Abstract
Retinoid X receptors (RXRs) are ligand-dependent nuclear receptors, which are activated by the potent agonist 9-cis retinoic acid (9cRA). 9cRA binds to the ligand binding domain (LBD) of RXRs, and recruits coactivator proteins for gene transcription. Using isothermal titration calorimetry, the binding of a 13-mer coactivator peptide, GRIP-1, to the hRXRα-LBD homodimer complex containing 9cRA (hRXRα-LBD:9cRA:GRIP-1) is reported between 20° and 37 °C. ΔG is temperature independent (−8.5 kcal/mol), and GRIP-1 binding is driven by ΔH (−9.2 kcal/mol) at 25 °C. ΔCp is large and negative (−401 cal/mol-K). The crystal structure of hRXRα-LBD:9cRA:GRIP-1 is reported at 2.05 Å. When the structures of hRXRα-LBD:9cRA:GRIP-1 and hRXRα-LBD:9cRA (1FBY) homodimers are compared, E453 and E456 on helix 12 bury and form ionic interactions with GRIP-1. R302 on helix 4 realigns to form new salt bridges to both E453 and E456. F277 (helix 3), F437 (helix 11), and F450 (helix 12) move toward the hydrophobic interior. The changes in the near-UV spectrum at 260 nm of the hRXRα-LBD:9cRA:GRIP-1 support this structural change. Helix 11 tilts toward helix 12 by ≈ 1 Å modifying the ring conformation of 9cRA. Hydrogen-deuterium exchange mass spectroscopy indicate GRIP-1 binding to hRXRα-LBD:9cRA significantly decreases the exchange rates for peptides containing helices 3 (F277), 4 (R302), 11 (F437) and 12 (E453; E456). The structural changes and loss of dynamics of the GRIP-1 bound structure are used to interpret the energetics of coactivator peptide binding to the agonist-bound hRXRα-LBD.
Nuclear receptors (NRs) are an important class of signaling molecules that bind lipophilic ligands and lead to ligand-induced gene transcription (1, 2). NRs are tightly regulated in a ligand and coregulator dependent mechanism. Class II NRs such as peroxisome proliferator-activated receptors (PPARs), liver oxysterol receptors (LXRs), retinoic acid receptors (RARs), and the vitamin D receptor (VDR) are active as heterodimers in complex with the retinoid X receptors (RXRα, RXRβ, and RXRγ) (3). RXRs also form transcriptionally silent tetramers which convert to transcriptionally active homodimers or heterodimers in the presence of agonist (4). There are many RXR-signaling pathways, which control cellular proliferation, differentiation and growth in epithelial tissue as well as maintain proper lipid and glucose homeostasis (5, 6). 9-cis-retinoic acid (9cRA, Figure 1A) is a potent ligand for RXRs and RARs even though its role as a high affinity agonist in vivo is questioned (7, 8). 9cRA is approved clinically for the topical treatment of Kaposi's sarcoma, but it displays significant toxicity with oral administration to humans (9). To reduce the side-effects of the pan-agonist 9cRA, agonists selective for RXRs over RARs have been designed and evaluated. Targretin, an RXR-selective agonists approved for the oral treatment of cutaneous T-cell lymphoma, displays significantly fewer dose limiting toxicities than 9cRA (7, 10). To understand how agonists enhance RXR-signaling at the molecular level requires an understanding of how agonists induce structural and/or dynamical changes in RXRs to recruit coactivators and displace corepressors.
Figure 1.
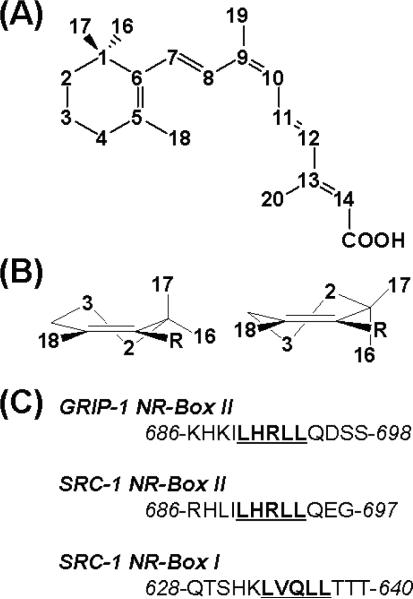
(A) The structure and numbering scheme of 9-cis-retinoic acid. (B) The two half-chair conformations of the trimethylcyclohexenyl ring of 9cRA are shown: the pseudo-axial orientation of the C-17 methyl group in the C-2 down conformation (left), and the pseudo-equatorial orientation of C-17 methyl in the C-2 up conformation (right). (C) The peptide sequences of GRIP-1 NR-Box I and SRC-1 NR-Box I and II coactivator peptides are shown.
The structural domains of RXRs include the N-terminal A/B domain, a highly conserved DNA-binding domain (DBD), a flexible hinge region, and a ligand-binding domain (LBD) that includes a ligand-dependant activation function-2 (AF-2) site and the dimer interface. Helix 12 (H12) of the LBD contains many core residues needed for ligand-dependent induction of transcription. Indeed when residues on this helix are mutated or the helix is truncated, the mutant RXR loses its capability to induce transcription, but mutant RXRs still bind 9cRA and heterodimerize (11). Normal development is significantly impaired in mice expressing mutant RXRα with H12 truncated (11, 12). In the crystal structure of hRXRα-LBD homodimer reported by Moras et al., H12 is extended from the rest of the LBD in the absence of ligand (13). Potent agonists like 9cRA bind to hRXRα-LBD homodimers and induce a substantial conformational change to H12 and other helices in this domain. Both H11 and H12 in the structure of the homodimer complex bound to 9cRA (hRXRα-LBD:9cRA) are folded to a compact structure and interact with the core of the hRXRα-LBD (14). Moras and coworkers proposed the mouse trap model (15), which suggests a potent agonist-bound NR complex is sufficient to lock H12 into a single conformation ready for coactivator binding. However, the residues on H12 make no direct interaction with the RXR agonist. The most direct contact occurs between the hydrophobic trimethylcyclohexenyl ring of 9cRA (Figure 1B) and hydrophobic residues of H11. The x-ray structure of hRXRα-LBD:9cRA homodimer without coactivator supports the view that 9cRA is sufficient to induce H12 to change conformation and form a cleft capable of binding the LxxLL motif contained in NR coactivator proteins/peptides. However, there are numerous solution studies indicating H12 in hRXRα-LBD homodimer complexes with agonists but without coactivator is dynamic and not fixed in a single conformation to the core of the LBD. Using hydrogen deuterium exchange coupled to mass spectrometry (HDX MS), the amide exchange rates of H12 in apo-hRXRα-LBD are fast and not significantly slowed when 9cRA is bound to this domain (16). Similar results are reported for other NRs (17). Li and coworkers used NMR to establish hRXRα-LBD:9cRA without coactivator peptide exists as a dynamic ensemble of conformations (18). These studies are consistent with fluorescent anisotropic measurement of dyes labeling the carboxyl terminus of hRXRα-LBD (19).
Signaling through NRs is influenced by not only the presence of agonist but also by the type and abundance of coactivators and corepressors in a cell (20, 21). Cells dosed with potent agonists lose their capability to induce transcription as the abundance of coactivators decreases or corepressors increases (21, 22). In most models of ligand-induced gene transcription of NRs, the coactivator binds to the surface of the holo-LBD, which has a hydrophobic pocket stable and ready to recognize the LxxLL motif and anchored by flanking residues constituting the charge-clamp (23). While this may be a reasonable description of the binding of coactivators to NRs containing agonists that make direct interactions with H12 (and thus directly stabilizing the structure of the coactivator binding site), this may not be a valid structural model to describe the recruitment of coactivators to the surface of NRs like RXRs. Without direct contact between the agonist and H12 residues, RXR-agonists must communicate their presence by changing the conformation and/or dynamics of residues in the ligand binding pocket (LBP) to enhance binding of coactivators. In this report we reveal these interactions by determining the structure of hRXRα-LBD:9cRA bound to the coactivator peptide GRIP-1 and we compare this structure to hRXRα-LBD:9cRA without coactivator (24). These structural studies are complemented by HDX MS, solution spectroscopy, and a thermodynamic analysis of the binding of the coactivator peptide GRIP-1 to hRXRα-LBD homodimers containing 9cRA. A model emerges for how GRIP-1 binds to the agonist-bound hRXRα-LBD. Coactivator peptide binding induces structural changes in the LBD which significantly reduces the dynamics of residues that are important for coactivator binding as well as those that make no direct contact with the coactivator but are near to the LBP of the agonist.
MATERIALS AND METHODS
Materials
9cRA was provided by Dr Grubbs at UAB, and its structure and purity was confirmed by NMR and LC-MS. A 13-mer peptide derived from residues 686 to 698 of the glucocorticoid receptor interacting protein-1 (GRIP-1) was synthesized by AnaSpec Inc. (KHKILHRLLQDSS) with a molecular weight of 1575.9 Da (sequence in Figure 1C). The concentration of peptide was determined by 1H NMR spiked with a known concentration of tryptophan. The 1D 1H NMR spectrum of the peptide was obtained with long delays between pulses to ensure accurate proton integration. The methyl peaks of the peptide were integrated and compared to the downfield resonances of tryptophan.
Protein Expression and Purification
The hRXRα-LBD (T223–T462) expression vector was kindly provided by Dr Ellen Li of Washington University in St Louis (18). Over expression and purification of the His6-tagged hRXRα-LBD fusion protein were performed as described by Egea and Moras (25) with a few modifications. BL21-(DE3) Escherichia coli bacteria (Invitrogen, Carlsbad, CA) containing the expression plasmid for hRXRα-LBD were grown in Luria broth (LB) medium at 20 °C, and protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside when the OD600 reached 0.6. After 18 h, the cells were harvested by 30-min centrifugation with a speed of 2500 rpm and resuspended in buffer A (20 mM Tris-HCL, 500 mM NaCl, pH 8.0). Cells were lysed by a French press under a pressure of 1500 psi and then centrifuged at 25000 rpm for 30 min. The clarified supernatant containing the His6-tagged hRXRα-LBD was loaded on a Ni-chelating column (GE health, Piscataway, NJ), washed with buffer A (20 mM Tris, 500 mM NaCl, 10 mM imidazole, pH 8.0, 30 CV) and then eluted with buffer B containing 300 mM imidazole, 500 mM NaCl, 20 mM Tris, pH 8.0. The eluted hRXRα-LBD fractions were dialyzed in 2 L buffer C (50 mM NaCl, 10 mM Tris, 0.5 mM EDTA, 2 mM DTT, pH 8.0) for at least 6 h. Human α-thrombin (Novagen, Madison, WI) was added at 4 °C and allowed to hydrolyze the His6-tag for 16 h. An additional Ni-chelating purification was used to separate hRXRα-LBD from any remaining His-tagged protein. The hRXRα-LBD, which did not bind the Ni-chelating column, was separated by a HiLoad Superdex 75 gel filtration column (GE health, Piscataway, NJ) at 4 °C in buffer C at 1.0 mL/min, and the fractions containing hRXRα-LBD homodimers were pooled. SDS-PAGE and MALDI mass spectrometry were used to establish a purity of >97% and mass of the monomers (m/z=26433.1 Da), and native-PAGE confirmed the oligomeric state of the pooled fractions were hRXRα-LBD homodimers.
