The histone deacetylase HDAC4 associates with the nuclear pore complex component Nup155 to modulate gene expression during cardiomyocyte hypertrophy.
Abstract
Class IIa histone deacetylases (HDACs) can modulate chromatin architecture and transcriptional activity, thereby participating in the regulation of cellular responses such as cardiomyocyte hypertrophy. However, the target genes of class IIa HDACs that control inducible cardiac growth and the broader mechanisms whereby these deacetylases modulate locus-specific gene expression within chromatin remain a mystery. Here, we used genome-wide promoter occupancy analysis, expression profiling, and primary cell validation to identify direct class IIa HDAC4 targets in cardiomyocytes. Simultaneously, we identified nucleoporin155 (Nup155) as an HDAC4-interacting protein. Mechanistically, we show that HDAC4 modulated the association of identified target genes with nucleoporins through interaction with Nup155. Moreover, a truncated mutant of Nup155 that cannot bind HDAC4 suppressed HDAC4-induced gene expression patterns and chromatin–nucleoporin association, suggesting that Nup155-mediated localization was required for HDAC4’s effect on gene expression. We thus propose a novel mechanism of action for HDAC4, suggesting it can function to dynamically regulate gene expression through changes in chromatin–nucleoporin association.
Introduction
In response to various disease-causing stimuli the adult myocardium undergoes hypertrophic growth (Dorn et al., 2003). Class IIa histone deacetylases (HDACs) are signal-regulated effectors of gene expression that modulate chromatin structure and can suppress the hypertrophic growth of cardiomyocytes (McKinsey et al., 2000; Zhang et al., 2002; Backs et al., 2006; Ago et al., 2008). Class IIa HDACs—HDAC4, HDAC5, HDAC7, and HDAC9—are unique among the four groups of HDACs and contain a large N-terminal regulatory domain that is subject to phosphorylation as a means of inducibly controlling their nuclear egress to permit gene activation (Verdin et al., 2003; Ago et al., 2008; Haberland et al., 2009). Although class IIa HDACs can bind myocyte enhancer factor-2 (MEF2) in mediating transcriptional repression of select genes (Zhang et al., 2002), their target loci in vivo and the mechanisms whereby they integrate their actions over these endogenous loci in cardiomyocyte nuclei remain unknown.
The nuclear envelope is a highly specialized membrane that surrounds the eukaryotic cell nucleus and provides attachment sites for the lamina, nuclear pore complex (NPC), and chromatin (Hetzer et al., 2005). The NPC is a large protein assembly composed of multiple copies of roughly 30 distinct nucleoporins (NUPs) that regulates the trafficking of macromolecules between the nucleoplasm and cytosol but also provides anchoring sites for chromatin (Lim et al., 2008). Inactive heterochromatin is often partitioned against the inner face of the nuclear membrane in association with the nuclear lamina, while active gene regions and euchromatin may be associated with NUPs at the periphery, or more centrally in the nucleus (Akhtar and Gasser, 2007). Although mechanisms regulating chromatin occupancy within these domains are poorly defined, NUP–chromatin association can be modified by acetylation/deacetylation. Indeed, general class I and class II HDAC blockade by trichostatin A (TSA) induced the association of differentially expressed genes with NUPs at the NPC (Brown et al., 2008). It was also recently discovered that the association of chromatin with NUPs is important for gene expression (Vaquerizas et al., 2010). However, the regulation of these processes, direct binding partners, and the acetyltransferases or deacetylases involved are unknown. Here, we show a novel paradigm whereby select gene loci are subject to inducible regulation through class IIa HDAC binding to nucleoporin 155 (Nup155), which can modulate the hypertrophic growth program in cardiomyocytes.
Results and discussion
To investigate the mechanisms whereby class IIa HDACs regulate the growth of cardiac myocytes we first identified HDAC4 target genes. HDAC4 was mildly overexpressed in primary cultures of neonatal rat ventricular cardiomyocytes (NRVMs) using recombinant adenoviruses expressing HDAC4 or control β-galactosidase, after which RNA was collected for transcript profiling. The array expression data showed that 1,130 genes were changed with HDAC4 overexpression, of which 815 genes were down-regulated and 315 up-regulated (Fig. S1). Gene-grouping analysis using DAVID (Dennis et al., 2003; Huang et al., 2009) showed that HDAC4-repressed genes were enriched for cardiac transcripts, such as myofilament genes (12.3-fold enrichment, benjamini P < 2.2E-14), and heart contraction genes (11.4-fold enrichment, benjamini P < 1.8E-7). Interestingly, we also observed a significant enrichment for Ca2+ ion homeostasis genes (5.8-fold enrichment, benjamini P < 3.3E-6), a cardiac gene group not previously shown to be regulated by class IIa HDACs. These data suggest that HDAC4 represses specific gene groups associated with cardiac function and hypertrophy.
To build off the HDAC4 array screening approach and further refine the list of potential direct HDAC4 target genes in cardiomyocytes we performed a genome-wide occupancy mapping analysis in vivo using the DamID technique (Vogel et al., 2007). The approach consisted of expressing an HDAC4-adenine methyltransferase (Dam, from Escherichia coli) fusion protein to tag occupied DNA sequences by methylation in vivo. Methylated DNA fragments were then amplified, labeled, and hybridized to genomic arrays. We performed the DamID screen with both quiescent and hypertrophic cardiomyocytes using lentiviral vectors and a weak promoter to produce low levels of expression to guard against nonspecific methylation (Guelen et al., 2008). The screen identified 6,434 genomic targets under quiescent conditions, 4,961 targets under hypertrophic conditions, and 2,751 targets present under both conditions (Fig. S1). Thus, more loci were identified under quiescent/repressed conditions, and for loci that were present under both conditions; the peak ratio (HDAC4-Dam/Dam) was significantly higher under quiescent conditions than under hypertrophic stimulation (1.16 ± 0.001-fold increase; P < 0.0001). Corroborating these data, the number of loci with multiple peaks was significantly higher under quiescent conditions (33.8 vs. 29%; P < 0.0001). These data support the paradigm that class IIa HDACs are mostly repressive under quiescent conditions and that hypertrophic stimuli relieve this inhibition (Chang et al., 2004).
