Abstract
We evaluated whether MYC, TP53, and chromosome 17 copy-number alterations occur in ACP02, ACP03, and AGP01 gastric cancer cell lines and in their tumor counterpart. Fluorescence in situ hybridization for MYC and TP53 genes and for chromosome 17 was applied in the 6th, 12th, 60th, and 85th passages of the cell lines and in their parental primary tumors. We observed that three and four MYC signals were the most common alterations in gastric cell lines and tumors. ACP02 presented cells with two copies of chr17 and loss of one copy of TP53 more frequently than ACP03 and AGP01. Only ACP03 and AGP01 presented clonal chr17 trisomy with three or two TP53 copies. The frequency of MYC gain, TP53 loss, and chromosome 17 trisomy seems to increase in gastric cell lines compared to their parental tumors. Our findings reveal that these cell lines retain, in vitro, the genetic alterations presented in their parental primary tumors.
1. Introduction
Chromosomal instability is characterized by changes in chromosome copy number (aneuploidy) and alterations in chromosomal regions, which may induce oncogene activation, tumor suppressor gene inactivation, or both. Chromosomal instability is one of the two major genomic instability pathways observed in gastric cancer (GC) [1], the fourth most frequent type of cancer and second most frequent cause of cancer mortality worldwide [2]. However, due to the difficulty in obtaining high-quality chromosome preparations in these neoplasias, the evaluation of chromosomal alterations is complicated [3–7].
Cell lines derived from human cancers are useful in order to understand the chromosomal alterations and other molecular alterations in the carcinogenesis process. Cell lines are also a useful tool for the study of anticancer treatments in vitro and in animal xenograft models. However, the process of cell line immortalization has been implicated as a source of cytogenetic changes, and growth passages have been associated with random genomic instability [8–11]. Given their importance as models, it is important to understand in which way and to what degree cell lines grown under artificial conditions reflect their parental in vivo genetic architecture [8].
Our research group previously established three GC cell lines from tumor samples of individuals from Northern Brazil. ACP02 cell line was established from a diffuse-type GC, and ACP03 and AGP01 were from an intestinal-type GC. These cell lines exhibited a composite karyotype with several clonal chromosomal alterations. All these cell lines presented chromosome 8 trisomy—where MYC oncogene is located—and deletion of chromosome arm 17p, which includes the TP53 tumor suppressor locus. Moreover, chromosome 17 (chr17) trisomy in ACP03 and AGP01 cell lines was detected [12].
Here, we analyzed the number of MYC, TP53, and chr17 copies in 6th (short-duration culture), 12th (long-duration culture), 60th and 85th passages of ACP02, ACP03 and AGP01 cell lines, and in their parental primary tumors. We aim to evaluate whether these gene/chromosomal alterations occur in cell lines and in their tumor counterpart, as well as whether multiple passage growth leads to a difference in the frequency of these alterations.
2. Materials and Methods
FISH was performed on recently made slides from methanol/acetic acid fixed cells of four passages (6th, 12th, 60th, and 85th) of ACP02, ACP03, and AGP01 cell lines, as previously described [13]. FISH was also applied on nuclei isolated from parental primary tumors as previously reported [14]. Tumor samples were obtained from João de Barros Barreto University Hospital (HUJBB) in Pará State, Brazil.
To determine MYC gene copy number, cells were hybridized with a rhodamine-labeled probe (Chromotrax, USA) for MYC gene region (8q24.1-q24.2). To determine the chromosome 17 and TP53 copy numbers, cells were hybridized using a dual-color direct labeled probe (Qbiogene, USA) specific for chr17 α-satellite and for TP53 gene region, and labeled with fluorescein and rhodamine, respectively. Nuclei were counterstained with DAPI/antifade (Chemicon, USA). Fluorescence was detected using an Olympus BX41 fluorescence microscope with DAPI/FITC/TRICT filters (Olympus, Japan), and signals were analyzed using FISHView of Applied Spectral Imaging image analysis system (ASI Ldt., Israel). For each cell line or tumor, 200 interphase nuclei were analyzed and scored using the criteria of Hopman et al. [15]. To avoid misinterpretation due to technical error, gastric mucosal tissue (nonneoplastic) and normal lymphocyte nuclei were used as control.
