Abstract
Sensing of environmental signals is often mediated by G-protein coupled receptors and their cognate heterotrimeric G-proteins. In Trichoderma reesei (Hypocrea jecorina) the signals transmitted via the G-protein alpha subunits GNA1 and GNA3 cause considerable modulation of cellulase transcript levels and the extent of this adjustment is dependent on the light status. We therefore intended to elucidate the underlying mechanism connecting light response and heterotrimeric G-protein signaling.
Analysis of double mutant strains showed that constitutive activation of GNA1 or GNA3 in the absence of the PAS/LOV domain protein ENVOY (ENV1) leads to the phenotype of constitutive G-alpha activation in darkness. In light, however the deletion-phenotype of Δenv1 was observed with respect to growth, conidiation and cellulase gene transcription. Additionally deletion of env1 causes decreased intracellular cAMP accumulation, even upon constitutive activation of GNA1 or GNA3. While supplementation of cAMP caused an even more severe growth phenotype of all strains lacking env1 in light, addition of the phosphodiesterase inhibitor caffeine rescued the growth phenotype of these strains.
ENV1 is consequently suggested to connect the light response pathway with nutrient signaling by the heterotrimeric G-protein cascade by adjusting transcript levels of gna1 and gna3 and action on cAMP levels – presumably through inhibition of a phosphodiesterase.
Keywords: Trichoderma reesei, Hypocrea jecorina, Heterotrimeric G-proteins, cAMP pathway, Cellulase gene expression, Light response
1. Introduction
The adaptation of fungi to their environment is crucial for successful competition in their natural habitat. In this process, they respond to the availability of nutrients, light or the presence of a potential mating partner or competitor. Such key signals are received and transmitted via a complex machinery of signal transduction pathways, one of the most important among them being heterotrimeric G-protein signaling (Lengeler et al., 2000). G-protein signaling is widespread in mammalians as well as in fungi (Lafon et al., 2006; Li et al., 2007; Offermanns, 2003). In fungi, this signaling cascade is involved in many important processes, such as reproduction, virulence, stress responses and development (Hsueh et al., 2007; Ivey et al., 2002; Kays and Borkovich, 2004; Segers and Nuss, 2003; Yang et al., 2002). The main route of G-protein signaling is well studied (for review see Hamm, 1998; Li et al., 2007). G-proteins are composed of three subunits: alpha, beta and gamma. When the G-proteins are activated, a conformational change leads to the exchange of the G-alpha bound GDP to GTP. The associated beta and gamma subunits dissociate as a dimer. The free alpha subunit, as well as the betagamma-dimer are now able to bind and influence downstream factors. The genome of Trichoderma reesei comprises three G-alpha subunits (GNA1, GNA2 and GNA3), one beta and one gamma subunit (Schmoll, 2008). The number of different G-alpha subunits allows for many possibilities of combinations and hence response to various signals (Dohlman and Thorner, 1997). Interestingly, the transmitted signals do not necessarily lead to isolated or independent outputs: for example in Cryptococcus neoformans two G-alpha subunits were shown to act in opposite directions with respect to initiation of mating (Hsueh et al., 2007). The most fascinating element in this pathway is the fine-tuning mechanism, which allows the fungi to react immediately and appropriately to changes in the environment. This effect can be established by different proteins, which interact with G-proteins, such as the RGS domain proteins (Regulators of G-protein Signaling; Dohlman and Thorner, 1997) or other factors influencing abundance and/or activity of G-proteins (Li et al., 2007). Many of the factors which are involved in the regulation of G-protein signaling remain unknown.
Recently it was shown that in the biotechnological workhorse T. reesei (Hypocrea jecorina), the heterotrimeric G-protein pathway impacts light dependent transmission of the cellulose signal and hence regulation of cellulase gene expression (Schmoll et al., 2009, 2010; Seibel et al., 2009). Since the rise in energy costs and the increased awareness of the imminent climate change at least in part caused by utilization of fossil fuels, development of more sustainable fuels, i.e. the so-called biofuels has become a focus of research. With respect to second generation biofuels derived from agricultural waste products (Rubin, 2008; Sticklen, 2008), T. reesei plays an important role as one of the most prolific producers of a highly efficient cellulase mixture (Schuster and Schmoll, 2010). In spite of its great potential to degrade plant material (cellulose and hemicellulose), the genome only comprises seven genes for cellulases (Martinez et al., 2008). Nevertheless, this fungus is known as a successful colonizer in its natural habitat, which may also be due to optimized use of resources by fine-tuning of the physiological response to the presence of nutrients in the environment. In this respect, the finding that ENVOY impacts the production of cellulases in a light dependent way (Schmoll et al., 2005) warrants further investigation in order to elucidate the underlying mechanism.
The modulation of cellulase gene transcription in T. reesei by light was found after screening for novel signaling factors involved in regulation of this process, which surprisingly identified a signaling factor (later named ENVOY for messenger) related to the light signaling machinery (Schmoll et al., 2004, 2005). ENVOY is a PAS/LOV domain protein, related to the Neurospora crassa photoreceptor VIVID (Brunner and Kaldi, 2008; Heintzen et al., 2001), but not conserved in all ascomycetes and thus far are absent outside of the ascomycete lineage (Schmoll et al., 2005, 2010; Idnurm et al., 2010; Rodriguez-Romero et al., 2010). PAS (PER, ARNT, SIM) domains are sensory domains and able to convey a protein/protein interaction (Taylor and Zhulin, 1999). “LOV” stands for sensing of light, oxygen and voltage (Cheng et al., 2003; Crosson et al., 2002) and LOV domains are characteristic for a subfamily of the PAS superfamily. Previously, it was shown that ENVOY is subject to an autoregulatory feedback loop (Schmoll et al., 2005). Moreover, constitutive activation of the Gα subunit GNA1 enhances gna1 transcription levels upon growth on glycerol, hence suggesting a feedback loop (Seibel et al., 2009) and the constitutive activation of GNA3 leads to an increase in transcript levels of gna3 on cellulose as carbon source in light. Interestingly, the light regulatory protein ENVOY turned out to influence the transcript levels of gna3 (Schmoll et al., 2009).
Therefore the aim of this study was to investigate the underlying interplay of signal transduction pathways in order to understand the regulatory network constituting this fine-tuning mechanism between nutrient signaling and light response. The following study identified ENVOY as a central regulatory element in the fine-tuning mechanism of G-protein signaling.
2. Materials and methods
2.1. Strains, plasmids and culture conditions
T. reesei wild-type strains QM9414 (ATCC 26921) and TU-6 (ATCC MYA-256; uridine auxotroph; (Gruber et al., 1990)), Δenv1 (Castellanos et al., 2010), GNA1QL (Seibel et al., 2009) and GNA3QL (Schmoll et al., 2009) strains were used in the present study. In the latter strains the Q-L mutation causes constitutive activation of the respective G-alpha subunit. Unless otherwise noted, all strains including those constructed for this study (see below) were grown on malt extract agar. For cultivations of the uridine auxotrophic strain TU-6, the medium was supplemented with 10 mM uridine (Merck, Darmstadt, Germany).
