measured using flowcytometry. B) Epifluorescent images of the untreated GFP̄ and GFP+ cells as well as SKOV3/GFP cells treated with biopolymers in complex with GFP-siRNA. C) Knockdown of BCL2 protein expression in SKOV3/BCL2 cancer cells that overexpress BCL2. BCL2-siRNA was complexed with FDT to knockdown the BCL2 expression. i) Knockdown was detected by westernblot analysis using anti-BCL2 antibody. Lane 1: BCL2 protein (positive control); Lane 2: FDT/BCL2-siRNA (1:1 ratio); Lane 3: FDT/BCL2-siRNA (1:5 ratio); Lane 4: scrambled-siRNA (1:5 ratio); ii) GAPDH control.
Based on the results from the abovementioned studies, the potential use of the biopolymer FDT in BCL2 (pro-survival protein) knockdown by delivering BCL2-siRNA was also examined. It was observed that FDT has the ability to ablate the expression of BCL2 in SKOV3/BCL2 cell line Figure 6C. As a result, SKOV-3 cells could become more susceptible to apoptosis that is induced by drugs or therapeutic genes.
Use of biopolymers for combination gene and siRNA therapy
The ability of one biopolymeric platform to deliver different therapeutic nucleic acids and result in significant cell killing was evaluated by transfecting SKOV-3 cancer cells with BCL2-siRNA and pSR39 gene. The results demonstrated that FDNT/pSR39 complexes in combination with prodrug ganciclovir (GCV) are able to kill SKOV-3 cells significantly (*t-test, p<0.05) (Figure 7). In comparison to cells treated with FDNT/pSR39 complexes plus GCV, maximum killing efficiency was observed when FDNT/pSR39 plus GCV was combined with FDT/BCL2-siRNA (*t-test, p<0.05). No other control group resulted in statistically significant cell killing in comparison to the untreated cells.
Figure 7.
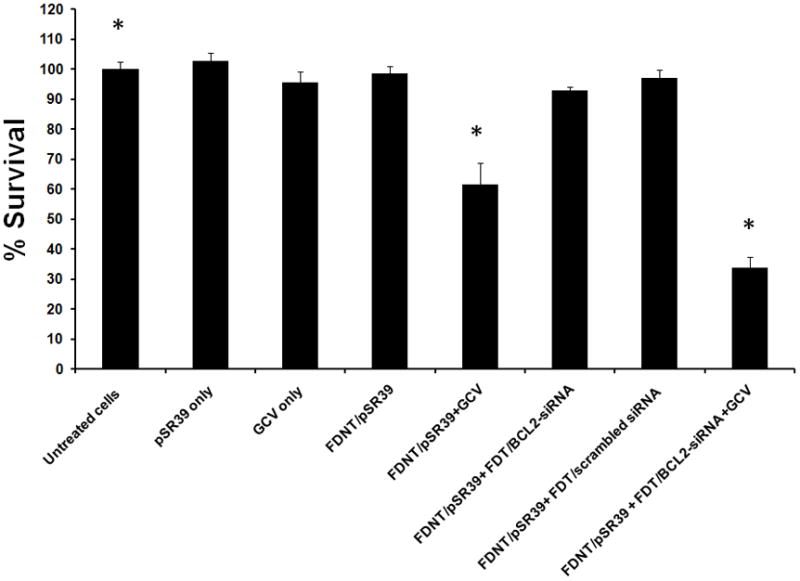
Evaluation of cell killing efficiency by biopolymers in complex with therapeutic nucleic acids. Cells were transfected with two test groups (FDNT/pSR39 plus GCV; and FDNT/pSR39 plus GCV + FDT/BCL2-siRNA) and six control groups. The cell survival was assessed at day 8 with WST-1 cell toxicity assay.
DISCUSSION
In the past decade, since the discovery of RNA interference (RNAi), significant amounts of money have been invested in the therapeutic application of gene silencing in humans [15]. Concurrently, significant efforts have been made in therapeutic application of genes to cure various diseases. Despite early successes, however, the widespread use of RNAi and gene therapeutics for disease prevention and treatment has not been achieved. Future success requires the development of nucleic acid delivery systems that are safe, efficient, targeted and cost-effective. One of the major impediments to the effective siRNA and pDNA delivery is the inability to efficiently localize such molecules to their site of action. While various gene delivery systems have been designed for nucleic acid delivery, there has been no report on the design and development of a vector that is tailor-made specifically for either siRNA or pDNA. To address this issue, development of a class of biomaterials is required that will allow precise correlation of structure with function. One class of biomaterials that provides the possibility of performing such precise structure/activity relationship studies is genetically engineered polymers. Using recombinant techniques, novel polymers can be synthesized with precise compositions, molecular weights, sterotacticity and specified functions. Compared to synthetic methods of polymer production, the principal advantages are (i) monodisperse material, (ii) full control over polymer architecture at the molecular level, (iii) precise covalent attachment of functional moieties (e.g. targeting motifs), (iv) programmability via amino acid sequence, and from a manufacturing standpoint, (v) elimination of the conjugation steps [9]. In 1997, Dan Urry was among the first to discuss the possibility of developing biopolymers that can be programmed via their amino acid sequences to perform self-guided functions [16]. Here, we demonstrate that multifunctional biopolymers can be designed where amino acid sequence could dictate not only their function but localization site.
