Abstract
Background
The novel ability to epigenetically reprogram somatic cells into induced pluripotent stem cells through the exogenous expression of transcription promises to revolutionize the study of human diseases.
Objective
Here we report on the generation of 25 induced pluripotent stem cell lines from 6 patients with various forms of Primary Immunodeficiencies, affecting adaptive and/or innate immunity.
Methods
Patients’ dermal fibroblasts were reprogrammed by expression of four transcription factors, OCT4, SOX2, KLF4, and c-MYC using a single excisable polycistronic lentiviral vector.
Results
Induced pluripotent stem cells derived from patients with primary immunodeficiencies show a stemness profile that is comparable to that observed in human embryonic stem cells. Following in vitro differentiation into embryoid bodies, pluripotency of the patient-derived indiced pluripotent stem cells lines was demonstrated by expression of genes characteristic of each of the three embryonic layers. We have confirmed the patient-specific origin of the induced pluripotent stem cell lines, and ascertained maintenance of karyotypic integrity.
Conclusion
By providing a limitless source of diseased stem cells that can be differentiated into various cell types in vitro, the repository of induced pluripotent stem cell lines from patients with primary immunodeficiencies represents a unique resource to investigate the pathophysiology of hematopoietic and extra-hematopoietic manifestations of these diseases, and may assist in the development of novel therapeutic approaches based on gene correction.
Keywords: Primary Immunodeficiency, Induced Pluripotent Stem Cells, Reprogramming
Introduction
Primary immunodeficiencies (PIDs) comprise over 150 distinct disorders of immune system development and/or function1. Dissection of the cellular pathophysiology of PIDs has been largely based on in vitro studies using patient-derived cells and on analysis of suitable animal models. While largely successful, both of these approaches have important inherent limitations. In particular, many forms of PID are rare, severe and affect predominantly infants and young children. In these cases, access to biological specimens from affected patients may be problematic. Furthermore, there is significant heterogeneity of clinical and immunological phenotype among patients with different mutations in the same gene, but limited information is available on this diversity at the cellular level2. Finally, studies that aim to define the cellular pathophysiology of human PIDs are usually performed on blood samples, occasionally on the bone marrow, rarely on lymphoid tissues (thymus, lymph nodes, spleen) and almost never on non hematopoietic tissues, yet many forms of PID also include extra-immune manifestations1, 3. This is the case for immunodeficiency syndromes characterized by multi-system developmental defects (such as DiGeorge syndrome4 and cartilage hair hypoplasia5), broad expression of the disease-specific gene (as in defects of DNA repair6, NEMO deficiency7, hyper-IgE syndrome due to STAT3 deficiency8, 9, and adenosine deaminase deficiency10) or tissue-specific susceptibility to infections (as in herpes simplex encephalitis11–13).
On the other hand, while murine models of PID have provided key insights, they also carry significant inherent limitations because of differences in immune system development and function between mice and humans and the relative lack of phenotypic variability and heterogeneity of mutations in murine models as compared to PIDs in humans.
Following the demonstration in 2006 by Takahashi and Yamanaka that mouse fibroblasts can be reprogrammed into induced pluripotent stem cells (iPSC) through transient forced expression of defined transcription factors14, generation of iPSCs from terminally differentiated human cells has been recently reported15–17. Similar to embryonic stem cells, these cells hold the unique potential to differentiate into various tissue cell types, including neurons18–25, cardiomyocytes26–28, hepatic cells29–31, gastrointestinal cells32, thymic epithelial cells33, hematopoietic cells34, 35 and many others36–41. Furthermore, iPSCs have also been used to correct genetic disorders in mice following gene targeting and homologous recombination42, 43.
Over the last ten years, we have established an extended repository of fibroblast cell lines from patients with various forms of PIDs. This repository is also representative of the diversity of the clinical and immunological phenotype that is associated with different mutations occurring in the same gene. Using this collection of fibroblast cell lines, we now report on the successful generation and characterization of a series of PID-specific iPSCs that may serve as the foundation for future studies of disease pathophysiology and gene correction.
Materials and methods
Patients
Dermal fibroblast samples were obtained from 6 PID patients carrying mutations in different genes as detailed in Table I. Informed consent was obtained from a parent or a guardian. Study protocols were approved by Children’s Hospital Boston Institutional Review Board.
Table I.