For Northern Blot analysis and qRT-PCR experiments, strains were grown in 1 L Erlenmayer shake flasks in constant darkness (DD), constant light (LL, 25 μmol photons m−2 s−1; 1800 lux) or pregrown for 24 h in darkness and thereafter exposed to light (DL) at 28 °C on a rotary shaker (200 rpm). Harvesting of dark grown cultures was done under safe-red-light (darkroom lamp, Philips PF712E, red, E27, 15 W). For liquid culture Mandels–Andreotti minimal medium (Mandels and Andreotti, 1978) was used, supplemented with 0.1% (w/v) peptone to induce germination and with 1% (w/v) carbon source, i.e. glycerol (Merck, Darmstadt, Germany) or microcrystalline cellulose (# 1402; SERVA, Heidelberg, Germany), as indicated at the respective experiments. For inoculation 109 conidia per liter were used. For cAMP measurements strains were grown on Mandels–Andreotti medium, supplemented with 0.1% (w/v) peptone, 2% (w/v) agar–agar and 1% (w/v) of carboxymethylcellulose sodium salt (Roth, Karlsruhe, Germany) in constant light. Escherichia coli JM109 was used for the propagation of vector molecules and DNA manipulations (Yanisch-Perron et al., 1985).
2.2. Construction of T. reesei GNA1QL and GNA3QL strains lacking env1
For deletion of env1 in GNA1QL and GNA3QL strains, plasmid pDELENV2 (Castellanos et al., 2010) conferring hygromycin B resistance was used. The deletion cassette was released from pDELENV2 by restriction enzymes Acc65I and BamHI. This linear fragment was used for the transformation of protoplasts of strains GNA1QL and GNA3QL (Gruber et al., 1990). Transformants were selected on plates containing 50 μg/mL hygromycin B (Roth, Karlsruhe, Germany). Stable transformants were obtained after two rounds of single spore isolation. Fungal DNA was isolated from transformants by standard protocols. To check for successful deletion, we performed two different control PCRs, using primers csfDEL5F and csfDEL3R as well as ENVSP1F and ENVNEU2R (for primer sequences see Table 1). These primers bind outside the deleted region, which enables verification of replacement of the env1 gene with the hygromycin resistance cassette. Successful deletion, as expected, resulted in a significantly longer fragment than amplification from wild-type DNA as well as disappearance of the shorter wild-type fragment. These results were confirmed by Southern blotting (data not shown).
Table 1.
Oligonucleotides used in this study.
| Gene | Fragment length | Primer | Sequence |
|---|---|---|---|
| 18s rRNA | 297 bp | 18SRF | 5′ GGTGGAGTGATTTGTCTG 3′ |
| 18SRR | 5′ CTTACTAGGGATTCCTCG 3′ | ||
| cbh1 | 1264 bp | CBH1SF | 5′ TCGGCCTGCACTCTCCAATC 3′ |
| CBH1SR | 5′ TGGAGTCCAGCCACAGCATG 3′ | ||
| env1 | 794 bp | ENVGST1F | 5′ TGAATTCCAATGCTCGAAGCACCAATC 3′ |
| ENVGST1R | 5′ ATCTCGAGATCTATAGCTCAGCCTGGAG 3′ | ||
| 1119 bp for wild-type | ENVSP1F | 5′ TCCCTGGATCTGGATACG 3′ | |
| 3056 bp for env1 deletion mutant | ENVNEU2R | 5′ CTGGCGTGGTATTTCTCTGAC 3′ | |
| 2740 bp for wild-type | csfDEL5F | 5′ ATGGTACCGATGATGTCCAGCCTTC 3′ | |
| 5140 bp for env1 deletion mutant | csfDEL5R | 5′ ATCTCGAGTCGTAACTAGGCATAGAACTG 3′ | |
| 143 bp | RTenv1F | 5′ CATTGACCTTGGCCCTCTC 3′ | |
| RTenv1R | 5′ GACAGTTTCGACCCATGATCTC 3′ | ||
| rpl6e | 110 bp | RTL6eF1 | 5′ GATACGTCATCGCCACCTCC 3′ |
| RTL6eR1 | 5′ CTTCTCCTTGGCCTTCTCG 3′ | ||
| gna1 | 133 bp | RTgna1F | 5′ CACCACCATCCTCTTCCTG 3′ |
| RTgna1R | 5′ CGTCTTGATGAACCACCTG 3′ | ||
| gna3 | 93 bp | RTgna3F | 5′ CTCACACAAGCCACCGACAC 3′ |
| RTgna3R | 5′ ATGCCCGAATCCTTGAGC 3′ | ||
| 1068 bp | GNA3CDNA1F | 5′ AACGTCGTCACTCTCGTC 3′ | |
| GNA3CDNA1R | 5′ ATGGGAAAGAACGTGATG 3′ | ||
| acy1 | 159 bp | RTAcyF1 | 5′ CGCCGCATGGACTACTTTG 3′ |
| RTAcyR1 | 5′ CGACCCGGTGGATACATTTC 3′ |
2.3. Nucleic acid isolation and manipulation
Fungal mycelia were harvested by filtration, briefly washed with tap water, frozen and ground in liquid nitrogen. Extraction of genomic DNA was performed as described previously (Schmoll et al., 2004). Total RNA for Northern blots was isolated by the guanidinium thiocyanate method (Chomczynski and Sacchi, 1987). Standard methods (Sambrook et al., 1989) were used for electrophoresis, blotting and hybridization of nucleic acids. Northern blot analysis was performed as described previously (Sambrook et al., 1989; Schmoll et al., 2004). Twenty micrograms of total RNA were loaded onto the gel and blotted. The following probes were used: for 18S rRNA hybridization a probe using primers 18SRF and 18SRR (for all primer sequences see Table 1), amplifying a 297 bp fragment and a 1264 bp cbh1 fragment, generated by PCR amplification with primers cbh1S1F and cbh1S1R.
2.4. RNA preparation and reverse transcription for quantitative RT-PCR
Fungal mycelia were harvested by filtration, briefly washed with tap water and frozen in liquid nitrogen. For extraction of total RNA, buffers and columns of the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) were used. Mycelium was added to 800 μl ice-cold buffer RLC with 10 μL/mL 2-mercaptoethanol (Merck) and 12 mm3 0.5 mm Zirconia/Silica Beads (Biospec, Bartlesvielle, USA). The mycelium was ground using a RETSCH MM301 Ball Mill (Retsch, Haan, Germany), two rounds for 30 s (frequency 30 1/s) and left on ice between the two cycles for 30 s. The tubes were incubated at 56 °C for three minutes and then the whole content was transferred to a QIAshredder spin column. Thereafter the procedure as recommended by the manufacturer was followed. Concentration was measured using a Nanodrop ND-1000 spectrophotometer (PEQLAB, Erlangen, Germany).
The obtained total RNA was digested by DNase I (Fermentas, Vilnius, Lithuania) and purified with the RNeasy Mini Kit (QIAGEN) according to the manufacturers’ instructions. Prior to preparation of cDNA, the RNA quality was evaluated by the Experion System (Bio-Rad, Hercules, USA). The Experion RNA StdSens Analysis Kit (Bio-Rad) was used and the samples were treated as recommended by the manufacturer.
