Abstract
Aggregation of Cu, Zn superoxide dismutase (SOD1) is implicated in Amyotrophic Lateral Sclerosis (ALS). Glutathionylation and phosphorylation of SOD1 is omnipresent in the human body, even in healthy individuals, and has been shown to increase SOD1 dimer dissociation, which is the first step on the pathway toward SOD1 aggregation. We find that post-translational modification of SOD1, especially glutathionylation, promotes dimer dissociation. We discover an intermediate state in the pathway to dissociation, a conformational change that involves a “loosening” of the β-barrels and a loss or shift of dimer interface interactions. In modified SOD1, this intermediate state is stabilized as compared to unmodified SOD1. The presence of post-translational modifications could explain the environmental factors involved in the speed of disease progression. Because post-translational modifications such as glutathionylation are often induced by oxidative stress, post-translational modification of SOD1 could be a factor in the occurrence of sporadic cases of ALS, which make up 90% of all cases of the disease.
Introduction
The homodimeric metal-binding enzyme Cu, Zn superoxide dismutase (SOD1) is implicated in Amyotrophic Lateral Sclerosis (ALS). Ten percent of ALS cases are familial (fALS), and 20% of these fALS cases are attributed to mutations in SOD11. Since the initial identification in 1993 of 11 different SOD1 mutants in patients with ALS2, researchers have linked over 100 point mutations throughout the length of this protein to fALS (http://alsod.iop.kcl.ac.uk).
Structurally, SOD1 is a cytosolic enzyme made up of two monomers of 153 residues each. Each monomer folds into a β-barrel formed of eight anti-parallel strands configured in a Greek key formation3. Each monomer has two major loops, the electrostatic and zinc-binding loops, coordinated by the β-barrels4. These loops are coordinated by a zinc ion, which makes a large contribution to the stability of the dimer5,6. The dimer is also stabilized by a disulfide bond between cysteines 57 and 146, which connect the zinc-binding loop to the C-terminal β-strand7–9. The enzymatic activity of SOD1 is enabled by a copper ion, which dismutates superoxide ion to molecular oxygen and hydrogen peroxide10,11.
Although the exact mechanism of the progressive motor neuron death characteristic of ALS is still unknown, there is evidence that aggregates of SOD1 are the cytotoxic species12,13. The first step towards SOD1 protein aggregation is hypothesized to be the dissociation of SOD1 dimer into monomer, followed by metal loss and unfolding5,6,14–22. Mutations in SOD1 decrease the stability of the dimeric species, causing dimer dissociation and unfolding22,23. Interestingly, those mutations having the largest effect on folding and thermodynamic stability are located on the surface of the protein, and not in the hydrophobic core24. Moreover, because intra-protein interactions are highly networked in SOD1, mutations to residues spread throughout the enzyme, and not only those near the dimer interface, may affect dissociation25. Other factors that destabilize the dimer interface include the loss of metals and dissolution of the disulfide bond26, which are both promoted in SOD1 mutants. The resulting apo-monomer is the reactive species in protein aggregation of both wild type and mutant SOD122,27,28, due to drastically reduced stability and partial unfolding26. However, the molecular details of progression along this aggregation pathway are still unclear.
Recently, Wilcox et al. reported post-translational modifications present in SOD1 purified from human erythrocytes29. The authors reported phosphorylation at Thr-2 and glutathionylation at Cys-111, both sites near the dimer interface. Wilcox et al. hypothesized that these post-translational modifications promote monomer formation, which leads to aggregation. They found a three-fold increase in monomer formation for post-translationally modified species as compared to unmodified species when measured by size-exclusion chromatography. In an activity assay, modified species exhibited a two-fold increase in Kd. This increase does not seem large, but such a change results in a 70% increase in the monomer population, which significantly increases the frequency of aggregation nucleation events29. Post-translational modifications present a possible link between familial and sporadic ALS, since they are present ubiquitously in the human population, although in varying amounts29–31. However, the molecular mechanism of dimer destabilization by post-translational modifications, glutathionylation in particular, is not yet understood.
The large size of glutathione and the location of both modifications at the dimer interface suggest a steric explanation of the dimer destabilization observed by Wilcox et al. To gain structural insight into the causes of increased dimer dissociation, we perform all-atom discrete molecular dynamics32 simulations of modified and unmodified forms of the wild type and mutant monomer and dimer species. We utilize the replica exchange method33 to increase the sampling efficiency of our simulations for thermodynamic calculations. We find that the processes of dimer dissociation and monomer unfolding are highly coupled, and that modification reduces but does not eliminate the coupled nature of the reaction. We discover an unfolding intermediate structure common to wild type and mutant unmodified SOD1 dimer that precedes dimer dissociation in the unfolding pathway. Post-translational modification shifts the intermediate state to a lower energy, and decreases the potential energy gap (ΔE) between the native-like and dissociated states. In order to identify structural differences between modified and unmodified forms of SOD1 species, we also perform constant temperature simulations at a constant temperature so as to avoid low- and high- temperature structural aberrations. We discover differences in dimer interface contacts as a result of post-translational modification, which could explain the increased dimer dissociation of the post-translationally modified species.
