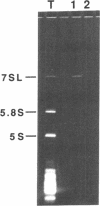
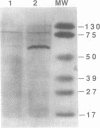
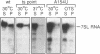
Abstract
Mammalian signal recognition particle (SRP), a complex of six polypeptides and one 7SL RNA molecule, is required for targeting nascent presecretory proteins to the endoplasmic reticulum (ER). Earlier work identified a Schizosaccharomyces pombe homolog of human SRP RNA and showed that it is a component of a particle similar in size and biochemical properties to mammalian SRP. The recent cloning of the gene encoding a fission yeast protein homologous to Srp54p has made possible further characterization of the subunit structure, subcellular distribution, and assembly of fission yeast SRP. S. pombe SRP RNA and Srp54p co-sediment on a sucrose velocity gradient and coimmunoprecipitate, indicating that they reside in the same complex. In vitro assays demonstrate that fission yeast Srp54p binds under stringent conditions to E. coli SRP RNA, which consists essentially of domain IV, but not to the full-length cognate RNA nor to an RNA in which domain III has been deleted in an effort to mirror the structure of bacterial homologs. Moreover, the association of S. pombe Srp54p with SRP RNA in vivo is disrupted by conditional mutations not only in domain IV, which contains its binding site, but in domains I and III, suggesting that the particle may assemble cooperatively. The growth defects conferred by mutations throughout SRP RNA can be suppressed by overexpression of Srp54p, and the degree to which growth is restored correlates inversely with the severity of the reduction in protein binding. Conditional mutations in SRP RNA also reduce its sedimentation with the ribosome/membrane pellet during cell fractionation. Finally, immunoprecipitation under native conditions of an SRP-enriched fraction from [35S]-labeled fission yeast cells suggests that five additional polypeptides are complexed with Srp54p; each of these proteins is similar in size to a constituent of mammalian SRP, implying that the subunit structure of this ribonucleoprotein is conserved over vast evolutionary distances.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein H. D., Poritz M. A., Strub K., Hoben P. J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989 Aug 10;340(6233):482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Brennwald P., Liao X., Holm K., Porter G., Wise J. A. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol Cell Biol. 1988 Apr;8(4):1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Wise J. A. A homologous cell-free system for studying protein translocation across the endoplasmic reticulum membrane in fission yeast. Yeast. 1994 Feb;10(2):159–172. doi: 10.1002/yea.320100204. [DOI] [PubMed] [Google Scholar]
- Brown S., Thon G., Tolentino E. Genetic selection and DNA sequences of 4.5S RNA homologs. J Bacteriol. 1989 Dec;171(12):6517–6520. doi: 10.1128/jb.171.12.6517-6520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Franklin A. E., Hoffman N. E. Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem. 1993 Oct 15;268(29):22175–22180. [PubMed] [Google Scholar]
- Gilmore R., Blobel G. Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell. 1983 Dec;35(3 Pt 2):677–685. doi: 10.1016/0092-8674(83)90100-9. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982 Nov;95(2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud B., Salminen A., Walworth N. C., Novick P. J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988 Jun 3;53(5):753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Hann B. C., Poritz M. A., Walter P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kD subunit of the signal recognition particle that in S. cerevisiae is essential for growth. J Cell Biol. 1989 Dec;109(6 Pt 2):3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B. C., Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991 Oct 4;67(1):131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Herz J., Flint N., Stanley K., Frank R., Dobberstein B. The 68 kDa protein of signal recognition particle contains a glycine-rich region also found in certain RNA-binding proteins. FEBS Lett. 1990 Dec 10;276(1-2):103–107. doi: 10.1016/0014-5793(90)80518-n. [DOI] [PubMed] [Google Scholar]
- Honda K., Nakamura K., Nishiguchi M., Yamane K. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol. 1993 Aug;175(15):4885–4894. doi: 10.1128/jb.175.15.4885-4894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak F., Walter P., Johnson A. E. Fluorescence-detected assembly of the signal recognition particle: binding of the two SRP protein heterodimers to SRP RNA is noncooperative. Biochemistry. 1992 Jun 30;31(25):5830–5840. doi: 10.1021/bi00140a019. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lacy M. J., Voss E. W., Jr A modified method to induce immune polyclonal ascites fluid in BALB/c mice using Sp2/0-Ag14 cells. J Immunol Methods. 1986 Mar 13;87(2):169–177. doi: 10.1016/0022-1759(86)90527-2. [DOI] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Y., Reddy R., Henning D., Epstein P., Busch H. Nucleotide sequence of 7 S RNA. Homology to Alu DNA and La 4.5 S RNA. J Biol Chem. 1982 May 10;257(9):5136–5142. [PubMed] [Google Scholar]
- Liao X. B., Brennwald P., Wise J. A. Genetic analysis of Schizosaccharomyces pombe 7SL RNA: a structural motif that includes a conserved tetranucleotide loop is important for function. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4137–4141. doi: 10.1073/pnas.86.11.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Selinger D., Althoff S., Chiang A., Hamilton D., Ma M., Wise J. A. Random mutagenesis of Schizosaccharomyces pombe SRP RNA: lethal and conditional lesions cluster in presumptive protein binding sites. Nucleic Acids Res. 1992 Apr 11;20(7):1607–1615. doi: 10.1093/nar/20.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom J. T., Chu B., Belanger F. C. Isolation and characterization of an Arabidopsis thaliana gene for the 54 kDa subunit of the signal recognition particle. Plant Mol Biol. 1993 Dec;23(6):1265–1272. doi: 10.1007/BF00042359. [DOI] [PubMed] [Google Scholar]