Crystallization of hRXRα-LBD:9cRA:GRIP-1 Ternary Complex
Prior to forming the ternary complex, the affinity of hRXRα-LBD for 9cRA was measured using fluorescence quenching at 334 nm. The Kd of 9cRA binding to hRXRα-LBD was determined to be 14 ± 3 nM (data not shown) which agrees with the binding constant of 13 nM reported by Moras, et al (13). The ternary complex was prepared by adding 4-fold excess of 9cRA in methanol into a solution of hRXRα-LBD homodimers followed by adding 4-fold excess of GRIP-1. The protein complex solution was then concentrated to 10–15 mg/mL by centrifugation using Millipore Amicon Ultra-15 5000 MWCO centrifugal filter units. Crystals of hRXRα-LBD:9cRA:GRIP-1 complexes were obtained at 22 °C by use of a vapor diffusion method in hanging drops (24). All sample manipulations and the crystallization were performed under dimmed red light. Reservoir solutions contained 4–20% PEG4000, 4–16% glycerol, 0.1M Bis-Tris, pH7.0. Crystals of the hRXRα-LBD:9cRA:GRIP-1 complexes with suitable diffraction size preferred lower PEG4000 (4–10%) and glycerol (4–8%) concentration. These crystals belonged to the tetragonal space group P43212. Two monomers were present in each asymmetric unit. The crystals had two shapes. One with tetrahedral shape diffracted while the second with an elliptical shape did not diffract.
Data Collection, Structure Determination and Refinement
Crystals were flash frozen in liquid nitrogen using a reservoir cryoprotectant solution containing glycerol. The crystals were placed in stepwise increasing concentrations of glycerol (10%, 12.5%, 15%, 17.5%, 20.0%, 22.5%, and 25.0%) in order to avoid any damages to the crystals due to a sharp concentration change of glycerol. Diffraction data were collected using a Rigaku IV+ with a rotating copper anode. Diffraction data were processed using the HKL2000 program (26). The structure was solved by molecular replacement using the MOLPREP program in the CCP4 suite (27). The structure of hRXRα-LBD:DHA complex (PDB ID: 1MV9) with its ligand deleted was used as a search model. The ligand file and its associated topology and parameter files were created by the PRODRG program (28). The molecular replacement solution was refined in the Crystallography & NMR System (CNS) software (29) and interactive sessions of model building were performed in Quanta (Molecular Simulations Inc., Burlington, MA). The ligand was clearly identified by its electron density map. The protein and ligand files were merged in Quanta and the structure of hRXRα-LBD:9cRA:GRIP-1 complex was further refined in CNS. The refined structure of hRXRα-LBD:9cRA:GRIP-1 complex had an R factor of 20.5% and a free R of 24.2%. The complete summary of data for this structure is given in Table 1.
Table 1.
Crystallographic data-collection and refinement statistics for hRXRα-LBD:9cRA:GRIP-1 complex*
| Unit Cell parameters (Å, °) | a=65.83, b=65.83, c=1 11.86 |
| α=90, β=90, γ=90 | |
| Space group | P43212 |
| Resolution (last shell) (Å) | 50–2.05 (2.12–2.05) |
| No. of total/unique reflections | 229000/16156 |
| Rmerge (last shell) (%) | 0.071 (0.324) |
| Redundancy (last shell) | 14.2 (14.1) |
| Completeness (last shell) (%) | 100.0 (99.9) |
| I/sigma (last shell) | 13.2 (9.3) |
| Refinement | |
| No. of Residues | 213 |
| No. of protein atoms | 1791 |
| No. of water molecules | 144 |
| No. of ligand atoms | 22 |
| Rcryst (%) | 20.5 |
| Rfree (%) | 24.2 |
| R.M.S. Deviation from ideality | |
| Bond length (Å) | 0.0057 |
| Bond angles (°) | 1.135 |
| Ramachandran plot, residues in Most favored regions (%) | 94.98 |
| Additional allowed regions (%) | 3.20 |
| Generously allowed regions (%) | 1.82 |
| Average B factor (Å2) | 30.85 |
| Nonhydrogen protein atoms (Å2) | 29.78 |
| Nonhydrogen ligand atoms (Å2) | 39.79 |
| Water molecules (Å2) | 37.72 |
Analysis of hRXRα-LBD Contacts with 9cRA or GRIP-1 Peptide
The interactions between hRXRα-LBD and 9cRA or the interactions between the surface of the hRXRα-LBD and GRIP-1 peptide were determined using the program Ligand-Protein Contacts (LPC) (30) (Weizmann Institute of Science; http://bip.weizmann.ac.il/oca-bin/lpccsu). LPCs with an atom-to-atom distance less than 4.2 Å or a van der Waals surface area greater than 8 Å2 were examined (14). Contacts of Structural Units (CSU) (30) were also used to analyze interactions between GRIP-1 and hRXRα-LBD:9cRA. The volume of the LBP was calculated using VOIDOO (31) from Uppsala Software Factory of Sweden, and a readable format of LBP was created by the program MAPMAN (32).
Hydrogen Deuterium Exchange Experiments
Deuterium content was determined by comparing the molecular masses of undeuterated (control) and deuterated peptic peptides. For control experiments, a 10-fold dilution of purified hRXRα LBD homodimer (50 μM, 50 pmol) was incubated at 25 °C in buffer C in the presence and absence of ligand (500 μM in methanol). Ligand was added at 10-fold excess of protein to ensure saturation; an equal volume of methanol was used for unbound experiments (1 μL in 11 μL total deuterium on-exchange reaction). Hydrogen deuterium exchange experiments were initiated by a 10-fold dilution of hRXRα-LBD homodimer in the same buffer described above made with D2O. Time points were initiated upon addition of protein, samples taken at 15 s, 30 s, 120 s, 900 s, and 3600 s performed in triplicate. Deuterium on-exchange reactions were quenched with 1% (v/v) formic acid (final pH = 2.5) and immediately flash frozen in liquid nitrogen. Samples were stored at −80 °C until time of analysis.
Peptide Sequencing
Proteolytic digestion allowed regional monitoring of deuterium incorporation. Identification of hRXRα-LBD peptides was achieved by use of positive ion electrospray ionization (ESI) liquid chromatography (LC) tandem mass spectrometry (MS/MS) on a hybrid linear quadrupole ion trap 7 Tesla Fourier transform ion cyclotron resonance mass spectrometer (LTQ FT) (Thermo Fisher Scientific, San Jose, CA). Pilot experiments optimized protein digestion conditions (described below) and identified by data-dependent tandem LC MS/MS as previously described (33). Precursor ions were identified by use of the TurboSEQUEST algorithm available from Bioworks 3.2 software (Thermo Electron) filtered with a peptide mass accuracy limit of 5 ppm. All peptide ion assignments were verified by manual sequencing.
FT-ICR MS of HDX Samples and HDX Data Analysis
ESI LC MS analysis of HDX samples were performed under conditions optimized to preserve exchange information (0 °C, pH 2.5) (34). At the time of analysis, samples were rapidly thawed for 30 s at ambient temperature. Upon addition of pepsin (10 μL, 0.68 mg/mL, 0.05% formic acid) samples were immediately injected into a manual six port valve held in the load position and allowed to digest for 60 s in a 20 μL loop under ice bath conditions. Peptic peptides were then loaded onto a desalting C8 phase trap column (Michrom Bioresources, Inc.) and eluted with a rapid 5 m 5–95% (v/v) acetonitrile gradient, flow rate 50 μL/min as previously described (33). To determine the levels of deuterium incorporation for the peptic peptides, raw data files were converted to MxZML format and submitted for processing by The Deuterator data processing software (35)
Isothermal Titration Calorimetry Measurement of GRIP-1 Binding to hRXRα-LBD
ITC experiments were performed on a Microcal VP-Isothermal titration calorimeter (36). The concentration of hRXRα-LBD homodimers was determined by the absorbance at 280 nm using an extinction coefficient of 16.9 mM−1 cm−1 per hRXRα-LBD monomer. 9cRA was dissolved in degassed DMSO and added to the hRXRα-LBD solution at a ratio of 2:1 (retinoid:hRXRα-LBD monomer). Each titration experiment consisted of 30 injections of 8 μL of GRIP-1 peptide (0.4–1.5 mM) into the sample cell containing 1.34 mL of hRXRα-LBD:9cRA (40–60 μM) in buffer C at 20°, 25°, 30° and 37° C. Background reaction enthalpy was determined from injection of buffer into protein or from each peptide into buffer. The background enthalpy values were subtracted from the raw titration data prior to curve fitting. Titration curves were fit to a single-site binding model provided by the ORIGIN 7 software (Microcal, Northampton, MA). The C value (C = PTKa, where Ka is the association constant and PT is the total protein concentration) was kept in the range of 30–60 to optimize Ka measurements (37). The heat capacity change for GRIP-1 binding was determined by measuring the enthalpy change at three temperatures and fitting the corrected enthalpies versus temperature by linear regression.
Far-UV CD Spectroscopy
The CD spectra of hRXRα-LBD homodimers (5 μM) were obtained with a Jasco J-815 spectropolarimeter in buffer C at 25 °C. Spectra were acquired in a 0.1-cm cell every 1 nm with a 1 nm bandwidth and a 4 s response. The spectra were baseline-corrected by subtraction of the buffer spectrum. The CD spectra of hRXRα-LBD:9cRA homodimer complex (prepared with 4-fold excess of 9cRA to achieve > 99% saturation) was measured with or without 20 μM of GRIP-1 (37). The far-UV CD spectra were analyzed using CDSSTR (38) and SELCON3 (39) programs. A solution of hRXRα-LBD:9cRA homodimers (5 μM) was titrated with 0.5 mM of GRIP-1 solution.
