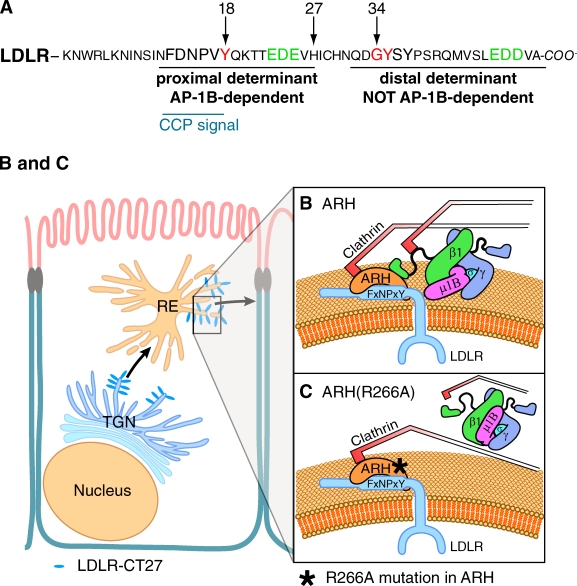
The low-density lipoprotein receptor (LDLR) doesn’t directly bind AP-1B; however, it relies on this clathrin adaptor for basolateral exocytosis. Identification of the autosomal recessive hypercholesterolemia protein (ARH) as a link between LDLR and AP-1B explains this apparent discrepancy and provides a model for how other endocytic proteins may contribute to endosomal recycling.
Abstract
The autosomal recessive hypercholesterolemia protein (ARH) is well known for its role in clathrin-mediated endocytosis of low-density lipoprotein receptors (LDLRs). During uptake, ARH directly binds to the FxNPxY signal in the cytoplasmic tail of LDLR. Interestingly, the same FxNPxY motif is used in basolateral exocytosis of LDLR from recycling endosomes (REs), which is facilitated by the epithelial-specific clathrin adaptor AP-1B. However, AP-1B directly interacts with neither the FxNPxY motif nor the second more distally located YxxØ sorting motif of LDLR. Here, we show that ARH colocalizes and cooperates with AP-1B in REs. Knockdown of ARH in polarized epithelial cells leads to specific apical missorting of truncated LDLR, which encodes only the FxNPxY motif (LDLR-CT27). Moreover, a mutation in ARH designed to disrupt the interaction of ARH with AP-1B specifically abrogates exocytosis of LDLR-CT27. We conclude that in addition to its role in endocytosis, ARH cooperates with AP-1B in basolateral exocytosis of LDLR from REs.
Introduction
Columnar epithelial cells polarize their plasma membrane into apical and basolateral domains, each with their own distinct assortment of transmembrane proteins and lipids (Martin-Belmonte and Mostov, 2008). Polarized sorting of newly synthesized or recycling receptors takes place either at the TGN or in recycling endosomes (REs) dependent on specific sorting determinants (Mellman and Nelson, 2008; Fölsch et al., 2009). Whereas apical sorting information is often decoded in a protein’s ectodomain or transmembrane anchor, sorting to the basolateral membrane is frequently facilitated by tyrosine- or dileucine-based sorting signals encoded in the cytoplasmic tails of transmembrane proteins. These cytoplasmic tail signals are recognized by cytosolic adaptor proteins and are generally cis dominant over apical sorting information (Rodriguez-Boulan et al., 2005; Fölsch, 2008). Tyrosine-based sorting signals conform to either FxNPxY or YxxØ consensus sequences.