Patients and mutations
| Disease Phenotype | Gene | Mutation | Reference |
|---|---|---|---|
| SCID | RAG1 | c.1228C>T; c.2332C>T | unpublished |
| Leaky SCID | RAG1 | c.1180C>T; c.1180C>T | unpublished |
| OS | RAG1 | c.256-257del; c.2164G>A | 59 |
| HSE1 | STAT1 | c.1928_1929 insA; c.1928_1929 insA | 60 |
| HSE2 | TLR3 | c.1660C>T; c.2236G>T | Guo Y et al., submitted |
| CHH | RMRP | c. 27G>A; c. 27G>A | unpublished |
SCID: Severe combined immunodeficiency; OS: Omenn syndrome; HSE: Herpes Simplex Encephalitis; CHH: Cartilage Hair Hypoplasia; RAG1: Recombinase Activating Gene 1; STAT1: Signal Transducer and Activator of Transcription 1; TLR3: Toll-like Recepetor 3; RMRP: RNA component of the Mitochondrial RNA Processing endoribunuclease
Cell lines and culture
A previously reported human iPSC line 17, obtained by reprogramming dermal fibroblasts with retroviral vectors encoding SOX2, OCT4, KLF4 and c-MYC transcription factors, was used as an internal control.
Patient and healthy control fibroblasts were maintained in DMEM (high glucose and L-glutamine) media containing 10% FBS, 1 mM L-glutamine and penicillin/streptomycin (hFib media).
iPSCs were maintained in human embryonic stem (hES) cells medium, composed of DMEM/F12 (Invitrogen, Carlsbad, CA) containing 20% KOSR (Invitrogen), 10 ng/ml basic Fibroblast Growth Factor (bFGF, Gemini Bio-Products, West Sacramento, CA), 1 mM L-glutamine, 100 μM non-essential amino acids, 100 μM 2-β-mercaptoethanol, and penicillin/streptomycin. The cells were co-cultured on CF1 irradiated mouse embryonic fibroblasts (iMEFs, Globalstem Inc, Rockville, MD) as previously described16, 17. Expansion and splitting of the iPSC colonies was done by either mechanical passage or by the use of collagenase, as previously described44.
iPSC differentiation into embryoid bodies (EB) was achieved by transferring iPSC colonies into low-adhesion plates free of feeder cells and using a bFGF-free hES medium as previously described16, 17, 45.
Lentiviral reprogramming vector production
Lentiviruses containing the polycistronic lentiviral vector STEMCCA–LoxP previously described46–49 were produced using a five-plasmid transfection system as previously described 50.
Reprogramming of fibroblasts and human iPSC generation
Fibroblasts were infected with the lentiviral reprogramming vector in hFib media supplemented with 5 μg/ml protamine sulphate (Sigma) for 24 hours. After 72 hours, cells were transferred onto iMEFs in hES media. iPSCs colonies with ES-like morphology started to appear after 3 to 5 weeks. Colonies were picked, sub-cloned and expanded by mechanical transfer into new plates containing fresh and adhered iMEFs, as previously described16. Several clones were derived and characterized from each fibroblast line (Table E1 in the Online Repository).
Immunohistochemistry
iPSCs colonies were stained for OCT4, NANOG, TRA-1-60, TRA-1-81, SSEA3 and SSEA4 as previously described45. Images were acquired with a Pathway 435 bioimager equipped with a 10x objective (BD Biosciences, San Jose, CA).
Quantitative real-time PCR (qPCR)
mirVana™ RNA isolation kit (Ambion) was used for total RNA extraction and reverse transcription preformed using qScript cDNA supermix (Quanta Biosciences, Gaithersburg, MD), according to manufacturer’s instructions.
Real-time qPCR in PowerSYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) was performed on an AB 7500 Real-Time PCR system (Applied Biosystems). Primer sequences used for amplifying OCT4, SOX2, NANOG, REX1, GDF3, hTERT, KLF4, cMYC, RUNX, AFP, GATA4, Brachyury, NESTIN, NCAM and β-ACTIN were as previously described16, 17, 45. Results were normalized to β-actin (ACTB) gene expression and relative expression calculated by the ddCT method relative to expression levels in the individual parent cell lines using SDSv1 Software.
Mutation Analysis
Genomic DNA was isolated from dermal fibroblasts and iPSCs using the QiAMP DNA Kit (Qiagen, Valencia, CA). Genes known to be mutated in the patients were amplified by PCR as previously described, using primer sets as detailed in Table E2 in the Online Repository. Amplified products were purified using QIAquick PCR purification kit (Qiagen) and sequenced by the DF/HCC DNA Sequencing Facility. Sequences were analyzed using the Sequencher 4.8 software (Gene Codes Corporation).
In some of the compound heterozygote patients, PCR products were cloned using the TOPO TA Cloning® Kit with pCR®2.1-TOPO® vector (Invitrogen). Cloning products were amplified in competent bacteria, purified (QiaPrep miniprep kit, Qiagen) and later sequenced as described above.