Near-UV-Vis Absorption Spectra
Near-UV absorbance spectra were collected using a Cary 100 Conc UV-Vis Spectrometer (Varian) or by converting the dynode voltage of a Jasco J-815 spectropolarimeter to absorbance. 9cRA (4.2 mM in methanol) was added to 30 μM of hRXRα-LBD homodimers in buffer C to achieve 95% saturation based on the Kd of 14 nM. The spectra (average of 4 scans) of hRXRα-LBD homodimers or hRXRα-LBD:9cRA homodimers were measured between 250 and 450 nm in 0.5 nm increments with an averaging time of 4 s using a 1-cm quartz cell. The difference spectrum between hRXRα-LBD:9cRA:GRIP-1 and hRXRα-LBD:9cRA homodimers was measured in buffer C at 25 °C. Identical preparations of hRXRα-LBD:9cRA were placed in sample and reference cells to establish a baseline. Then, 54 μL of a 7.0 mM GRIP-1 solution in buffer C was added to 3.0 mL of the protein (2.5-fold excess) in the sample cell, an equal volume of buffer was added to the reference cell, and the difference spectrum was obtained as an average of 4 scans.
The near-UV-Vis difference spectrum was simulated by first measuring the near UV-Vis spectra of the aromatic amino acids in buffer C. Using Microsoft Excel, the λmax and εmax of the component amino acid spectra were summed according to aromatic amino acid composition of hRXRα-LBD: εcalc = 2*εmax(W) + 4*εmax(Y) +10* εmax(F) (40). The sum of square of error (SSE) between calculated and observed spectra were minimized to fit the hRXRα-LBD homodimer near-UV spectrum. Next, the absorption spectrum of 9cRA bound to hRXRα-LBD was obtained by subtracting the spectrum of hRXRα-LBD:9cRA from the apo-spectrum. Gaussian functions were used to fit the two π-π* transitions from 9cRA (trans and cis bands). (Different bandwidths were used for the long-wavelength and short-wavelength side of the asymmetric absorption band centered near 334 nm.) The calculated absorption spectrum of hRXRα-LBD:9cRA homodimers was obtained by optimizing the extinction coefficients of the aromatic residues and the 9cRA transitions. The calculated spectrum of hRXRα-LBD:9cRA:GRIP-1 was fit to the experimental difference spectrum by nonlinear optimization of the extinction coefficients for the aromatic residues to minimize the SSE between calculated and measured spectra.
RESULTS
Binding of GRIP-1 Peptide to hRXRα-LBD:9cRA
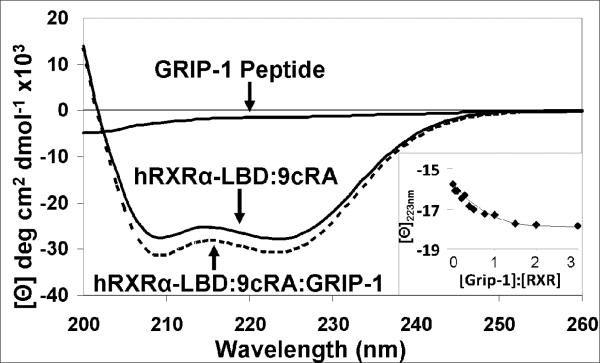
Vogel et al demonstrated the overexpression of peptides corresponding to GRIP-1 (Figure 1C) significantly enhanced RXRα-mediated gene expression when 9cRA was present (41). The 13-mer GRIP-1 NR-Box II peptide was also used to facilitate the crystallization of hRXRα-LBD homodimers (24) and used in NMR experiments on hRXRα-LBD homodimers (37). Based on these previous studies, this peptide sequence was chosen for our studies. The far-UV CD spectrum of the GRIP-1 peptide was measured in solution at pH 8. As displayed in Figure 2, the CD spectrum was near baseline with a small negative band appeared at wavelengths less than 210 nm. In contrast the far-UV CD spectrum of the hRXRα-LBD:9cRA homodimer displayed significant negative bands at 209 and 223 nm, consistent with high helical content (calculated helical content was 65% versus 61% from x-ray studies). When GRIP-1 was added in 3-fold excess, the intensity of negative bands at 209 and 223 nm increased in magnitude by −1850 deg-cm2-decimol−1, which is consistent with induced helical structure of GRIP-1 bound to hRXRα-LBD:9cRA or increased helical secondary structure of hRXRα-LBD. The spectroscopic change was used to measure the binding affinity of GRIP-1 to hRXRα-LBD:9cRA homodimers. A titration of GRIP-1 produced a binding isotherm which was fit to a Kd of ≈ 0.9 ± 0.1 μM at 25 °C (see inset Figure 2).
Figure 2.
Far-UV CD spectrum of hRXRα-LBD:9cRA homodimers in the absence (Solid) and presence (Dashed) of 3-fold excess GRIP-1 coactivator peptide. The far-UV CD spectrum of GRIP-1 coactivator peptide in solution is presented. INSET: The change in mean molar ellipticity at 223 nm for the titration of GRIP-1 into hRXRα-LBD:9cRA homodimers.
The energetics of GRIP-1 binding to hRXRα-LBD:9cRA homodimers were measured by ITC using similar conditions to those used in the CD experiment. The binding of GRIP-1 to hRXRα-LBD:9cRA was nearly stoichiometric for each GRIP-1 binding site on the hRXRα-LBD (Table 2). The isotherms were fit to a Kd of 0.55 ± 0.02 μM at 25 °C. Isotherms for GRIP-1 binding to hRXRα-LBD:9cRA were also measured using ITC at three other temperatures with little change in Kd (Table 2). At 25 °C, the GRIP-1 binding was driven by enthalpy with little entropy contribution. At 37 °C, even though the free energy of binding was essentially temperature independent, GRIP-1 binding was highly exothermic to overcome a significant entropy contribution opposing coactivator peptide binding. The heat capacity change (ΔCp) for GRIP-1 binding was determined from the temperature dependence of ΔH at 20, 25 and 30 °C. The heat capacity change was large and negative (ΔCp = −401 ± 18 cal-mol−1-K−1). To judge if buffer ionization significantly affected the binding enthalpy, these experiments were repeated in HEPES and phosphate buffer at pH 8. No significant change in enthalpy was observed (Supporting Information Figure S2).
Table 2.
Thermodynamic Parameters for the Binding of GRIP-1 Coactivator Peptide to hRXRα-LBD:9cRA Homodimers.a
| Temperature | Kd | ΔH | −TΔS | ΔG | n | ΔCp |
|---|---|---|---|---|---|---|
| (°C) | (μM) | (kcal/mol) | (kcal/mol) | (kcal/mol) | cal mol−1 K−1 | |
| 20 | 0.47±0.02 | −6.78±0.02 | −1.71±0.03 | −8.49±0.02 | 0.98 | |
| 25 | 0.55±0.02 | −9.15±0.03 | 0.61±0.02 | −8.54±0.03 | 0.92 | −401±18 |
| 30 | 0.72±0.01 | −11.13±0.03 | 2.61±0.03 | −8.52±0.03 | 0.95 | |
| 37 | 1.00±0.05 | −13.65±0.04 | 5.11±0.04 | −8.53±0.08 | 0.93 | |
The ITC data were fit to a single-site binding model (assuming independent binding sites for GRIP-1 on each hRXRα-LBD monomer). The ITC experiments were repeated three times for 20°. 25° and 30 °C. The enthalpies for these temperatures were used to calculate ΔCp.
X-Ray Crystal Structure of hRXRα-LBD:9cRA:GRIP-1 Complex
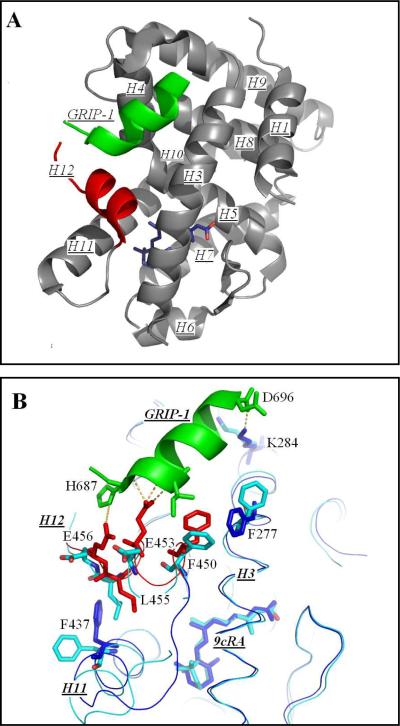
In order to identify structural changes from the binding of a coactivator to a NR homodimer, the hRXRα-LBD:9cRA homodimer was crystallized with the GRIP-1 peptide. The x-ray structure was determined to 2.05 Å resolution using the molecular replacement method. The asymmetric unit contained two monomers with GRIP-1 bound to each. The tertiary structure of each monomer displayed a very similar α-helical sandwich structure found in most NR LBDs (Figure 3A). Residues for 11 out of the 12 helices were resolved (H2 missing). GRIP-1 adopted a two-turn helix and sat in a cleft formed by H3, H4 and H12 residues (Figure 3A). This structure was compared directly to the hRXRα-LBD:9cRA complex without coactivator peptide (PDB ID: 1FBY) (14). The backbone atoms of helices that did not undergo large conformation changes upon GRIP-1 binding (H5; H6; H7; H8; H9; H10 and the beta structure, 75 residues in total) were overlaid. The RMSD of the 229 residues of the peptide backbone was 0.157 Å. GRIP-1 binding induced tertiary structural changes which occurred near the GRIP-1 binding site (H3, H4, and H12 residues) but also extended to H11 residues that interacted with the trimethylcyclohexenyl ring of 9cRA.
Figure 3.