We next crossed the expression data with the genomic occupancy data to identify dynamically regulated HDAC4 targets. Of the 1,130 genes whose expression was significantly changed by HDAC4, 372 genes were identified as direct targets only under quiescence, 321 under hypertrophic conditions, and 221 under both conditions with the DamID screen (Fig. S1; data deposited in public database, see Materials and methods). These direct targets from the combined dataset displayed even higher enrichment scores for the contraction and the Ca2+ ion homeostasis gene groups (Fig. S1, chi square; P < 0.02), indicating that HDAC4 specifically controls select functional gene groups in the heart. We also performed a search for evolutionary conserved motifs in the promoters (−2,000 to +500 bp) of the HDAC4 target genes. However, multiple analyses did not reveal a conserved DNA sequence motif, suggesting that HDAC4 can function more broadly and is not restricted to conserved transcription factor binding sites.
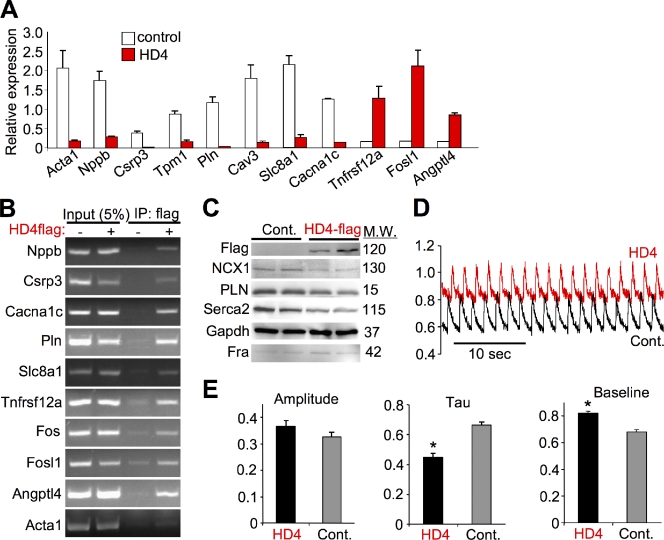
We next experimentally validated the DamID and expression analysis results for the selected gene groups. Quantitative RT-PCR across 11 selected genes showed that the HDAC4 targets were either significantly repressed or activated, consistent with the expression arrays following expression of HDAC4 (Fig. 1 A). We also analyzed several of the direct HDAC4 target genes by chromatin immunoprecipitation (ChIP) for their promoter regions, which validated the DamID screen and showed occupancy of HDAC4 within 10 selected gene regions corresponding to their promoters (Fig. 1 B, band sizes given in Materials and methods). Changes in gene expression through HDAC4 also correlated with changes in protein levels of key Ca2+-handling genes (Fig. 1 C). We observed a reduction in expression of the Na+/Ca2+ exchanger (NCX1), phospholamban (PLN), and the sarcoplasmic reticulum Ca2+ ATPase (Serca2), a profile that should alter Ca2+ handling in myocytes. Indeed, we directly measured intracellular Ca2+ concentration and handling in stimulated cardiomyocytes using a fluorescent Ca2+ indicator (Fig. 1, D and E). Expression of HDAC4 resulted in a significant increase in baseline Ca2+ levels and a significant decrease in the transient relaxation time constant. Thus, HDAC4 acts in an orchestrated manner to alter the expression of several Ca2+-handling genes, validating the expression and promoter occupancy screen with a functional correlate.
Figure 1.
Validation of the expression and occupancy screens. (A) Validation of the transcription array profiling using normalized quantitative RT-PCR in NRVM at baseline or with HDAC4 (HD4) adenoviral-mediated overexpression showing HDAC4 repressed and activated transcripts. Gapdh was used as a control (P < 0.05 for all transcripts, n = 4). (B) Validation of the DamID screen with ChIP assays in NRVM overexpressing HDAC4-flag (adenoviral-mediated overexpression) followed by flag IP and PCR for the promoters of the indicated genes. Input represents 5% of chromatin before IP. (C) Western blot from control (β-galactosidase containing adenovirus) or adeno-HDAC4-flag infected NRVMs showing a decrease in several Ca2+ handling proteins and increased Fosl1 product Fra. Molecular weights in kD are shown. (D) Recordings of total cytoplasmic Ca2+ fluxing using the fluorescent Ca2+ probe Indo-1 in NRVMs paced at 0.5 Hz. Expression of HDAC4 resulted in an increase in baseline Ca2+ and a decrease in relaxation time (tau). (E) Quantitative analysis of the Ca2+ transient amplitude, tau, and baseline diastolic Ca2+ levels (*, P < 0.01; n = 18).