Karyotype analysis
Karyotyping and G-banding were performed as described (see: Supplementary Methods in the Online Repository) in a blinded fashion by Cell Line Genetics, Madison, WI.
Results
We have established a repository of dermal fibroblast cell lines from more than 60 patients with various forms of PID, that are representative of defects in various components of the immune system. This repository of PID-specific fibroblast lines has been used to establish a pipeline for the systematic production of PID-specific iPSCs.
The initial cohort of iPSCs was derived from, TLR3 deficiency), immune deficiency associated with systemic disorders (cartilage hair hypoplasia, CHH), and early defects in T and B cell development (RAG1 mutations) (Table I). For the latter, we sought to derive iPSCs from patients with a different clinical and immunological phenotype (SCID, leaky SCID, Omenn syndrome) associated with null or hypomorphic mutations in the same gene (RAG1). In parallel, iPSCs were also derived from healthy control fibroblasts.
Various strategies have been previously described for reprogramming somatic cells to pluripotency. We have made use of an excisable, human stem cell cassette (STEMCCA)-containing single polycistronic lentiviral vector, that allows transduction of four reprogramming factors, OCT4, SOX2, KLF4, and c-MYC (Figure E1 in the Online Repository). Following infection with the STEMCCA lentivirus, patient- and control-derived fibroblasts were maintained under stringent human ES cell supporting culture conditions as previously described16, 17, 45. After 3–5 weeks of culture, ES-like colonies emerged and were picked and expanded. Several clones were derived from each of the fibroblast line (Table E1 in the Online Repository).
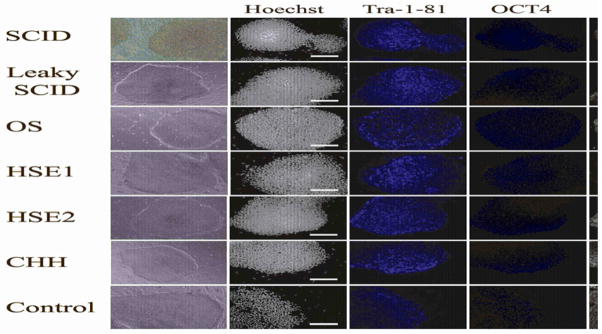
Human iPSC colonies were initially selected based on similarity to hES colonies, with discrete and compact colony morphology (Figure 1, left column). The selected colonies were then expanded and studied for expression of stemness markers, including Tra-1–81, Tra-1–60, OCT4, NANOG, SSEA3, and SSEA4. As shown in Figure 1, all iPSC colonies demonstrated uniform expression of these pluripotency markers.
Figure 1. PID-specific iPSC express markers of pluripotency.
PID-specific iPSC have morphology similar to hES cells (left column) when grown in coculture with iMEFs and express pluripotency markers including Tra-1–81, OCT4, SSEA4, SSEA3, NANOG and Tra-1-60 as shown demonstrated by immunohistochemistry. Nuclear staining with Hoechst 33342 is shown in the second and sixth columns to indicate the total cell content per image.
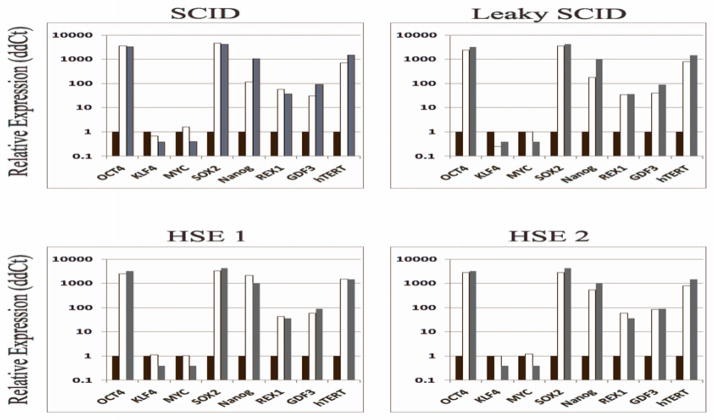
Expression of pluripotency-related genes was also evaluated by quantitative RT-PCR. As compared to the parental fibroblast lines, patient- (and control-) derived iPSCs demonstrated robust expression of pluripotency-associated genes such as OCT4, SOX2, NANOG, REX1, GDF3, and hTERT (Figure 2). As an internal control of pluripotency gene expression profile, we used a previously reported control iPSC line17 that had been generated using 4 retroviral vectors, each of which contained one of the four reprogramming factors. A similar profile of gene expression was demonstrated in the newly generated patient- and control-derived iPSCs and in reference iPSCs.