The RNA quality index (RQI) provides information about RNA integrity. An RQI factor of 1 means fragmented and degraded RNA, whereas an RQI of 10 means an intact, non-fragmented RNA sample. The RQI factor shows higher reproducibility and sensitivity than the corresponding RIN (RNA integrity number) factor. The importance of an intact RNA sample of high quality, which is used for qPCR, is crucial for reliable transcription analysis. Using RNA samples of different integrity can result in up to 7-fold transcript values (Fleige and Pfaffl, 2006; Imbeaud et al., 2005). The cut-off value for samples with proper quality is recommended to be above RIN-factor 5 (Fleige and Pfaffl, 2006; Imbeaud et al., 2005) and the threshold for the experiments in this study was set to RQI > 7. Samples with lower quality were not used in the experiments.
Five microgram of total RNA were reverse-transcribed using RevertAid-H− First Strand cDNA Synthesis Kit (Fermentas) and oligo-d(T)-primers for cDNA synthesis following the instruction manual. qRT-PCR was performed using the iQ SYBR Green supermix (Bio-Rad) and the IQ5 ICycler system (Bio-Rad). For data analysis the Pfaffl-method was used (Pfaffl, 2001). Experiments were done in triplicates from two independent biological replicates.
Primers for qRT-PCR analysis and the respective product length are given in table 1. For our studies under changing light conditions it was crucial to find a reliable reference gene for qRT-PCR in T. reesei. A gene (Protein ID 21658; T. reesei genome database v2.0), which is related to the ribosomal protein L6 of N. crassa (NCU02707) was used for normalization of the qRT-PCR data. The expression of the N. crassa gene was shown to be non-clock and non-temperature regulated (Nowrousian et al., 2003) and was used as reference gene for qRT-PCR analysis of wc-1 and phytochrome genes in N. crassa (Froehlich et al., 2005; Lee et al., 2003).
Reliability of the chosen reference gene, which we denominated rpl6e, was compared with two different commonly used reference genes, namely glyceraldehyde-3-phosphate dehydrogenase (gpdh) and translation elongation factor 1 alpha (tef1) with the two different algorithms of the software packages NormFinder (Andersen et al., 2004) and geNORM (Vandesompele et al., 2002). NormFinder ranks gpdh as the best reference gene before rpl6e, but since gpdh is reported to be clock regulated on a circadian basis in N. crassa (Shinohara et al., 1998) and mRNA expression levels vary during photoinduced conidiation in Trichoderma harzianum (Puyesky et al., 1997), it would not be suitable for light/darkness experiments. The geNORM software put rpl6e in first position together with gpdh as best reference genes. The corresponding primers for amplification of the rpl6e fragment are given in table 1.
2.5. cAMP and phosphodiesterase inhibitor assays
For cAMP measurements the Direct cAMP enzyme immunoassay kit (Sigma–Aldrich, USA) was used. Strains were cultured on Mandels–Andreotti minimal medium with carboxymethyl cellulose as carbon source (1% w/v) in constant light (2000 lux). After 6 days of cultivation, the mycelium was frozen in liquid nitrogen and ground to fine powder. Samples were diluted with 10 volumes 0.1 M HCl and centrifuged at 25 °C for 10 min at 700g. The supernatant was used for cAMP measurements performed according to the manufacturer’s instruction.
For testing the effect of inhibition of phosphodiesterase, caffeine and theophylline (both Sigma–Aldrich) were used in the concentrations given with the experiments.
3. Results
3.1. env1, gna1 and gna3 form a regulatory network in signal transmission
Since the G-protein alpha subunit gene gna3 was shown to be regulated both by light and ENV1 (Schmoll et al., 2009) and an influence of light on the regulatory output of both GNA1 and GNA3 was reported (Schmoll et al., 2009; Seibel et al., 2009) the hierarchy of their regulatory interactions should be established.
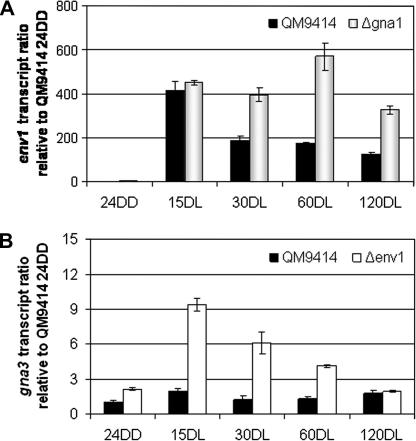
As a basis for our further experiments the extent of the light response of these genes in wild-type and mutant strains was first quantified on glycerol by qRT-PCR. The transcript levels of env1 are very low in darkness but upon illumination, env1 transcript abundance increased 400-fold compared to the dark levels of the wild-type QM9414 with a peak 15 min after start of illumination. In the mutant strain Δgna1, env1 transcript levels showed a comparable response to light as in the wild-type, however, the levels remained elevated beyond 15 min in Δgna1 (Fig. 1A). Hence the dampening of env1 levels after the initially strong response likely involves the function of GNA1, which has a negative effect on env1 transcription and its absence enhances or prolongs light response of env1.
Fig. 1.
Regulatory interactions between gna1, gna3 and env1. (A) Transcript levels of env1 in a gna1 deletion mutant (Δgna1) compared to wild-type QM9414 as analyzed by qRT-PCR. (B) Transcript levels of gna3 in an env1 deletion mutant (Δenv1) compared to wild-type QM9414. Data are given relative to wild-type strain QM9414 in darkness (24DD). Strains were cultivated on Mandels–Andreotti medium with 1% (w/v) glycerol in darkness for 24 h (24DD) and then exposed to constant light for 15 (15DL), 30 (30DL), 60 (60DL) and 120 min (120DL), respectively.
Analysis of the influence of ENV1 on transcription of gna3 had revealed a negative effect of ENV1 (Schmoll et al., 2009). This dampening effect of ENV1 on light response of gna3 was up to 5-fold. (Fig. 1B) These results suggest a hierarchically organized regulatory cascade of gna1, env1 and gna3.
3.2. Both gna1 and gna3 show positive feedback upon activation of their gene products
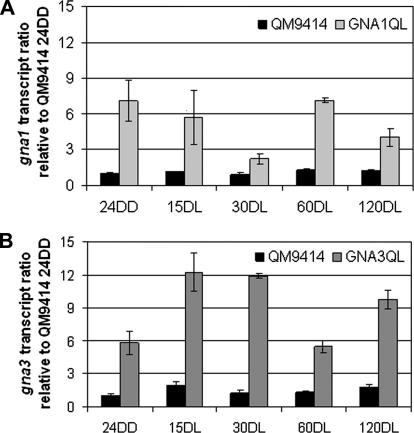
For both G-alpha subunits (GNA1 and GNA3), earlier data indicated positive feedback upon activation on glycerol (gna1) or cellulose (gna3), which is assumed to be operative upon activation of the respective G-alpha subunit (Schmoll et al., 2009; Seibel et al., 2009). Since for gna1 this feedback was carbon source dependent (Seibel et al., 2009), a different result for gna3 on glycerol (instead of cellulose) could not be excluded. Therefore, transcription of both G-alpha subunit genes was analyzed under equal conditions (growth on glycerol, in darkness and after illumination). Transcript quantification of gna1 and gna3 by qRT-PCR revealed a 5- to 10-fold up-regulation of transcript abundance of gna1 or gna3 upon constitutive activation of the respective G-alpha subunit in GNA1QL (Fig. 2A) and GNA3QL (Fig. 2B). These strains comprise additional alleles for expression of constitutively activated versions of GNA1 and GNA3 (Schmoll et al., 2009; Seibel et al., 2009). These results show that both G-protein alpha subunits are subject to positive feedback loops upon growth on glycerol, which of course must involve further components, most importantly transcription factors.