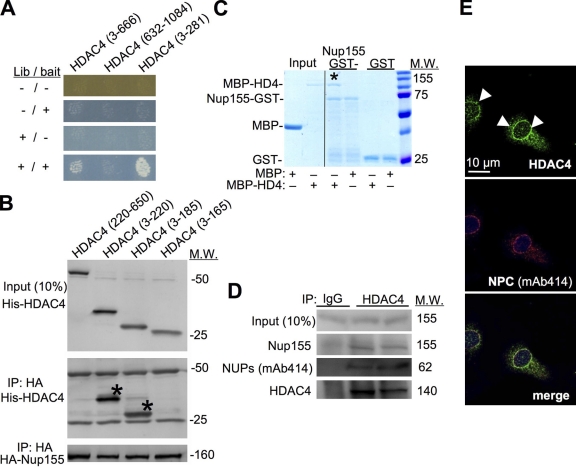
To investigate the transcriptional mechanisms whereby HDAC4 might selectively control gene expression in the heart we performed a modified yeast two-hybrid screen, termed the Ras recruitment system (RRS; Aronheim, 2004). We used the N-terminal regulatory domain of HDAC4 (aa 3–666) as the bait with a heart cDNA library to identify prey plasmids that specifically interacted. This screen identified the C terminus of the Nup155 (aa 886–1391) as an HDAC4 binding partner. Examination in yeast showed that an HDAC4 N-terminal sequence (aa 3–281) was sufficient to bind Nup155 C terminus (Fig. 2 A). Coimmunoprecipitation (coIP) in mammalian cells using HA-tagged full-length Nup155 and His-tagged HDAC4 fragments showed that peptides containing amino acid 3–220 and 3–185 of HDAC4 were sufficient for the interaction, whereas a 3–165 fragment was not, suggesting that the interacting domain in HDAC4 is between amino acids 165 and 185 (Fig. 2 B). This domain in HDAC4 also mediates an interaction with MEF2 and HDAC1 (Chan et al., 2003). Finally, recombinant GST was fused to the C terminus of Nup155 (aa 886–1391) and used to assess interaction with the recombinant HDAC4 N terminus (aa 3–628) fused to the maltose-binding protein (MBP). This analysis showed that GST-Nup155 was unable to bind MBP alone, although it did bind MBP-HDAC4 recombinant protein (Fig. 2 C, asterisk). GST alone did not bind MBP-HDAC4.
Figure 2.
HDAC4 partners with Nup155 and associates with the NUPs. (A) Yeast two-hybrid growth assay with baits containing either the N terminus (aa 3–281 or 3–666) or C terminus (aa 632–1084) of HDAC4 (HD4) cotransfected with the library plasmid, which identified the Nup155 C terminus. Inducible promoters on library and bait plasmids allows differential activation of expression (−/+), showing growth and therefore interaction between Nup155 and both N-terminal constructs of HDAC4, but not with the C-terminal construct. (B) Western blots for the indicated proteins from mammalian cells transfected with the indicated constructs. For the bottom two blots, HA-Nup155 was immunoprecipitated (IP) followed by Western blotting for His-HDAC4 fragments or HA-Nup155. The asterisks show the fragments that interact. Input represents 10% of protein before IP. (C) Coomassie gel showing interaction between the C terminus of Nup155 as a GST fusion protein and the N terminus of HDAC4 as an MBP fusion protein. The asterisk shows the pull-down of MBP-HDAC4 with GST-Nup155 from the GST binding column. Input protein is shown on the left. (D) Western blots from NRVMs after endogenous HDAC4 immunoprecipitation (IP) or control IgG to investigate Nup155 and nucleoporin (mAb414) interaction. (E) Immunofluorescent staining of NRVMs for endogenous HDAC4 (green, arrowheads), the NUPs (mAb414, red), and DNA (ToPro3, blue) showing partial colocalization of HDAC4 with NUPs.
Coimmunoprecipitation experiments showed that endogenous HDAC4 interacted with endogenous Nup155 in extracts from primary cultures of NRVMs (Fig. 2 D). We used the well-validated mAb414 monoclonal antibody raised against the entire rat nuclear pore, which recognizes several NUPs but not Nup155 (Davis and Blobel, 1986), to show that other NUPs (bands at 62 and 107 kD) may be part of this complex (Fig. 2 D, and unpublished data). Remarkably, immunofluorescent staining of cardiomyocytes showed that endogenous HDAC4 is enriched at the nuclear periphery, and partially colocalized with the mAb414 antibody (Fig. 2 E). Interestingly, HDAC5 was not able to bind Nup155 (unpublished data), suggesting greater specificity for HDAC4. Collectively, these data suggest that Nup155 and HDAC4 can interact in conjunction with other NUPs, especially at the nuclear periphery where the NPC is located.
Global HDAC inhibition was previously shown to alter chromatin association with the NUPs for several gene loci (Brown et al., 2008), and a study in Drosophila cells showed that mAb414-positive NUPs were found to associate with sites of active transcription (Capelson et al., 2010). Therefore, we hypothesized that HDAC4 could modify the association of select genomic loci with the NUPs, and that the interaction between HDAC4 and Nup155 was required for this change in association. To test this hypothesis we initially performed ChIP assays using the mAb414 antibody in NRVMs, with or without expression of HDAC4 and TSA treatment. Many of the identified HDAC4 target genes indeed displayed some degree of association with the NUPs. Remarkably, as we hypothesized, overexpression of HDAC4 decreased the association of many target loci with the NUPs, and the addition of TSA restored this association (Fig. 3 A). Interestingly, several HDAC4 targets showed a reversed association pattern. Loci like Pln, Fos, Tnfrsf12a, and Csrp3 displayed low association with NUPs at baseline but overexpression of HDAC4 enhanced the association, which was reversed with TSA (Fig. 3 A). Although this differential behavior of specific loci has been described in yeast (Casolari et al., 2004) and mammalian cells (Brown et al., 2008), the reasons for it are unknown. The reversal of the dissociation by TSA shows that the process is acetylation/deacetylation dependent, although the enzymes involved, such as association with class I HDACs, and the recipients of the acetyl group, are unknown. Thus, our results identify a mechanism whereby HDAC4 can modify chromatin association of select gene loci with NUPs.
Figure 3.