Results
Thermodynamics of the dimer dissociation and monomer unfolding transition
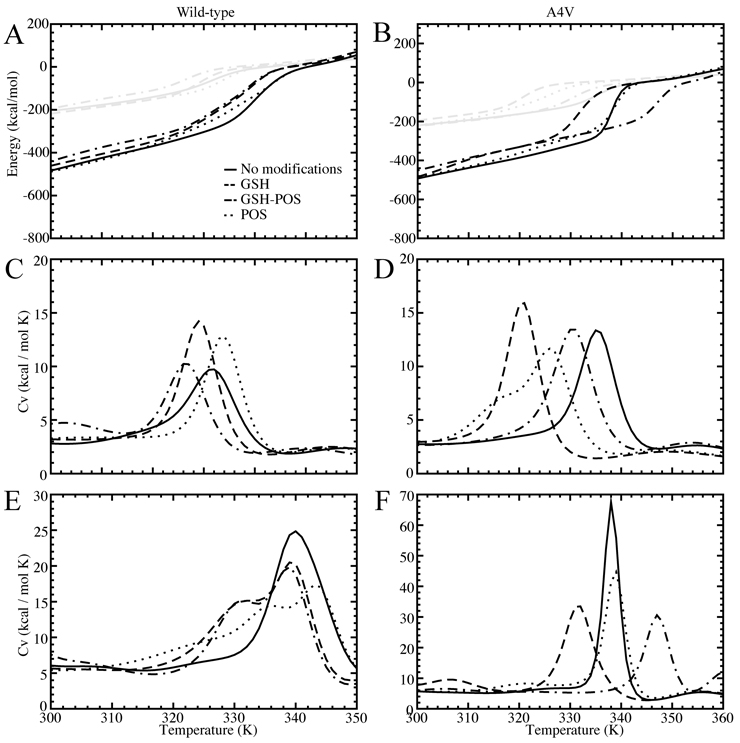
Based on replica exchange simulations, we analyze folding thermodynamics using the Weighted Histogram Analysis Method (WHAM34). WHAM incorporates multiple simulation trajectories with overlapping energetic sampling. The density of states is calculated self-consistently by combining histograms from these multiple trajectories, and thereby thermodynamic parameters, such as specific heat, can be computed. Because we do not observe multiple unfolding and refolding transitions in our simulations due to insufficient sampling, we do not expect to obtain a fully quantitative free energy landscape for the various species of SOD1. However, we expect to gain insight to the stability of both the monomer and dimer species by using the unfolding transition temperature, which corresponds to the major peak in specific heat26 (Figure 1, Supplementary Figure 1). Specific heat is a measure of the amount of energy necessary to increase the temperature (kinetic energy) of the protein. Hence, the peak in specific heat corresponds to a transition between energetic states, where energy is devoted to raising the potential as opposed to the kinetic energy of the complex. Monomer species exhibit one significant thermodynamic transition, corresponding to monomer unfolding (Figure 1, Supplementary Figure 1). Dimer species also exhibit one major transition, with the transition temperature significantly shifted to higher temperature as compared to the corresponding monomer species. Because the SOD1 dimer species feature one major peak rather than two peaks, corresponding to dimer dissociation and monomer unfolding, we conclude that dimer dissociation and monomer unfolding are highly coupled processes. The strong interactions in the dimer interface stabilize the individual monomers by shifting the unfolding transition to a higher temperature, keeping the dimer associated and folded at temperatures higher than the monomer unfolding temperature.
Figure 1. Thermodynamics.
Comparison of wild type (first column) and mutant A4V (second column) modified and unmodified SOD1. (A),(B): Potential energy of the SOD1 complex according to simulation temperature. Monomer curves are shown in gray, dimer curves in black. (C),(D): Specific heat as a function of temperature for SOD1 monomer. (E),(F): Specific heat as a function of temperature for SOD1 dimer. Legends shown for wild type are also relevant for A4V.
We observe that, in wild type and all mutant variants except A4V and I112T, modification causes a decrease in the coupled nature of the dissociation/unfolding transition. All unmodified species exhibit a single peak in specific heat, representing a single transition, but modified species feature a shoulder, representing a partial decoupling of the two processes (Figure 1, Supplementary Figure 1).
We find that modifications destabilize the dimer by shifting the transition temperature to a lower temperature. However, the effect of specific modification types on the dimer transition temperature varies between mutants (Supplementary Table 1). The exceptions to the destabilization trend are A4V, where the glutathionylated-phosphorylated dimer is stabilized in comparison with the unmodified dimer, and G93A, where all modified species are stabilized with respect to the unmodified form (Figure 1, Supplementary Figure 1).
We observe in dimer species that the maximum value of the major specific heat peak (corresponding to the energy input needed to raise the complex temperature by 1°C during the dimer dissociation/unfolding transition) is highest in the unmodified species, as opposed to in the various modified species (Figure 1, Supplementary Figure 1), with the exceptions of G93A and I112T. The absolute value of the specific heat indicates the cooperativity of the phase transition. Therefore, this observation suggests that the dissociation/unfolding transitions are more cooperative in the unmodified species, and that modified species exhibit a more gradual melting transition from associated to dissociated states.