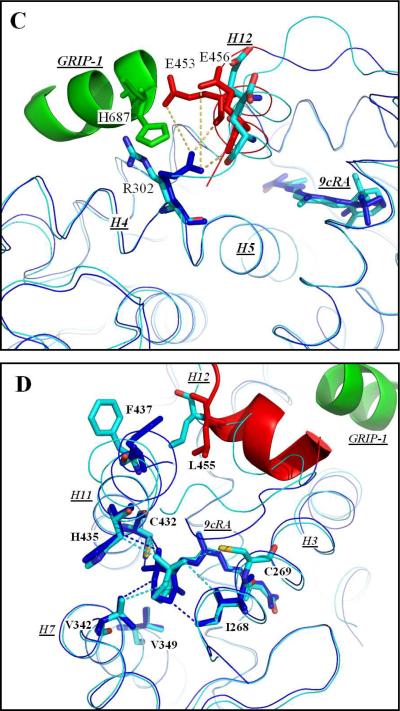
The structure of hRXRα-LBD homodimers bound to 9cRA and to GRIP-1. (A) the tertiary conformation of one monomer of the hRXRα-LBD homodimer containing 9cRA (Dark Blue) and coactivator peptide GRIP-1 (Green). Helices 1 through H11 are numbered (H2 missing) and shown in Grey. H12 is colored Red. The amino acid sequences of the helices are: H1 (P231-E243), H3 (D263-R285), H4 (L294-A303), H5 (G304-V320), H6 (R334-S339), H7 (V342-M360), H8 (D363-F376), H9 (N385-K407), H10 (R414-L419), H11 (L422-G443), and H12 (T449-L455). (B) A comparison of hRXRα-LBD:9cRA:GRIP-1 (PDBID-3OAP) and hRXRα-LBD:9cRA (PDBID-1FBY). The structure of hRXRα-LBD:9cRA:GRIP-1 is displayed in Dark Blue for all residues except for H12 which are shown in Red. The structure of hRXRα-LBD:9cRA is displayed in light blue. Significant changes occur for F450, E453, and E456 on H12 (Red), and for F437 on H11 and F277 on H3 (Dark Blue). Yellow dashes refer to new ionic or hydrogen bonding interactions. (C) The structural changes of R302 on H4 (Dark Blue) and E453 and E456 on H12 (Red). New ionic interactions/hydrogen bonds are shown in Yellow. (D) The structural change of the trimethylcyclohexenyl ring of 9cRA in hRXRα-LBD:9cRA:GRIP-1 (Dark Blue) and hRXRα-LBD:9cRA (Light Blue). The trimethylcyclohexenyl ring inverts from one half-chair conformation to the other. Hydrophobic interactions with H11 residues are shown in Dark Blue dashes for the GRIP-1 bound structure and in Light Blue dashes for the structure without GRIP-1.
Residues on H3, H4 and H12 of hRXRα-LBD form the coactivator binding site on nuclear receptors (23). From structural studies on coactivator binding to hRXRα-LBD homodimers using other rexinoids (20, 42) or to hRXRα-LBD:9cRA heterodimers (43–45), coactivators bind to the surface of nuclear receptors by interacting with both hydrophobic and hydrophilic residues. As reported by Nolte et al (23), the LxxLL motif of SRC-1 NR-Box I and NR-Box II (sequences compared in Figure 1C) were positioned in the PPARγ LBD binding site by charge clamps. The N-terminal of SRC-1 (K632; L633 of NR-Box I; and K688, I689 and L690 of NR-Box II) formed a charge clamp with E471. Similarly, the C-terminal of SRC-1 (L636, T639 on NR-Box I; L693, L694 on NR-Box II) formed another charge clamp with K301. For the hRXRα-LBD:9cRA:GRIP-1 homodimer presented here, two charge clamps were also observed at the N- and C-terminal ends of GRIP-1 (Figure 3B). E453 on H12 was solvent exposed in hRXRα-LBD:9cRA without coactivator peptide (1FBY). Upon GRIP-1 binding, the carboxylate group of E453 moved toward GRIP-1 and formed 3 hydrogen bonds with the N-H amides of K688, I689, and L690 (2.8 to 3.2 Å). The alkyl ammonium side-chain of K284 of H3 moved from a solvent-exposed environment to one that formed strong hydrogen bonds to GRIP-1. This residue formed the second charge clamp with the amide carbonyl oxygen atoms of L693 and D696 of GRIP-1 (≈ 2.5 Å). In addition to these ionic interactions, the carboxylate of E456 on H12 formed a charge-charge interaction with the imidazole ring nitrogen of H687 of GRIP-1 (4.3 Å). This salt bridge was not observed in holo-hRXRα-LBD/hPPARγ-LBD heterodimers bound to SRC-1 (1FM6) (Supporting Information Figure S3).
In each monomer of the hRXRα-LBD:9cRA:GRIP-1 homodimer, three leucine residues (L690, L693, L694) in LxxLL motif of GRIP-1 and I689 at the -1 position in the LxxLL motif made hydrophobic contacts with several residues on H3 (F277, V280, and K284), H4 (L294, Q297, V298, L301, R302), and H12 (T449, F450) of hRXRα-LBD. The hydrophobic surface area contacts between GRIP-1 and hRXRα-LBD were significant (33.7 Å2 for I689; 115.4 Å2 for L690; 84 Å2 for L693; 118 Å2 for L694). In addition to these hydrophobic interactions, significant hydrophobic interactions occurred between I689 and T449 and F450 on H12 and with F277 on H3. (Table S1 of the Supporting Information summarizes closest atom-to-atom distances between GRIP-1 residues and hRXRα-LBD, and the total van der Waals contact surface area). The phenyl group of F450 on H12 was in van der Waals contact with the sec-butyl and isobutyl side chains of I689, L690 and L693 of GRIP-1. F450 changed conformations upon GRIP-1 binding, and moved from a solvent exposed environment toward the interface of the hydrophobic residues of the amphipathic GRIP-1 helix (Figure 3B). F277 on H3 rotated from a surface-exposed environment toward the interior of the protein to form new hydrophobic interactions with L694 of GRIP-1. The two phenyl rings of F277 and F450 were oriented at a 72° dihedral angle, and the distance between the two phenyl ring centroids was 5.3 Å (Figure 3B), suggestive of a Caro-H – π interaction (46).
The polar side-chain of R302 on H4 protruded into the solvent in the hRXRα-LBD:9cRA structure without GRIP-1. In the structure reported here, GRIP-1 binding to hRXRα-LBD:9cRA induced R302 to flip ~90° toward H12 in order to avoid steric interaction with the coactivator peptide (Figure 3C). The guanidinium nitrogen atoms of R302 were coordinated to the carboxylate oxygens of E453 (5.54 Å) and E456 (6.42 Å) on H12, consistent with the formation of two salt bridges. Additionally, the guanidinium group of R302 and the carbonyl oxygens of E453 (3.13 Å) and E456 (2.74 Å) were in close contact and consistent with two hydrogen bonds formed between the nitrogen atoms on R302 and the amide backbone of GRIP-1.
GRIP-1 Binding Induces Structural Changes in the LBP
Three residues on H12 (F450, E453 and E456) and two residues on H3 (F277, K284) were solvent exposed in the hRXRα-LBD:9cRA homodimer, but were found to interact with GRIP-1 in the hRXRα-LBD:9cRA:GRIP-1 homodimer. The carboxyl end of H12 moved 2 Å toward GRIP-1. In response to this change, the position of H11 was moved closer to H12. The phenyl ring of F437 on H11 rotated from a solvent exposed environment toward L455 on H12 (Figure 3D). H11 residues interact with 9cRA near the trimethylcyclohexenyl ring. The change in the orientation of H11 in the GRIP-1 bound structure influenced the structure of 9cRA in the LBP. When coactivator was not bound to hRXRα-LBD:9cRA, the C-16 methyl of 9cRA had contact with C269 (28 Å2) and with I268 (7 Å2) on H3. Smaller contacts occurred for C-16 methyl with residues H435 and L436 on H11. The C-17 methyl group made contacts with H11 residues: C432 (10 Å2), H435 (32 Å2), and F439 (5 Å2). In the GRIP-1 bound structure, the C-16 methyl group of 9cRA did not interact with H3 residues (C269 and I268), but it formed new interactions to the H11 residues, C432 (23 Å2) and H435 (13 Å2). The contacts between the C-17 methyl on 9cRA and H435 on H11 were reduced. However, the contact between C-17 and C432 on H11 increased by 18 Å2, and a new contact appeared between C-17 with I345 of H7 (20 Å2). Changes occurred in the interactions for other carbons in the trimethylcyclohexenyl ring of 9cRA and residues in the LBP. The C-18 methyl group lost contact between V349 on H3 and C432 on H11 in the GRIP-1 bound structure. C-2 formed new interacts with V342 on H7 (15 Å2) and F439 on H11 (9 Å2), while the contacts between C-3 and C-4 and H3 and H7 residues were weakened. Due to these significant changes in LBP contacts with 9cRA, the conformation of trimethylcyclohexenyl ring switched from one half-chair conformation (with C-2 pointed toward H7 and C-3 pointed toward H11) to the alternate half-chair conformation (Figure 1B; Figure 3D). In the GRIP-1 bound structure, the surface area of contact of 9cRA with the H11 residues increased from 118 Å2 to 140 Å2.
The change in trimethylcyclohexenyl ring interactions with H3, H7 and H11 residues also altered the orientation of the retinoid ring to the tetraene chain of 9cRA. The C6–C7 torsional angle of 9cRA was −70 ° without GRIP-1 and decreased to −20° with GRIP-1 (data not shown). The L-shaped conformation was still present with GRIP-1. The interactions of the methyl-substituted tetraene chain of 9cRA with residues in the LBP were substantial for both structures, and the differences in contacts between these residues were small (< 8 Å2) for all carbons except for the C-19 and C-20 methyl groups and the C-15 carboxylate. In the GRIP-1 bound structure, the C-19 methyl of 9cRA formed a stronger contact with L436 on H11 (increased by 18 Å2), while the contact between C-20 methyl and I268 was reduced by 14 Å2. A small change was observed in the interaction of the C-15 carboxylate group of 9cRA with residues A271, R316 and L326 and A327. C-15 moved 0.6 Å away from R316 in the structure with GRIP-1 bound. The surface area contacts between C-15 and R316, A327, and L326 decreased by about 30% in the GRIP-1 bound structure.