Fig. 2.
Analysis of feedback regulation of G-alpha subunit gene transcription upon activation of the respective gene product. (A) gna1 transcript levels as analyzed by qRT-PCR in wild-type QM9414 and GNA1QL, which expresses constitutively activated GNA1. (B) gna3 transcript levels in wild-type QM9414 and GNA1QL, which expresses constitutively activated GNA3. Increased transcript abundance in strains expressing a constitutively activated G-alpha subunit indicate positive feedback regulation upon activation of the respective G-protein. Data are given relative to wild-type strain QM9414 in darkness (24DD). Strains were cultivated on Mandels–Andreotti medium with 1% (w/v) glycerol in darkness for 24 h (24DD) and then exposed to constant light for 15 (15DL), 30 (30DL), 60 (60DL) and 120 min (120DL), respectively.
3.3. Deletion of env1 in GNA1QL and GNA3QL reveals a central position for ENV1
In order to determine the position of ENV1 in the presumed regulatory cascade, the entire reading frame of env1 in the genomes of the mutant strains GNA1QL and GNA3QL was deleted, resulting in the double mutant strains GNA1QLE and GNA3QLE, with the suffix “E” representing the lack of ENV1.
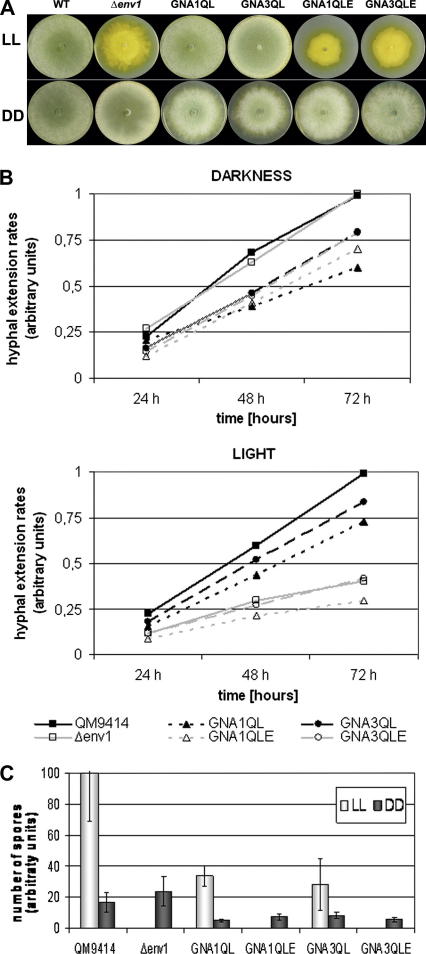
The severe growth phenotype of the env1 mutant in light, i.e. slower growth, hardly any aerial hyphae and lack of conidiation (Castellanos et al., 2010; Schmoll et al., 2005) was observed in GNA3QLE, which could be expected from its presumed position downstream of env1 in the cascade. Surprisingly, the same phenotype of retarded growth in light was also observed for GNA1QLE (Fig. 3A). However, this epistatic effect of env1 deletion was light dependent: both GNA1QLE and GNA3QLE showed the same phenotype as GNA1QL and GNA3QL in darkness, while the Δenv1 mutant shows wild-type behaviour in darkness. Consequently GNA1 and GNA3 are epistatic to ENV1 in darkness. We conclude that ENV1 is a crucial checkpoint in signal transmission by GNA1 and GNA3 in light, but does not interfere with heterotrimeric G-protein signaling in darkness.
Fig. 3.
Characteristics of wild-type, strains expressing constitutively activated GNA1 or GNA3 (GNA1QL and GNA3QL), Δenv1 and double mutants lacking env1 (GNA1QLE and GNA3QLE) in light and darkness. (A) Growth phenotypes of wild-type and mutants on malt extract medium in light and darkness. Strains were grown on malt extract agar plates in constant light or constant darkness for 4 days at 28 °C. (B) Hyphal extension rates upon growth on malt extract medium in light and darkness. Extension rates are given relative to QM9414 after 24, 48 or 72 h. (C) Conidiation upon growth on malt extract medium in light and darkness. Cultures were grown on malt extract agar plates in constant light or constant darkness. Number of spores is given relative to wild-type QM9414 upon growth in light.
To confirm these data in detail growth (hyphal extension rates; Fig. 3B) as well as sporulation (Fig. 3C) were analyzed in light and darkness. As expected, the analyses of the double mutants (GNA1QLE and GNA3QLE) in darkness show similar results as their corresponding parental strains GNA1QL and GNA3QL, while the results for Δenv1 resemble those for the wild-type QM9414 in darkness. In light however, GNA1QLE and GNA3QLE again clearly show the Δenv1 phenotype both with respect to hyphal extension rates and sporulation. Consequently, ENV1 overrules G-protein signaling by GNA1 and GNA3, despite constitutive activation of these G-alpha subunits, with respect to growth and sporulation in light, but does not interfere with this pathway in darkness.
3.4. ENV1 positively acts on the feedback loop of gna1 but not gna3
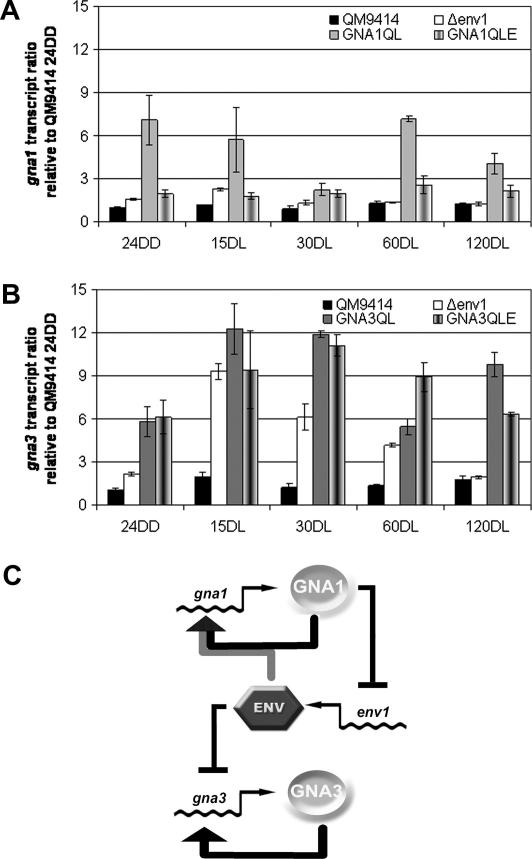
This obvious function of ENV1 as a checkpoint in heterotrimeric G-protein signaling led to the question, whether this impact may be exerted via interference of ENV1 with the previously identified feedback regulation of gna1 and gna3.
Consequently, transcript levels of gna1 and gna3 were measured in response to light during growth on glycerol as carbon source in the parent strain QM9414, in the mutant strains expressing constitutively activated G-alpha subunits (GNA1QL and GNA3QL), in the double mutants (GNA1QLE and GNA3QLE) and in Δenv1. While deletion of env1 significantly reduced the extent of positive feedback regulation of gna1 (lowered transcript levels in GNA1QLE as compared to GNA1QL; Fig. 4A), only minor differences between transcript levels of gna3 in GNA3QL and GNA3QLE can be observed (Fig. 4B). Remarkably, these effects were detectable both in light and darkness. Therefore we conclude that ENV1 interferes with the gna1-feedback loop, but not with the gna3-feedback loop (Fig. 4C). Moreover, these data show that the organization of the regulatory interactions between env1, gna1 and gna3 is not strictly hierarchical, but rather functions via interconnected feedback cycles.