HDAC4 binds and regulates the association of specific chromatin loci with NUPs. (A) ChIP assay in NRVM with the NUP (mAb414) antibody showing association of loci with NUPs with or without HDAC4, with or without TSA treatment. Input represents amplification of 5% of chromatin before IP. (B) Immunofluorescent staining of NRVMs for tagged full-length Nup155 or truncated Nup155 (Nup155ΔC) showing similar localization (arrowheads). Troponin (Tropo.) staining verified myocyte identity. (C) Immunofluorescent staining of NRVM for endogenous HDAC4 (HD4, green), the NUPs (mAb414, red), and DNA (ToPro3, blue) at control or after Nup155ΔC expression. The arrowheads show localization of HDAC4 at baseline, which is lost with Nup155ΔC expression, yet the NUPs maintain their peripheral location (red). (D) ChIP assay in NRVMs with mAb414 antibody during control (βgal, −), HDAC4, or HDAC4 + Nup155ΔC overexpression. (E) ChIP assay as in D, except NRVMs were infected with an adenovirus containing activated CaMKII, which reversed HDAC4 actions.
Next we wanted to verify that the association with nucleoporins is required for HDAC4 action on gene expression. Unfortunately, knockdown of Nup155 resulted in cardiomyocyte death (unpublished data). To circumvent this problem we constructed a truncated Nup155 mutant, referred to as Nup155ΔC (aa 1–886), which lacks the HDAC4-interacting C-terminal domain. Analysis in cardiomyocytes showed that this truncation mutant is able to localize in a similar manner to full-length Nup155 (Fig. 3 B, arrowheads). Importantly, overexpression of Nup155ΔC did not seem to result in overt nuclear pore transport defects or otherwise compromise the health of cultured cells (Fig. S2). Overexpression of Nup155ΔC by adenoviral-mediated gene transfer in NRVMs also did not result in major alterations in other NUPs as assessed with m414 or Nup153 antibody (unpublished data).
Remarkably, adenoviral overexpression of Nup155ΔC replaced endogenous full-length Nup155 and now no longer permitted endogenous HDAC4 to associate with nucleoporins (Fig. 3 C and Fig. S3). Careful analysis and quantification showed that ∼30% of the HDAC4 colocalized with NUPs (mAb414) at baseline, and overexpression of Nup155ΔC reduced this association to only 3% (Fig. S3). More importantly, ChIP assays using immunoprecipitated NUPs showed that although HDAC4 is able to modify the association of selected loci with NUPs, coexpression of Nup155ΔC abolished the effect and restored NUPs association (Fig. 3 D). Similarly, coexpression of activated calmodulin-dependent kinase II (CaMKII), which can shuttle HDAC4 out of the nucleus (Haberland et al., 2009), abolished the effect of HDAC4 and restored association with nucleoporins (Fig. 3 E). Thus, recruitment of HDAC4 by Nup155 binding is required for HDAC4’s ability to displace chromatin from the NUPs.
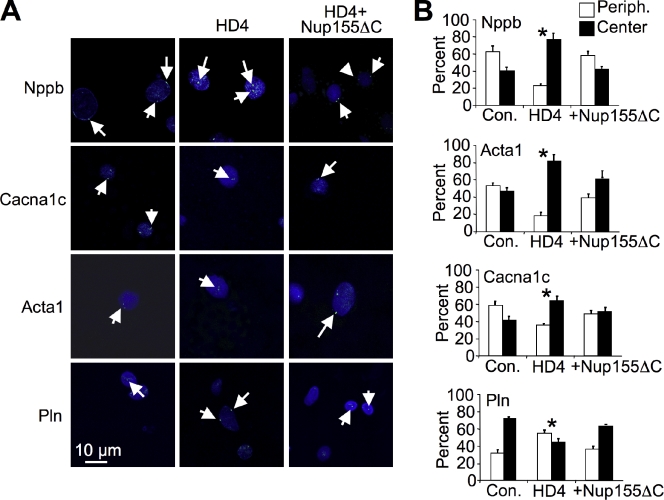
To further verify dynamic association between NUPs and HDAC4 on select target genes in NRVMs we performed fluorescent in-situ hybridization (FISH) combined with confocal microscopy and quantified the results in both a blinded and unblinded manner, producing similar results (Fig. 4, A and B). This analysis showed predominant peripheral nuclear localization of the Nppb, Acta1, Cacna1c loci during control conditions, a shift to a more central nuclear position under HDAC4 (HD4) overexpression, and reversal of the HDAC4 effect by expression of Nup155ΔC (Fig. 4, A and B). In contrast, the Pln locus demonstrates a reverse pattern. The Nlrx1 locus was used as a control, which showed no significant changes in intranuclear location (not depicted). These results corroborate the ChIP experiments and support the dynamic movement of specific loci to the nuclear periphery, the ability of HDAC4 to modify this balance, and the requirement of HDAC4–Nup155 interaction for this mechanism.
Figure 4.
HDAC4 regulates spatial chromatin organization. (A) Confocal FISH of selected loci (green dots, arrows) with nuclear Topro3 counterstain (blue) in NRVMs showing predominant peripheral nuclear localization of the Nppb, Acta1, Cacna1c during control conditions, a shift to a more central nuclear position under HDAC4 (HD4) overexpression, and reversal of the HDAC4 effect by expression of Nup155ΔC. In contrast, the Pln locus demonstrates a reverse pattern. (B) Quantitative summary of the FISH data showing the percentage of peripheral nuclear FISH signals (white bar) versus central nuclear signals (black bar). *, P < 0.001; significance of HDAC4-expressing vs. control and HDAC4+ Nup155ΔC, n = 250.