Figure 2. PID-specific iPSC show a gene expression panel similar to reference iPSCs derived from a healthy control.
Quantitative real-time PCR (RT-PCR) assay for gene expression of OCT4, KLF4, MYC, SOX2, NANOG, REX1, GDF3, and hTERT was performed in iPSCs derived from healthy contriol and from patients with PID, and compared to the pattern observed in a previously established control human iPSC line, obtained by reprogramming using 4 different retroviral vectors17. Quantitative RT-PCR reactions were normalized against β-actin (ACTB). Expression was calculated using the ddCT method relative to expression levels in the individual parent fibroblast cell lines for the normal and PID-specific iPSC or normal control human fibroblasts for the human ES cells.
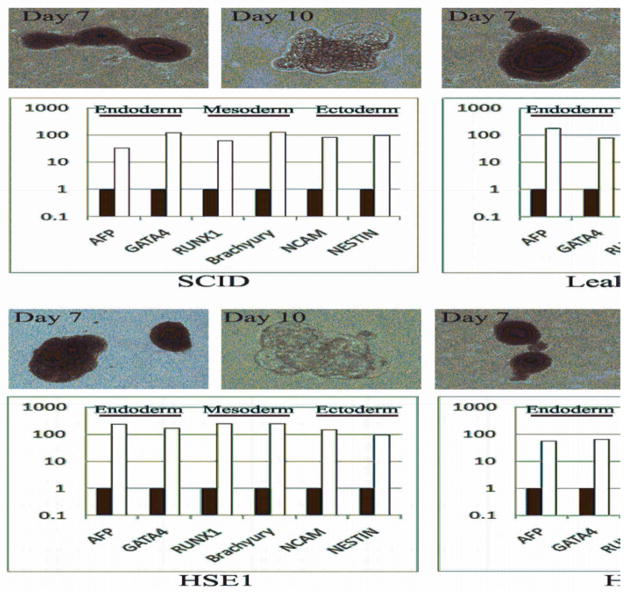
To confirm pluripotency and the ability to support multilineage differentiation, the PID-specific iPSC lines were allowed to differentiate in vitro into embryoid bodies as previously described17, 45. Similar to what was previously shown for hES, tight clusters of differentiating cells formed by day 7, and later cavitated, becoming cystic, by day 10 (Figure 3). Both PID-specific and control iPSCs showed expression of markers of all three embryonic germ layers (ectoderm, mesoderm, and endoderm) (Figure 3), thus confirming their ability to develop along multiple lineages.
Figure 3. Differentiation of PID-specific iPSCs reveals lineage-specific gene expression.
PID-specific iPSC were allowed to differentiate into embryoid bodies (EB) by culture in a bFGF-free hES medium and without co-culture with feeder cells. Robust formation of tight and well-formed cell clusters was detected by day 7, that became cystic by day 10 (upper row in each cell-specific panel). Quantitative RT-PCR gene expression analysis of the derived EB after 10 days shows increased expression of lineage-specific markers from each of the three embryonic germ layers, including: AFP and GATA4 (endoderm), RUNX1 and Brachyury (mesoderm), NCAM and NESTIN (ectoderm). Quantitative RT-PCR reactions were normalized against β-actin (ACTB). Expression was calculated using the ddCT method relative to expression levels in undifferentiated iPSCs. Black and white bars identify undifferentiated iPSCs and day 10 EB, respectively.
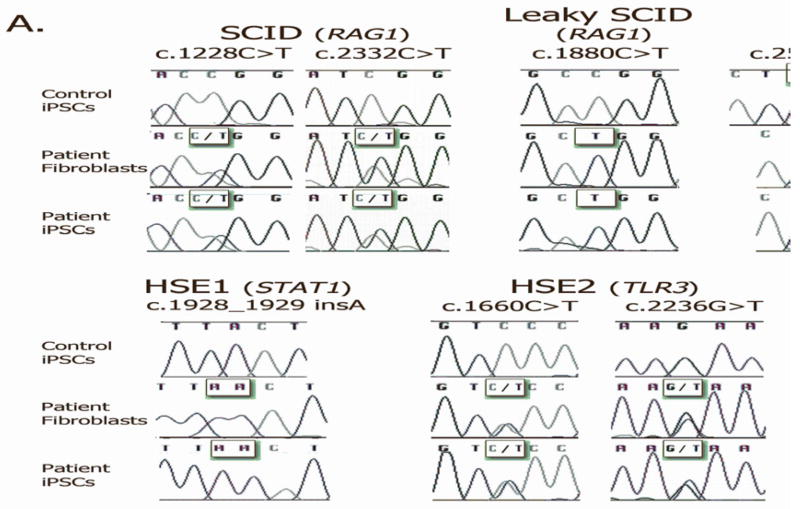
To confirm patient-specific origin, we analyzed each of the iPSCs and the parental fibroblast lines for the specific gene mutation(s) identified in each patient. In all cases, genetic identity was observed between patient-derived fibroblasts and iPSCs (Figure 4A).