For statistical analysis, the MYC, TP53, or chr17 copy numbers were compared among cell lines using Kruskal-Wallis nonparametric test followed by Games-Howell posthoc test. Friedman test followed by Wilcoxon posthoc test with Bonferroni correction were used to evaluate differences among primary tumor and their cell line passages. In all analyses, the confidence interval was 95%, and P values less than .05 were considered significant.
3. Results
3.1. MYC
Two signals for MYC probe were observed in 99.5% of peripheral blood lymphocytes and in 97% of normal gastric cells (Figure 1(a)). Table 1 shows the frequency of MYC signals in cell lines and parental tumors. In primary tumor samples, three MYC signals were the most frequent alteration, ranging from 39%–45% of cells (Figure 1(b)). In the 6th and 12th passages, 3 MYC signals were the most frequent alteration (about 40%) followed by 4 signals (about 28%). In the 60th passage of cell lines, 3 and 4 MYC signals were commonly observed. In the 85th passage, the most frequent alteration was the presence of 4 signals for MYC probe, ranging from 33.5%–42% of cells. Five or more MYC copies by cells were also observed in all cell lines and parental tumors. High amplification of MYC was detected as clonal alteration mainly in cell lines (Figure 1(c)).
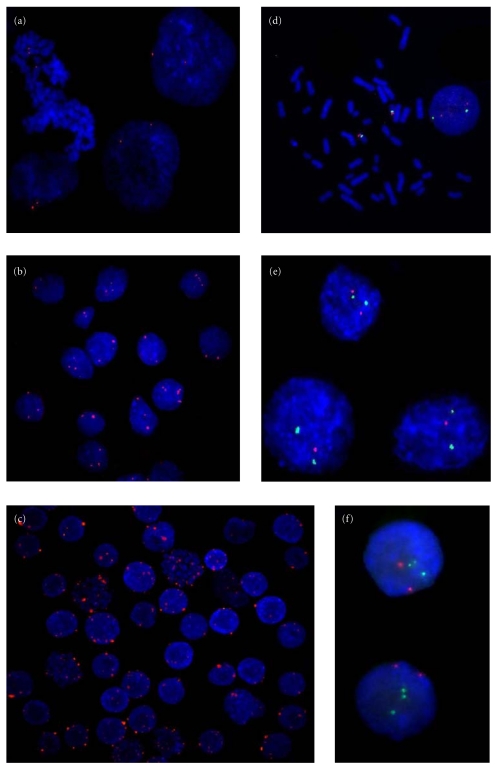
Figure 1.
Fluorescence in situ hybridization assay. (a) Interphase nuclei presenting two MYC signals from normal gastric mucosa; (b) interphase nuclei presenting 2–5 MYC signals from ACP02 parental primary tumor; (c) interphase nuclei presenting MYC signal number alterations, including high amplification, from the 85th passage of AGP01 cell line; (d) interphase and metaphase cells presenting two copies of chr17/TP53 from lymphocytes control, with the green spots representing the 17 centromere probe and the red representing the TP53 gene probe; (e) interphase nuclei presenting two signals of chr17 and two or one TP53 signal(s) from ACP02 parental primary tumor; (f) interphase nuclei presenting three signals for chr17 and two TP53 signals from the 85th passage of ACP03 cell line.
Table 1.
FISH analysis of MYC copy number in gastric cancer cell lines, in their parental primary tumors and in control samples.