Fig. 4.
The role of ENVOY in feedback regulation of gna1 and gna3. Increased transcript abundance in strains expressing a constitutively activated G-alpha subunit indicates positive feedback regulation upon activation of the respective G-protein. Abolishment of this effect upon deletion of env1 shows an involvement of ENVOY in establishing this feedback. Data are given relative to wild-type strain QM9414 in darkness (24DD). (A) Comparison between feedback regulation of gna1 upon constitutive activation of GNA1 (in GNA1QL) and after deletion of env1 (double mutant GNA1QLE). (B) Comparison between feedback regulation of gna3 upon constitutive activation of GNA3 and after deletion of env1 (double mutant GNA1QLE). Strains were cultivated on Mandels–Andreotti medium with 1% (w/v) glycerol in darkness for 24 h (24DD) and then exposed to constant light for 15 (15DL), 30 (30DL), 60 (60DL) and 120 min (120DL), respectively. (C) Model for the regulatory interaction between ENVOY and the G-protein alpha subunits GNA1 and GNA3. ENVOY is negatively regulated by GNA1 and itself regulates gna3. Additionally, ENVOY is necessary for a fully operational GNA1 feedback loop.
3.5. ENVOY overrules G-protein signaling with respect to cellulase gene expression
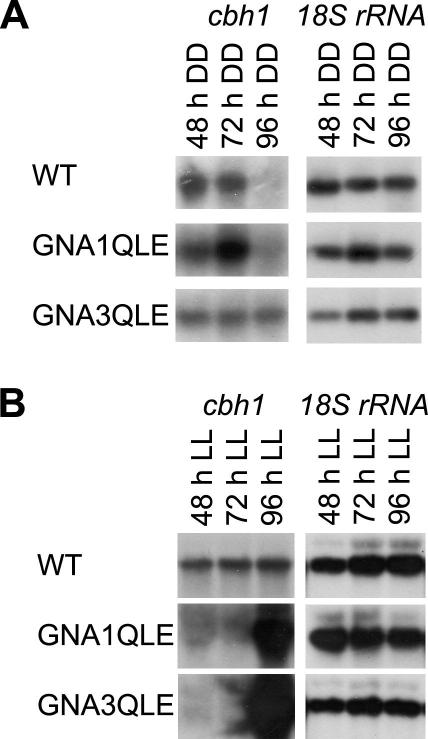
It was shown previously that GNA1, GNA3 and ENV1 influence cellulase gene expression, and in every case, light dependent alterations in their regulatory impact were observed. In darkness, constitutive activation of GNA1 or GNA3 causes minor or no alteration of cellulase transcript levels, whereas in Δenv1 the initially strong transcription of the major cellobiohydrolase gene cel7a (cbh1) at 48 h, significantly drops at 72 and 96 h upon growth on cellulose (Castellanos et al., 2010; Schmoll et al., 2005, 2009; Seibel et al., 2009). Analysis of GNA1QLE and GNA3QLE under these conditions only shows minor alterations compared to the wild-type (Fig. 5A), hence resembling their parental strains, but they clearly do not show the characteristics of the env1-deletion strain. In contrast to these data, our earlier studies showed that upon light exposure cellulase transcript levels are significantly elevated in both strains bearing the alleles for expression of constitutively activated GNA1 or GNA3 (up to 10-fold) and in Δenv1 induction of cellulase transcription is delayed and only starts at 96 h, thus resulting in very characteristic patterns for this strain (Castellanos et al., 2010; Schmoll et al., 2005, 2009; Seibel et al., 2009). Analysis of the double mutants GNA1QLE and GNA3QLE in light on cellulose clearly showed that transcription patterns in these strains resemble the easily distinguishable pattern for Δenv1 with the onset of detectable cellulase transcription only after 96 h (Fig. 5B). Hence also with respect to regulation of cellulase gene expression, ENV1 overrules G-protein signaling via GNA1 and GNA3 in light.
Fig. 5.
The influence of ENVOY on regulation of cellulase gene transcription by GNA1 and GNA3. Northern Blot for analysis of cbh1 transcription in wild-type (WT) and the double mutants GNA1QLE and GNA3QLE. Strains were grown in constant darkness (A) or constant light (B) on Mandels–Andreotti medium with 1% (w/v) microcrystalline cellulose as carbon source. 18S rRNA was used as a control.
3.6. ENVOY intervenes in the cAMP pathway
The second messenger cyclic adenosine monophosphate (cAMP) influences cellulase gene expression (Sestak and Farkas, 1993) and its intracellular levels are responsive to light (Farkas et al., 1990). Moreover, recent studies showed that GNA3 strongly regulates cAMP levels, while GNA1 has only a moderate influence (Schmoll et al., 2009; Seibel et al., 2009).
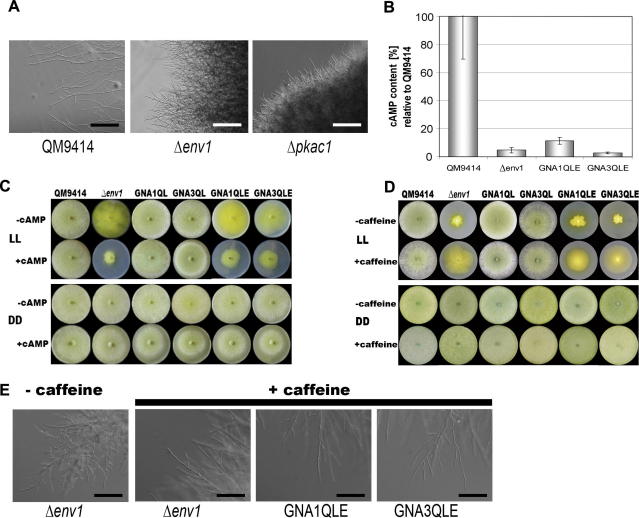
One of the main elements in the cAMP pathway is protein kinase A, which is activated by cAMP and in turn phosphorylates downstream targets (D’Souza et al., 2001). Mutations in the catalytic subunit of this kinase result in a severe growth phenotype (Banno et al., 2005; Saudohar et al., 2002; Shimizu and Keller, 2001), which resembles that seen in Δenv1 in light (Castellanos et al., 2010; Schmoll et al., 2005). Microscopic analysis of Δenv1 and a deletion mutant in the catalytic subunit of cAMP-dependent protein kinase A (PKAC1), Δpkac1, (Schuster et al., 2011), grown in light showed that both mutants no longer show hyphal avoidance or form leading hyphae in the peripheral hyphal zone (Fig. 6A). Based on these data from us and others we reasoned that ENV1 may intervene in regulation of cAMP levels.
Fig. 6.
