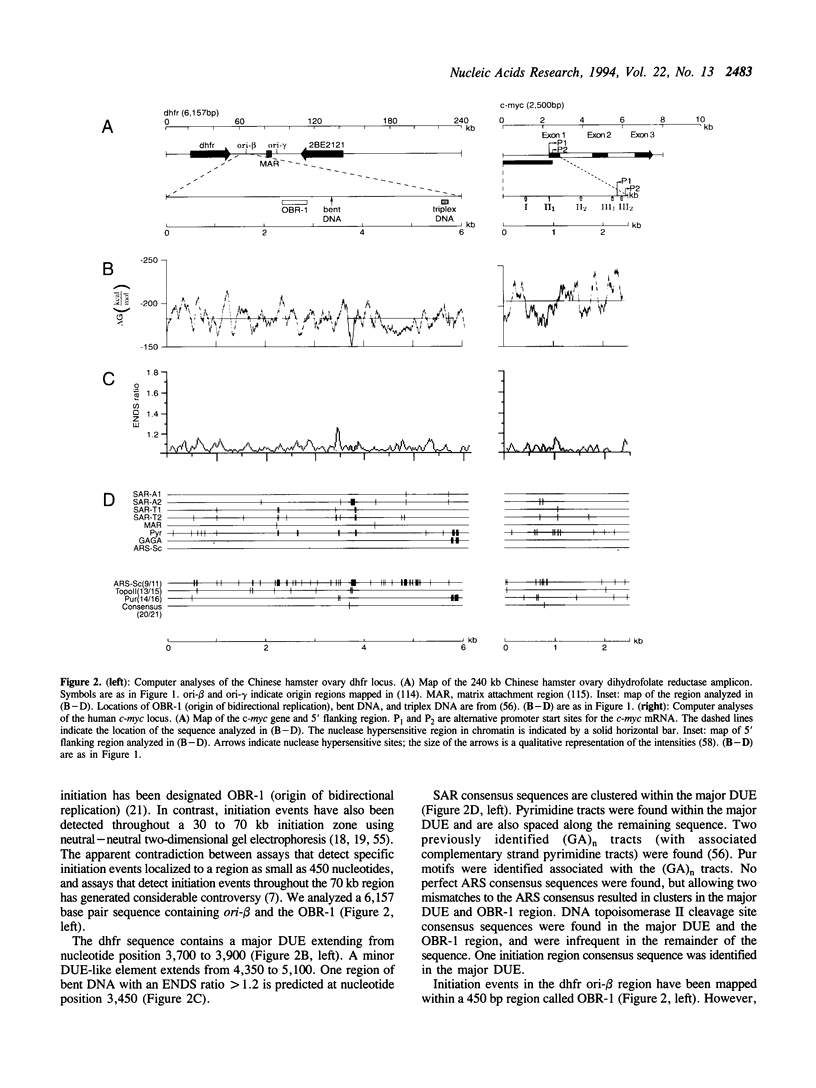
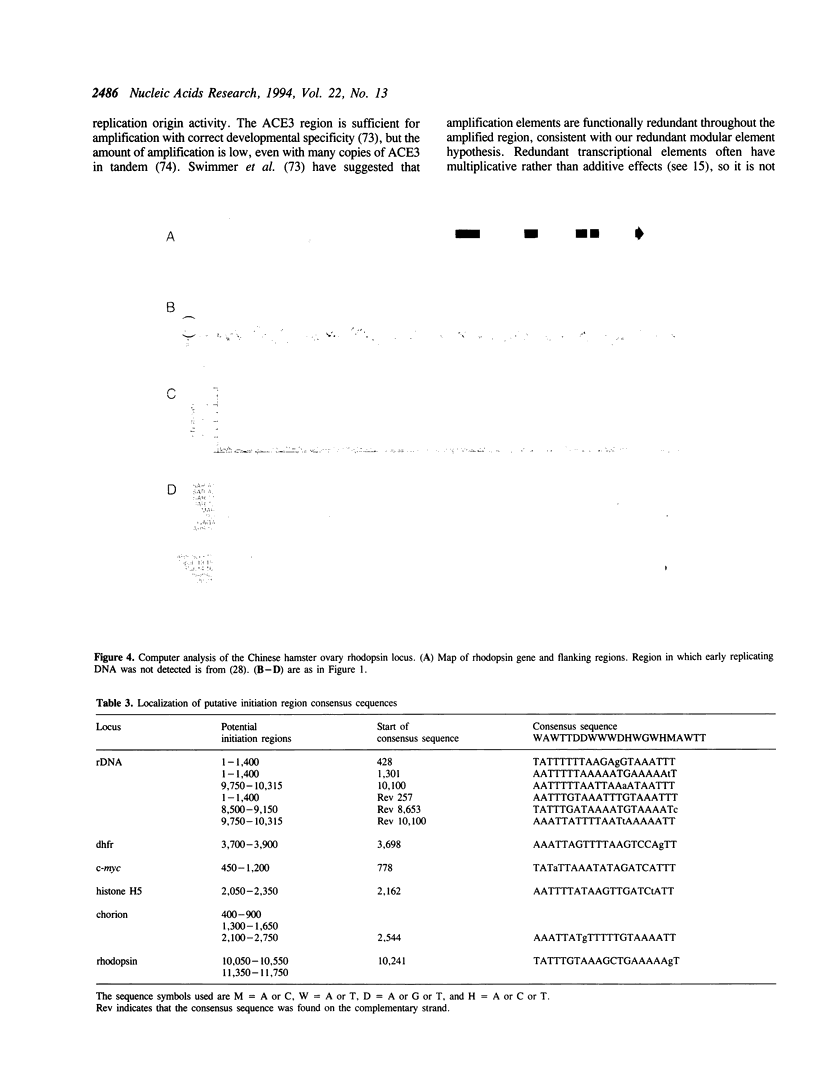
Abstract
We have postulated that chromosomal replication origin regions in eukaryotes have in common clusters of certain modular sequence elements (Benbow, Zhao, and Larson, BioEssays 14, 661-670, 1992). In this study, computer analyses of DNA sequences from six origin regions showed that each contained one or more potential initiation regions consisting of a putative DUE (DNA unwinding element) aligned with clusters of SAR (scaffold associated region), and ARS (autonomously replicating sequence) consensus sequences, and pyrimidine tracts. The replication origins analyzed were from the following loci: Tetrahymena thermophila macronuclear rDNA gene, Chinese hamster ovary dihydrofolate reductase amplicon, human c-myc proto-oncogene, chicken histone H5 gene, Drosophila melanogaster chorion gene cluster on the third chromosome, and Chinese hamster ovary rhodopsin gene. The locations of putative initiation regions identified by the computer analyses were compared with published data obtained using diverse methods to map initiation sites. For at least four loci, the potential initiation regions identified by sequence analysis aligned with previously mapped initiation events. A consensus DNA sequence, WAWTTDDWWWDHWGWHMAWTT, was found within the potential initiation regions in every case. An additional 35 kb of combined flanking sequences from the six loci were also analyzed, but no additional copies of this consensus sequence were found.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Affolter M., Ruiz-Carrillo A. Transcription unit of the chicken histone H5 gene and mapping of H5 pre-mRNA sequences. J Biol Chem. 1986 Sep 5;261(25):11496–11502. [PubMed] [Google Scholar]
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C. T., Amaldi F., Buongiorno-Nardelli M. Electron microscopic analysis of replicating DNA of sea urchin embryos. Cell. 1978 Nov;15(3):1095–1107. doi: 10.1016/0092-8674(78)90293-3. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zhao J., Larson D. D. On the nature of origins of DNA replication in eukaryotes. Bioessays. 1992 Oct;14(10):661–670. doi: 10.1002/bies.950141004. [DOI] [PubMed] [Google Scholar]
- Bergemann A. D., Johnson E. M. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992 Mar;12(3):1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann A. D., Ma Z. W., Johnson E. M. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol. 1992 Dec;12(12):5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992 Jan 10;255(5041):195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Bonven B., Westergaard O. DNase I hypersensitive regions correlate with a site-specific endogenous nuclease activity on the r-chromatin of Tetrahymena. Nucleic Acids Res. 1982 Dec 11;10(23):7593–7608. doi: 10.1093/nar/10.23.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan P. M., Dayton A. I. A specific replication origin in the chromosomal rDNA of Lytechinus variegatus. Nature. 1982 Sep 30;299(5882):453–456. doi: 10.1038/299453a0. [DOI] [PubMed] [Google Scholar]