| Nuclei exhibiting MYC signals, no. (%)a | |||||||
|---|---|---|---|---|---|---|---|
| 1 signal | 2 signals | 3 signals | 4 signals | ≥5 signals | HA | ||
| ACP02 | Parental primary tumor | 0 (0) | 57 (28.5) | 78 (39) | 57 (28.5) | 8 (4) | 0 (0) |
| 6th passage | 1 (0.5) | 40 (20) | 84 (42) | 62 (31) | 13 (6.5) | 0(0) | |
| 12th passage | 0 (0) | 34 (17) | 88 (44) | 63 (31.5) | 15 (7.5) | 0(0) | |
| 60th passage | 0 (0) | 6 (3) | 83 (41.5) | 79 (39.5) | 24 (12.5) | 7 (3.5) | |
| 85th passage | 2 (1) | 8 (4) | 65 (32.5) | 84 (42) | 30 (15) | 11 (5.5) | |
| ACP03 | Parental primary tumor | 0 (0) | 39 (19.5) | 84 (42) | 41 (20.5) | 23 (11.5) | 13 (6.5) |
| 6th passage | 0(0) | 33 (16.5) | 89 (44.5) | 40 (20) | 24 (12) | 14 (7) | |
| 12th passage | 1 (0.5) | 19 (9.5) | 85 (42.5) | 51 (25.5) | 29 (14.5) | 15 (7.5) | |
| 60th passage | 1 (0.5) | 8 (4) | 71 (35.5) | 63 (31.5) | 39 (19.5) | 18 (9) | |
| 85th passage | 0 (0) | 6 (3) | 47 (23.5) | 69 (34.5) | 49 (24.5) | 29 (14.5) | |
| AGP01 | Parental primary tumor | 0 (0) | 47 (23.5) | 90 (45) | 37 (18.5) | 17 (8.5) | 9 (4.5) |
| 6th passage | 4 (2) | 12 (6) | 69 (34.5) | 59 (29.5) | 36 (18) | 20 (10) | |
| 12th passage | 1 (0.5) | 13 (6.5) | 70 (35) | 56 (28) | 33 (16.5) | 27 (13.5) | |
| 60th passage | 0 (0) | 2 (1) | 64 (32) | 63 (31.5) | 41 (20.5) | 30 (15) | |
| 85th passage | 1 (0.5) | 1 (0.5) | 45 (22.5) | 67 (33.5) | 49 (24.5) | 37 (18.5) | |
| Control | Normal stomach tissue | 4 (2) | 194 (97) | 2 (1) | 0 (0) | 0 (0) | 0 (0) |
| Lymphocytes | 1 (0.5) | 199 (99.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
aThe FISH analysis was performed on 200 nuclei. HA: high amplification.
Statistical analysis revealed that the frequency of cells with high MYC amplification was significantly different among ACP02, ACP03, and AGP01 cell lines (χ2 = 9.206, df = 2, P = .01, by Kruskal-Wallis test). The Games-Howell posthoc analyses demonstrated that the ACP02, cell line, and parental tumor, presented a lower frequency of high MYC amplification cells than AGP01 (P = .019) and ACP03 (P = .014).
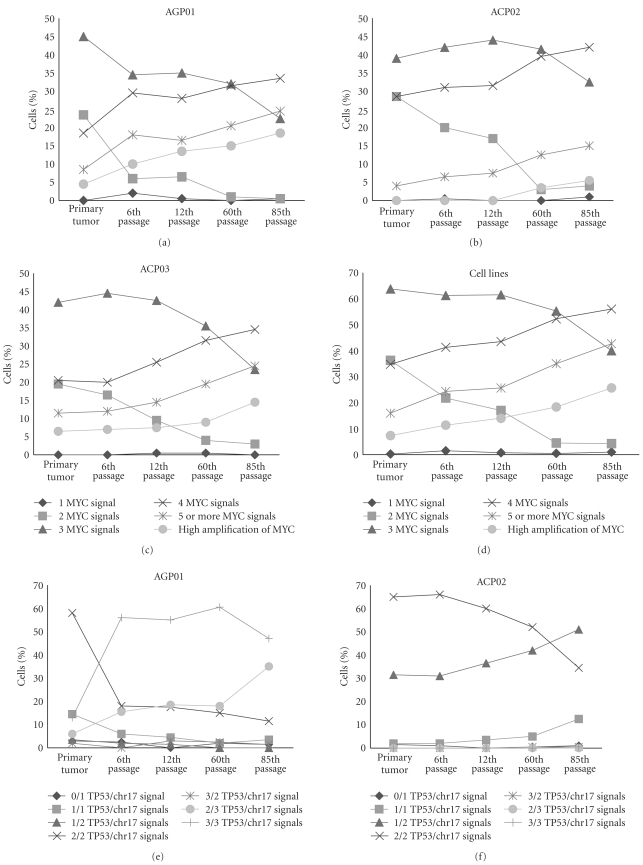
Concerning the MYC signal frequency during culture process, we observed that the frequency of cells with 2 (χ2 = 10.933, df = 4, P = .027, by Friedman test), 4 (χ2 = 10.667, df = 4, P = .031, by Friedman Test), 5 or more (χ2 = 11.467, df = 4, P = .022, by Friedman test), and high amplification (χ2 = 11.429, df = 4, P = .022, by Friedman test) MYC copies were significantly different among parental tumor and their passages. However, the posthoc analysis by Wilcoxon test with Bonferroni correction did not reveal any significant difference, probably due to the gradual alterations among passages (Figures 2(a), 2(b), 2(c), 2(d)).