- Lingelbach K., Zwieb C., Webb J. R., Marshallsay C., Hoben P. J., Walter P., Dobberstein B. Isolation and characterization of a cDNA clone encoding the 19 kDa protein of signal recognition particle (SRP): expression and binding to 7SL RNA. Nucleic Acids Res. 1988 Oct 25;16(20):9431–9442. doi: 10.1093/nar/16.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H., High S., Römisch K., Ashford A. J., Dobberstein B. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992 Apr;11(4):1543–1551. doi: 10.1002/j.1460-2075.1992.tb05199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H., Prehn S., Ashford A. J., Remus M., Frank R., Dobberstein B. Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J Cell Biol. 1993 Jun;121(5):977–985. doi: 10.1083/jcb.121.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. D., Wilhelm H., Gierasch L., Gilmore R., Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993 Nov 25;366(6453):351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Siegel V., Hansen W., Walter P. Small ribonucleoproteins in Schizosaccharomyces pombe and Yarrowia lipolytica homologous to signal recognition particle. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4315–4319. doi: 10.1073/pnas.85.12.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz M. A., Strub K., Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988 Oct 7;55(1):4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Ribes V., Dehoux P., Tollervey D. 7SL RNA from Schizosaccharomyces pombe is encoded by a single copy essential gene. EMBO J. 1988 Jan;7(1):231–237. doi: 10.1002/j.1460-2075.1988.tb02804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990 Nov 2;63(3):591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989 Aug 10;340(6233):478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Lingelbach K., Gausepohl H., Dobberstein B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1990 Nov;111(5 Pt 1):1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T. A Mycoplasma protein homologous to mammalian SRP54 recognizes a highly conserved domain of SRP RNA. Nucleic Acids Res. 1992 Nov 11;20(21):5763–5770. doi: 10.1093/nar/20.21.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D., Brennwald P., Liao X., Wise J. A. Identification of RNA sequences and structural elements required for assembly of fission yeast SRP54 protein with signal recognition particle RNA. Mol Cell Biol. 1993 Mar;13(3):1353–1362. doi: 10.1128/mcb.13.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D., Liao X., Wise J. A. Functional interchangeability of the structurally similar tetranucleotide loops GAAA and UUCG in fission yeast signal recognition particle RNA. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5409–5413. doi: 10.1073/pnas.90.12.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA "footprinting". Proc Natl Acad Sci U S A. 1988 Mar;85(6):1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988 Jan 15;52(1):39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol. 1985 Jun;100(6):1913–1921. doi: 10.1083/jcb.100.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986 Mar 6;320(6057):81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Hewitt E. W. The S. cerevisiae SEC65 gene encodes a component of yeast signal recognition particle with homology to human SRP19. Nature. 1992 Apr 9;356(6369):534–537. doi: 10.1038/356534a0. [DOI] [PubMed] [Google Scholar]
- Strub K., Moss J., Walter P. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol Cell Biol. 1991 Aug;11(8):3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol Cell Biol. 1990 Feb;10(2):777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Walter P. Isolation of a cDNA clone of the 14-kDa subunit of the signal recognition particle by cross-hybridization of differently primed polymerase chain reactions. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9747–9751. doi: 10.1073/pnas.86.24.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck J. C., Lempicki R. A., Toschka H. Y., Erdmann V. A., Fournier M. J. Escherichia coli 4.5S RNA gene function can be complemented by heterologous bacterial RNA genes. J Bacteriol. 1990 Mar;172(3):1284–1288. doi: 10.1128/jb.172.3.1284-1288.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck J. C., Vogel D. W., Ulbrich N., Erdmann V. A. The Bacillus subtilis scRNA is related to the 4.5S RNA from Escherichia coli. Nucleic Acids Res. 1988 Mar 25;16(6):2719–2719. doi: 10.1093/nar/16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983 Sep;34(2):525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle: a ribonucleoprotein required for cotranslational translocation of proteins, isolation and properties. Methods Enzymol. 1983;96:682–691. doi: 10.1016/s0076-6879(83)96057-3. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Subcellular distribution of signal recognition particle and 7SL-RNA determined with polypeptide-specific antibodies and complementary DNA probe. J Cell Biol. 1983 Dec;97(6):1693–1699. doi: 10.1083/jcb.97.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G., Dobberstein B. Protein transfer across microsomal membranes reassembled from separated membrane components. Nature. 1978 Jun 15;273(5663):569–571. doi: 10.1038/273569a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H., Luirink J., Tollervey D. Evolutionary conserved nucleotides within the E.coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 1992 Nov 25;20(22):5919–5925. doi: 10.1093/nar/20.22.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D., Bernstein H. D., Johnson A. E., Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990 Dec;9(13):4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. Interaction of protein SRP19 with signal recognition particle RNA lacking individual RNA-helices. Nucleic Acids Res. 1991 Jun 11;19(11):2955–2960. doi: 10.1093/nar/19.11.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]