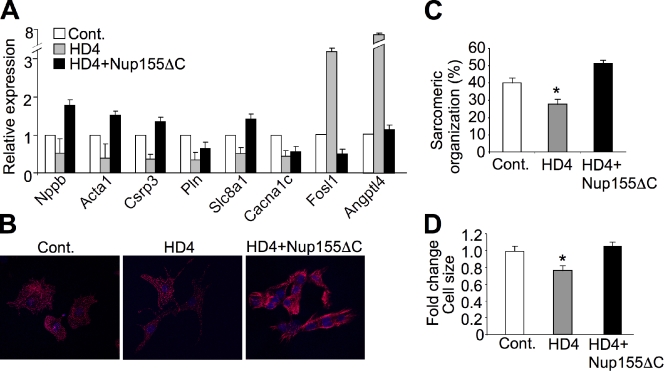
Consistent with the changes described above, detailed mRNA expression analysis using qRT-PCR showed that overexpression of Nup155ΔC reversed the expression pattern of HDAC4 for many of its target genes (Fig. 5 A). Functionally, these changes in gene expression correlated with alterations in cardiomyocyte hypertrophy. Indeed, although overexpression of HDAC4 reduced sarcomeric organization at baseline in quiescent NRVMs, expression of the Nup155ΔC mutant induced organization of sarcomeres and induced an increase in cell size, likely by relieving HDAC-dependent repression of select genes involved in myocyte growth (Fig. 5, B–D).
Figure 5.
Non-HDAC4 binding Nup155ΔC reverses HDAC4 gene expression patterns and alters the growth response of NRVMs. (A) Quantitative qRT-PCR in NRVMs (normalized to Gapdh) shows that combined expression of Nup155ΔC reverses the HDAC4 expression pattern for the indicated transcripts. (B) Immunofluorescent staining of NRVM for sarcomeric α-actinin (red) with Adβgal (control), AdHDAC4 (HD4), or AdHDAC4 + AdNup155ΔC overexpression. The data show that Nup155ΔC expression increased sarcomeric organization associated with hypertrophy, which is quantified in C (*, P < 0.001; n = 50). (D) HDAC4 overexpression also reduced myocyte size in culture, which was restored by expression of Nup155ΔC (*, P < 0.01).
Collectively, we identified clusters of sarcomeric genes and Ca2+-handling genes that are directly repressed by HDAC4 and showed that HDAC4 exists in a complex with Nup155 and mAb414-positive NUPs, and that HDAC4 modified the association of these chromatin loci with the NUPs to control gene expression and hypertrophy. The association of these loci with the NUPs may not be locus- or sequence-specific, but rather the activation of functional gene groups may induce their association with the NUPs. HDAC4 appears to be affixed to the NUPs at rest in quiescent cardiomyocytes, which might function as a general inhibitory mechanism to quench gene expression of selected target loci. Upon agonist or mitogen stimulation, class IIa HDACs are exported from the nucleus, thereby enhancing chromatin–NUP association, gene transcription, and activation of otherwise repressed loci, now poised for rapid expression. Interestingly, NUPs were shown to interact with and regulate genes in the nucleoplasm, independent of the nuclear periphery (Capelson et al., 2010; Kalverda et al., 2010), and some NUPs are highly dynamic and rapidly shuttle between the NPC and the nucleoplasm (Hou and Corces, 2010). Therefore, although we did not observe it directly in our system, it is possible that HDAC4 can modify the association between chromatin and NUPs in the nucleoplasm or on the NPC at the periphery.
Materials and methods
Cell culture
Neonatal cardiomyocytes were isolated from 1–3-d-old Sprague Dawley rat pups using the Neonatal Cardiomyocyte Isolation System (Worthington) according to the manufacturer’s instructions. In brief, hearts were rinsed with ice-cold HBSS and trypsinized for 16 h at 4°C. Hearts were incubated with trypsin inhibitor at 37°C, and then incubated for 60 min with a collagenase solution at 37°C with gentle mixing. The cell pellet was triturated and resuspended in M199 media (Invitrogen) containing 10% fetal calf serum. Cells were then preplated for 1 h to deplete culture of fibroblasts, and nonadherent cells were plated on gelatin-coated culture dishes. After plating, cells were washed twice with serum-free or serum-containing culture media, and grown for 24–72 h.
Adenoviral and lentiviral vector production
The Gateway system (Invitrogen) was used to generate pEntry clones for adenoviral and lentiviral vector production. Adenoviral vectors were generated as described previously (Xu et al., 2006). NRVMs were infected for 2 h at 10 multiples of infection; cells were washed twice and incubated for 24–48 h. Lentiviral vectors were produced by the Viral Vector Core at the Translational Core Laboratories, Cincinnati Children’s Hospital Research Foundation (Cincinnati, OH). Lentiviral infection was performed overnight with addition of hexadimethrine bromide; cells were washed twice and incubated for 24–48 h.
Ras recruitment system (RRS) yeast two-hybrid
The RRS screen was performed as described previously (Aronheim, 2004). In brief, the bait was a hybrid protein with the mammalian activated Ras protein lacking its farnesylation CAAX box fused to the human HDAC4 N terminus (aa 3–666). The bait plasmid was cotransfected into Cdc25-2 yeast cells with a myristoylated heart cDNA library (CryoTrap; Agilent Technologies). The expression of the cDNA library is designed under the control of the Gal1-inducible promoter, while the expression of the bait is controlled by a Met-off inducible promoter. Plates were incubated for 7 d at the permissive temperature of 24°C and were subsequently replica plated onto inductive medium and incubated at the restrictive temperature of 36°C. Colonies that exhibit efficient growth were selected and grown on appropriate glucose plates for 2 d. Subsequently, galactose and methionine dependency was assayed by replica plating at the restrictive temperature of 36°C. Colonies that exhibited efficient cell growth and full dependency were further analyzed. Plasmid DNA was extracted and the library plasmid identified by sequencing. Identified library plasmids were reintroduced into Cdc25-2 cells with either the specific (HDAC4 aa 3–666 or aa 3–281) or nonspecific baits (HDAC4 aa 632-end) and the interaction was reexamined using the galactose and methionine dependency test at the restrictive temperature of 36°C.