Regulation of cAMP levels by ENVOY. (A) Microscopic analysis of QM9414, Δpkac1 and Δenv1. Scale bar = 100 μm. (B) cAMP content in QM9414, Δenv1, GNA1QLE and GNA3QLE. cAMP content is given in relation to QM9414. (C) Phenotype of wild-type and mutants upon growth on malt extract medium in the presence (+cAMP) or absence (−cAMP) of 2 mM (final concentration) cAMP in the medium in light and darkness. Strains were grown for 5 days at 28 °C. (D) Phenotype of wild-type and mutants upon growth on malt extract medium in the presence (+caffeine) or absence (−caffeine) of 2 mM (final concentration) caffeine in the medium in light and darkness. Strains were grown for 3 days at 28 °C. (E) Microscopic investigation of the effect of 2 mM caffeine on strains lacking env1. Scale bar = 50 μm.
To test the hypothesis that cAMP is involved in ENVOY function we determined the intracellular cAMP content of the Δenv1 mutant strains (Δenv1, GNA1QLE, GNA3QLE) in light and compared them to wild-type cAMP levels. Indeed we found that the intracellular cAMP levels are drastically (by roughly 90% compared to wild-type) decreased in all strains lacking the function of ENV1 (Fig. 6B). Even the strong increase in cAMP levels of GNA3QL in light, which is more than 5-fold, (Schmoll et al., 2009) does not occur in the absence of ENV1. Consequently, these findings show a positive correlation between the availability of ENV1, hyphal growth and cAMP levels in light and again confirm the key regulatory position of this protein.
From these data one could assume that the reduced growth rate and reduced conidiation in Δenv1 could be due to the decrease in cAMP levels. This in turn could be caused by decreased activity of adenylate cyclase. Especially the finding that adenylate cyclase is activated by light (Kolarova et al., 1992) would hint to an activating function of ENVOY on this enzyme. Therefore, we supplemented the growth media of wild-type and mutant strains with 2 mM cAMP. In the wild-type or the mutant strains expressing constitutively activated G-alpha subunits, growth and conidiation were not influenced by this treatment, neither in light nor in darkness (Fig. 6C). We want to note here that the phenotypes shown here were recorded at a time when the respective effect on growth was most obvious (i.e. after 3, 4 or 5 days). Controls of course showed equal characteristics throughout the experiments.
In the mutant strains expressing constitutively activated GNA1 or GNA3 but lacking env1 (GNA1QLE and GNA3QLE), growth in light is even more retarded than without addition of exogenous cAMP. Therefore, the decreased cAMP levels are unlikely to be caused by a lower activity of adenylate cyclase in the mutants. This conclusion is also corroborated by a qRT-PCR analysis of acy1, encoding adenylate cyclase, which revealed no significant influence of env1 on transcription of acy1 (data not shown). It should be noted that the exogenous addition of cAMP has no effect on the Δenv1 mutant strains in darkness.
An alternative explanation for the lower cAMP levels in the strains lacking ENV1 could be that instead of adenylate cyclase, which would increase cAMP levels, the actual target of ENV1 in light might be one or both of the two phosphodiesterases (Schmoll, 2008) of T. reesei, since phosphodiesterases are known to be activated by cAMP (Hicks et al., 2005; Wang et al., 2005). We consequently tested whether the addition of the phosphodiesterase inhibitor caffeine, which is also effective in T. reesei (Scott and Solomon, 1973; Sestak and Farkas, 1993) (2 mM final concentration) would rescue the growth phenotype of strains lacking ENV1 in light. Indeed enhanced growth in the presence of caffeine was observed. Complete rescue of the wild-type phenotype, however, would likely require optimization of caffeine concentrations, although also functions of caffeine aside from inhibition of phosphodiesterase might be involved. (Fig. 6D). Similar results were obtained with theophylline (5 mM final concentration; data not shown), which also acts as inhibitor of phosphodiesterases. We therefore hypothesize that ENV1 negatively influences phosphodiesterase activity in light, which leads to increased activity in the absence of ENV1 and hence to decreased intracellular cAMP levels. As with the other processes we studied, the effect of ENV1 occurs in light. In this respect, these results indicate that for all regulatory targets of protein kinase A/cAMP light dependent regulation should be taken into consideration.
4. Discussion
Signal transduction via heterotrimeric G-proteins impacts numerous physiological phenomena and a plethora of genes triggering the underlying processes. Because of earlier findings that two G-alpha subunits exert their function in a light dependent manner (Schmoll et al., 2009; Seibel et al., 2009), we were interested in how heterotrimeric G-protein signaling and light response are linked and whether the respective interconnection is organized hierarchically or cooperatively.
Intriguingly, it became very clear that ENVOY is an important key regulator in light-dependent signal transmission via G-alpha subunits. It was especially surprising that ENVOY interferes with the heterotrimeric G-protein pathway at the level of adjustment of cAMP abundance in the cell. cAMP and its role in signal transduction has been studied for decades. This secondary messenger regulates many physiological processes because of its function as activator of cAMP-dependent protein kinase A (PKA). This kinase is an important regulatory component for a plethora or target proteins, including those responsible for regulation of metabolic pathways and exerts its function by phosphorylation (D’Souza and Heitman, 2001; Walsh and Van Patten, 1994). It modulates activity or function of factors binding to DNA motifs within the promotor regions of cAMP inducible genes (Lalli and Sassone-Corsi, 1994). Known regulatory outputs of cAMP/PKA mediated signaling are for example regulation growth (Adachi and Hamer, 1998; Banno et al., 2005; Saudohar et al., 2002; Shimizu and Keller, 2001), spore germination (Fillinger et al., 2002), sporulation (Friedl et al., 2008), virulence (D’Souza et al., 2001), endoglucanase expression (Sestak and Farkas, 1993) or the circadian clock (Hsueh et al., 2007).
The intracellular cAMP amount is directly regulated by two enzymes: the adenylate cyclase, which generates cAMP and phosphodiesterases (PDE), which degrades cAMP (Houslay and Adams, 2003). However, the steady state levels of intracellular cAMP are subject to a sophisticated fine-tuning mechanism: PDE and PKA establish a negative feedback loop (Hicks et al., 2005; Wang et al., 2005), which seems to be targeted by ENV1 via negative impact on PDE in light.
It was shown that the activity of adenylate cyclase, and therefore the amount of cAMP, is regulated by G-alpha subunits in several fungi (Alspaugh et al., 2002; Bölker, 1998; Chen et al., 1996; Gao and Nuss, 1996; Ivey et al., 2002; Kays et al., 2000) and also in T. reesei (Schmoll et al., 2009; Seibel et al., 2009). Data obtained in this study suggest that ENVOY acts on phosphodiesterase, which via altered cAMP levels in turn impacts cAMP-dependent protein kinase A (PKA). This connection hence indicates a widespread effect of the action of ENVOY.
A recent study on the role of ENVOY in T. reesei revealed that this protein not only has functions in light response but influences several other pathways, including carbon source utilization (Schuster et al., 2007). The involvement of ENVOY in signal transmission via G-proteins and the downstream cAMP/protein kinase A pathway provides a reasonable explanation for this broad function and highlights the importance of light for the signals transmitted via these pathways. Adjustment of an appropriate steady state level of cAMP in light and darkness seems to be one important function of the interplay between ENVOY and G-protein signaling. This interplay is supported by an earlier study, which revealed that adenylate cyclase is activated by light, but phosphodiesterase is not (Kolarova et al., 1992) – which in view of the presented data could due to a paramount function, i.e. negative action of ENVOY.