In monomer species, the unmodified form often exhibits the lowest specific heat peak for monomer unfolding (Figure 1, Supplementary Figure 1), which implies interactions between the modification molecules and the monomer, although these interactions do not affect the stability of the monomer. We expect this effect to some extent because of the slightly larger size of the modified system. In agreement with recent experimental studies by Redler et al. (Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted), we find that modification does not exhibit a large effect on monomer stability, although we observe a slight destabilization in A4V monomer (Figure 1, Supplementary Figure 1, Supplementary Table 1).
Energetic states near the dissociation/unfolding transition
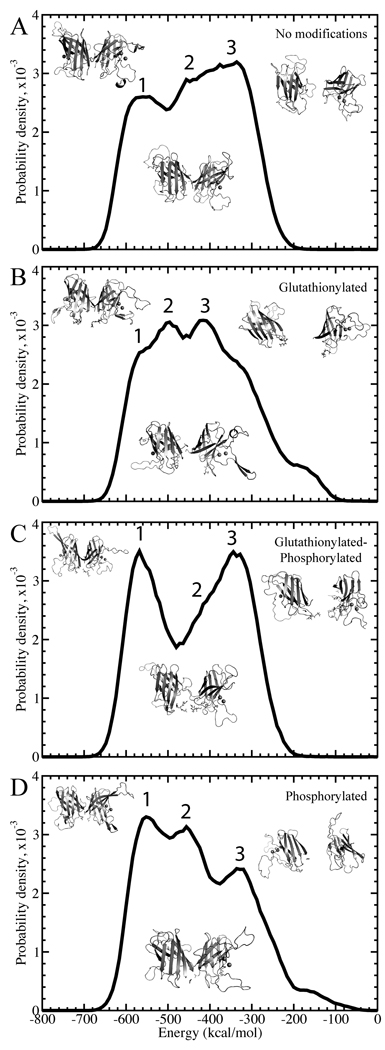
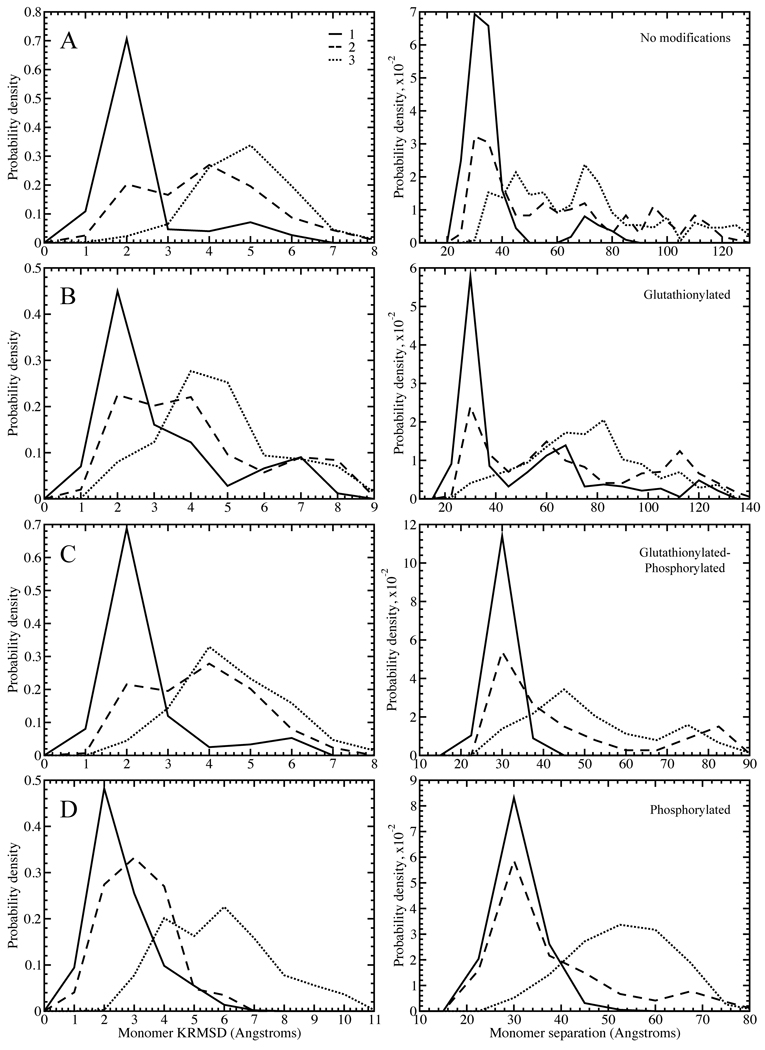
In order to study the more subtle transitions between native-like states that may explain increased dissociation in modified species, it is necessary to examine the potential energy distributions of structures in the mid-range temperature region of the replica exchange simulations, 275–325 Kelvin, just reaching the lower bound of the dissociation/unfolding transition (Figure 2, Supplementary Figures 2–6). These structures should include populations of native-like, dissociated, and any intermediate states. All energies discussed are potential energies of the system. In each system, we observe the existence of at least three low-energy states with Gaussian-like peaks or shoulders in the energy distribution. Because of the large size and close spacing of the surrounding populations, the distributions of some states as plotted on the energy coordinate (e.g. the second state in Figure 2C) appear as shoulders. However, deconvolution of the peaks based on potential energies and subsequent clustering of structures within the energy window of each peak, allows for the clear separation of these structural states. These low energy states correspond to the native state and the early excitation states in the dissociation/unfolding process. Because the goal of this study is to uncover the effect of post-translational modifications on the dissociation process, we focus on these first three states by fitting each distribution to a trimodal Gaussian function and identifying the three lowest energy states present for each species. The first state is a low-energy state with a native-like structure; the second has undergone a conformational change that can be characterized as a loosening of the β-barrels and/or a slight movement outwards of the two centers of mass (Figure 4, Supplementary Figures 7–11, Supplementary Table 2); and the third is the dissociated state, with partial unfolding due to the coupled nature of the dissociation and unfolding processes.
Figure 2. Deciphering the states for wild type.
Distributions of total potential energies are shown with a solid curve. Potential energy is sampled throughout the simulations, at temperatures of 0.55–0.65 kcal/mol•kB, for (A) unmodified, (B), glutathionylated, (C) glutathionylated-phosphorylated, and (D) phosphorylated cases of SOD1. The three Gaussian-like curves are highlighted for each species. Representative structures for each of the three states in each species are shown to the right of each plot.
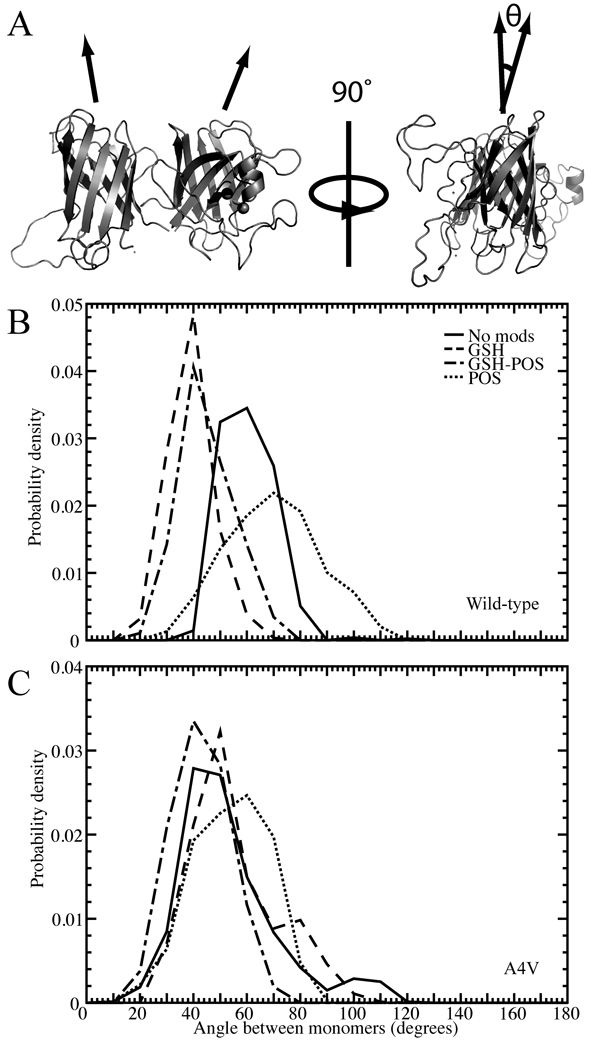
Figure 4. Structural effects of modification on the angle between monomers in the SOD1 dimer.
(A) Depiction of the angle, θ, measured between monomers in the SOD1 dimer. Distribution of angle between monomers in constant temperature populations of modified and unmodified (B) wild type and (C) A4V dimer. Legend shown for (B) is also relevant for (C).