Near-UV-Vis Spectroscopic Studies
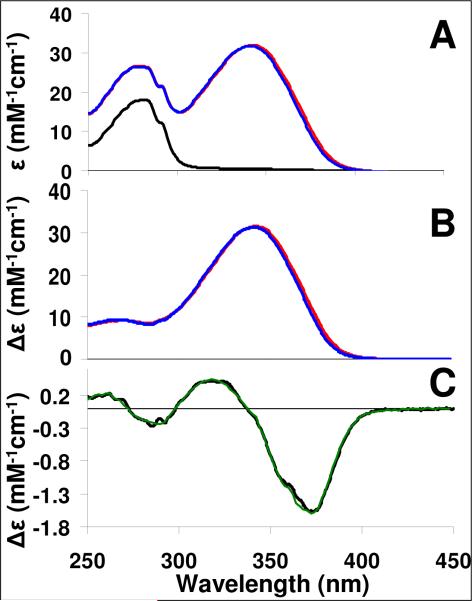
In order to provide evidence that the structural changes observed in the x-ray structures occurred in solution, the near-UV spectrum of hRXRα-LBD:9cRA homodimers was measured in the presence and absence of GRIP-1. 9cRA was added to apo-hRXRα-LBD to achieve 95% saturation. At these protein concentrations, the amount of free 9cRA was less than 1% based on our measured Kd of 14 nM. The near-UV spectrum of hRXRα-LBD:9cRA displayed two intense absorbance bands with λmax values at 334 and 280 nm (Figure 4A). The spectrum of 9cRA was revealed by subtracting the spectrum of apo-hRXRα-LBD from the spectrum of hRXRα-LBD:9cRA (Figure 4B). In the subtracted absorption spectrum, a cis band arising from 9cRA was revealed at 267 nm. In order to evaluate the spectral differences due to the addition of GRIP-1, the difference spectrum was measured between hRXRα-LBD:9cRA with GRIP-1 (2.5 fold excess) and without GRIP-1 (Figure 4C). In this spectrum, the long-wavelength π-π* of 9cRA shifted to the blue by about 1 nm, resulting in a difference spectrum containing a negative band centered at 370.5 nm, a positive band at 322.5 nm, and a cross-over at 342 nm. In addition to these spectral changes, the near-UV spectrum in the 280 nm range also changed. Negative bands appeared at 285 and 292 nm, and a positive band was centered at 259 (with less intense maxima at 254-nm and 265-nm). Second derivative spectra of the near-UV region (250–300 nm) demonstrated the wavelength maxima of the near-UV absorbance (280-nm band) did not change when GRIP-1 was added. This indicated the observed spectral changes in the near-UV region were due to changes in the extinction coefficients of the transitions. In contrast, the second derivative data clearly showed a 1 nm shift to the blue for the 334-nm band of 9cRA when GRIP-1 was added.
Figure 4.
(A) Near UV-Vis spectrum of apo-hRXRα-LBD homodimer (black), hRXRα-LBD:9cRA complex in the absence of GRIP-1 (Red), and hRXRα-LBD:9cRA:GRIP-1 complex (Blue). (B) Difference UV-Vis spectrum of hRXRα-LBD:9cRA complex (Red) or the hRXRα-LBD:9cRA:GRIP-1 complex (Blue) minus the spectrum of apo-hRXRα-LBD. (C). Difference UV-Vis spectrum of hRXRα-LBD:9cRA:GRIP-1 minus the spectrum of hRXRα-LBD:9cRA (Black) versus the computer-fitted difference spectrum (Green).
A simulated near-UV region difference spectrum was constructed to gain further insight into our difference measurement. Using the spectra for apo-hRXRα-LBD and hRXRα-LBD:9cRA, the simulated near-UV-Vis difference spectrum was calculated for hRXRα-LBD:9cRA and hRXRα-LBD:9cRA:GRIP-1 by summing the constituent spectra as detailed in the methods section. The simulated difference spectrum agreed remarkably well with the experimentally measured difference spectrum (Figure 4C). The best-fit simulation yielded three notable changes in the near-UV-Vis absorption spectra of hRXRα-LBD:9cRA upon GRIP-1 binding: (a) the long-wavelength π-π* of 9cRA was shifted to the blue by ~1 nm without a change in εmax, (b) the εmax of tryptophan residues and tyrosine were decreased by 1% without change in wavelength maximum, and (c) the εmax of phenylalanine residues were increased by 22% without change in wavelength maximum. Solvent effects on the intensity and wavelength position of phenylalanine absorption demonstrated nonpolar solvents increased the extinction coefficient of the phenylalanine relative to polar ones (47). The change in solvent exposed surface area was calculated for the R-groups of each phenylalanine residue from the two crystal structures. The solvent exposed surface area of three phenylalanines, F437, F450 and F453, decreased by 18%, 71% and 36%, respectively, with little change for other aromatic residues (Figure 3B). The phenyl ring of F450 was nearly completely buried from solvent by GRIP-1. The 285-nm and 292-nm absorbance were attributed primarily to tryptophan absorbance. Two tryptophan residues are present in hRXRα-LBD. W305 is near 9cRA, 3 Å from the C-19 methyl group, and W282 is located on H3 8 Å away. Even though both tryptophan residues were completely buried in both x-ray structures, the C-19 methyl of 9cRA moved about 0.5 Å away (and displayed 7 Å2 less contact) with the indole ring of W305 in the GRIP-1 bound structure.
HDX MS analysis of hRXRα-LBD complexes
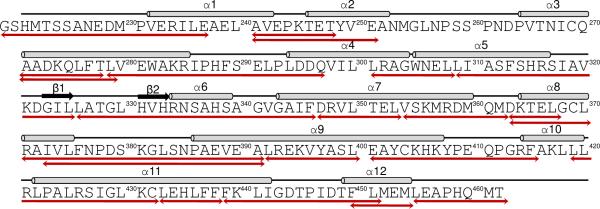
The in-solution dynamics of these hRXRα-LBD complexes in the presence and absence of the GRIP-1 coactivator peptide were further analyzed by hydrogen deuterium exchange coupled with high-resolution mass spectrometry (HDX MS). The backbone amide hydrogen exchange for deuterium at variable rates were measured for four hRXRα-LBD homodimer complexes: hRXRα-LBD, hRXRα-LBD:GRIP-1, hRXRα-LBD:9cRA, and hRXRα-LBD:9cRA:GRIP-1. The preparations were pepsin digested prior to mass spectrometry analysis, which allowed regional assessment of hydrogen-deuterium exchange events. Thirty-eight peptic peptides comprising 88% of hRXRα-LBD were identified with high confidence (< 2 ppm) by FT MSMS. Deuterium incorporation for all peptides was calculated by monitoring centroid mass shifts across a series of time points for the four different homodimer preparations. Deuterium incorporation increased as a function of time for hRXRα-LBD in all observed states. The averaged differences in deuterium incorporation percentages for five on-exchange time points prepared in triplicate (15 s, 30 s, 120 s, 900 s, 3600 s) relative to apo-hRXRα-LBD are shown in Table 3 for 24 of the observed peptides from the three different preparations (apo-hRXRα-LBD:GRIP-1; hRXRα-LBD:9cRA; hRXRα-LBD:9cRA:GRIP-1). The remaining 14 observed overlapping peptides (Supplemental Table S4) served to corroborate these 24 peptides in Table 3. Figure 5 overlays the 24 peptides in Table 3 onto the sequence of hRXRα-LBD. A negative percentage value represents a reduction in the number of hydrogen deuterium exchange events compared to the number observed for the unbound domain, which is an indication of an increased level of protection for that region. Values shaded in increasing intensities of blue indicate increasing levels of protection for each peptide. Our data clearly demonstrated that 9cRA and/or GRIP-1 binding caused region-specific reductions in deuterium incorporation. Out of those 38 peptides, 9 peptides representing 23% of the domain showed no significant change in deuterium incorporation levels (<5%) across all complexes (shaded in gray in Table 3).
Table 3.
Averaged differences in deuterium incorporation percentages for twenty-four hRXRα-LBD peptic peptides relative to apo-hRXRα-LBD for three conditions.
| Peptide Sequence | Secondary Structure | hRXRα-LBD | hRXRα-LBD:GRIP-1 | hRXRα-LBD:9cRA | hRXRα-LBD:9cRA: GRIP-1 |
|---|---|---|---|---|---|
| G219SHMTSSANEDMPVERILE237 | H1 | 73% | −5 | −3 | −5 |
| A241VEPKTET248 | H1 | 100% | −1 | 0 | 1 |
| A241VEPKTETYVE251 | H1 | 100% | −8 | −3 | −11 |
| A271ADKQLFT278 | H3 | 99% | −14 | −15 | −30 |
| A271ADKQLFTL279 | H3 | 100% | −9 | −28 | −39 |
| L279VEWAKRIPHFSELPLDDQ297 | H3 | 30% | −2 | −1 | −2 |
| L301RAGWNEL308 | H4 | 37% | −11 | −12 | −20 |
| L309IASFSHRSIAVKDGIL325 | H5 | 57% | −5 | −24 | −27 |
| L326ATGLHVHRNSAHSAGVGAIF346 | Beta | 65% | −5 | −9 | −12 |
| D347RVLTEL353 | H7 | 48% | −5 | −31 | −35 |
| V354SKMRDMQM362 | H7 | 69% | −8 | −31 | −38 |
| D363KTEL367 | H8 | 41% | −8 | −6 | −6 |
| D363KTELGCL370 | H8 | 20% | −4 | 0 | −1 |
| R371AIVLFNPDSKGLSNPAEVEA391 | H8 | 50% | −4 | 1 | −3 |
| I373VLFNPDSKGLSNPAEVEA391 | H8 | 53% | −4 | 0 | −5 |
| L392REKVYASL400 | H9 | 3% | −1 | −1 | 0 |
| E401AYCKHKYPEQPGRF415 | H9 | 40% | −2 | 1 | −2 |
| L419LRLPALRSIGLKC432 | H10 | 86% | −6 | −37 | −47 |
| L419LRLPALRSIGLKCLEHLFF438 | H10 | 91% | −9 | −26 | −37 |
| P423ALRSIGLKCLEHLFF438 | H11 | 88% | −8 | −25 | −37 |
| L433EHLFF438 | H11 | 80% | −8 | −1 | −10 |
| F439KLIGDTPIDTFL451 | H11 | 88% | −1 | −3 | −5 |
| F450LMEM454 | H12 | 92% | −17 | 4 | −14 |
| L455EAPHQMT462 | CTERM | 84% | −3 | −2 | −6 |
Values reflect differences in percent deuterium incorporation for each peptic peptide averaged across five time points (15 s, 30 s, 120 s, 900 s, 3600 s) relative to apo-hRXRα-LBD (column 3). Negative values indicate an increased level of protection for that peptide under the indicated conditions and are color coded in increasing shades of blue. Differences <5% are considered insignificant (shown in grey). Peptides are listed from N- to C- terminus, holo structural element indicated in column 2. Observed hRXRα-LBD amino acid contacts with 9cRA are in red. Observed hRXRα-LBD amino acid contacts with GRIP1 are underlined.