To achieve the objective, two multifunctional biopolymers were designed one with NLS and the other without (Figure 1). Using genetic engineering techniques, they were cloned and produced in E.coli expression system and isolated to high purity using nickel column chromatography. Both purified biopolymers were used to complex with pEGFP to form particles with sizes less than 150nm which would make them suitable for receptor-mediated endocytosis [17]. These nanoparticles were then used to transfect SKOV-3 ovarian cancer cells which overexpress HER2 on their surfaces [18]. It was observed that the cells that were transfected with vector FDNT had significantly higher gene expression in comparison to the cells that were transfected with FDT (Figure 3). This observation was somewhat expected as the site of action for plasmid DNA is the cell nucleus and vector FDNT has the major necessary tools to facilitate its localization in the nucleus. To make a direct observation of the trafficking of the nanoparticles, we utilized real-time live cell imaging technique. This technique has previously been described in detail by Hanes group [19, 20]. Vectors FDNT and FDT were complexed with Cy3 labeled pEGFP and used to transfect cells. The trafficking of the nanoparticles was observed in a single SKOV-3 cell at a time using an epifluorescent microscope for a period of 80 minutes (Figure 4). Our observations reaffirmed our previous findings that the presence of NLS in the biopolymer structure is necessary for microtubule mediated translocation of genetic materials toward nucleus [14]. To examine whether the labeled nanoparticles could internalize and not merely attach to the outer membrane, z-stack images were obtained (Figure 5). Observation of bright red signals in the midst sections illustrated that the nanoparticles were in fact inside the cells.
To further examine the localization site of FDNT versus FDT, we changed the payload to siRNA where the site of action is cytoplasmic. From the cell transfection studies with pEGFP and imaging studies we understood that FDT/pDNA nanoparticles were inefficient in reaching the cell nucleus and localized in the cytoplasm. Therefore, we hypothesized that FDT is a suitable vehicle for siRNA delivery. To test the hypothesis, we used GFP-siRNA as a model gene silencer to demonstrate the localization of FDT in the cytoplasm. FDNT biopolymer was used as control to demonstrate that presence of NLS in the structure is detrimental to the localization of siRNA in the cytoplasm. Unmodified GFP-siRNA was purchased from Ambion and used as a gene silencer to evaluate the localization of the biopolymers in cytoplasm. It is noteworthy that for true evaluation of the efficiency of siRNA delivery systems, unmodified siRNA needs to be used. Several companies such as Dharmacon (Lafayette, CO) have modified the siRNA structure by attaching lipids or other molecules to enhance its internalization into the cells. Such modified siRNA are not suitable for the evaluation of the efficiency of the delivery systems as they themselves can enter the cells without the help of a carrier. In this study, we purchased unmodified siRNA from Ambion to ensure that siRNA by itself cannot enter the cells. For the evaluation of the gene expression knockdown, we also engineered a SKOV-3 cell line that stably expresses GFP (SKOV3/GFP).
Both FDT and FDNT biopolymers were complexed with GFP-siRNA and used to knockdown GFP expression in SKOV3/GFP cells. Significant GFP expression knockdown indicated that vector FDT is the more suitable carrier for siRNA delivery (Figure 6A and B). To examine the potential use of FDT in cancer therapy, we delivered BCL2-siRNA to SKOV-3 cancer cells to knockdown one of their most prominent pro-survival signaling pathway. When under stress (e.g., chemotherapy or gene therapy), cancer cells have the capacity to up-regulate production of BCL2 protein which make them more resistant to apoptosis [21]. Under normal conditions, the basal level of BCL2 production in SKOV-3 cells is low and hard to detect. Therefore, we engineered and used a SKOV-3 cell line that overexpresses BCL2 (SKOV3/BCL2) in order to evaluate the efficiency of the FDT biopolymer to knockdown BCL2 expression. Both biopolymers were complexed with BCL2-siRNA and used to transfect SKOV3/BCL2 cells. The results showed that only FDT was able to ablate the expression of BCL2, whereas FDNT was not effective (Figure 6C). This finding not only was in agreement with our GFP-siRNA observations, but also indicated the potential use of the FDT vector in siRNA delivery for cancer therapy. To examine this potential, we combined BCL2-siRNA delivery with thymidine kinase gene delivery.