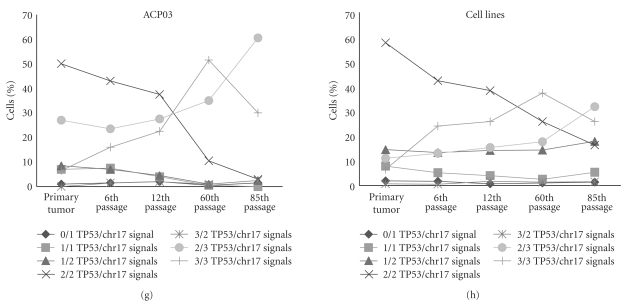
Figure 2.
Distribution of cells according to (a) MYC signals in AGP01 parental tumor and cell line passages; (b) MYC signals in ACP02 parental tumor and cell line passages; (c) MYC signals in ACP03 parental tumor and cell line passages; (d) mean of MYC signals of AGP01, ACP02, and ACP03 parental tumor and cell line passages; (e) TP53/chr17 signals in AGP01 parental tumor and cell line passages; (f) TP53/chr17 signals in ACP02 parental tumor and cell line passages; (g) TP53/chr17 signals in ACP03 parental tumor and cell line passages; (h) mean of TP53/chr17 signals of AGP01, ACP02, and ACP03 parental tumor and cell line passages.
3.2. Chr17/TP53
Two signals for chr17 and TP53 were observed in about 97% of control cells (Figure 1(d)). Table 2 shows the frequency of chr17/TP53 signals in cell lines from the 6th, 12th, 60th, and 85th passages and tumor samples.
Table 2.
FISH analysis of chr17/TP53 copy number in gastric cancer cell lines, in their parental primary tumors and in control samples.
| Nuclei exhibiting chr17/TP53 signals, no. (%)a | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1/0 signal | 1/1 signals | 2/1 signals | 2/2 signals | 2/3 signals | 3/2 signals | 3/3 signals | ||
| ACP02 | Parental primary tumor | 3 (1.5) | 4 (2) | 63 (31.5) | 130 (65) | 0 (0) | 0 (0) | 0 (0) |
| 6th passage | 2 (1) | 4 (2) | 62 (31) | 132 (66) | 0 (0) | 0 (0) | 0 (0) | |
| 12th passage | 0 (0) | 7 (3.5) | 73 (36.5) | 120 (60) | 0 (0) | 0 (0) | 0 (0) | |
| 60th passage | 1 (0.5) | 10 (5) | 84 (42) | 104 (52) | 1 (0.5) | 0 (0) | 0 (0) | |
| 85th passage | 2 (1) | 25 (12.5) | 102 (51) | 69 (34.5) | 1 (0.5) | 0 (0) | 1 (0.5) | |
| ACP03 | Parental primary tumor | 2 (1) | 14 (7) | 17 (8.5) | 100 (50) | 0 (0) | 54 (27) | 13 (6.5) |
| 6th passage | 3 (1.5) | 15 (7.5) | 14 (7) | 86 (43) | 3 (1.5) | 47 (23.5) | 32 (16) | |
| 12th passage | 4 (2) | 8 (4) | 9 (4.5) | 75 (37.5) | 4 (2) | 55 (27.5) | 45 (22.5) | |
| 60th passage | 1 (0.5) | 1 (0.5) | 2 (1) | 21 (10.5) | 2 (1) | 70 (35) | 103 (51.5) | |
| 85th passage | 3 (1.5) | 0 (0) | 5 (2.5) | 6 (3) | 5 (2.5) | 121 (60.5) | 60 (30) | |
| AGP01 | Parental primary tumor | 6 (3) | 29 (14.5) | 7 (3.5) | 116 (58) | 4 (2) | 12 (6) | 26 (13) |
| 6th passage | 5 (2.5) | 12 (6) | 4 (2) | 36 (18) | 0 (0) | 31 (15.5) | 112 (56) | |
| 12th passage | 0 (0) | 9 (4.5) | 3 (1.5) | 35 (17.5) | 6 (3) | 37 (18.5) | 110 (55) | |
| 60th passage | 4 (2) | 4 (2) | 0 (0) | 30 (15) | 5 (2.5) | 36 (18) | 121 (60.5) | |
| 85th passage | 3 (1.5) | 7 (3.5) | 0 (0) | 23 (11.5) | 3 (1.5) | 70 (35) | 94 (47) | |
| Control | Normal stomach tissue | 0 (0) | 4 (2) | 1 (0.5) | 193 (96.5) | 0 (0) | 0 (0) | 2 (1) |
| Lymphocytes | 0 (0) | 3 (1.5) | 1 (0.5) | 195 (97.5) | 1 (0.5) | 0 (0) | 0 (0) | |
aThe FISH analysis was performed on 200 nuclei.