- Bozzoni I., Baldari C. T., Amaldi F., Buongiorno-Nardelli M. Replication of ribosomal DNA in Xenopus laevis. Eur J Biochem. 1981 Sep 1;118(3):585–590. doi: 10.1111/j.1432-1033.1981.tb05559.x. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brown J. A., Holmes S. G., Smith M. M. The chromatin structure of Saccharomyces cerevisiae autonomously replicating sequences changes during the cell division cycle. Mol Cell Biol. 1991 Oct;11(10):5301–5311. doi: 10.1128/mcb.11.10.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986 Jan 28;25(2):441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Wu J., Sogo J. M., Nallaseth F. S., DePamphilis M. L. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991 Dec;10(13):4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Carminati J. L., Johnston C. G., Orr-Weaver T. L. The Drosophila ACE3 chorion element autonomously induces amplification. Mol Cell Biol. 1992 May;12(5):2444–2453. doi: 10.1128/mcb.12.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challoner P. B., Amin A. A., Pearlman R. E., Blackburn E. H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985 Apr 11;13(7):2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Yuen M. H., Garrard W. T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987 Apr 15;262(11):5394–5397. [PubMed] [Google Scholar]
- D'Anna J. A., Church V. L., Gale J. M., Tobey R. A. DNA contents of replication without DNA density labeling. Anal Biochem. 1990 May 15;187(1):1–9. doi: 10.1016/0003-2697(90)90409-3. [DOI] [PubMed] [Google Scholar]
- Dailey L., Caddle M. S., Heintz N., Heintz N. H. Purification of RIP60 and RIP100, mammalian proteins with origin-specific DNA-binding and ATP-dependent DNA helicase activities. Mol Cell Biol. 1990 Dec;10(12):6225–6235. doi: 10.1128/mcb.10.12.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication in metazoan chromosomes. J Biol Chem. 1993 Jan 5;268(1):1–4. [PubMed] [Google Scholar]
- Delidakis C., Kafatos F. C. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989 Mar;8(3):891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C., Kafatos F. C. Amplification of a chorion gene cluster in Drosophila is subject to multiple cis-regulatory elements and to long-range position effects. J Mol Biol. 1987 Sep 5;197(1):11–26. doi: 10.1016/0022-2836(87)90605-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992 Sep;12(9):3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991 Aug;11(8):3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Wenink P. W., Poddighe J. Permanent attachment of replication origins to the nuclear matrix in BHK-cells. Nucleic Acids Res. 1986 Apr 25;14(8):3241–3249. doi: 10.1093/nar/14.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J., Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990 Dec 11;18(23):6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale J. M., Tobey R. A., D'Anna J. A. Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J Mol Biol. 1992 Mar 20;224(2):343–358. doi: 10.1016/0022-2836(92)90999-z. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Amati B. B., Cardenas M. E., Hofmann J. F. Studies on scaffold attachment sites and their relation to genome function. Int Rev Cytol. 1989;119:57–96. doi: 10.1016/s0074-7696(08)60649-x. [DOI] [PubMed] [Google Scholar]