Herpes simplex virus thymidine kinase (HSV-TK) is a widely used suicide gene that in conjunction with prodrug ganciclovir (GCV) results in cell killing by selectively converting nontoxic prodrugs to highly toxic metabolites. In the case of HSV-TK/GCV, the viral enzyme phosphorylates a nucleotide analog resulting in analog incorporation during DNA replication, halting the replication process [22]. HSV-TK/GCV also demonstrates a Bystander affect meaning that cells not directly transfected with the gene may also be affected due to the transfer of the cytotoxic metabolites [23]. This offers an advantage in systems with less efficient transfection levels as fewer cells need to be transfected to see a substantial effect. In this study, we used the SR39 gene which is a mutant HSV-TK and more selective to GCV than wild-type [24]. The rationale behind the choice of BCL2 knockdown in combination with pSR39 is that expression of thymidine kinase gene in transfected cells in combination with GCV will cause significant stress in those cells. As a result, transfected cells would up-regulate the pro-survival BCL2 signaling pathway to resist apoptosis. Subsequent transfection of the cells with BCL2-siRNA would inhibit such defense mechanism and render the cells more responsive to suicide gene therapy. Published data by others also reiterates the potential use of this approach in combination gene and siRNA cancer therapy [25].
This approach in cancer therapy was evaluated using FDNT/pSR39 complexes plus GCV in combination with FDT/BCL2-siRNA complexes. Using a cell toxicity assay, we evaluated the synergistic cell killing effects of combination siRNA and suicide gene delivery (Figure 7). In comparison to untreated cells, we observed statistically significant cell killing with FDNT/pSR39 plus GCV (*t-test, p<0.05). As discussed above, cells that undergo stress may up-regulate BCL2 pathway to resist apoptosis. Therefore, by knocking down the BCL2 expression, we expected to observe significantly higher cell killing efficiency. In comparison to the cell killing efficiency of FDNT/pSR39 plus GCV, we observed significantly higher cell death in cells that were treated with FDT/BCL2-siRNA in combination with FDNT/pSR39 plus GCV (*t-test, p<0.05).
The significance of these studies is that one biopolymeric platform can be tailor-made with minimum costs to deliver different types of nucleic acids to their site of action for maximum therapeutic outcome. These proof-of-concept studies justify the use of animals in the next steps to evaluate the potential application of such biopolymers to establish preliminary efficacy and toxicity in vivo.
CONCLUSIONS
One of the most important features of using biopolymers for gene delivery is the enormous possibility for recombinant engineering that facilitates accurate structure/activity relationship studies. As a result, the rate limiting steps to non-viral gene transfer can be studied with higher degree of accuracy and precision. These biopolymers allow full control over their architecture at the molecular level; hence, customizable and programmable via their amino acid sequences. The results of this study demonstrate that biopolymers can be designed to target and deliver nucleic acids to their site of action inside the cells.
Supplementary Material
Western Blot analysis (left panel) and SDS-PAGE (right panel) of the purified FDT biopolymer. The expected molecular weight for FDT is 18,210 daltons. Lane 1: lysate; Lane 2: flow through; Lane 3: protein marker; Lane 4: purified FDT biopolymer.