Figure 4. Patient origin and chromosomal integrity of the PID-specific iPSCs.
A) PCR amplification followed by DNA sequencing of genomic DNA derived from the PID-specific iPSCs and their parental fibroblasts was performed using specific primers corresponding to the disease-causing mutations for each of the lines, and demonstrated that the PID-specific iPSC lines carry the same disease-causing mutations as their parental fibroblasts. In the case of the first allele (c.256-257del) of the RAG1-mutated patient with Omenn syndrome, genetic identity between patient-derived iPSCs and fibroblasts was demonstrated upon cloning and sequencing of the specific product.
B) PID-specific iPSCs were analyzed for chromosomal integrity by G-banding karyotyping.
It has been previously reported that reprogramming of somatic cells to pluripotency and prolonged culture of hES cells may result in clonal somatic chromosomal aberrations47, 51, 52. We tested representative PID-specific and control-derived iPSC lines for karyotypic integrity. At least one line was analyzed for each patient-specific iPSCs (Table E1 in the Online Repository). For each line assayed, cytogenic analysis was performed on 20 G-banded metaphases. All but one of the various PID-specific iPSC lines that were analyzed demonstrated normal karyotype (Figure 4B). In one of the iPSC lines derived from a patient with RAG1-deficient SCID, 2 cells out of the 20 analyzed carried a trisomy of chromosome 7 (47,XX +7) and hence may represent a clonal chromosomal aberration. Two other lines derived from the same patient demonstrated a normal karyotype.
Discussion
The field of iPSCs is rapidly growing as novel reprogramming strategies and protocols that allow differentiation of iPSC into various cell types become available. In spite of this progress, variable efficiency of the nuclear reprogramming process, incomplete maintenance of iPSC stemness profile, failure to achieve transgene silencing, and integration-dependent effects on endogenous genes expression remain significant challenges53–55.
To generate iPSCs from patients with various forms of PID, we have chosen to use a single lentiviral vector expressing the four common reprogramming factors OCT4, SOX2, KLF4, and c-MYC that are coded on a single polycistronic cassette. Using this vector, we have succeeded in generating multiple iPSC clones for each of the fibroblast cell lines that we have infected. A very high efficiency of reprogramming had been also observed with a murine version of the STEMCCA cassette48. Another potential advantage of this vector is that the STEMCCA cassette is flanked by two inverted LoxP sites, thus permitting excision of the lentiviral sequences by transient expression of the Cre recombinase46, 47. This strategy limits the residual genetic signature that is left following the reprogramming process to a minimum, and removes all reprogramming transgenes.
We have demonstrated that the PID-specific iPSCs generated with this approach exhibit a robust stemness gene expression profile. Furthermore, iPSC lines expressed TRA-1-60, a marker that has been shown to identify fully reprogrammed cells, capable of pluripotency50. In keeping with this, when cultured under appropriate conditions and allowed to differentiate into EB, iPSC lines expressed genes specific for each of the three embryonic layers. Nuclear reprogramming is thought to result from transient expression of the OCT4, SOX2, KLF4, and c-MYC transgenes, followed by induction of endogenous genes while the transgenes are silenced. Comparison of the pattern of expression of OCT4, SOX2, KLF4, and c-MYC genes in our series of PID-specific iPSCs is consistent with this notion. In particular, KLF4 and c-MYC were expressed at similar levels in patient-derived iPSCs, their parental fibroblasts, and in reference iPSC cells, whereas iPSCs maintained high levels of expression of SOX2 and OCT4. Since transcription of all four transgenes contained in the STEMCCA cassette is under control by the same promoter, these data suggest silencing of the transgenes, and differential induction of the endogenous genes. This has been recently confirmed using a modified version of the STEMCCA lentiviral vector also containing an m-Cherry reporter gene46.
Clonal chromosomal aberrations have been previously reported in aged hES cells51, 52. Maintenance of karyotypic integrity is an important feature, when considering using patient-derived iPSCs to study the pathophysiology of the disease at the cellular level. With one single exception, all PID-specific iPSC lines tested retained a normal karyotype, demonstrating that genomic integrity is generally maintained at this level of resolution after reprogramming. However, assessment of genomic integrity remains an important test that should be performed on all newly generated iPSCs.