- Gaudette M. F., Benbow R. M. Replication forks are underrepresented in chromosomal DNA of Xenopus laevis embryos. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5953–5957. doi: 10.1073/pnas.83.16.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J. L., Dijkwel P. A., Vaughn J. P. Initiation of replication in the Chinese hamster dihydrofolate reductase domain. Chromosoma. 1992;102(1 Suppl):S17–S23. doi: 10.1007/BF02451781. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L. Mammalian origins of replication. Bioessays. 1992 Oct;14(10):651–659. doi: 10.1002/bies.950141002. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L., Vaughn J. P., Dijkwel P. A., Leu T. H., Ma C. Origins of replication: timing and chromosomal position. Curr Opin Cell Biol. 1991 Jun;3(3):414–421. doi: 10.1016/0955-0674(91)90068-a. [DOI] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Heck M. M., Spradling A. C. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990 Apr;110(4):903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H., Dailey L., Held P., Heintz N. Eukaryotic replication origins as promoters of bidirectional DNA synthesis. Trends Genet. 1992 Nov;8(11):376–381. doi: 10.1016/0168-9525(92)90298-i. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H. Transcription factors and the control of DNA replication. Curr Opin Cell Biol. 1992 Jun;4(3):459–467. doi: 10.1016/0955-0674(92)90012-2. [DOI] [PubMed] [Google Scholar]
- Held P. G., Heintz N. H. Eukaryotic replication origins. Biochim Biophys Acta. 1992 Apr 6;1130(3):235–246. doi: 10.1016/0167-4781(92)90435-3. [DOI] [PubMed] [Google Scholar]
- Hines P. J., Benbow R. M. Initiation of replication at specific origins in DNA molecules microinjected into unfertilized eggs of the frog Xenopus laevis. Cell. 1982 Sep;30(2):459–468. doi: 10.1016/0092-8674(82)90243-4. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Sudo T., Yoshida M., Kubota H., Ueyama H. In vitro replication of recombinant plasmids carrying chromosomal segments of Xenopus laevis. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3697–3701. doi: 10.1073/pnas.79.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Méchali M. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 1992 Apr 11;20(7):1463–1469. doi: 10.1093/nar/20.7.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Okazaki T., Itani T., Ogata M., Sato Y., Ariga H. An initiation site of DNA replication with transcriptional enhancer activity present upstream of the c-myc gene. EMBO J. 1988 Oct;7(10):3135–3142. doi: 10.1002/j.1460-2075.1988.tb03180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Ghandour G. Comparative statistics for DNA and protein sequences: single sequence analysis. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5800–5804. doi: 10.1073/pnas.82.17.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Natale D. A., Eddy M. J. Stable DNA unwinding, not "breathing," accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 1983 Feb 11;11(3):619–627. doi: 10.1093/nar/11.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Calos M. P. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol Cell Biol. 1991 Mar;11(3):1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Leffak M. DNA topology of the ordered chromatin domain 5' to the human c-myc gene. Nucleic Acids Res. 1989 Apr 11;17(7):2819–2833. doi: 10.1093/nar/17.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S., Zannis-Hadjopoulos M. Classes of autonomously replicating sequences are found among early-replicating monkey DNA. Biochim Biophys Acta. 1991 Feb 16;1088(2):234–244. doi: 10.1016/0167-4781(91)90059-u. [DOI] [PubMed] [Google Scholar]