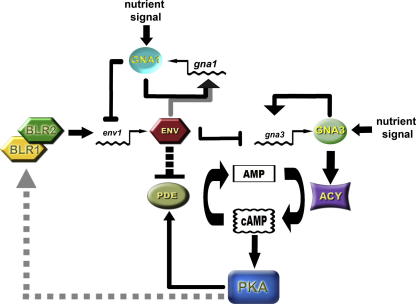
Due to its obvious impact on the cAMP pathway, a model for the regulatory interference of ENVOY with the G-protein pathway can be designed (Fig. 7): ENVOY interferes with the autoregulatory feedback loop of GNA1, but not GNA3. On the one hand this indicates that ENVOY can adjust the relevance of the GNA1 signal by modifying the abundance of the respective message (gna1 levels). On the other hand, the regulatory circuit involving env1, gna3 and cAMP levels would also ensure the control of the relevance of the GNA3 signal. Although deletion of env1 would result in higher levels of gna3, the positive effect of GNA3 on cAMP levels (Schmoll et al., 2009) should initiate an even stronger activation of PDE as is caused by deletion of env1 and hence this regulatory wing is unlikely to counteract the effect of activated PDE.
Fig. 7.
Model for the regulatory interaction of ENVOY with G-protein signaling and cAMP signaling. In env1 deletion mutants, phosphodiesterase activity (PDE) is not dampened and cAMP levels drop due to deregulated degradation of cAMP to AMP by PDE. If exogenous cAMP is added to the medium, PDE activity is increased even further and cannot not be kept at lower levels by ENVOY in the env1 deletion mutants in light. ENVOY can modify the regulatory output of GNA1 by interfering with its feedback cycle. ENVOY interferes with feedback of GNA3 via regulation of gna3 transcript abundance and additionally impacts the GNA3 output-pathway by altering cAMP levels via PDE. The broken line shows the hypothesized feedback of cAMP levels via PKA on the photoreceptors.
Taken together, ENVOY can influence cAMP levels via its action on transcript levels of G-alpha subunits, although their effect is dependent on activation by environmental signals and termination of signal transmission – for example by Regulators of G-protein Signaling (RGS). Hence ENVOY regulates the supply of signal transmitters for a certain signal, and thus sets its relevance for the cell upon reception, i.e. activation of the G-protein. Additionally, ENVOY can still dampen the output of the heterotrimeric G-protein cascade in terms of cAMP by alleviating its negative effect on PDE, which would decrease elevated cAMP levels despite positive effects of for example GNA3.
The surprisingly close relationship of ENVOY with cAMP signaling raises another interesting possibility: since the cAMP-dependent protein kinase A acts as a priming kinase in phosphorylation of the N. crassa photoreceptors WC-1 and WC-2 hence inhibiting their activity (Huang et al., 2007), ENVOY might act on the photoreceptors via regulation of cAMP levels. In N. crassa, the homologue of ENVOY, VIVID (encoded by vvd), was shown to function by physical interaction with WC-1 (Chen et al., 2010; Hunt et al., 2010; Malzahn et al., 2010). However, neither gna1 nor gna3 were found to be direct targets of the white collar complex (WCC) in this fungus (Smith et al., 2010), though due to the flat hierarchical network of regulation by WCC (Smith et al., 2010), an impact of this complex on regulation of gna3 and possibly gna1 cannot be excluded. Earlier studies had revealed that caffeine lengthens the free running period of N. crassa (Feldman, 1975) and was later on confirmed to have a series of clock-related effects. It was also assumed that cAMP may play a role in the effects seen on the vvd mutant in light (Brody et al., 2010; Lakin-Thomas and Brody, 1985). Although the circadian clock of T. reesei has not been studied so far, the data on the role of cAMP in the vvd mutant of N. crassa is in agreement with the hypothesis that the link between cAMP signaling and the clock is established by ENVOY via PDE.
The interrelationship with at least two G-alpha subunits suggests an even more significant role of ENVOY in coordination and prioritizing of signals: by previous integration of nutrient signals and possible additional environmental signals into the respective message at this stage, the T. reesei counterpart of the White collar complex (WCC) could be modified according to nutrient signals and again act on env1 (Fig. 7). Thereby an efficient fine-tuning mechanism for integration of light and nutrient signals could be explained. Hence the relevance of the light signal under the current environmental conditions would be adjusted and vice versa. In support of this hypothesis, an influence of the carbon source on the difference between growth in light and darkness was shown long ago, included even in a review from the 1960s (Carlile, 1965), and later studies confirmed this finding in more detail as well as a plethora of effects of light on metabolic processes (Tisch and Schmoll, 2010).
It seems that ENVOY – possibly in coordination with other factors – could be responsible for adjusting the response of an output-pathway according to the relevance of the respective signal in light and darkness. In this respect, the finding that the N. crassa homologue of env1, vvd, acts as a universal brake once transcription is turned on by light-activated WCC (Chen et al., 2009) additionally strengthens the central function of ENVOY (and presumably VIVID) in distributing resources to certain pathways in response to light. The rationale behind the interconnection of this signal transduction pathway with ENVOY is in agreement with the fact that this protein has many regulatory targets, on which it exerts both positive as well as negative influence (Schuster et al., 2007). Chen and coworkers (Chen et al., 2009) found a correlation between the timing of induction of gene transcription in response to light and the underlying biological process. This tight regulation of light dependent processes requires assessment of signal relevance and interpretation for best use of given resources and competition in the natural habitat. It will be interesting to learn how further signaling pathways feed into this system and whether the target of ENVOY in darkness is a different one than in light.
Acknowledgments
We want to thank Gabriela Gremel and Caroline Kotlowski for preliminary experimental work for this study. Our work was supported by grants from the Austrian Science Fund (FWF) to MS: P21072 and V152-B20. MS is recipient of an Elise Richter fellowship of the Austrian Science Fund (FWF). DT is recipient of a DOC fFORTE fellowship of the Austrian Academy of Sciences at the Institute of Chemical Engineering, Vienna University of Technology.
Contributor Information
Doris Tisch, Email: doris.tisch@tuwien.ac.at.
Christian P. Kubicek, Email: ckubicek@mail.tuwien.ac.at.
Monika Schmoll, Email: monika.schmoll@tuwien.ac.at.