Coimmunoprecipitation
Cultured cells were washed with ice-cold phosphate-buffered sodium (PBS) buffer and lysed in buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% NP-40. 400 µg of protein was immunoprecipitated with anti-HA, HDAC4, or IgG antibodies and protein A/G agarose beads (Santa Cruz Biotechnology, Inc.). Beads were washed three times with buffer and suspended in 30 µl of Laemmli buffer. Proteins were separated on an SDS gel, blotted, and detected with primary anti-His antibody (Cell Signaling Technology), HA antibody (Cell Signaling Technology), Nup155 antibody (custom made from YenZym Antibodies, LLC.), nucleoporin mAb414 antibody (Covance), and HDAC4 antibody (Abcam). Secondary alkaline phosphatase–conjugated antibodies were also used (Santa Cruz Biotechnology, Inc.).
Recombinant protein pull-down
cDNA constructs encoding recombinant GST-Nup155 (aa 886–1391) and MBP-HDAC4 (aa 3–628) in pDest15 and pKM596 (Chen et al., 2009) vectors were generated using the Gateway system (Invitrogen) according to the manufacturer’s instructions in BL21 E. coli. Bacteria were induced at OD600 0.4–0.6 with 0.5 mM IPTG and grown for 5 h at 30°C (MBP fusion proteins) or overnight at room temperature (GST fusion proteins). Bacterial pellets were resuspended in lysis buffer (150 mM Nacl, 100 mM, Tris pH 8, and 1% Triton), sonicated, and cleared by centrifugation. Recombinant MBP fusion proteins were purified on amylose resins (New England Biolabs, Inc.) according to the manufacturer’s instructions, washed, and eluted with 10 mM maltose. Recombinant GST fusion proteins were immobilized on glutathione–Sepharose beads (GE healthcare) and washed with 10x volume of lysis buffer. The beads were then incubated with either MBP or MBP fused to HDAC4 N terminus. After washing three times in lysis buffer, the beads were suspended in Laemmli buffer and boiled. Proteins were resolved on an acrylamide gel and stained with Coomassie brilliant blue.
Immunofluorescent staining and analysis
NRVMs were plated on glass slides, infected with adenoviral vectors encoding β-galactosidase, Flag-HDAC4, Nup155-V5, or Nup155ΔC-V5, and grown for 24–48 h. Cells were fixed in 4% paraformaldehyde in PBS, with or without preextraction with 0.1% Triton in PBS for 2 min. Cells were permeabilized with 0.5% Triton in PBS and blocked in 5% goat serum in PBS. Incubation with primary antibodies was performed in 3% goat serum containing PBS at 4°C overnight. Primary antibodies used were anti-HDAC4 (Abcam), anti-nucleoporin mAb414 (Covance), anti-V5 epitope (Abcam), and anti-sarcomeric α-actinin (Sigma-Aldrich). Alexa Fluor 488– or 568–conjugated secondary antibodies were used for detection with a confocal microscope (Invitrogen). Nuclei were counterstained with To-Pro-3 (Invitrogen). Imaging was performed using a laser-scanning confocal microscope (PCM 2000; Nikon) equipped with Plan Apochromat 20x 0.75 NA and Plan Apochromat water immersion 60x 1.2 NA objectives (Nikon) at room temperature. Images were acquired using Simple PCI software (version 4.0; C-Imaging Systems) and arranged for display using Photoshop CS (Adobe) and PowerPoint (Microsoft) without processing.
For quantification of sarcomeric organization and cell size, NRVMs were stained with sarcomeric α-actinin (Sigma-Aldrich) antibody and imaged using a confocal microscope. The ratio of signal area above a threshold level to cell surface area was calculated using ImageJ 1.38, and used as a measure of sarcomeric content and organization.
Chromatin immunoprecipitation
NRVMs were infected with adenoviral vectors encoding β-galactosidase, flag-HDAC4, and Nup155ΔC. Chromatin immunoprecipitation was performed using the EZ-Chip kit (Millipore) according to the manufacturer’s instructions. Antibodies for immunoprecipitation were anti-flag (Sigma-Aldrich) or anti-nucleoporin mAb414 (Covance). PCR was performed using promoter-specific primer pairs as follows (size in base pairs, the amplified band is shown in parenthesis): Acta1 (218) 5′-AGCTGTTCCCATTTGGAGTG-3′, 5′-CCCAATATGAATGCCCAAAG-3′; Angptl4 (196) 5′-CACCCACCTGTAAACCCAAC-3′, 5′-CTCCAATTGCTTCTGGCTCT-3′; Cacna1c (120) 5′-CGAGCACGTGTTCTGCCAGTGT-3′, 5′-TGCAGGAGGTGCCATGCTGT-3′; Fos (170) 5′-CGGTGTGTAAGGCAGTTTCA-3′, 5′-GCTCAGCCTTTCTCCTCCAT-3′; Fosl1 (174) 5′-TGTCGCTCTAATTGTCCGTG-3′, 5′-GAGAATATGGAGCGCGAAAG-3′; Jph2 (184) 5′-GGCAAGGAAATGGAGTCAAA-3′, 5′-GAACTCCATTGGTCTGGGAA-3′; Mybpc3 (143) 5′-CCCCTGCTGCTCCCAGGCTA-3′, 5′-CGATGGGCCCAGCCTCCTCT-3′; Nppb (100) 5′-CCAATACGGGTGGGGCACGG-3′, 5′-GGGTGGGAGTGTGTGGGGGA-3′; Obscn (98) 5′-AGCCCCTGCCACTCGCTCTT-3′, 5′-GGCCCACCCTGCAAGAAGCA-3′; Pln (198) 5′-GTTGTACGAGAGCAAAGCCC-3′, 5′-CATCCCATCAGTTGCCAATA-3′; Slc8a1 (104) 5′-AGAGAACCAGGGGCCTGCAA-3′, 5′-TCCTTGGTGTGGTGCAAGGCA-3′; Tnfrsf12a (185) 5′-CAGACCTGTCTTCATTCGCA-3′, 5′-TCTCTCTGGGAGGAGCAGAG-3′; Tnni3 (192) 5′-TGGACTCAGCATTCTGGTTG-3′, 5′-TCCGAGCTCTTCATCACCTT-3′; Trdn (90) 5′-AGCCTGGAGAGTCCCTGTGGC-3′, 5′-TTCTGCACCCACCATGACTGAGA-3′.