Figure 5.
HDX MS peptides. The 24 HDX MS peptides in Table 3 are mapped onto the hRXRα-LBD protein sequence. Structural features of the hRXRα-LBD are shown above the sequence. Arrows below the sequence correspond to the observed HDX MS peptides in Table 3. An additional 14 overlapping peptides that were observed are provided in Supplemental Table S4.
Dynamic Changes Induced by GRIP-1 Coactivator Peptide Binding to hRXRα-LBD:9cRA
HDX MS of hRXRα-LBD was performed in the presence of 10-fold excess of 9cRA (>99% saturation). Ligand binding elicited the greatest suppression in deuterium incorporation in peptides with residues directly involved or surrounding the ligand binding pocket (H3, H5, H7, H10 and H11). Every peptide containing residues directly involved in contact with 9cRA demonstrated a significant reduction in deuterium incorporation in the presence of 9cRA. Peptide L419-C432 (H10-H11) demonstrated the largest degree of protection (36%) and contained one reside (C432) that is directly involved in 9cRA binding. Surprisingly, peptide V354-M362 (H7) had no residues that directly contacted 9cRA, but it displayed a significant reduction in deuterium incorporation with 9cRA binding.
To investigate the dynamical effects of the binding of the coactivator peptide GRIP-1, we performed HDX MS of hRXRα-LBD:9cRA in the presence of 3-fold excess of GRIP-1 (>95% saturation based on Kd of 1μM). Compared to the HDX MS analysis of the hRXRα-LBD:9cRA complex, GRIP-1 binding resulted in further suppressions in deuterium incorporation for many of the same peptides (Table 3). For several of these peptides, there was an additional 5–12% reduction in deuterium incorporation that included H3, H4, H7, H10, and H11.. There were four additional peptides which also had significantly reduced deuterium incorporation profiles that were distinct from the hRXRα-LBD:9cRA complex. The peptide L433-F438 (H11) had little change in deuterium incorporation in the presence of 9cRA alone, but it was significantly altered upon the addition of GRIP-1 to the hRXRα-LBD:9cRA complex. In the plot of deuterium incorporation versus time for this peptide, the exchange of deuterium for apo-hRXRα-LBD homodimer complex was fast and reached >80 % deuterium incorporation by 90 s (Supporting Information Figure S4 C.). The addition of 9cRA did not change the rate of deuterium incorporation for this region, but addition of GRIP-1 induced a significant decrease in the early time points. This reflected an altered level of solvent accessibility for the C-terminal end of H11 when GRIP-1 was bound to hRXRα-LBD:9cRA. This same trend was also observed for the N-terminal region of the hRXRα-LBD spanning F450-T462 (H12). In ours and previous HDX MS analysis (16), H12 remained a fast exchanging region with unaltered rates of incorporation in both apo-hRXRα-LBD and hRXRα-LBD:9cRA homodimer complexes. However, our current results demonstrate the addition of GRIP-1 did significantly altered the solvent accessibility of H12.
In the GRIP-1 bound state, a significant decrease occurred in the deuterium incorporation of peptides located outside of the LBP or the GRIP-1 binding site. The protection of two peptides from H3, which include F277, was increased in the GRIP-1 bound structure. The x-ray structures demonstrated tertiary structural changes for F277 on H3 interacting with F450 on H12, which was supported by the near-UV difference spectra (Figure 4). Consistent with these changes, HDX MS peptides including F277 (A271-T278, A271-L279) and another containing F450 (F450-M454) displayed a decrease in deuterium incorporation indicating further stabilization of the AF-2 recognition helix upon coactivator peptide binding. This latter peptide also included E453, which was essential for formation of the charge clamp between several residues in the amino end of GRIP-1 and H12. Another region of the LBD which changed dynamics due to GRIP-1 binding was in H10 and H11. Four peptides in this region showed substantial decreases in deuterium incorporation. These helices contained several residues that interacted with the trimethylcyclohexenyl ring of 9cRA (C432; H435 and L436). Additionally they contained F437 on H11, which was observed to move from a water exposed environment (without GRIP-1 bound) toward L455 on H11 (Figure 3 B).
HDX MS of apo-hRXRα-LBD:GRIP-1 complexes
HDX MS of apo-hRXRα-LBD in the presence of GRIP-1 was performed for complete analysis (Table 3). Our results demonstrated that the presence of GRIP-1 caused hRXRα-LBD homodimers to undergo differential structural dynamics even in the absence of ligand. While reductions in deuterium incorporation were considerably smaller compared to the complexes discussed above, several peptides in the same regions within the hRXRα-LBD were altered. This included the H11 peptide L433-F438 and the N-terminal peptide F450-M454 that were only altered when GRIP-1 was present. These results suggested that the interactions between apo-hRXRα-LBD homodimers and GRIP-1 were similar (yet not as strong) to those observed between GRIP-1 and hRXRα-LBD:9cRA.
DISCUSSION
Using several methods, we provide in this study important structural, dynamical and thermodynamic information on the binding of a coactivator peptide, GRIP-1, to the LBD of hRXRα homodimers containing the potent agonist 9cRA. Upon binding, the 13-residue GRIP-1 adopts a helical structure involving 10-residues of its sequence centered on the ILHRLL hydrophobic motif. The GRIP-1 peptide binds stoichiometrically to each monomer of the hRXRα-LBD homodimer, with a Kd ≈1 μM (ΔG ~ −8.5 kcal/mol). Even though there are significant interactions between the ILxxLL motif on GRIP-1 and the hydrophobic residues in H3, H4 and H12 of hRXRα-LBD, the binding of GRIP-1 to hRXRα-LBD is strongly driven by enthalpy especially at 37 °C. Burying the ILxxLL sequence to a hydrophobic pocket on the surface of hRXRα-LBD:9cRA certainly is entropically favorable, but this favorable entropy must be compensated by other entropically unfavorable events. The large negative enthalpy contribution to the binding free energy is a result of many factors. An N-terminal charge clamp is observed between four N-H residues of the peptide bonds of H687, K688, I689, L690 on GRIP-1 residues and E453 on H12 (Figure 3 B and C), and a C-terminal charge clamp is present between D696 on GRIP-1 and K284 on H3 (Figure 3 B). These interactions were nearly identical to those reported for other NRs and coactivator peptides (24, 48). In addition to these electrostatic interactions, a new ionic interaction was identified in this study. E456, which is close to E453 on H12, forms an ionic interaction with H687 on GRIP-1 (Figure 3 B). This interaction is not observed in hPPARγ-LBD:SRC-1 complexes (23). R302 on H4 also changes its side chain conformation upon GRIP-1 binding. Two salt bridges and two hydrogen bonds between this residue on H4 with E453 and E456 on H12 locks in these two helices (Figure 3 C yellow dashes) (49).. Our HDX MS data verified that GRIP-1 binding produces a significant decrease in HD exchange rates of peptides containing R302 (H4/H5) and E456 (H12) (Table 3 L301-L308 and L450-T454). Previous mutational studies indicate R302 is an important residue in preventing the binding of corepressor SMRT-ID2 and promoting the recruitment of coactivator (50).
GRIP-1 binding to hRXRα-LBD:9cRA alters the tertiary structure of the NR LBD. Most significantly the phenyl rings of three phenylalanine residues (F277, F437, F450) on H3, H11 and H12 bury and become less solvent exposed (Figure 3 B). The near-UV difference spectral studies (Figure 4) support that this is not consequence of different crystallization conditions reported here for the hRXRα-LBD:9cRA:GRIP-1 homodimers versus those of Moras and coworkers for the hRXRα-LBD:9cRA without coactivator peptide (14). The conformational changes in hRXRα-LBD:9cRA upon GRIP-1 binding are also reflected in the dynamics of this region of the structure. Our HDX MS results indicate the N-H exchange rates are slowed for H3 and H11 peptides containing these phenylalanine residues (Table 3). These findings support the concept that GRIP-1 binding to the NR LBD surface stabilizes not only its interface, but also the secondary structure one layer inward toward the retinoid binding site. Noy and coworkers previously demonstrated F437 on H11 is an important residue in communicating transcriptional activation (51). This study supports the role F437 plays in the transmission of the conformational change toward the LBP of the NR LBD. The helical axis of H11 containing F437 on the carboxy-end moves toward the GRIP-1 binding site when coactivator peptide is bound to the NR LBD. The movement is small but it is significant enough to alter the conformation of the binding pocket of the retinoid especially near the trimethylcyclohexenyl ring. The ring of 9cRA flips between alternate half-chair conformations (C-2' up in hRXRα-LBD:9cRA; C-2' down in hRXRα-LBD:9cRA:GRIP-1). Numerous ligand-protein contacts change for the 9cRA especially for the gem-dimethyl groups, C-16 and C-17. The number of surface area contacts between trimethylcyclohexenyl ring atoms and H11 residues increase (Table S2 of the Supporting Information). The HDX MS data is consistent with these structural changes; three peptides containing F437 become significantly more protected when GRIP-1 is bound (Table 3). Significantly, the smallest of these overlapping peptides spanning L433-H438 was protected only when GRIP-1 is bound. Taken together, these data reflect that the dynamics of H11 residues are reduced due to two factors: increased hydrophobic interactions with 9cRA and increased hydrophobic/hydrophilic interactions with residues that form the GRIP-1 binding site. And these data establish a structural model for how retinoid agonists interact with the LBP residues on H11 to positively aid in recruitment of coactivators.