- Larson D. D., Blackburn E. H., Yaeger P. C., Orias E. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986 Oct 24;47(2):229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- Leffak M., James C. D. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol Cell Biol. 1989 Feb;9(2):586–593. doi: 10.1128/mcb.9.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. Activation of a mammalian origin of replication by chromosomal rearrangement. Mol Cell Biol. 1992 Jun;12(6):2804–2812. doi: 10.1128/mcb.12.6.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J., Spradling A. DNA sequence of a 3.8 kilobase pair region controlling Drosophila chorion gene amplification. Chromosoma. 1985;92(2):136–142. doi: 10.1007/BF00328465. [DOI] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. The two faces of higher eukaryotic DNA replication origins. Cell. 1990 Sep 7;62(5):845–847. doi: 10.1016/0092-8674(90)90258-g. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Mahbubani H. M., Paull T., Elder J. K., Blow J. J. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992 Apr 11;20(7):1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhinney C., Leffak M. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 1990 Mar 11;18(5):1233–1242. doi: 10.1093/nar/18.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli G., Baldari C. T., Carri M. T., Di Cello G., Buongiorno-Nardelli M. An electron microscope study of chromosomal DNA replication in different eukaryotic systems. Exp Cell Res. 1982 Jan;137(1):127–140. doi: 10.1016/0014-4827(82)90015-5. [DOI] [PubMed] [Google Scholar]
- Natale D. A., Schubert A. E., Kowalski D. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles E. G., Cunningham K., Jain R. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription termination region. J Biol Chem. 1981 Dec 25;256(24):12857–12860. [PubMed] [Google Scholar]
- Orr-Weaver T. L. Drosophila chorion genes: cracking the eggshell's secrets. Bioessays. 1991 Mar;13(3):97–105. doi: 10.1002/bies.950130302. [DOI] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr, Beyer A. L. Visualization of Drosophila melanogaster chorion genes undergoing amplification. Mol Cell Biol. 1988 Jul;8(7):2811–2821. doi: 10.1128/mcb.8.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr Novel amplification and transcriptional activity of chorion genes in Drosophila melanogaster follicle cells. Cell. 1983 Jun;33(2):543–553. doi: 10.1016/0092-8674(83)90435-x. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Rao B. S., Zannis-Hadjopoulos M., Price G. B., Reitman M., Martin R. G. Sequence similarities among monkey ori-enriched (ors) fragments. Gene. 1990 Mar 15;87(2):233–242. doi: 10.1016/0378-1119(90)90307-d. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Affolter M., Renaud J. Genomic organization of the genes coding for the six main histones of the chicken: complete sequence of the H5 gene. J Mol Biol. 1983 Nov 15;170(4):843–859. doi: 10.1016/s0022-2836(83)80191-0. [DOI] [PubMed] [Google Scholar]