References
- Adachi K., Hamer J.E. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell. 1998;10:1361–1374. doi: 10.1105/tpc.10.8.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh J.A. Adenylyl cyclase functions downstream of the G-alpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C.L. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Banno S. A catalytic subunit of cyclic AMP-dependent protein kinase, PKAC-1, regulates asexual differentiation in Neurospora crassa. Genes Genet. Syst. 2005;80:25–34. doi: 10.1266/ggs.80.25. [DOI] [PubMed] [Google Scholar]
- Bölker M. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 1998;25:143–156. doi: 10.1006/fgbi.1998.1102. [DOI] [PubMed] [Google Scholar]
- Brody S. Circadian rhythms in Neurospora crassa: downstream effectors. Fungal Genet. Biol. 2010;47:159–168. doi: 10.1016/j.fgb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Brunner M., Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- Carlile M.J. The photobiology of fungi. Annu. Rev. Plant Physiol. 1965;16:175–202. [Google Scholar]
- Castellanos F. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet. Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Chen B. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA. 1996;93:7996–8000. doi: 10.1073/pnas.93.15.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc. Natl. Acad. Sci. USA. 2010;107:16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA. 2003;100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crosson S. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2002;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- D’Souza C.A. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C.A., Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Dohlman H.G., Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J. Biol. Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- Farkas V. Biochemical and physiological changes during photoinduced conidiation and derepression of cellulase synthesis in Trichoderma. In: Kubicek C.P., editor. Trichoderma reesei Cellulase: Biochemistry, Genetics, Physiology and Application. Graham House; Cambridge, UK: 1990. pp. 139–155. [Google Scholar]
- Feldman J.F. Circadian periodicity a Neurospora: alteration by inhibitors of cyclic AMP phosphodiesterase. Science. 1975;190:789–790. doi: 10.1126/science.173018. [DOI] [PubMed] [Google Scholar]
- Fillinger S. CAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2002;44:1001–1016. doi: 10.1046/j.1365-2958.2002.02933.x. [DOI] [PubMed] [Google Scholar]
- Fleige S., Pfaffl M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Friedl M.A. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl. Environ. Microbiol. 2008;74:245–250. doi: 10.1128/AEM.02068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A.C. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell. 2005;4:2140–2152. doi: 10.1128/EC.4.12.2140-2152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Nuss D. Distinct roles for two G protein alpha subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA. 1996;93:14122–14127. doi: 10.1073/pnas.93.24.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber F. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 1990;18:71–76. doi: 10.1007/BF00321118. [DOI] [PubMed] [Google Scholar]
- Hamm H.E. The many faces of G protein signaling. J. Biol. Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- Heintzen C. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Hicks J.K. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell. 2005;4:1971–1981. doi: 10.1128/EC.4.12.1971-1981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay M.D., Adams D.R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y.P. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol. Biol. Cell. 2007;18:3237–3249. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.M. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc. Natl. Acad. Sci. USA. 2010;107:16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet. Biol. 2010;47:881–892. doi: 10.1016/j.fgb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeaud S. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey F.D. Shared and independent roles for a G-alpha(i) protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot. Cell. 2002;1:634–642. doi: 10.1128/EC.1.4.634-642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays A.M., Borkovich K.A. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G alpha proteins. Genetics. 2004;166:1229–1240. doi: 10.1534/genetics.166.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays A.M. Regulation of conidiation and adenylyl cyclase levels by the G-alpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 2000;20:7693–7705. doi: 10.1128/mcb.20.20.7693-7705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarova N. Light-activated adenyl cyclase from Trichoderma viride. FEMS Microbiol. Lett. 1992;72:275–278. doi: 10.1016/0378-1097(92)90474-3. [DOI] [PubMed] [Google Scholar]
- Lafon A. G-protein and cAMP-mediated signaling in Aspergilli: a genomic perspective. Fungal Genet. Biol. 2006;43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P.L., Brody S. Circadian rhythms in Neurospora crassa: interactions between clock mutations. Genetics. 1985;109:49–66. doi: 10.1093/genetics/109.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli E., Sassone-Corsi P. Signal transduction and gene regulation: the nuclear response to cAMP. J. Biol. Chem. 1994;269:17359–17362. [PubMed] [Google Scholar]
- Lee K. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics. 2003;163:103–114. doi: 10.1093/genetics/163.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler K.B. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 2007;61:423–452. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- Malzahn E. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell. 2010;142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Mandels M., Andreotti R. Problems and challenges in the cellulose to cellulase fermentation. Proc. Biochem. 1978;13:6–13. [Google Scholar]
- Martinez D. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat. Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Nowrousian M. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns S. G-proteins as transducers in transmembrane signalling. Prog. Biophys. Mol. Biol. 2003;83:101–130. doi: 10.1016/s0079-6107(03)00052-x. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyesky M. Glyceraldehyde-3-phosphate dehydrogenase expression in Trichoderma harzianum is repressed during conidiation and mycoparasitism. Microbiology. 1997;143:3157–3164. doi: 10.1099/00221287-143-10-3157. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Romero J. Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol. 2010;64:585–610. doi: 10.1146/annurev.micro.112408.134000. [DOI] [PubMed] [Google Scholar]
- Rubin E.M. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- Sambrook J. Cold Spring Harbour Laboratory Press; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Saudohar M. Cyclic AMP-dependent protein kinase is involved in morphogenesis of Aspergillus niger. Microbiology. 2002;148:2635–2645. doi: 10.1099/00221287-148-8-2635. [DOI] [PubMed] [Google Scholar]
- Schmoll M. The information highways of a biotechnological workhorse – signal transduction in Hypocrea jecorina. BMC Genom. 2008;9:430. doi: 10.1186/1471-2164-9-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M. The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot. Cell. 2009;8:410–420. doi: 10.1128/EC.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M. Trichoderma in the light of day – physiology and development. Fungal Genet. Biol. 2010;47:909–916. doi: 10.1016/j.fgb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M. Cloning of genes expressed early during cellulase induction in Hypocrea jecorina by a rapid subtraction hybridization approach. Fungal Genet. Biol. 2004;41:877–887. doi: 10.1016/j.fgb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Schuster A. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genom. 2007;8:449. doi: 10.1186/1471-2164-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A., Schmoll M. Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 2010;87:787–799. doi: 10.1007/s00253-010-2632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, A. et al., 2011. Targets of the cAMP signaling pathway in Trichoderma reesei, in preparation.
- Scott W.A., Solomon B. Cyclic 3′,5′-AMP phosphodiesterase of Neurospora crassa. Biochem. Biophys. Res. Commun. 1973;53:1024–1030. doi: 10.1016/0006-291x(73)90194-0. [DOI] [PubMed] [Google Scholar]
- Segers G.C., Nuss D.L. Constitutively activated Galpha negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 2003;38:198–208. doi: 10.1016/s1087-1845(02)00534-0. [DOI] [PubMed] [Google Scholar]
- Seibel C. Light-dependent roles of the G-protein alpha subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei) BMC Biol. 2009;7:58. doi: 10.1186/1741-7007-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestak S., Farkas V. Metabolic regulation of endoglucanase synthesis in Trichoderma reesei: participation of cyclic AMP and glucose-6-phosphate. Can. J. Microbiol. 1993;39:342–347. doi: 10.1139/m93-048. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Keller N.P. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M.L. Glyceraldehyde-3-phosphate dehydrogenase is regulated on a daily basis by the circadian clock. J. Biol. Chem. 1998;273:446–452. doi: 10.1074/jbc.273.1.446. [DOI] [PubMed] [Google Scholar]
- Smith K.M. Transcription factors in light and circadian clock signaling networks revealed by genome wide mapping of direct targets for Neurospora white collar complex. Eukaryot. Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticklen M.B. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat. Rev. Genet. 2008;9:433–443. doi: 10.1038/nrg2336. [DOI] [PubMed] [Google Scholar]
- Taylor B.L., Zhulin I.B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch D., Schmoll M. Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D.A., Van Patten S.M. Multiple pathway signal transduction by the cAMP-dependent protein kinase. FASEB J. 1994;8:1227–1236. doi: 10.1096/fasebj.8.15.8001734. [DOI] [PubMed] [Google Scholar]
- Wang L. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics. 2005;171:1523–1533. doi: 10.1534/genetics.105.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. A G-protein beta subunit required for sexual and vegetative development and maintenance of normal G alpha protein levels in Neurospora crassa. Eukaryot. Cell. 2002;1:378–390. doi: 10.1128/EC.1.3.378-390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]