The HDX MS studies reveal the helical structure of hRXRα-LBD:9cRA:GRIP-1 homodimers is fluctuating less and is more stable than hRXRα-LBD:9cRA. As presented in this study, the addition of GRIP-1 reduces the overall number of deuterium incorporated compared to data from hRXRα-LBD:9cRA without coactivator (Table 3). The HDX MS protection factors reported here for apo-hRXRα-LBD and for hRXRα-LBD:9cRA without coactivator are very similar to studies done by Deinzer and coworkers (16). These studies each reveal that the H11 peptide (on the carboxyl end starting from L433) and the H12 peptide (F450-M454) are exchanging rapidly with solvent in both apo- and holo-complexes, even though the x-ray structure reported by Moras and coworkers reveal a compact structure with H11 and H12 folded into an active conformation. NMR solution measurements on hRXRα-LBD:9cRA homodimers also support the dynamical nature of the carboxyl terminal helices of hRXRα-LBD. Even with 9cRA present in the binding site, the NMR signals from H11 residues are undetectable (18). Many residues on H11 (I428–T449) including the three phenylalanine residues identified by Noy and coworkers (F437; F438 and F439) cannot be resolved (51). HDX MS studies support this finding especially for residues in the carboxy end of this sequence. 9cRA binding induces repositioning of H11, and is a key structural change needed for the movement of H12 into the active conformation. The HDX MS analysis demonstrates a significant reduction in deuterium incorporation for peptides from H12 only when GRIP-1 is bound. While the changes in H10 and H11 peptides are apparent in table 3, comparison of the overlapping HDX MS peptides that span this region in % deuterium vs. time plots provides more insight into the dynamical changes occurring in this region (Supplemental Figure S4 C and D). The deuterium incorporation of H11 peptide L419-F438 is reduced by >20% following 15 s of deuterium incorporation in the presence of 9cRA alone. Addition of GRIP-1 to the complex further reduces the incorporation an additional 10 % for the longer time points (2 min, 15 min, and 1 h).. The P423-F438 H11 peptide follows this same pattern. In contrast, the overlapping H11 C-terminal peptide (L433-F438) is different. The addition of 9cRA slightly increases the deuterium incorporation for the longer time points (2 min and 15 min) indicating a slight increase in dynamics for this portion of H11 after repositioning upon binding 9cRA. The resulting average difference is small (Table 3). Upon binding of GRIP-1 this peptide is reduced 10% in deuterium incorporation. In the case of H12, the influence in deuterium incorporation is exclusively from GRIP-1 binding (Table 3 and Supplemental S4 D). The analysis of the HDX MS data indicates the carboxyl end of H11 and H12 of hRXRα-LBD:9cRA are much more dynamic in conformation (or multiple conformations). Then H11 and H12 are considerably stabilized when coactivator peptide is bound suggesting a move towards a “single” conformation.
A comparison of the x-ray crystal structures also reveals GRIP-1 binding induces the formation of a Caro-H-π interaction between residues F277 and F450. A dramatic reduction in deuterium incorporation is found for peptide F450-M454 (H12 peptide) when GRIP-1 is bound. Additionally H3 and H4 peptides are also significantly protected when GRIP-1 is bound. Deuterium incorporation for peptide A271-L279 (H3) is reduced by greater than 25% following one hour of exchange in the presence of GRIP-1 (Supplemental Figure S4 A). This peptide contains four residues involved in ligand or coactivator binding. Peptide L301-L308 (H4) shows 20% reduction in deuterium incorporation upon the addition of GRIP-1 compared to 12% reduction in the presence of 9cRA alone (Table 3). This observation is supported by the x-ray crystal structure reported here which reveals the involvement of residue L301 in coactivator binding. The reduced deuterium incorporation of H3, H4 and H11 peptides indicates the structure is less dynamical in solution, which is in good agreement with structural changes found in x-ray crystal structures that identify new interactions involving residues in these peptides. Further the HDX MS studies reveals regions of hRXRα-LBD which are protected from deuterium incorporation in the presence of GRIP-1 (e.g., peptide L419-C432 in H10) even though the structure of this region of holo-complex did not change significantly in the presence or absence of GRIP-1.
X-ray crystal structures, spectroscopic studies and HDX MS measurements provide structural and dynamical views of GRIP-1 binding to hRXRα-LBD. The ITC titrations establish GRIP-1 binding to hRXRα-LBD:9cRA is driven by a large negative enthalpy change with a large negative heat capacity change (−401 ± 18 cal/mol-K, Table 2) at pH 8. The ΔCp value we measure for GRIP-1 binding to hRXRα-LBD:9cRA homodimers is significantly larger than that measured for SRC-1 coactivator peptide binding to hRXRα-LBD:9cRA heterodimers with CAR at pH 7.2 (52). Our ΔCp is similar in magnitude to that found for SCR-1 binding to apo-hRXRα-LBD:CAR heterodimers (36). The largest contribution to ΔCp from ligand binding is a result of the changes of the surface area between the ligand-bound complex and the surfaces of unbound protein and free ligand (53). Polar and nonpolar atoms contribute differently. Burying nonpolar atoms lead to a negative ΔCp while polar atoms that lose contact with water result in a positive ΔCp. Burying the hydrophobic residues of ILxxLL or hydrophobic residues (e.g., F277; F437; F450) leads to a large negative term in ΔCp, while burying the hydrophilic residues of the peptide (K686; H687: K688) and the residues involved in the charge clamp on H12 (E453; E456) opposes this trend. The large negative ΔCp is consistent with the importance of the hydrophobic forces involved with GRIP-1 recognition of hRXRα-LBD:9cRA.
Why is GRIP-1 binding to hRXRα-LBD not entropically driven given the amphipathic nature of GRIP-1 and obvious burial of nonpolar residues on the surface of hRXRα-LBD? In the absence of ionization effects for ligand binding as demonstrated here and with SRC-1 binding to RXR:CAR heterodimers (52), the most significant contributions to the entropy change of binding arise from three terms: 1. ΔSsolv, the increase in entropy from burial of hydrophobic groups in the complex away from the aqueous solvent; 2. ΔSrt, the decrease in entropy from loss of rotational and translational degrees of freedom in the complex; and 3. ΔSconf, the decrease in entropy from reduction of conformational freedom in the complex relative to the free peptide and hRXRα-LBD (54).
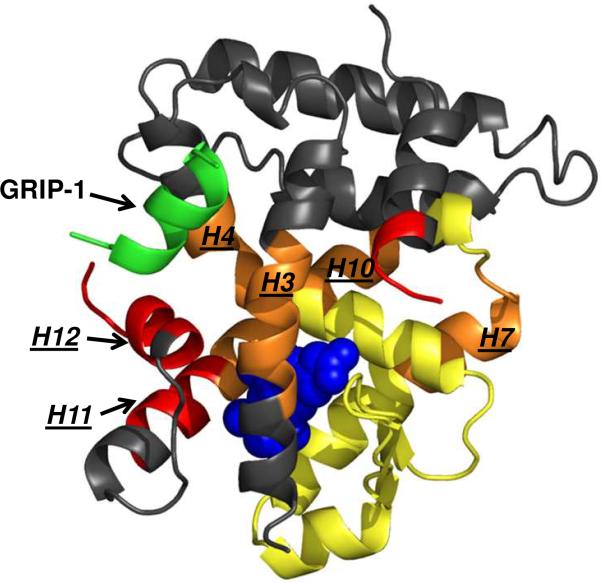
The ΔSsolv can be estimated by ΔCp ln(T/385), which is +102 cal-mol−1 K−1 at 25 °C. Estimates for ΔSrt for 1:1 binding are −8 cal-mol−1 K−1 (54). The contributions of ΔSconf are more difficult to estimate. However, Gomez and Freire (52) established parameters for amino acids based on the work of Lee et al (55). These authors successfully calculated the binding energetics (including the binding entropy) of a small peptide inhibitor (pepstain A) to endothiapepsin (Table 2) (52). Using a similar approach and their published parameters, we estimate no more than a −50 cal-mol−1 K−1 change in entropy for an unstructured GRIP-1 in solution to adopt a single helical conformation bound to hRXRα-LBD (freezing the polypeptide backbone), and for the loss of entropy due to the burial of side chains from both GRIP-1 and hRXRα-LBD on the surface of the protein. The negative entropy terms (ΔSrt, ΔSconf) are only about 50% of the solvation entropy term suggesting that GRIP-1 binding should also be strongly entropically driven. However, the ΔSconf term may be significantly underestimated given the data provided in the HDX MS exchange studies. As presented in Table 3, GRIP-1 binding produces significant protection of amide nitrogens in regions beyond the immediate coactivator peptide binding site (peptides in H3, H5, H7, H10, H11 and the beta strands). Figure 6 highlights this by mapping the changes in observed in the HDX MS studies onto the structure. Upon 9cRA binding, several observed peptides around the ligand binding pocket of hRXRα-LBD become significantly more protected from deuterium incorporation (10–37% change relative to apo-hRXRα-LBD, highlighted in yellow and orange) as previously reported by Yan et. al (16). When GRIP-1 (green) is added to the complex, a subset of these same regions (those in orange) become further protected or stabilized (>7% additional change relative to hRXRα-LBD:9cRA) demonstrating the allosteric influence of GRIP-1 binding on the ligand binding pocket helices. Finally, the HDX MS analysis of the hRXRα-LBD:9cRA:GRIP-1 complex clearly identified peptides including H11 and H12 (in red) that were not protected (remained dynamic) by 9cRA addition but did become more protected when GRIP-1 was added to hRXRα-LBD:9cRA. This increased protection is expected to result in a reduced mobility and increased stability of the helices of the hRXRα-LBD involved in GRIP-1 binding and between the coactivator binding site and the LBP. This loss of mobility would significantly add to the negative entropy term ΔSconf. Thus the most consistent picture of GRIP-1 binding to hRXRα-LBDs involves a small change in the conformation of the LBD resulting in a significant decrease in the dynamics of the domain.
Figure 6.
HDX MS analyses mapped onto hRXRα-LBD:9cRA:GRIP-1. Regions within the hRXRα-LBD that do not change deuterium incorporation with 9cRA or 9cRA + GRIP-1 are colored in grey. Upon 9cRA binding, several HDX MS monitored peptides (yellow and orange) around the ligand binding pocket of hRXRα-LBD become significantly more protected from deuterium incorporation (10–37% change relative to hRXRα-LBD, see Table 3). When GRIP-1 (green) is added to the complex, a subset of these same regions (in orange, H3, H4, H10, and H7) become further protected or stabilized (>7% additional change relative to hRXRα-LBD:9cRA) demonstrating the stabilizing affect of GRIP-1 binding on the hRXRα-LBD beyond the coactivator binding site. Signficantly, several peptides that are part of H12 and the C-terminal end of H11 (in red) were not protected (remained dynamic) by 9cRA addition but then did show protection when GRIP-1 is added to the hRXRα-LBD:9cRA complex.