Low-density lipoprotein receptor (LDLR) encodes two different types of basolateral sorting signals in its cytoplasmic tail (see Fig. 5 A). The proximal signal is an FxNPxY motif that is co-linear with the endocytic motif, ensuring incorporation of LDLR into clathrin-coated pits, followed by a cluster of negative charges (Matter and Mellman, 1994). The distal signal is a noncanonical YxxØ motif and comprises the amino acids GYSY again followed by a cluster of negative charges (see Fig. 5 A; Matter and Mellman, 1994; Koivisto et al., 2001). Here, we use two well-established LDLR mutant proteins in which either the proximal or the distal signal is inactivated (Matter et al., 1992). LDLR-CT27 was truncated after amino acid 27 of LDLR’s cytoplasmic tail and contains only the proximal FxNPxY motif (see Fig. 5 A). LDLR(Y18A) contains a tyrosine-to-alanine mutation at position 18 that disrupts the FxNPxY motif while maintaining the distal sorting determinant (see Fig. 5 A).
Figure 5.
Model of ARH interactions with AP-1B and LDLR in REs. (A) Schematic depicting amino acids of the cytoplasmic tail of LDLR. Clathrin-coated pit (CCP) and proximal and distal targeting determinants are underlined. Red residues are critically important for either proximal or distal signal. Green residues denote clusters of acidic residues that are part of the sorting determinants. Arrows denote the position of amino acid residues in the cytoplasmic tail of LDLR. (B and C) Model depicting biosynthetic sorting of LDLR-CT27 from the TGN into REs. Here, LDLR-CT27 engages ARH and AP-1B for basolateral exocytosis (B). In cells expressing ARH(R266A), the interaction between ARH and AP-1B is broken and exocytosis of LDLR-CT27 is stalled (C).
YxxØ sorting motifs are recognized by the medium subunits of heterotetrameric clathrin adaptor protein (AP) complexes, AP-1 through AP-4 (Bonifacino and Traub, 2003). They are each composed of two large subunits (γ, α, δ, or ε, and β1–β4), one medium subunit (µ1–µ4), and one small subunit (σ1–σ4; Boehm and Bonifacino, 2001; Brodsky et al., 2001). The medium subunits recognize YxxØ motifs (Owen and Evans, 1998), and the ear domains of the large subunits interact with accessory proteins and clathrin (Edeling et al., 2006). AP-2 plays a role in clathrin-mediated endocytosis at the plasma membrane, and AP-1, AP-3, and AP-4 facilitate cargo sorting at the TGN or endosomes (Nakatsu and Ohno, 2003).
Epithelial cells co-express AP-1A and AP-1B, which differ only in the incorporation of their respective medium subunits µ1A or the tissue-specific µ1B (Fölsch et al., 1999; Ohno et al., 1999). Despite this close homology, AP-1A and AP-1B are functionally distinct (Fölsch et al., 2003). Whereas AP-1A localizes primarily at the TGN and functions in sorting to the endosomal/lysosomal system, AP-1B is localized in REs and operates in basolateral sorting of cargos with YxxØ motifs, including LDLR (Fölsch, 2005). Curiously, none of LDLR’s basolateral sorting determinants directly interacts with µ1B (Fields et al., 2007). Instead, the distal GYSY signal interacts weakly with µ2 and µ4 (Fields et al., 2007). Accordingly, basolateral sorting of LDLR(Y18A) is not dependent on AP-1B (Fields et al., 2007) but may rely on AP-4 (Simmen et al., 2002). In contrast, LDLR’s proximal FxNPxY motif failed to interact with any adaptor µ chain, though LDLR-CT27 depends on AP-1B for basolateral sorting (see Fig. 5 A; Fields et al., 2007).