The amount of differentiation between the states in partial unfolding or separation of the monomers varies with mutant and modification type. Although in some glutathionylated species, namely A4V-GSH, G37R-GSH, G93A-GSH-Pi, H46R-GSH-Pi, and I112T-GSH, the representative structure of the intermediate energetic state is dissociated (Supplementary Figures 2–6), we observe from the distributions of monomer separation and monomer unfolding that, statistically, the intermediate state as a whole is still distinct from the dissociated state (Supplementary Figures 7–11). In G93A, the unmodified species exhibits an intermediate state that maintains very few dimer interface contacts, and so has a larger tendency to be dissociated than the intermediate states of other mutant variants. However, modification restores the intermediate state to a form with similar characteristics to the other mutants. We also observe that in the phosphorylated species of G37R, G37R-GSH-Pi and G37R-Pi, phosphorylation causes the third state to have a larger population of associated structures than is seen in other species. However, as in the previously discussed case, the second and third states are still distinct when characterized by their distributions of monomer separation and unfolding among all structures (Supplementary Figure 8).
In general, we find that modification increases dissociation by decreasing the potential energy gap between the native-like and the dissociated states, as opposed to only destabilizing the native-like state. In most mutant variants, the native-like state, which occurs at approximately −550 kcal/mol but varying across mutant variants, does not undergo a significant energetic change upon modification (Figure 2, Supplementary Figures 2–6, Supplementary Table 3). However, the intermediate state population is shifted to a lower energy in modified species, with exceptions in some phosphorylated species: wild type-GSH-Pi, G37R-Pi, H46R-GSH-Pi, and I112T-Pi. Stabilization of this dissociation/unfolding intermediate increases the probability of the protein to misfold and dissociate. In many of the modified species, the third (dissociated) state is also stabilized as compared with the third state in the unmodified species, with exceptions in wild type phosphorylated species, all A4V modified species, G93A-GSH-Pi, and I112T-GSH. In general, we find that the overall ΔE between the native-like and the dissociated populations is decreased in modified cases as compared to unmodified, with exceptions in wild type-GSH-Pi, all modified species of A4V, G93A-GSH-Pi, and I112T-GSH (Figure 2, Supplementary Figures 2–6, Supplementary Table 3).
Structural changes induced by post-translational modifications
Modification affects the interactions between the two monomers, causing structural rearrangement. The two monomers of SOD1 can be represented as cylinders that, instead of being oriented in a parallel fashion, positioned at a torsional angle to one another (Figure 4A). In constant temperature simulations, we note that this torsional angle is affected by modification, which occurs near the dimer interface, but the effect is different for different mutants (Figure 4, Supplementary Figure 12). These differences are potentially due to variation between the mutants in their dimer interface configuration, and how they are affected by the addition of the modification molecule (Supplementary Figures 13–18).
We observe a major difference in dimer interface interactions between wild type and the various mutant SOD1 in constant temperature simulations. Where wild type exhibits monomer-monomer interactions occurring between residues located throughout the length of the protein sequence, mutant SOD1 monomer-monomer interactions almost exclusively involve a terminus from at least one of the monomers, with the exception of the structurally wild type-like G93A. This property is manifested in the interface contact maps of each species (Supplementary Figures 13–18): wild type has significant interactions present in the center of the map, whereas in the various mutant SOD1 the center of the map is blank. It may be for this reason that in the wild type enzyme only, modifications introduce an asymmetry in monomer-monomer interactions. For instance, in wild type-GSH we observe that interactions near 60A-115B (A and B representing the two monomers) have disappeared when compared to the unmodified, but 115A-60B interactions remain (Supplementary Figure 13B). Also, in some residues one monomer may see only a shift in interface contacts, losing interaction frequency in one residue but gaining it in an adjacent residue, while the other monomer loses contacts with no compensating gain. This compromise may introduce strain into the structure that leads to the increased dissociation seen experimentally29(Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted).
In contrast to wild type, mutant SOD1 is affected symmetrically in the two monomers by modification. In A4V, we find a loss of interactions in the dimer interface of the glutathionylated species, and in the phosphorylated species we observe increased interface contacts near the C-termini and decreased or shifting interactions elsewhere (Supplementary Figure 14). In the G37R mutant, we find a shifting in contacts in the C-termini, implying a structural movement in that area (Supplementary Figure 15). In G93A, we observe an overall increase in interface contacts upon modification, implying that the introduction of the modification molecules induces the formation of non-native interactions in the interface (Supplementary Figure 16). H46R loses contacts in all areas of the interface upon modification, although in the phosphorylated species the interactions in the N- and C-termini shift rather than be lost (Supplementary Figure 17), which can be seen in added contacts directly adjacent to the lost ones. In I112T, most losses of contacts are accompanied by a gain in the residues directly adjacent (Supplementary Figure 18), which would imply a shift in the location of the dimer interface and so a shift in its make-up.
Cumulatively, these results imply a shift in the composition and position of the dimer interface upon modification. A change in the identity, even if not the number, of dimer interface contacts could result in the loss of monomer-monomer binding affinity and increased dimer dissociation. In a parallel experimental study, Redler et al. have found a shift towards the monomer population in wild type and mutant A4V SOD1, and an increase in the rate of monomer formation in mutant I112T SOD1 (Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted), which would suggest that the contact loss seen in wild type-GSH and A4V-GSH (Supplementary Figures 13 and 14) results in a loss of monomer-monomer binding affinity, while the overall shift in contacts seen in I112T-GSH (Supplementary Figure 14) results in an increased koff for the dissociation reaction (Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted).