In summary, our study integrates x-ray crystallography with dynamical HDX MS spectrometry to understand the energetic effects of the binding of the GRIP-1 coactivator peptide to the hRXRα-LBD:9cRA complex. Our structural and dynamical studies of the complex with and without coactivator peptide clearly exhibit a translational movement of H11 and H12 toward GRIP-1 and the formation of new hydrophobic and hydrophilic interactions to stabilize the fluctuating complex. The binding of coactivator peptide to the hRXRα-LBD:9cRA complex induces effects which propagate to the terminal end of the retinoid agonist and are mediated by residues at the interface between agonist and coactivator peptide (especially on H11). While ligand-mediated NR activation and coactivator recruitment is a well-studied process, the structural features that allow a range of NR-specific ligands to produce a variety of responses in different tissues is still not well defined (an agonist in one tissue versus an antagonist in another). The recruitment of coactivator (and replacement of corepressors) is an essential question to address when evaluating ligand-induced gene expression and biological responses. This work draws attention to the important structural and dynamical changes that occur in one example of NR LBD recruitment of coactivators.
Supplementary Material
Acknowledgement
We thank Dr Ellen Li (Washington University in St. Louis) for providing the protein expression vector, and Dr David Graves' laboratory for training and use of the ITC. We also appreciate critical reading and helpful comments provided by Drs Christie Brouillette, David Graves and Bingdong Sha. Dr Sha's efforts to continue to train and assist Gang Xia in the methods of crystallography and structural analysis are especially appreciated in view of the unfortunate death of our colleague and our close friend Dr Craig D. Smith.
This work supported by grants from the National Institutes of Health (NIH) 2 P50 CA089019 (DDM),5 P50 CA089019 (MBR), RR17261 (MBR) and the Komen Foundation(BCTR 20000690) (DDM)
Abbreviations
- hRXRα-LBD
human Retinoid X Receptor alpha-Ligand Binding Domain
- 9cRA
9-cis Retinoic Acid
- GRIP-1
Glucocorticoid Receptor Interacting Protein-1
- hRXRα-LBD:9cRA
human Retinoid X Receptor alpha-Ligand Binding Domain homodimers bound to 9cRA
- hRXRα-LBD:9cRA:GRIP-1
human Retinoid X Receptor alpha-Ligand Binding Domain homodimers bound to 9cRA and GRIP-1
- HDX MS
Hydrogen/Deuterium Exchanging Mass Spectroscopy
- LBP
Ligand Binding Pocket
- LPC
Ligand-Protein Contacts
- NR
Nuclear receptor
- H
helix
Footnotes
The atomic coordinates and structure factors (code 3OAP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Supporting Information Available The ITC measurements of GRIP-1 coactivator peptide binding to hRXRα-LBD:9cRA at 20, 25, 30, and 37 °C are presented in Figure S1, and the ITC measurement of -1 coactivator peptide binding to hRXRα-LBD:9cRA at 25 °C in HEPES Buffer is presented in Figure S2. The interaction between E456 of RXR and H688 of GRIP-1 observed in our crystal structure is presented in Figure S3. The deuterium incorporation measurements of all peptide fragments are provided in Figure S4. The contact measurements between hRXRα-LBD and GRIP-1 coactivator peptide or 9-cis-retinoic acid are provided in Table S1 and Table S2, respectively. The optimized extinction coefficients used in our near-UV-Vis difference simulation in Figure 4 are presented in Table S3. Table S4 provides the complete list of 38 HDX MS peptides and their observed differences in the various complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- (2).Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- (3).Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dong D, Noy N. Heterodimer formation by retinoid X receptor: regulation by ligands and by the receptor's self-association properties. Biochemistry. 1998;37:10691–10700. doi: 10.1021/bi980561r. [DOI] [PubMed] [Google Scholar]
- (5).Svensson S, Ostberg T, Jacobsson M, Norstrom C, Stefansson K, Hallen D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. Embo J. 2003;22:4625–46233. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, Jr., McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- (8).Wolf G. Is 9-cis-retinoic acid the endogenous ligand for the retinoic acid-X receptor? Nutr Rev. 2006;64:532–538. doi: 10.1111/j.1753-4887.2006.tb00186.x. [DOI] [PubMed] [Google Scholar]
- (9).Lawrence JA, Adamson PC, Caruso R, Chow C, Kleiner D, Murphy RF, Venzon DJ, Shovlin M, Noone M, Merino M, Cowan KH, Kaiser M, O'Shaughnessy J, Zujewski J. Phase I clinical trial of alitretinoin and tamoxifen in breast cancer patients: toxicity, pharmacokinetic, and biomarker evaluations. J Clin Oncol. 2001;19:2754–2763. doi: 10.1200/JCO.2001.19.10.2754. [DOI] [PubMed] [Google Scholar]
- (10).Miller VA, Rigas JR, Benedetti FM, Verret AL, Tong WP, Kris MG, Gill GM, Loewen GR, Truglia JA, Ulm EH, Warrell RP., Jr. Initial clinical trial of the retinoid receptor pan agonist 9-cis retinoic acid. Clin Cancer Res. 1996;2:471–475. [PubMed] [Google Scholar]
- (11).Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. Embo J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mascrez B, Mark M, Dierich A, Ghyselinck NB, Kastner P, Chambon P. The RXRalpha ligand-dependent activation function 2 (AF-2) is important for mouse development. Development. 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- (13).Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- (14).Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. Embo J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- (16).Yan X, Broderick D, Leid ME, Schimerlik MI, Deinzer ML. Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry. 2004;43:909–917. doi: 10.1021/bi030183c. [DOI] [PubMed] [Google Scholar]
- (17).Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- (18).Lu J, Cistola DP, Li E. Analysis of ligand binding and protein dynamics of human retinoid X receptor alpha ligand-binding domain by nuclear magnetic resonance. Biochemistry. 2006;45:1629–1639. doi: 10.1021/bi051474j. [DOI] [PubMed] [Google Scholar]
- (19).Nahoum V, Perez E, Germain P, Rodriguez-Barrios F, Manzo F, Kammerer S, Lemaire G, Hirsch O, Royer CA, Gronemeyer H, de Lera AR, Bourguet W. Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc Natl Acad Sci U S A. 2007;104:17323–17328. doi: 10.1073/pnas.0705356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu Z, Auboeuf D, Wong J, Chen JD, Tsai SY, Tsai MJ, O'Malley BW. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc Natl Acad Sci U S A. 2002;99:7940–7944. doi: 10.1073/pnas.122225699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Phelan CA, Weaver JM, Steger DJ, Joshi S, Maslany JT, Collins JL, Zuercher WJ, Willson TM, Walker M, Jaye M, Lazar MA. Selective partial agonism of liver X receptor alpha is related to differential corepressor recruitment. Mol Endocrinol. 2008;22:2241–2249. doi: 10.1210/me.2008-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- (24).Egea PF, Mitschler A, Moras D. Molecular recognition of agonist ligands by RXRs. Mol Endocrinol. 2002;16:987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- (25).Egea PF, Moras D. Purification and crystallization of the human RXRalpha ligand-binding domain-9-cisRA complex. Acta Crystallogr D Biol Crystallogr. 2001;57:434–437. doi: 10.1107/s0907444901000385. [DOI] [PubMed] [Google Scholar]
- (26).Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. Academic Press Inc; San Diego: 1997. Processing of X-ray diffraction data collected in oscillation mode; p. 307. [DOI] [PubMed] [Google Scholar]
- (27).Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D Biol CrystallogrD. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- (28).Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- (29).Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- (30).Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- (31).Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- (32).Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- (33).Kang S, Poliakov A, Sexton J, Renfrow MB, Prevelige PE., Jr. Probing conserved helical modules of portal complexes by mass spectrometry-based hydrogen/deuterium exchange. J Mol Biol. 2008;381:772–784. doi: 10.1016/j.jmb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- (35).Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wright E, Vincent J, Fernandez EJ. Thermodynamic characterization of the interaction between CAR-RXR and SRC-1 peptide by isothermal titration calorimetry. Biochemistry. 2007;46:862–870. doi: 10.1021/bi061627i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lu J, Chen M, Dekoster GT, Cistola DP, Li E. The RXRalpha C-terminus T462 is a NMR sensor for coactivator peptide binding. Biochem Biophys Res Commun. 2008;366:932–937. doi: 10.1016/j.bbrc.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- (39).Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- (40).Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. Embo J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Love JD, Gooch JT, Benko S, Li C, Nagy L, Chatterjee VK, Evans RM, Schwabe JW. The structural basis for the specificity of retinoid-X receptor-selective agonists: new insights into the role of helix H12. J Biol Chem. 2002;277:11385–11391. doi: 10.1074/jbc.M110869200. [DOI] [PubMed] [Google Scholar]
- (43).Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pogenberg V, Guichou JF, Vivat-Hannah V, Kammerer S, Perez E, Germain P, de Lera AR, Gronemeyer H, Royer CA, Bourguet W. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280:1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- (45).Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- (46).Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- (47).Donovan JW. Changes in ultraviolet absorption produced by alteration of protein conformation. J Biol Chem. 1969;244:1961–1967. [PubMed] [Google Scholar]
- (48).Gampe RT, Jr., Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- (49).Lu J, Dawson MI, Hu QY, Xia Z, Dambacher JD, Ye M, Zhang XK, Li E. The effect of antagonists on the conformational exchange of the retinoid X receptor alpha ligand-binding domain. Magn Reson Chem. 2009;47:1071–1080. doi: 10.1002/mrc.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ghosh JC, Yang X, Zhang A, Lambert MH, Li H, Xu HE, Chen JD. Interactions that determine the assembly of a retinoid X receptor/corepressor complex. Proc Natl Acad Sci U S A. 2002;99:5842–5847. doi: 10.1073/pnas.092043399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lee WY, Noy N. Interactions of RXR with coactivators are differentially mediated by helix 11 of the receptor's ligand binding domain. Biochemistry. 2002;41:2500–2508. doi: 10.1021/bi011764+. [DOI] [PubMed] [Google Scholar]
- (52).Gomez J, Freire E. Thermodynamic mapping of the inhibitor site of the aspartic protease endothiapepsin. J Mol Biol. 1995;252:337–350. doi: 10.1006/jmbi.1995.0501. [DOI] [PubMed] [Google Scholar]
- (53).Baker BM, Murphy KP. Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 1998;295:294–315. doi: 10.1016/s0076-6879(98)95045-5. [DOI] [PubMed] [Google Scholar]
- (54).Lee KH, Xie D, Freire E, Amzel LM. Estimation of changes in side chain configurational entropy in binding and folding: general methods and application to helix formation. Proteins. 1994;20:68–84. doi: 10.1002/prot.340200108. [DOI] [PubMed] [Google Scholar]
- (55).Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.