The frequency of cells with 2 signals for chr17 and 2 signals for TP53 was significantly different among ACP02, ACP03, and AGP01 cell lines (χ2 = 6, df = 2, P = .05, by Kruskal-Wallis test). The Games-Howell posthoc analyses demonstrated that the ACP02, cell line, and parental tumor presented a higher number of this cell type than AGP01. ACP02 also presented a higher frequency of cells with two copies of chr17 than AGP01 (P = .003, by Games-Howell posthoc analysis) and ACP03 (P = .009, by Games-Howell posthoc analysis).
The frequency of cells with 2/1 (χ2 = 10.839, df = 2, P = .004, by Kruskal-Wallis test), 2/3 (χ2 = 11.423, df = 2, P = .003, by Kruskal-Wallis test), and 3/3 (χ2 = 10.691, df = 2, P = .005, by Kruskal-Wallis test) copies of chr17/TP53 was significantly different among cell lines. The Games-Howell posthoc analyses demonstrated that the ACP02 presented a higher frequency of cells with 2 copies of chr17 and loss of TP53 than AGP01 (P = .001) and ACP03 (P = .001). In ACP02, the most common alteration observed was the loss of one copy of TP53 (Figure 1(e)).
Only ACP03 and AGP01 presented clonal chr17 trisomy with two or three copies of TP53. Thus, the Games-Howell posthoc analysis among cell lines also demonstrated that AGP01 and ACP03 presented a higher frequency of cells with chr17 trisomy with 2 TP53 copies than ACP02 (P = .035 and P = .15, resp.; Figure 1(f)). AGP01 also presented a higher frequency of cells with chr17 trisomy and 3 TP53 copies than ACP02 (P = .013).
Concerning the chr17 and TP53 alterations during culture process, we observed that the frequency of cells with two copies of chr17/TP53 (χ2 = 11.467, df = 4, P = .022, by Friedman test), two chr17 copies (χ2 = 10.373, df = 4, P = .035, by Friedman test), and chr17 trisomy (χ2 = 10.4, df = 4, P = .034, by Friedman test) were significantly different among parental tumor and their passages. However, the posthoc analysis by Wilcoxon test with Bonferroni correction did not reveal any significant difference, probably due to the gradual alterations among passages (Figures 2(e), 2(f), 2(g), 2(h)).
4. Discussion
Fluorescence in situ hybridization (FISH) assay allows rapid detection of numerical genetic aberrations in interphase nuclei in tumor cells. FISH assay should be used to evaluate cell-to-cell heterogeneity in gene or loci copy number and detect small subpopulations of genetically aberrant cells [16]. Using FISH assay, our research group previously reported several frequent aneusomies in GC samples and cell lines from individuals of Northern Brazil, which suggests a genomic instability [7, 12, 13, 17–20]. Molecular cytogenetic studies have shown that gains at 3q, 7p, 7q, 8q, 13q, 17q, 20p, and 20q and losses at 4q, 9p, 17p, and 18q are recurrent chromosomal alterations in GC. (For a review, see [21].)