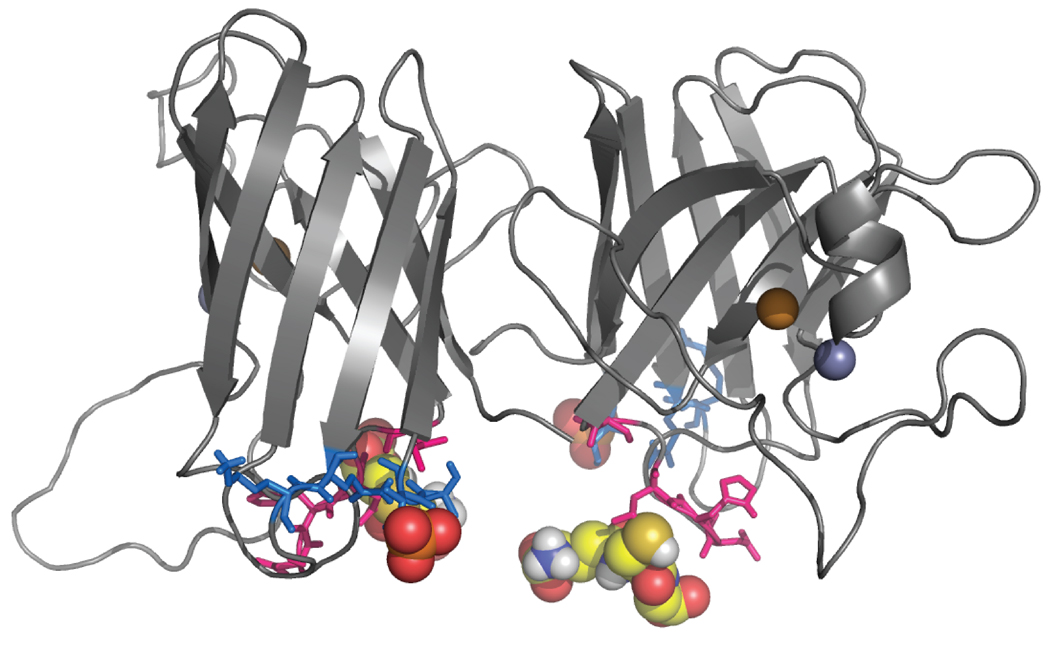
Predictably, we observe in constant temperature simulations that the modification molecules interact most frequently with the residues surrounding the modification site on their associated monomer (Supplementary Figures 19 and 20). In fact, the modification molecules interact almost exclusively with their associated monomer. Interactions with the opposing monomer or with the opposing modification molecule are comparatively infrequent and do not occur in all mutant variants. The locations of the interacting residues in small loop segments connecting the β-strands (Figure 5) could pull apart the β-barrels, which would result in the loss of crucial contacts and therefore dissociation.
Figure 5. Residues interacting with modifications.
Residues having the highest frequency of interaction with phosphate are shown in marine blue, and those with glutathione in hot pink. Modification molecules are shown as spheres.
Discussion
Our systematic replica exchange DMD simulations indicate a strong coupling of the dimer dissociation and monomer unfolding processes, which has important implications for overall protein stability and aggregation. Dimerization significantly stabilizes the folded monomer, which implies that contacts in the dimer interface contribute to the integrity of the monomer β-barrels. As these contacts are broken, the intra-monomer interactions maintaining the β-barrel formation of each monomer are also broken, causing a simultaneous dissociation and partial unfolding. The unfolding process is then more favorable to complete upon full dissociation.
Similarly, the wild type and mutant unmodified species in general dissociate and unfold more sharply with an increase in temperature than do the modified forms (Figure 1, Supplementary Figure 1). This finding implies that the shifting or gain of non-native contacts (Supplementary Figures 13–18) that occurs in modified species causes a loss of cooperativity in the interface and intra-monomer interactions. This loss of cooperativity allows some contacts to be lost much more frequently than others, whether due to a structural change or the steric interference of the modification molecules. A loss in interaction cooperativity may be manifest in the stabilization of the intermediate state, and also could be the reason behind the decreased potential energy gap between the native and dissociated states (Figure 2, Supplementary Figures 2–6, Supplementary Table 3). Interactions between the modifications and their associated monomer may also be responsible for this more drawn out dissociation interaction and loss of cooperativity; both modifications are located near the dimer interface (Figure 5), and their interactions with the monomers may disrupt or weaken native interface interactions. This effect could possibly be remedied by the introduction of additional interactions in the form of a drug that would bind, bridging the dimer interface and holding it together, as was found by Ray et al38. In addition, it may be possible to design a drug that would interfere with the modification of SOD1 in the first place, whether by occupation or blocking of the modification site or inhibition of the modification binding itself.
The results above indicate that the post-translational modifications glutathionylation and phosphorylation affect the energetic and structural properties of wild type and mutant SOD1. The effect observed varies both with mutant and with modification. This observation suggests that different types of modification have varied effects on the stability of dimer of the various genetic mutations, and for different reasons. For example, A4V dimer seems to be largely stabilized by modification, while the wild type dimer is destabilized by glutathionylation but appears to be “rescued” from this effect by phosphorylation. With the exceptions of A4V and I112T, glutathionylation in particular has a dramatic effect in decreasing the potential energy gap between the native-like state and the dissociated state. We infer from this that the presence of glutathione, a marker of oxidative stress in the cell39, would be detrimental in most types of familial ALS. This finding corroborates with reports that exercise40 and electrical stimulation41 of ALS model animals results in a more rapid and severe disease progression, since both of these would produce increased oxidative stress in cells and hence increased levels of glutathionylated protein42. An environmental factor such as oxidative stress could also help to explain the differences in disease progression between the various ALS-causative mutants.
Our simulation results indicate that post-translational modifications in wild type and mutant A4V SOD1 result into a net loss in the total number of interface contacts, and thus, a reduced binding energy. This result is consistent with the experimental observation of reduced dimer dissociation constant in modified wild type and A4V mutant (Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted). Our simulations of the I112T mutant also suggest that the modification induces a shift in the dimer interface, but no significant change in the total number of contacts, in contrast to the wild type and A4V mutant. This result suggests that modified I112T has a similar dimer dissociation constant to the unmodified species. Indeed, Redler et al. have shown that the equilibrium constant of I112T dimer dissociation is not affected by glutathionylation.
In this work, we study homo-modified species of wild type and mutant SOD1. However, it is possible that in vivo SOD1 may exhibit significant hetero-modified populations in addition to homo-modified species. Because of the means of experimental characterization of post-translational modifications from erythrocytes (mass spectrometry)29(Redler, R. L., K. C. Wilcox, E. A. Proctor, L. Fee, M. Caplow, and N. V. Dokholyan (2011) submitted), dimers are necessarily dissociated before measurement, and so it is impossible to determine whether dimers are hetero- or homo-modified in vivo using this method. The molecular mechanism of glutathionylation, as well as the kinase responsible for phosphorylation of SOD1, is still unknown, so it is unclear whether modification of the individual monomers is a cooperative or an independent process. However, even the presence of only one modification molecule near the dimer interface disrupts or changes monomer-monomer contacts and induces many of the same effects that we observe in homo-modified species.
On a further note, because glutathionylation decreases the potential energy gap between the native-like and dissociated states in wild type SOD1, and glutathionylation of SOD1 is present even in healthy individuals29, it is possible that glutathionylation caused by oxidative stress to motor neurons could be a factor in sporadic ALS. The late onset of ALS suggests that an environmental trigger could exist for both familial and sporadic cases. This possibility fits with the increased occurrence of sporadic ALS in athletes43 and soldiers44 as compared to the general population; both of these groups experience more extreme and frequent oxidative stress than does the average individual. Such environmental factors could possibly be counteracted with a drug or lifestyle decisions. Further investigation into the mechanism of post-translational modification in SOD1 could illuminate preventative measures against the observed increased dissociation and unfolding, and hence inhibit aggregation and disease.