Acknowledgments
This work was supported in part by the startup funds from the Washington State University to Hatefi and by the NIH biotechnology training fellowship (T32-GM008336) to B.F. Canine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SH, Kim SH, Park TG. Intracellular siRNA delivery system using polyelectrolyte complex micelles prepared from VEGF siRNA-PEG conjugate and cationic fusogenic peptide. Biochem Biophys Res Commun. 2007;357(2):511–516. doi: 10.1016/j.bbrc.2007.03.185. [DOI] [PubMed] [Google Scholar]
- 2.Garbuzenko OB, Saad M, Betigeri S, Zhang M, Vetcher AA, Soldatenkov VA, Reimer DC, Pozharov VP, Minko T. Intratracheal versus intravenous liposomal delivery of siRNA, antisense oligonucleotides and anticancer drug. Pharm Res. 2009;26(2):382–394. doi: 10.1007/s11095-008-9755-4. [DOI] [PubMed] [Google Scholar]
- 3.Inoue A, Sawata SY, Taira K. Molecular design and delivery of siRNA. J Drug Target. 2006;14(7):448–455. doi: 10.1080/10611860600845397. [DOI] [PubMed] [Google Scholar]
- 4.Meade BR, Dowdy SF. The road to therapeutic RNA interference (RNAi): Tackling the 800 pound siRNA delivery gorilla. Discov Med. 2009;8(43):253–256. [PubMed] [Google Scholar]
- 5.Wang Y, Saad M, Pakunlu RI, Khandare JJ, Garbuzenko OB, Vetcher AA, Soldatenkov VA, Pozharov VP, Minko T. Nonviral nanoscale-based delivery of antisense oligonucleotides targeted to hypoxia-inducible factor 1 alpha enhances the efficacy of chemotherapy in drug-resistant tumor. Clin Cancer Res. 2008;14(11):3607–3616. doi: 10.1158/1078-0432.CCR-07-2020. [DOI] [PubMed] [Google Scholar]
- 6.Akita H, Harashima H. Nonviral gene delivery. Contrib Nephrol. 2008;159:13–29. doi: 10.1159/000125560. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Kim KS, Liu D. Nonviral gene delivery: what we know and what is next. Aaps J. 2007;9(1):E92–104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louise C. Nonviral vectors. Methods Mol Biol. 2006;333:201–226. doi: 10.1385/1-59745-049-9:201. [DOI] [PubMed] [Google Scholar]
- 9.Canine BF, Hatefi A. Development of recombinant cationic polymers for gene therapy research. Adv Drug Deliv Rev. 2010 doi: 10.1016/j.addr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy HO, Wang Y, Mangipudi SS, Hatefi A. Advances with the use of bio-inspired vectors towards creation of artificial viruses. Expert Opin Drug Deliv. 2010;7(4):1–16. doi: 10.1517/17425240903579989. [DOI] [PubMed] [Google Scholar]
- 11.Mangipudi SS, Canine BF, Wang Y, Hatefi A. Development of a genetically engineered biomimetic vector for targeted gene transfer to breast cancer cells. Mol Pharm. 2009;6(4):1100–1109. doi: 10.1021/mp800251x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Canine BF, Hatefi A. HSV-TK/GCV Cancer Suicide Gene Therapy by a Designed Recombinant Multifunctional Vector. Nanomedicine. 2010 doi: 10.1016/j.nano.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Mangipudi SS, Canine BF, Hatefi A. A designer biomimetic vector with a chimeric architecture for targeted gene transfer. J Control Release. 2009;137:46–53. doi: 10.1016/j.jconrel.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canine BF, Wang Y, Hatefi A. Biosynthesis and characterization of a novel genetically engineered polymer for targeted gene transfer to cancer cells. J Control Release. 2009;138(3):188–196. doi: 10.1016/j.jconrel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J Phys Chem B. 1997;101(51):11007–11028. [Google Scholar]
- 17.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377(Pt 1):159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusnak DW, Alligood KJ, Mullin RJ, Spehar GM, Arenas-Elliott C, Martin AM, Degenhardt Y, Rudolph SK, Haws TF, Jr, Hudson-Curtis BL, Gilmer TM. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40(4):580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv Drug Deliv Rev. 2005;57(1):63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Sasi N, Hwang M, Jaboin J, Csiki I, Lu B. Regulated cell death pathways: new twists in modulation of BCL2 family function. Mol Cancer Ther. 2009;8(6):1421–1429. doi: 10.1158/1535-7163.MCT-08-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahan SD, Ireton GC, Knoeber C, Stoddard BL, Black ME. Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy. Protein Eng Des Sel. 2004;17(8):625–633. doi: 10.1093/protein/gzh074. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas TW, Read SB, Burrows FJ, Kruse CA. Suicide gene therapy with Herpes simplex virus thymidine kinase and ganciclovir is enhanced with connexins to improve gap junctions and bystander effects. Histol Histopathol. 2003;18(2):495–507. doi: 10.14670/HH-18.495. [DOI] [PubMed] [Google Scholar]
- 24.Black ME, Kokoris MS, Sabo P. Herpes simplex virus-1 thymidine kinase mutants created by semi-random sequence mutagenesis improve prodrug-mediated tumor cell killing. Cancer Res. 2001;61(7):3022–3026. [PubMed] [Google Scholar]
- 25.Hamel W, Magnelli L, Chiarugi VP, Israel MA. Herpes simplex virus thymidine kinase/ganciclovir-mediated apoptotic death of bystander cells. Cancer Res. 1996;56(12):2697–2702. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western Blot analysis (left panel) and SDS-PAGE (right panel) of the purified FDT biopolymer. The expected molecular weight for FDT is 18,210 daltons. Lane 1: lysate; Lane 2: flow through; Lane 3: protein marker; Lane 4: purified FDT biopolymer.