qRT-PCR
NRVMs were infected with adenoviral vectors encoding β-galactosidase or flag-HDAC4 and incubated in serum-free or 10% fetal bovine serum containing growth medium for 48 h. RNA was extracted using Trizol (Invitrogen). Reverse transcription (RT) reaction was performed using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) and random hexamer primers according to the manufacturer’s instructions. Quantitative real-time PCR was performed with sybr-green and taq polymerase master mix (Applied Biosystems). Data were normalized to the expression of GAPDH. Primers used were: Acta1 5′-TCGCGACCTTACTGACTACCTG-3′, 5′-GCTTCTCTTTGATGTCGCGC-3′; Nppb 5′-ACAATCCACGATGCAGAAGCT-3′, 5′-GGGCCTTGGTCCTTTGAGA-3′; Csrp3 5′-ATCAGAGAAGTGCCCACGATG-3′, 5′-GTAAGCCCTCCAAACCCAAT-3′; Pln 5′-TGACGATCACAGAAGCCAAG-3′, 5′-TGCCAGGAAGACAAAAGTAGG-3′; Cav3 5′-CCTCAAAATGGGAGTACATGG-3′, 5′-TGGTAGGCTGAGCAGTTTCC-3′; Slc8a1 5′-TTCCCTCTACCGTAATCAGCA-3′, 5′-ATTTCTGCAATGCGCCTCT-3′; Cacna1 5′-CATTCTGCTCAGTAGCATCTCC-3′, 5′-AACAATGTCAAAATAAAACAGAATGTG-3′; Tnfrsf12a 5′-TCGGGTTGGTGTTGATACG-3′, 5′-CCATGCACTTGTCGAGGTC-3′; Fos 5′-GCTCCCCTGTCAACACACA-3′, 5′-GACCAGAGTGGGCTGCAC-3′; Fosl1 5′-CCGACCAGGAGTCATACGAG-3′, 5′-TTCGGTTTCTGCACTTAGCA-3′; Angptl4 5′-TCTCCACCATTTTTGGTCAAC-3′, 5′-GTTCAGGCGTCTCTGAATCAC-3′.
Transcriptome analysis
NRVMs were infected with adenoviral vectors encoding β-galactosidase or flag-HDAC4, and incubated in serum-free or 10% fetal bovine serum containing growth medium for 48 h. RNA was extracted using the RNeasy Fibrous Tissue kit (QIAGEN) according to the manufacturer’s instructions. Expression pattern was analyzed using GeneChip Rat Gene 1.0 ST Array (Affymetrix) according to the manufacturer’s instructions. The Affymetrix Whole Transcript Sense Target Labeling Assay was used to create biotin-labeled sense-strand cDNA targets for hybridization to the Gene 1.0 ST Arrays. Hybridization cocktail for a single probe array contained 2.5 µg of fragmented cRNA, 50 pM Control Oligonucleotide B2 (Affymetrix), 20x Eukaryotic Hybridization Controls (1.5 pM bioB, 5 pM bioC, 25 pM bioD, 100 pM cre; Affymetrix), 7% DMSO, and 1x hybridization buffer. GeneChip Rat Gene 1.0 ST Array was hybridized at 45°C for 18 h in the GeneChip Hybridization Oven 640 (Affymetrix). Expression data has been submitted to GEO Datasets under accession no. GSE19771.
DamID mapping
The DamID vectors and protocol were a kind gift of B. van Steensel (Netherlands Cancer Institute, Amsterdam, The Netherlands). The DamID screen was performed using lentiviral vectors, as described previously (Guelen et al., 2008), in cultured neonatal rat cardiomyocytes in serum-free (SF) and in 10% fetal bovine serum (FCS) containing media for quiescence and hypertrophy, respectively. We used the Nimblegen tiling rat promoter arrays set (C4492001-00-01), based on genomic build RGSC3.4. Each feature on the array received a corresponding scaled log2 ratio of the input signals of the HDAC4-Dam and unfused Dam samples that were co-hybridized to the array. This log2 ratio was computed and scaled to center the ratio data at zero. Scaling was performed by subtracting the bi-weight mean for the log2 ratio values for all features on the array from each log2 ratio value. Peaks were detected by searching for four or more probes whose signals were above a cutoff value, ranging from 90% to 15%, using a 500-bp sliding window. The cutoff values were a percentage of a hypothetical maximum, calculated as the mean + six standard deviations. The ratio data were then randomized 20 times to evaluate the probability of “false positive” result. Each peak was then assigned a false discovery rate (FDR) score, based on the randomization. Analysis concentrated on peaks with FDR ≤0.1.
Analysis of evolutionary conserved DNA motifs
Promoters of HDAC4 target genes (−2,000 to +500 relative to the transcriptional start site) were extracted and crossed with genomic information of Homo sapiens, Rhesus macaca, Mus musculus, Bos taurus, and Canis familiaris to limit the analysis to evolutionary conserved sequences using GALAXY (Taylor et al., 2007). A search using MEME (Bailey and Elkan, 1994) and TOMTOM (Gupta et al., 2007) was used to identify conserved transcription factor–binding sites. In addition, a search for conserved transcription factor–binding sites was performed using GATHER (Chang and Nevins, 2006) and TRANSFAC databases.
Analysis of Ca2+ transients in cardiomyocytes
NRVMs plated on laminin-coated coverslips were loaded with 5 µmol/L Indo-1. Individual cells were excited at 350 nm and the fluorescence emission ratio (405/485) was collected every 20 ms using a 40x oil objective and a photomultiplier detection system (Photon Technology International). Cells were electrically stimulated (80 V, 5 ms, 0.5 Hz) using platinum electrodes. Ca2+ transient amplitudes were calculated by measuring the change in 405/485 fluorescence with each electrical pulse from 10 to 30 cells for each treatment. These Ca2+ measurements were conducted in normal rodent Ringer’s solution (in mM: 145 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, and 10 Hepes, pH 7.4).