Materials and Methods
All-atom Discrete Molecular Dynamics (DMD)
A complete description of the DMD algorithm can be found elsewhere45,46. Briefly, DMD utilizes discrete step function potentials to govern inter-atomic interactions, as opposed to the continuous potentials employed in traditional molecular dynamics (MD). The velocity of each atom is constant until a collision occurs at the step of the potential function, at which point the velocity of the atom changes instantaneously according to the laws of conservation of energy, momentum, and angular momentum. A collision table records all possible collisions between neighboring atoms, and the next collision is obtained by sorting these possible collisions. At each collision, only the colliding atoms have their positions and velocities updated, and the corresponding possible collisions of these two atoms are updated in the collision table. As a result, this innovation allows for a significant decrease in the number of calculations needed per simulation time step. Having fewer and faster calculations allows DMD to achieve a significant increase in sampling over MD.
In the all-atom protein model for DMD32, all heavy atoms and polar hydrogen atoms are explicitly represented. We represent bonded interactions by placing infinite square well constraints on bond lengths, bond angles, and dihedral angles. Non-bonded interactions are adapted from the Medusa force field47: Van der Waals interactions are modeled using the standard Lennard-Jones potential, while solvation interactions are modeled by the Lazaridis-Karplus solvation model48. Both potentials are discretized by multi-step square-well functions, characterized by the hard-core atom radius and a series of potential steps. We model hydrogen bonding interactions by the reaction algorithm49.
Systems studied
We perform constant volume DMD simulations with periodic boundary conditions for both monomer and dimer SOD1. We study disease-relevant mutants A4V, G37R, G93A, H46R, and I112T, as well as wild type SOD1. For each system of monomer or dimer and mutant or wild type, we study modifications consisting of glutathionylated (GSH), glutathionylated-phosphorylated (GSH-Pi), and phosphorylated (Pi), as well as unmodified structures, resulting in 48 systems total (6 (five mutants and wild type) × 2 (monomer or dimer) × 4 (modification types)). For simplicity, we refer to each mutation-modification combination (e.g., A4V-GSH-Pi, G93A-unmodified, wild type-Pi) as a species. For instance, “dimer species” refers to all mutation-modification combinations, in their dimerized form. For each species, we perform both replica exchange and constant temperature DMD simulations.
Modeling of modified SOD
We obtain structures of post-translationally modified mutant and wild type SOD1 using the known X-ray crystallographic structure of wild type SOD1 (PDBID: 1SPD) as a reference structure. We constrain glutathione and/or phosphate molecules to their respective SOD1 residues (Thr-2 for phosphate and Cys-111 for glutathione). We obtain parameters for bond length, angle, and dihedral constraints for glutathione, metals, and disulfide bonds from the CHARMM19 force field48. All dimers are homo-modified. We then mutate the wild type structure to generate each disease-relevant mutant (A4V, G37R, G93A, H46R, and I112T) using the Eris suite50,51. We make no structural adjustments to residues participating in metal-binding, glutathionylation, phosphorylation, or disulfide bond interactions, unless, as in the case of H46R, which coordinates the catalytic copper ion, they are necessary to the system of interest.
In order to minimize steric clashing between modifications and the SOD1 protein and to relax the SOD1 backbone, we employ an iterative relaxation and equilibration of each system utilizing all-atom DMD (described above). We perform three simulation iterations, each with a progressively lower heat exchange rate (0.2, 0.02, and 0.002 fs−1, respectively). Each iteration is performed for 50 picoseconds at a reduced unit temperature of 0.5 kcal/mol•kB (~251 K).
Replica exchange
In replica exchange simulations, we perform simulations of multiple copies of the same system in parallel at varying temperatures33. To fully explore protein conformational space, the upper end of the temperature range should be enough to unfold the protein. At given time intervals, replicas of neighboring temperatures exchange temperature values according to a Metropolis-based stochastic algorithm. We set the temperature exchange interval as 50 picoseconds. Exchange between replicas increases sampling efficiency in that energetic barriers can be more easily overcome, and in less time, with exposure to higher temperatures.
In the monomer species, twelve replicas were used for a simulation length of 100 nanoseconds, at temperatures of 0.50 (~252 K), 0.52 (~262 K), 0.54 (~272 K), 0.56 (~282 K), 0.58 (~292 K), 0.60 (~302 K), 0.62 (~312 K), 0.64 (~322 K), 0.66 (~332 K), 0.68 (~343 K), 0.70 (~353 K), and 0.72 (~363 K) kcal•mol. In the dimer species, sixteen replicas were used for a simulation length of 50 nanoseconds, at temperatures of 0.48 (~242 K), 0.495 (~249 K), 0.51 (~257 K), 0.525 (~264 K), 0.54 (~272 K), 0.555 (~280 K), 0.57 (~287 K), 0.585 (~295 K), 0.60 (~302 K), 0.615 (~310 K), 0.63 (~317 K), 0.645 (~325 K), 0.65 (~327 K), 0.67 (~337 K), 0.69 (~347 K), and 0.71 (~357 K) kcal/mol•kB. It is important to note that the temperatures used in MD simulations, including DMD simulations, often do not directly equate with physical temperatures. For example, the calculated melting temperature of wild type SOD1 dimer is different from the experimentally measured value of 93 °C (366 K)52. However, the simulations can be used to evaluate the relative changes in temperatures, which are relevant to our studies.
Replica trajectories were combined for the analysis of folding thermodynamics using Weighted Histogram Analysis Method (WHAM)34. In thermodynamics calculation, it is important to have sufficient sampling of the energetic sates. To test our sampling, we split our simulation trajectories in half and compare the distribution of energies for the two halves. Since we obtain similar results for both halves, we conclude that sampling was sufficient to reach equilibrium.
WHAM analysis of CV
We use the MMTSB tool53 to perform WHAM analysis of the replica exchange simulations. WHAM computes the density of states by combining histograms from overlapping simulation trajectories. Given the density of states ρ(E), the folding specific heat CV is computed at various temperatures according to the partition function, Z = ∫dE ρ(E)exp(−E/kT), where E denotes the potential energy.