Our research group has observed that chr8 trisomy, where MYC is located, is present in almost all gastric tumors and cell lines from our population by conventional and molecular cytogenetic analyses [7, 12, 13, 17, 19, 20, 22, 23]. We have previously described that ACP02, ACP03, and AGP01 at the 60th passage presented chr8 trisomy, as well as tetrasomy. In these cell lines, more than 5 signals of chr8 were observed in less than 5% of cells [12]. However, MYC copy number seems to be higher than the number of chr8 copies in our studies [19, 20, 23]. High MYC amplification has frequently been observed in primary tumors from our population, and we have also previously reported that MYC can be inserted into other chromosomes. The higher frequency of MYC high amplification in ACP03 and AGP01, originated from intestinal-type GC, than ACP02, originated from a diffuse-type GC, agrees with our previous observation in primary GC. Our group had observed that clonal high amplification of MYC is less frequent in diffuse-type than intestinal-type primary GC [19, 20, 23].
Thus, the presence of three or more copies of MYC, including gene high amplification, in all samples of the present study corroborates our previous observations in primary GC. Moreover, the frequency of MYC gain in advanced GC observed by FISH seems to be higher in our population than in East Asia, which ranges from 15.5% to 48% of cases [6, 24, 25]. MYC amplification has been suggested as the main mechanism for its deregulation in GC (see review [26]).
Concerning TP53/chr17 copies, we observed that ACP02, cell line, and its parental tumor, presented cells with two copies of chr17 and loss of one copy of TP53 more frequently than ACP03 and AGP01. This finding corroborates our previous study using dual-color FISH for chr17/TP53 in primary tumor samples, in which we observed that the frequency of cells with two chr17 and one TP53 signals was higher in the diffuse-type than in the intestinal-type GC [27].
We also observed that only ACP03 and AGP01 cells and parental tumors presented chr17 trisomy as clonal alteration, in agreement with the karyotype of these cell lines at 60th passage [12]. Although primary tumors of individuals from Northern Brazil present clonal crh17 trisomy or monosomy by FISH analysis [27], chr17 aneusomy is not the most frequent alteration within primary tumors of our population [7, 22, 28].
Moreover, we detected TP53 loss in all cell lines and primary tumors. TP53 somatic alteration is described in about 50% of human cancers, including GC [29]. Deletion of chromosome arm 17p was also observed in ACP02, ACP03, and AGP01 by conventional cytogenetic analysis [12]. In our population, TP53 deletion was previously observed in all analyzed primary tumor samples, despite Laurén's histopathologic types; therefore, corroborating the present study [27].
Although we did not observe any significant difference among parental tumors and their cell line passages after posthoc analysis, we were able to observe that the number of MYC signals is significantly higher in cell lines than primary tumors. We also observed a reduction in the number of nuclei with two signals for chr17/TP53 when we compared GC cell lines with their parental tumors, which reflects an increase of aneusomy cells.
Tumor samples are usually composed of a heterogeneous clonal population which can include nonneoplastic cells. The increased frequency of cells with MYC gain, TP53 loss, and chr17 trisomy in cell lines may be due to the selection of a subpopulation of cancer cells during the cell line establishment. Normal cells can divide only a limited number of times in vitro, because of replicative senescence. However, some tumor cells can become immortal and, therefore, they do not undergo senescence when cultured in vitro [30]. According to the Hayflick limit, the maximum number of passages that a normal cell attains before senescence is about 50 [31]. Here, we evaluated GC cell lines in the 60th and in 85th passages, which select cells with genomic alterations that are essential for cell survival and proliferation in vitro.
A meta-analysis of the studies using comparative genomic hybridization assay to evaluate chromosomal alterations in cancer cell lines and primary tumors demonstrated that, on average, the relatively large-scale copy number genetic aberrations seen in cell lines in vitro accurately reflect their parent histology [8]. In this meta-analysis study, it was also reported that MYC oncogene amplification seems to be more frequent in cell lines in several histologies, indicating that the deregulation of this gene may be acquired as part of cell immortalization or that their occurrence is selected when tumors are chosen for transformation. Therefore, we suggest that, in our GC cell lines, cells with MYC amplification are selected during the long culture process.