During clathrin-mediated endocytosis, the proximal FxNPxY motif of LDLR is recognized by co-adaptors autosomal recessive hypercholesterolemia protein (ARH), Dab2, or numb (Traub, 2009). Whereas both Dab2 and numb incorporate LDLR into nascent AP-2 vesicles through interaction with the α subunit (Traub, 2003), ARH interacts with the platform subdomain of β2, a binding interface that is conserved in β1 of AP-1 (He et al., 2002; Mishra et al., 2005; Keyel et al., 2008). Besides the β1/β2-binding domain, ARH has two additional well-characterized domains: a clathrin box and a phosphotyrosine-binding domain that interacts with FxNPxY motifs. Membrane recruitment of ARH is facilitated by binding to phosphorylated lipids such as phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2) in clathrin-coated pits at the plasma membrane (Mishra et al., 2002). We recently showed that REs in AP-1B–expressing epithelial cells are enriched in phosphatidylinositol 3,4,5-trisphosphate (PI[3,4,5]P3; Fields et al., 2010). Thus, the lipid environment in REs is similar to that found in clathrin-coated pits. Furthermore, ARH was shown to bind to AP-1 in vitro (Mishra et al., 2002). Therefore, we wondered whether ARH was the missing link needed for understanding basolateral exocytosis of LDLR. In this study, we provide strong evidence that ARH cooperates with AP-1B in basolateral exocytosis of LDLR from REs, finally providing a comprehensive mechanism for LDLR sorting in polarized epithelial cells. This study expands ARH’s function and opens up the field for future work aiming at assigning potential roles for other endocytic proteins in REs of polarized cells, perhaps including but not limited to β-arrestin and epsin 1, which like ARH bind to the β1/β2 platform subdomain (Owen et al., 2000; Traub, 2003; Mishra et al., 2005).
Results and discussion
ARH is expressed in epithelial cell lines
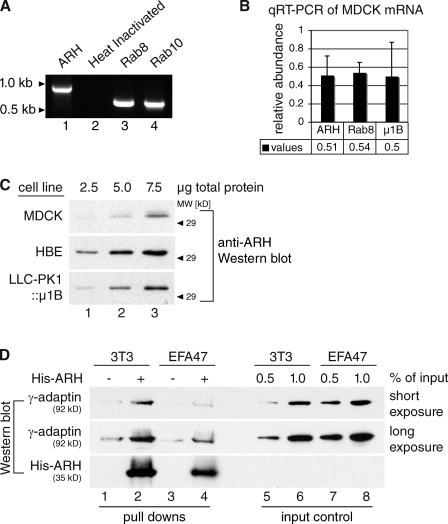
Using an antibody directed against rat ARH, a previous study detected only trace amounts of ARH in the most common model cell lines used to study polarized membrane trafficking, MDCK and LLC-PK1 (Nagai et al., 2003). Here, we reinvestigated the expression of ARH in columnar epithelial cells. First, we determined whether ARH is transcribed in MDCK cells by performing RT-PCR on isolated RNA. Transcripts for Rab8 and Rab10, small GTPases involved in polarized sorting, served as positive controls, and heat inactivation of the RT step served as a negative control. Indeed, ARH transcripts are present in MDCK cells (Fig. 1 A, lane 1). This was confirmed by using a quantitative real-time RT-PCR (qRT-PCR) set up to determine relative mRNA levels of ARH, Rab8, and µ1B normalized to the transcript levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Fig. 2 B).
Figure 1.
ARH is expressed in columnar epithelial cells and pulls down AP-1B. (A) RT-PCR was performed to amplify canine ARH, Rab8, or Rab10. Lane 2 shows a heat inactivation control for ARH amplification. (B) qRT-PCR was performed using one set of primers specific for canine ARH, Rab8, or μ1B. Transcript levels from at least three independent experiments done in triplicate were plotted in relation to a GAPDH control. Error bars indicate SD. (C) MDCK, HBE, and LLC-PK1::μ1B cell lysates were analyzed by SDS-PAGE and Western blotting using anti-ARH antibodies. (D) 3T3 (lanes 1 and 2) or EFA47 (lanes 3 and 4) cell lysates were incubated with purified His-ARH or NiNTA beads alone followed by SDS-PAGE and Western blotting using anti–γ-adaptin (shown are two different exposure times of the same blot) and anti-His antibodies. Lanes 5–8 show input controls for lanes 1–4.
Figure 2.