Identification of states
We calculate specific heat according to temperature using WHAM34. We calculate potential energy distributions over each simulation using intermediate temperatures (275–325 K) so as not to introduce structural aberrations in representative structures from low- and high-temperature conditions. Contributions from internal potential energy of the modification molecules are minimal in comparison to the potential energy of the system, therefore no adjustment is needed for comparisons of potential energy between modification systems. We fit each energy distribution with a trimodal Gaussian, corresponding to the three lowest energy peaks and/or shoulders in the distribution, and identify the energy of each state as the mean of its respective Gaussian function. Because the individual Gaussian functions corresponding to each state overlap significantly, and deconvolution with respect to individual structures is not possible, we identify representative structures by clustering structures whose energies fall within 1 kcal/mol around the mean of the respective Gaussian by RMSD using the OC suite54. Structures with energies in this range have a higher probability of belonging to the energetic state of interest, than of being a member of the tail of one of the other states. We choose RMSD cutoffs for clustering as the global maximum in a histogram of pairwise RMSDs of all structures clustered. We choose the centroid of the largest cluster as the representative structure for each state. The largest cluster is in all cases at least four times the weight of the next-largest cluster.
Measurement of angle between monomers
We define the radial axis vector for each monomer by creating vectors along each β-strand making up the approximately cylindrical β-barrel, and averaging over the components. We define all β-strand vectors to have the same directionality sign, and we length-normalize the vectors before averaging. The representative vector therefore has the three-dimensional average direction of all β-strand vectors in each monomer. In order for these calculations to be meaningful, structures must be both folded and associated. In order to eliminate those structures with monomer unfolding, for each simulation snapshot (snapshots recorded every 5 picoseconds) we calculate the aligned RMSD (Kabsch RMSD55, KRMSD) between the β-barrel alpha carbons of each monomer of the initial wild type-like structure (see Modeling of modified SOD1) and those of the corresponding monomer of the structural snapshot. We impose a KRMSD cutoff of 4 Å, which we choose based on the distribution of KRMSD values from all snapshots of all simulated species. To eliminate dissociated structures, we measure the distance between the centers of mass of the two monomers, which are calculated based on the alpha carbons in the β-barrels. We impose a cutoff of 35 Å, which we choose in the same manner as the cutoff for KRMSD. We retain those structures of each species that meet the criteria for both folded and associated, and measure the angle between the vectors characterizing the two monomers. The angle between the two vectors v1 and v2 is calculated as .
Interface contact maps
We define two residues as being in contact in the dimer interface if the two Cα atoms are within 10 Å of each other. At each simulation snapshot, we evaluate contacts between the two monomeric chains. The count between every pair of residues is normalized for the total number of simulation snapshots.
Modification hit maps
We define a molecule as being in contact with a residue or another molecule if any heavy atom in the molecule is within 4.5 Å of any heavy atom in the residue or second molecule. Contact counts are recorded and normalized similarly to in the interface contact maps described above.
Supplementary Material
Figure 3. Structural characterization of energetic populations in wild type SOD1.
Distributions of values for the monomer aligned KRMSD from the starting structure, used as a measure of β-barrel integrity and unfolding (left column), and the distance between monomer centers of mass, used as a measure of dissociation (right column). We show distributions for (A) unmodified, (B) glutathionylated, (C) glutathionylated-phosphorylated, and (D) phosphorylated wild type SOD1 for the first, second, and third energy populations described in Figure 2. Legend shown for (A) is relevant for all panels.
Acknowledgements
The authors would like to thank Dr. Michael Caplow, Rachel Redler, Dr. Kyle Wilcox, Dr. Lanette Fee, Pradeep Kota, and Srinivas Ramachandran for helpful discussions. This work was supported by the National Institutes of Health [grant numbers R01GM080742, ARRA supplements GM080742-03S1, GM066940-06S1]. E.A.P. was partially supported by the UNC Curriculum in Bioinformatics and Computational Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 3.Getzoff ED, Tainer JA, Stempien MM, Bell GI, Hallewell RA. Evolution of CuZn superoxide dismutase and the Greek key beta-barrel structural motif. Proteins. 1989;5:322–336. doi: 10.1002/prot.340050408. [DOI] [PubMed] [Google Scholar]
- 4.Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta. 2010;1804:245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayatekin C, Zitzewitz JA, Matthews CR. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J Mol Biol. 2008;384:540–555. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rumfeldt JA, Lepock JR, Meiering EM. Unfolding and folding kinetics of amyotrophic lateral sclerosis-associated mutant Cu,Zn superoxide dismutases. J Mol Biol. 2009;385:278–298. doi: 10.1016/j.jmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 8.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci U S A. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365:333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 10.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 11.Valentine JS, Hart PJ. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou SM, Wang HS, Komai K. Colocalization of NOS and SOD1 in neurofilament accumulation within motor neurons of amyotrophic lateral sclerosis: an immunohistochemical study. J Chem Neuroanat. 1996;10:249–258. doi: 10.1016/0891-0618(96)00137-8. [DOI] [PubMed] [Google Scholar]
- 13.Chou SM, Wang HS, Taniguchi A. Role of SOD-1 and nitric oxide/cyclic GMP cascade on neurofilament aggregation in ALS/MND. J Neurol Sci. 1996;139 Suppl:16–26. doi: 10.1016/0022-510x(96)00090-1. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg MJ, Tibell L, Oliveberg M. Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc Natl Acad Sci U S A. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stathopulos PB, Rumfeldt JA, Scholz GA, Irani RA, Frey HE, Hallewell RA, Lepock JR, Meiering EM. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc Natl Acad Sci U S A. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg MJ, Bystrom R, Boknas N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci U S A. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. J Mol Biol. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Vassall KA, Stathopulos PB, Rumfeldt JA, Lepock JR, Meiering EM. Equilibrium thermodynamic analysis of amyotrophic lateral sclerosis-associated mutant apo Cu,Zn superoxide dismutases. Biochemistry. 2006;45:7366–7379. doi: 10.1021/bi0600953. [DOI] [PubMed] [Google Scholar]