5. Conclusion
Our findings reveal that ACP02, ACP03, and AGP01 cell lines retain in vitro the genetic alterations presented in their parental primary tumors. Thus, these findings suggest that these cell lines are an interesting model to study GC biology as well as to evaluate new anticancer strategies.
Acknowledgments
This paper was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; MACS and RRB) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; MFL and DQC).
References
- 1.Ottini L, Falchetti M, Lupi R, et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Annals of Oncology. 2006;17(7):vii97–vii102. doi: 10.1093/annonc/mdl960. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. Ca-A Cancer Journal for Clinicians. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Ferti-Passantonopoulou AD, Panani AD, Vlachos JD, Raptis SA. Common cytogenetic findings in gastric cancer. Cancer Genetics and Cytogenetics. 1987;24(1):63–73. doi: 10.1016/0165-4608(87)90083-5. [DOI] [PubMed] [Google Scholar]
- 4.Xia JC, Lu S, Geng JS, Fu SB, Li P, Liu QZ. Direct chromosome analysis of ten primary gastric cancers. Cancer Genetics and Cytogenetics. 1998;102(1):88–90. doi: 10.1016/s0165-4608(97)00293-8. [DOI] [PubMed] [Google Scholar]
- 5.Ochi H, Douglass HO, Sandberg AA. Cytogenetic studies in primary gastric cancer. Cancer Genetics and Cytogenetics. 1986;22(4):295–307. doi: 10.1016/0165-4608(86)90022-1. [DOI] [PubMed] [Google Scholar]
- 6.Kitayama Y, Igarashi H, Sugimura H. Different vulnerability among chromosomes to numerical instability in gastric carcinogenesis: stage-dependent analysis by FISH with the use of microwave irradiation. Clinical Cancer Research. 2000;6(8):3139–3146. [PubMed] [Google Scholar]
- 7.Assumpcao PP, Ishak G, Chen ES, et al. Numerical aberrations of chromosome 8 detected by conventional cytogenetics and fluorescence in situ hybridization in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genetics and Cytogenetics. 2006;169(1):45–49. doi: 10.1016/j.cancergencyto.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Greshock J, Nathanson K, Martin AM, et al. Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Research. 2007;67(8):3594–3600. doi: 10.1158/0008-5472.CAN-06-3674. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y, Zhang H, Tsao SW, et al. Cytogenetic and molecular genetic characterization of immortalized human ovarian surface epithelial cell lines: consistent loss of chromosome 13 and amplification of chromosome 20. Gynecologic Oncology. 2004;92(1):183–191. doi: 10.1016/j.ygyno.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Ratsch SB, Gao Q, Srinivasan S, Wazer DE, Band V. Multiple genetic changes are required for efficient immortalization of different subtypes of normal human mammary epithelial cells. Radiation Research. 2001;155(1, part 2):143–150. doi: 10.1667/0033-7587(2001)155[0143:mgcarf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Meisner LF, Wu SQ, Christian BJ, Reznikoff CA. Cytogenetic instability with balanced chromosome changes in an SV40 transformed human uroepithelial cell line. Cancer Research. 1988;48(11):3215–3220. [PubMed] [Google Scholar]
- 12.Leal MF, Martins do Nascimento JL, da Silva CEA, et al. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genetics and Cytogenetics. 2009;195(1):85–91. doi: 10.1016/j.cancergencyto.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Costa Guimaraes A, Goncalves Quintana L, Ferreira Leal M, et al. Aneuploidy of chromosome 8 detected by fluorescence in situ hybridisation in ACP01 cell line gastric adenocarcinoma. Clinical and Experimental Medicine. 2006;6(3):129–133. doi: 10.1007/s10238-006-0108-5. [DOI] [PubMed] [Google Scholar]
- 14.Stamouli MI, Ferti AD, Panani AD, et al. Application of multiplex fluorescence in situ hybridization in the cytogenetic analysis of primary gastric carcinoma. Cancer Genetics and Cytogenetics. 2002;135(1):23–27. doi: 10.1016/s0165-4608(01)00635-5. [DOI] [PubMed] [Google Scholar]