Prior to this study, generation of iPSCs had been reported only for one type of PID, namely ADA deficiency45, using four retroviral vectors to allow transduction of the reprogramming factors. We have now shown that iPSCs can be generated with high efficiency from patients who suffer from various forms of PID that affect different arms of the immune system. However, it is possible that some forms of PID remain resistant to reprogramming. In particular, use of integrating vectors might fail to induce reprogramming in fibroblasts from patients with defects in DNA repair, because of toxicity and cell death associated with insertion of the vector. In this case, alternative strategies could be considered to generate iPSCs, such as delivery of the reprogramming factors through non integrating vectors, or transient correction of the cellular defect.
Generation of a repository of iPSCs from patients with various forms of PID provides unique research opportunities (Figure 5). In vitro generation of T lymphocytes has already been reported for hES cells cultured on stromal OP9-DL4 cells56–58. If a similar approach becomes available for human iPSCs, it would be possible to compare the cell-intrinsic potential of iPSCs carrying different mutations in the same gene to support T cell differentiation. For example, use of the three patient-derived iPSC lines with mutations in the RAG1 gene but with differing clinical phenotypes could provide a previously unforeseen experimental avenue to directly compare the efficiency and fidelity of human thymopoiesis.
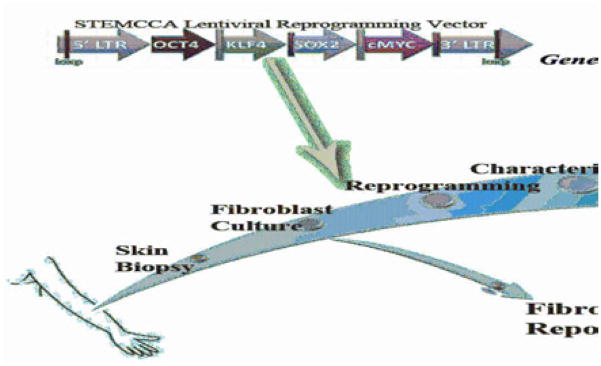
Figure 5.
Schematic representation of the process of producing PID-specific iPSCs and their possible use in disease characterization and gene correction.
Patient-derived iPSCs may also represent a unique tool to investigate in greater detail the pathophysiology of extra-hematopoietic manifestations associated with PIDs. Indeed, we have been able to differentiate iPSCs derived from patients with genetically-determined susceptibility to herpes simplex encephalitis into various mature cell types of the central nervous system, and by this mean dissect the cellular and molecular phenotype of the disease (Lafaille and Pessach et al., submitted). Similarly, it may become possible to study differentiation and function of thymic epithelial cells and heart cells starting from iPSCs from patients with DiGeorge syndrome, or inflammatory responses in various cell types obtained from iPSCs derived from patients with NEMO deficiency. Finally, iPSCs can be used as a limitless source of stem cells in which novel strategies to achieve gene correction may be tested. In particular, they share with ES cells a higher susceptibility to homologous recombination and thus represent a promising tool to study the ability of zinc-finger nucleases, meganucleases and sleeping-beauty transposons to mediate gene repair54,55.
In conclusion, we have reported on the successful generation and characterization of iPSCs from patients with various clinical PID phenotypes and underlying genotypes.
The iPSCs technology is still at its early days. Limitations and potential pitfalls of this approach include, among others, variability in the efficiency and validity of the reprogramming strategy, and the possible introduction of genomic abnormalities that may lead to increased tumorigenesis. These problems must be addressed before considering use of these cells in clinical settings. Nevertheless, it can be anticipated that this novel technology will provide new insights into the pathophysiology of PIDs, and facilitate development of novel and more effective forms of treatment for these disorders.
Clinical Implications.
Induced pluripotent stem cells derived from patients with primary immunodeficiencies represent a unique resource to study the pathophysiology and to develop novel therapeutic approaches for these disorders.
Acknowledgments
Source of funding: National Institutes of Health grants 1R03AI088352-01 and 1R21AI0898-10-01, March of Dimes (grant #6-FY10-282), Translation Research Program (all to L.D.N.), Harvard Catalyst Grant (to G.Q.D.)
This work was supported by the Manton Foundation (to L.D.N. and G.Q.D.) and by Fondazione “Angelo Nocivelli” (to S.G.).