- Ruiz J. C., Wahl G. M. Chromosomal destabilization during gene amplification. Mol Cell Biol. 1990 Jun;10(6):3056–3066. doi: 10.1128/mcb.10.6.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T., Ina S. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991 Jul 25;19(14):3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Spitzner J. R., Muller M. T. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988 Jun 24;16(12):5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Izuta S., Savoysky E., Sakurai T., Simbulan C., Tatebe M., Kojima K., Yoshida S. Deoxypyrimidine cluster mediates the priming by calf thymus DNA primase subunit. Biochem Mol Biol Int. 1993 Mar;29(4):645–652. [PubMed] [Google Scholar]
- Suzuki M., Savoysky E., Izuta S., Tatebe M., Okajima T., Yoshida S. RNA priming coupled with DNA synthesis on natural template by calf thymus DNA polymerase alpha-primase. Biochemistry. 1993 Nov 30;32(47):12782–12792. doi: 10.1021/bi00210a030. [DOI] [PubMed] [Google Scholar]
- Swimmer C., Delidakis C., Kafatos F. C. Amplification-control element ACE-3 is important but not essential for autosomal chorion gene amplification. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8823–8827. doi: 10.1073/pnas.86.22.8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J. P., Lindstrom Y. I., Leffak M. Opposite replication polarities of transcribed and nontranscribed histone H5 genes. Mol Cell Biol. 1988 Apr;8(4):1657–1663. doi: 10.1128/mcb.8.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2486–2490. doi: 10.1073/pnas.87.7.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J. D., Bronson E. C., Anderson J. N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991 Oct 11;19(19):5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., Burhans W. C., DePamphilis M. L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., DePamphilis M. L. Guide to identification of origins of DNA replication in eukaryotic cell chromosomes. Crit Rev Biochem Mol Biol. 1992;27(6):445–472. doi: 10.3109/10409239209082569. [DOI] [PubMed] [Google Scholar]
- Vassilev L., Johnson E. M. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol. 1990 Sep;10(9):4899–4904. doi: 10.1128/mcb.10.9.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990 Jun 15;61(6):1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Mullenders L. H., Hamlin J. L. Replication forks are associated with the nuclear matrix. Nucleic Acids Res. 1990 Apr 25;18(8):1965–1969. doi: 10.1093/nar/18.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger P. C., Orias E., Shaiu W. L., Larson D. D., Blackburn E. H. The replication advantage of a free linear rRNA gene is restored by somatic recombination in Tetrahymena thermophila. Mol Cell Biol. 1989 Feb;9(2):452–460. doi: 10.1128/mcb.9.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Hendrickson E. A., DePamphilis M. L. DNA primase-DNA polymerase alpha from simian cells. Modulation of RNA primer synthesis by ribonucleoside triphosphates. J Biol Chem. 1985 May 25;260(10):6254–6263. [PubMed] [Google Scholar]
- Yu G. L., Hasson M., Blackburn E. H. Circular ribosomal DNA plasmids transform Tetrahymena thermophila by homologous recombination with endogenous macronuclear ribosomal DNA. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5151–5155. doi: 10.1073/pnas.85.14.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Newlon C. S., Huberman J. A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]