Fluorescent in-situ hybridization (FISH)
NRVMs were plated on glass slides, infected with adenoviral vectors encoding β-galactosidase, Flag-HDAC4, or Nup155ΔC-V5, and grown for 24–48 h. Cells were pretreated with 0.8% sodium citrate and fixed in ice-cold 3:1 methanol/acetic acid. Slides were treated with protease solution (FISH specimen pretreatment reagent kit; Abbott Molecular) for 10 min at 37°C. Denaturation was performed with 70% formamide/2x SSC solution for 5 min at 70°C. Probes were generated using a nick translation reagent kit (Abbott Molecular) with Green-dUTP (Enzo Life Sciences) according to the manufacturer’s instructions, with bacterial artificial chromosome (BAC) clones as templates. Probes were suspended in buffer (cDenHyb; Insitus Biotechnologies), denaturated on the slides for 5 min at 75°C, and incubated overnight at 37°C. Washing was performed in 0.4x SSC/0.3% NP-40 for 2 min at 73°C and in 2x SSC/0.1% NP-40 for 1 min at room temperature. Nuclei were counterstained with To-Pro-3 (Invitrogen). Specimens were visualized using a confocal microscope.
Statistical analysis
Analysis of the DamID screen is described in Fig. S1. Data are presented as mean ± SEM. ANOVA was used for expression data, with P < 0.05 considered a statistically significant result. FISH data were quantified using χ2 test with P < 0.05 considered a statistically significant result.
Online supplemental material
Fig. S1 shows an analysis of HDAC4-induced expression changes, occupancy, and enrichment for functional groups. Fig. S2 shows that infection with AdNup155ΔC does not result in protein import and mRNA export defects. Fig. S3 shows and quantifies the partial colocalization of endogenous HDAC4 and mAb414 NUP immunosignals that are disrupted with AdNup155ΔC infection. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101046/DC1.
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Fondation Leducq, and the Howard Hughes Medical Institute (to J.D. Molkentin). I. Kehat was supported by a grant from the Human Frontiers Science Program and the Legacy Heritage Clinical Research Institute at Rambam Medical Center.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- HDAC
- histone deacetylase
- MBP
- maltose-binding protein
- NPC
- nuclear pore complex
- NRVM
- neonatal rat ventricular cardiomyocyte
- NUP
- nucleoporin
- TSA
- trichostatin A
References
- Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J.D., Vatner S.F., Sadoshima J. 2008. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 133:978–993 10.1016/j.cell.2008.04.041 [DOI] [PubMed] [Google Scholar]
- Akhtar A., Gasser S.M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Aronheim A. 2004. Ras signaling pathway for analysis of protein-protein interactions in yeast and mammalian cells. Methods Mol. Biol. 250:251–262 [DOI] [PubMed] [Google Scholar]
- Backs J., Song K., Bezprozvannaya S., Chang S., Olson E.N. 2006. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116:1853–1864 10.1172/JCI27438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- Brown C.R., Kennedy C.J., Delmar V.A., Forbes D.J., Silver P.A. 2008. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22:627–639 10.1101/gad.1632708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., Hetzer M.W. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Chan J.K., Sun L., Yang X.J., Zhu G., Wu Z. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515–23521 10.1074/jbc.M301922200 [DOI] [PubMed] [Google Scholar]
- Chang J.T., Nevins J.R. 2006. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 22:2926–2933 10.1093/bioinformatics/btl483 [DOI] [PubMed] [Google Scholar]
- Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467–8476 10.1128/MCB.24.19.8467-8476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37(Web Server issue):W305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I., Blobel G. 1986. Identification and characterization of a nuclear pore complex protein. Cell. 45:699–709 10.1016/0092-8674(86)90784-1 [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Dorn G.W., II, Robbins J., Sugden P.H. 2003. Phenotyping hypertrophy: eschew obfuscation. Circ. Res. 92:1171–1175 10.1161/01.RES.0000077012.11088.BC [DOI] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. 2007. Quantifying similarity between motifs. Genome Biol. 8:R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R.L., Olson E.N. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10:32–42 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W., Walther T.C., Mattaj I.W. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380 10.1146/annurev.cellbio.21.090704.151152 [DOI] [PubMed] [Google Scholar]
- Hou C., Corces V.G. 2010. Nups take leave of the nuclear envelope to regulate transcription. Cell. 140:306–308 10.1016/j.cell.2010.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140:360–371 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Lim R.Y., Ullman K.S., Fahrenkrog B. 2008. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 267:299–342 10.1016/S1937-6448(08)00632-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Olson E.N. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA. 97:14400–14405 10.1073/pnas.260501497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J., Schenck I., Blankenberg D., Nekrutenko A. 2007. Using galaxy to perform large-scale interactive data analyses. Curr. Protoc. Bioinformatics. Chapter 10:Unit 10 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas J.M., Suyama R., Kind J., Miura K., Luscombe N.M., Akhtar A. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6:e1000846 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H.G. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286–293 10.1016/S0168-9525(03)00073-8 [DOI] [PubMed] [Google Scholar]
- Vogel M.J., Peric-Hupkes D., van Steensel B. 2007. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2:1467–1478 10.1038/nprot.2007.148 [DOI] [PubMed] [Google Scholar]
- Xu J., Gong N.L., Bodi I., Aronow B.J., Backx P.H., Molkentin J.D. 2006. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 281:9152–9162 10.1074/jbc.M510217200 [DOI] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey T.A., Chang S., Antos C.L., Hill J.A., Olson E.N. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 110:479–488 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]