Abbreviations
- bFGF
basic fibroblast growth factor
- CHH
cartilage hair hypoplasia
- HSE
herpes simplex encephalitis
- iMEFs
irradiated mouse embryonic fibroblasts
- iPSC
induced pluripotent stem cells
- OS
Omenn syndrome
- PID
primary immunodeficiency
- SCID
severe combined immune deficiency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Notarangelo LD, Fischer A, Geha RS, Casanova JL, Chapel H, Conley ME, et al. Primary immunodeficiencies: 2009 update. J Allergy Clin Immunol. 2009;124:1161–78. doi: 10.1016/j.jaci.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pessach I, Walter J, Notarangelo LD. Recent advances in primary immunodeficiencies: identification of novel genetic defects and unanticipated phenotypes. Pediatr Res. 2009;65:3R–12R. doi: 10.1203/PDR.0b013e31819dbe1e. [DOI] [PubMed] [Google Scholar]
- 3.Notarangelo LD. Primary immunodeficiencies. J Allergy Clin Immunol. 125:S182–94. doi: 10.1016/j.jaci.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan KE. Chromosome 22q11.2 deletion syndrome: DiGeorge syndrome/velocardiofacial Syndrome Immunol Allergy. Clin North Am. 2008;28:353–66. doi: 10.1016/j.iac.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Notarangelo LD, Roifman CM, Giliani S. Cartilage-hair hypoplasia: molecular basis and heterogeneity of the immunological phenotype. Curr Opin Allergy Clin Immunol. 2008;8:534–9. doi: 10.1097/ACI.0b013e328310fe7d. [DOI] [PubMed] [Google Scholar]
- 6.Slatter MA, Gennery AR. Primary immunodeficiencies associated with DNA-repair disorders. Expert Rev Mol Med. 12:e9. doi: 10.1017/S1462399410001419. [DOI] [PubMed] [Google Scholar]
- 7.Hanson EP, Monaco-Shawver L, Solt LA, Madge LA, Banerjee PP, May MJ, et al. Hypomorphic nuclear factor-kappaB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol. 2008;122:1169–77. e16. doi: 10.1016/j.jaci.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 9.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 10.Sauer AV, Aiuti A. New insights into the pathogenesis of adenosine deaminase-severe combined immunodeficiency and progress in gene therapy. Curr Opin Allergy Clin Immunol. 2009;9:496–502. doi: 10.1097/ACI.0b013e3283327da5. [DOI] [PubMed] [Google Scholar]
- 11.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–12. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 12.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–7. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 13.Pérez de Diego R, Sancho-Shimizu V, Lorenzo L, Puel A, Plancoulaine S, Picard C, et al. Human TRAF3 Adaptor Molecule Deficiency Leads to Impaired Toll-like Receptor 3 Response and Susceptibility to Herpes Simplex Encephalitis Immunity. 2010 doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–8. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–6. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 17.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 18.Cai J, Yang M, Poremsky E, Kidd S, Schneider JS, Iacovitti L. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 19:1017–23. doi: 10.1089/scd.2009.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–21. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 21.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G, Chambers SM, Tomishima MJ, Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc. 5:688–701. doi: 10.1038/nprot.2010.35. [DOI] [PubMed] [Google Scholar]
- 23.Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–6. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nizzardo M, Simone C, Falcone M, Locatelli F, Riboldi G, Comi GP, et al. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol Life Sci. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swistowski A, Peng J, Liu Q, Mali P, Rao MS, Cheng L, et al. Efficient Generation of Functional Dopaminergic Neurons from Human Induced pluripotent Stem Cells under Defined Conditions. Stem Cells. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freund C, Davis RP, Gkatzis K, Ward-van Oostwaard D, Mummery CL. The first reported generation of human induced pluripotent stem cells (iPS cells) and iPS cell-derived cardiomyocytes in the Netherlands. Neth Heart J. 18:51–4. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–23. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 29.Gai H, Nguyen DM, Moon YJ, Aguila JR, Fink LM, Ward DC, et al. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 79:171–81. doi: 10.1016/j.diff.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Wang D, Qin J, Liu C, Zhang Q, Zhang X, et al. Generation of functional hepatocytes from mouse induced pluripotent stem cells. J Cell Physiol. 222:492–501. doi: 10.1002/jcp.22000. [DOI] [PubMed] [Google Scholar]
- 31.Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–42. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 32.Ueda T, Yamada T, Hokuto D, Koyama F, Kasuda S, Kanehiro H, et al. Generation of functional gut-like organ from mouse induced pluripotent stem cells. Biochem Biophys Res Commun. 391:38–42. doi: 10.1016/j.bbrc.2009.10.157. [DOI] [PubMed] [Google Scholar]