- 15.Hopman AHN, Ramaekers FCS, Raap AK, et al. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry. 1988;89(4):307–316. doi: 10.1007/BF00500631. [DOI] [PubMed] [Google Scholar]
- 16.Kallioniemi A, Visakorpi T, Karhu R, Pinkel D, Kallioniemi OP. Gene copy number analysis by fluorescence in situ hybridization and comparative genomic hybridization. Methods. 1996;9(1):113–121. doi: 10.1006/meth.1996.0015. [DOI] [PubMed] [Google Scholar]
- 17.Lima EM, Rissino JD, Harada ML, et al. Conventional cytogenetic characterization of a new cell line, ACP01, established from a primary human gastric tumor. Brazilian Journal of Medical and Biological Research. 2004;37(12):1831–1838. doi: 10.1590/s0100-879x2004001200008. [DOI] [PubMed] [Google Scholar]
- 18.Guimaraes AC, Lima EM, Khayat AS, et al. Interrelationships among chromosome aneuploidy, promoter hypermethylation, and protein expression of the CDKN2A gene in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genetics and Cytogenetics. 2007;179(1):45–51. doi: 10.1016/j.cancergencyto.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Calcagno DQ, Leal MF, Seabra AD, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World Journal of Gastroenterology. 2006;12(38):6207–6211. doi: 10.3748/wjg.v12.i38.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queiroz Calcagno D, Ferreira Leal M, Satomi Takeno S, et al. Aneuploidy of chromosome 8 and C-MYC amplification in individuals from northern Brazil with gastric adenocarcinoma. Anticancer Research. 2005;25(6 B):4069–4074. [PubMed] [Google Scholar]
- 21.Panani AD. Cytogenetic and molecular aspects of gastric cancer: clinical implications. Cancer Letters. 2008;266(2):99–115. doi: 10.1016/j.canlet.2008.02.053. [DOI] [PubMed] [Google Scholar]
- 22.Burbano R, Assumpcao PP, Ferreira Leal M, et al. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Research. 2006;26(4 B):2909–2914. [PubMed] [Google Scholar]
- 23.Calcagno DQ, Guimaraes AC, Leal MF, et al. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Research. 2009;29(7):2479–2483. [PubMed] [Google Scholar]
- 24.Koo SH, Kwon KC, Shin SY, et al. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence in situ hybridization studies. Cancer Genetics and Cytogenetics. 2000;117(2):97–103. doi: 10.1016/s0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- 25.Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Laboratory Investigation. 1998;78(9):1143–1153. [PubMed] [Google Scholar]
- 26.Calcagno DQ, Leal MF, Assumpcao PP, Smith MDAC, Burbano RR. MYC and gastric adenocarcinoma carcinogenesis. World Journal of Gastroenterology. 2008;14(39):5962–5968. doi: 10.3748/wjg.14.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khayat AS, Guimaraes AC, Calcagno DQ, et al. Interrelationship between TP53 gene deletion, protein expression and chromosome 17 aneusomy in gastric adenocarcinoma. BMC Gastroenterology. 2009;9, article 55 doi: 10.1186/1471-230X-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satomi Takeno S, Leal MF, Frias Lisboa LC, et al. Genomic alterations in diffuse-type gastric cancer as shown by high-resolution comparative genomic hybridization. Cancer Genetics and Cytogenetics. 2009;190(1):1–7. doi: 10.1016/j.cancergencyto.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Szymanska K, Hainaut P. TP53 and mutations in human cancer. Acta Biochimica Polonica. 2003;50(1):231–238. [PubMed] [Google Scholar]
- 30.Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21(3):477–484. doi: 10.1093/carcin/21.3.477. [DOI] [PubMed] [Google Scholar]
- 31.Rubin H. Promise and problems in relating cellular senescence in vitro to aging in vivo. Archives of Gerontology and Geriatrics. 2002;34(3):275–286. doi: 10.1016/s0167-4943(01)00221-7. [DOI] [PubMed] [Google Scholar]