- 19.Rumfeldt JA, Stathopulos PB, Chakrabarrty A, Lepock JR, Meiering EM. Mechanism and thermodynamics of guanidinium chloride-induced denaturation of ALSassociated mutant Cu,Zn superoxide dismutases. J Mol Biol. 2006;355:106–123. doi: 10.1016/j.jmb.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 20.Mulligan VK, Kerman A, Ho S, Chakrabartty A. Denaturational stress induces formation of zinc-deficient monomers of Cu,Zn superoxide dismutase: implications for pathogenesis in amyotrophic lateral sclerosis. J Mol Biol. 2008;383:424–436. doi: 10.1016/j.jmb.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Nordlund A, Leinartaite L, Saraboji K, Aisenbrey C, Grobner G, Zetterstrom P, Danielsson J, Logan DT, Oliveberg M. Functional features cause misfolding of the ALS-provoking enzyme SOD1. Proc Natl Acad Sci U S A. 2009;106:9667–9672. doi: 10.1073/pnas.0812046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khare SD, Caplow M, Dokholyan NV. FALS mutations in Cu, Zn superoxide dismutase destabilize the dimer and increase dimer dissociation propensity: a large-scale thermodynamic analysis. Amyloid. 2006;13:226–235. doi: 10.1080/13506120600960486. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez JA, Shaw BF, Durazo A, Sohn SH, Doucette PA, Nersissian AM, Faull KF, Eggers DK, Tiwari A, Hayward LJ, Valentine JS. Destabilization of apoprotein is insufficient to explain Cu,Zn-superoxide dismutase-linked ALS pathogenesis. Proc Natl Acad Sci U S A. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khare SD, Ding F, Dokholyan NV. Folding of Cu, Zn superoxide dismutase and familial amyotrophic lateral sclerosis. J Mol Biol. 2003;334:515–525. doi: 10.1016/j.jmb.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 25.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci U S A. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding F, Dokholyan NV. Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc Natl Acad Sci U S A. 2008;105:19696–19701. doi: 10.1073/pnas.0803266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–5504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 28.Chattopadhyay M, Durazo A, Sohn SH, Strong CD, Gralla EB, Whitelegge JP, Valentine JS. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc Natl Acad Sci U S A. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilcox KC, Zhou L, Jordon JK, Huang Y, Yu Y, Redler RL, Chen X, Caplow M, Dokholyan NV. Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J Biol Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi T, Kishikawa M, Miyazaki A, Shimizu A, Ogawa Y, Sakoda S, Ohi T, Shoji H. Simple and defined method to detect the SOD-1 mutants from patients with familial amyotrophic lateral sclerosis by mass spectrometry. J Neurosci Methods. 1998;81:41–44. doi: 10.1016/s0165-0270(98)00012-0. [DOI] [PubMed] [Google Scholar]
- 31.Marklund SL, Andersen PM, Forsgren L, Nilsson P, Ohlsson PI, Wikander G, Oberg A. Normal binding and reactivity of copper in mutant superoxide dismutase isolated from amyotrophic lateral sclerosis patients. J Neurochem. 1997;69:675–681. doi: 10.1046/j.1471-4159.1997.69020675.x. [DOI] [PubMed] [Google Scholar]
- 32.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with allatom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto Y. Generalized-ensemble algorithms: enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J Mol Graph Model. 2004;22:425–439. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman P. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. Journal of Computational Chemistry. 1992;13:1011–1021. [Google Scholar]
- 35.Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, Sohn SH, Durazo A, Faull KF, Gralla EB, Nersissian AM, Valentine JS. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 36.Strange RW, Yong CW, Smith W, Hasnain SS. Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase. Proc Natl Acad Sci U S A. 2007;104:10040–10044. doi: 10.1073/pnas.0703857104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons TJ, Nersissian A, Goto JJ, Zhu H, Gralla EB, Valentine JS. Metal ion reconstitution studies of yeast copper-zinc superoxide dismutase: the "phantom" subunit and the possible role of Lys7p. J Biol Inorg Chem. 1998;3:650–652. [Google Scholar]
- 38.Ray SS, Nowak RJ, Brown RH, Jr, Lansbury PT., Jr Small-moleculemediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proc Natl Acad Sci U S A. 2005;102:3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–324. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen A, Montes J, Mitsumoto H. The role of exercise in amyotrophic lateral sclerosis. Phys Med Rehabil Clin N Am. 2008;19:545–557. ix–x. doi: 10.1016/j.pmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Lepore AC, Tolmie C, O'Donnell J, Wright MC, Dejea C, Rauck B, Hoke A, Ignagni AR, Onders RP, Maragakis NJ. Peripheral hyperstimulation alters site of disease onset and course in SOD1 rats. Neurobiol Dis. 2010;39:252–264. doi: 10.1016/j.nbd.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers SK, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chio A, Traynor BJ, Swingler R, Mitchell D, Hardiman O, Mora G, Beghi E, Logroscino G. Amyotrophic lateral sclerosis and soccer: a different epidemiological approach strengthen the previous findings. J Neurol Sci. 2008;269:187–188. doi: 10.1016/j.jns.2007.10.005. author reply 188-9. [DOI] [PubMed] [Google Scholar]
- 44.Horner RD, Kamins KG, Feussner JR, Grambow SC, Hoff-Lindquist J, Harati Y, Mitsumoto H, Pascuzzi R, Spencer PS, Tim R, Howard D, Smith TC, Ryan MA, Coffman CJ, Kasarskis EJ. Occurrence of amyotrophic lateral sclerosis among Gulf War veterans. Neurology. 2003;61:742–749. doi: 10.1212/01.wnl.0000069922.32557.ca. [DOI] [PubMed] [Google Scholar]
- 45.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Karplus M. Folding thermodynamics of a model three-helix-bundle protein. Proc Natl Acad Sci U S A. 1997;94:c29–3232. doi: 10.1073/pnas.94.26.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 49.Ding F, Borreguero JM, Buldyrey SV, Stanley HE, Dokholyan NV. Mechanism for the alpha-helix to beta-hairpin transition. Proteins. 2003;53:220–228. doi: 10.1002/prot.10468. [DOI] [PubMed] [Google Scholar]
- 50.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 51.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 52.Galaleldeen A, Strange RW, Whitson LJ, Antonyuk SV, Narayana N, Taylor AB, Schuermann JP, Holloway SP, Hasnain SS, Hart PJ. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch Biochem Biophys. 2009;492:40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feig M, Karanicolas J, Brooks CL. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. Journal of Molecular Graphics & Modelling. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 54.Barton GJ. OC - A cluster analysis program. Scotland, UK: University of Dundee; 1993, 2002. [Google Scholar]
- 55.Kabsch W. Solution for Best Rotation to Relate 2 Sets of Vectors. Acta Crystallographica Section A. 1976;32:922–923. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.