- 33.Inami Y, Yoshikai T, Ito S, Nishio N, Suzuki H, Sakurai H, et al. Differentiation of induced pluripotent stem cells to thymic epithelial cells by phenotype. Immunol Cell Biol. doi: 10.1038/icb.2010.96. [DOI] [PubMed] [Google Scholar]
- 34.Choi KD, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–67. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengerke C, Grauer M, Niebuhr NI, Riedt T, Kanz L, Park IH, et al. Hematopoietic development from human induced pluripotent stem cells. Ann N Y Acad Sci. 2009;1176:219–27. doi: 10.1111/j.1749-6632.2009.04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–34. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 37.Karner E, Unger C, Cerny R, Ahrlund-Richter L, Ganss B, Dilber MS, et al. Differentiation of human embryonic stem cells into osteogenic or hematopoietic lineages: a dose-dependent effect of osterix over-expression. J Cell Physiol. 2009;218:323–33. doi: 10.1002/jcp.21605. [DOI] [PubMed] [Google Scholar]
- 38.Lei F, Haque R, Weiler L, Vrana KE, Song J. T lineage differentiation from induced pluripotent stem cells. Cell Immunol. 2009;260:1–5. doi: 10.1016/j.cellimm.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Bronson S, Niyibizi C. Derivation of murine induced pluripotent stem cells (iPS) and assessment of their differentiation toward osteogenic lineage. J Cell Biochem. 109:643–52. doi: 10.1002/jcb.22440. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno Y, Chang H, Umeda K, Niwa A, Iwasa T, Awaya T, et al. Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. FASEB J. 24:2245–53. doi: 10.1096/fj.09-137174. [DOI] [PubMed] [Google Scholar]
- 41.Morizane R, Monkawa T, Itoh H. Differentiation of murine embryonic stem and induced pluripotent stem cells to renal lineage in vitro. Biochem Biophys Res Commun. 2009;390:1334–9. doi: 10.1016/j.bbrc.2009.10.148. [DOI] [PubMed] [Google Scholar]
- 42.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 43.Xu D, Alipio Z, Fink LM, Adcock DM, Yang J, Ward DC, et al. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci U S A. 2009;106:808–13. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lerou PH, Yabuuchi A, Huo H, Miller JD, Boyer LF, Schlaeger TM, et al. Derivation and maintenance of human embryonic stem cells from poor-quality in vitro fertilization embryos. Nat Protoc. 2008;3:923–33. doi: 10.1038/nprot.2008.60. [DOI] [PubMed] [Google Scholar]
- 45.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somers A, JCJ, Sommer CA, Ford CC, Mills JA, Ying L, et al. Generation of transgene-free lung-disease specific human iPS cells using a single excisable lentiviral stem cell cassette. Stem cell. 2010 doi: 10.1002/stem.495. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, et al. Excision of Reprogramming Transgenes Improves the Differentiation Potential of iPS Cells Generated with a Single Excisable Vector. Stem Cells. 2009;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–9. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staerk J, Dawlaty M, Gao Q, Maetzel D, Hanna J, Sommer C, et al. Reprogramming of Human Peripheral Blood Cells to Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;7:20–4. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–11. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buzzard JJ, Gough NM, Crook JM, Colman A. Karyotype of human ES cells during extended culture. Nat Biotechnol. 2004;22:381–2. doi: 10.1038/nbt0404-381. author reply 2. [DOI] [PubMed] [Google Scholar]
- 52.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–4. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 53.Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 120:51–9. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nat Rev Mol Cell Biol. 2008;9:725–9. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 55.Rolletschek A, Wobus AM. Induced human pluripotent stem cells: promises and open questions. Biol Chem. 2009;390:845–9. doi: 10.1515/BC.2009.103. [DOI] [PubMed] [Google Scholar]
- 56.Dervovic D, Zuniga-Pflucker JC. Positive selection of T cells, an in vitro view. Semin Immunol. 2010;22:276–86. doi: 10.1016/j.smim.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Mohtashami M, Shah DK, Nakase H, Kianizad K, Petrie HT, Zuniga-Pflucker JC. Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. J Immunol. 2010;185:867–76. doi: 10.4049/jimmunol.1000782. [DOI] [PubMed] [Google Scholar]
- 58.Sultana DA, Bell JJ, Zlotoff DA, De Obaldia ME, Bhandoola A. Eliciting the T cell fate with Notch. Semin Immunol. 2010;22:254–60. doi: 10.1016/j.smim.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cassani B, Poliani PL, Moratto D, Sobacchi C, Marrella V, Imperatori L, et al. Defect of regulatory T cells in patients with Omenn syndrome. J Allergy Clin Immunol. 125:209–16. doi: 10.1016/j.jaci.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176:5078–83. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]