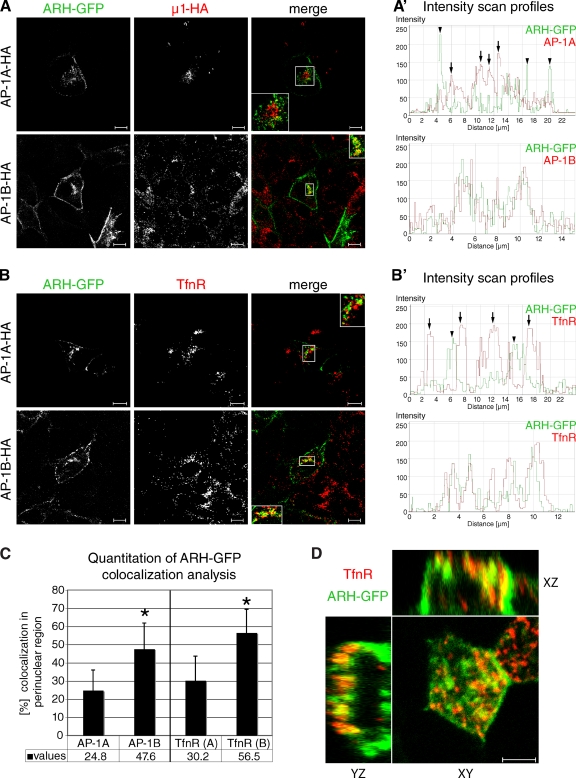
ARH localization in TfnR-positive REs depends on AP-1B. LLC-PK1::μ1A-HA and LLC-PK1::μ1B-HA cells were grown on coverslips and infected with defective adenoviruses encoding ARH-GFP. After 24 h, cells were fixed and stained with anti-HA (A) or anti-TfnR (B) antibodies. Specimens were analyzed by confocal microscopy and representative images are shown. Insets show 2× magnifications of the boxed regions. A′ and B′ show representative fluorescence intensity profiles through a region in the boxed area where noncoincidental peaks are marked by arrowheads (ARH-GFP) or arrows (AP-1A or TfnR). Bars, 10 µm. (C) For quantitation, confocal raw data were analyzed using Volocity software to determine the degree of colocalization between ARH-GFP and AP-1A (n = 64) or AP-1B (n = 71) as well as TfnR in LLC-PK1::µ1A-HA (TfnR[A], n = 54) or LLC-PK1::µ1B-HA cells (TfnR[B], n = 60). Data represent mean values from at least three independent experiments, error bars indicate SD. *, P < 0.0001. (D) Filter-grown MDCK cells were infected with defective adenoviruses encoding ARH-GFP. 24 h later, cells were stained for TfnR. Specimens were analyzed by confocal microscopy and representative xy, xz, and yz images are shown. Bar, 5 µm.
Finally, we investigated ARH protein levels in several columnar epithelial cell lines. Indeed, a polyclonal antibody that was raised against human ARH detected ARH in MDCK cells (see also Cui et al., 2010) as well as in human bronchial epithelial (HBE) cells and LLC-PK1 cells that were stably transfected with µ1B (LLC-PK1::µ1B; Fig. 1 C).
ARH pulls down AP-1B in vitro
Mishra et al. (2002) have found that ARH precipitated AP-1A and AP-2 from brain and liver lysates. To investigate whether ARH might also precipitate AP-1B, we used affinity purified His6-tagged ARH (His-ARH) to pull down AP-1B from EFA47 cell lysates. EFA47 cells are murine µ1A−/− embryonic fibroblasts that express µ1B exogenously so that AP-1B is the only AP-1 complex present (Fölsch et al., 2001; Eskelinen et al., 2002). Lysates from 3T3 fibroblasts served as a positive control for AP-1A.
We found that ARH can pull down AP-1A and AP-1B (Fig. 1 D), and thus could potentially cooperate with both AP-1 complexes in vivo.
ARH localizes in REs of AP-1B–positive epithelial cells
To cooperate, ARH and AP-1B would have to colocalize in REs. To test this, we used LLC-PK1 cells stably expressing HA-tagged µ1B (LLC-PK1::µ1B-HA) or HA-tagged µ1A as a control (LLC-PK1::µ1A-HA; Fölsch et al., 2001). Because LLC-PK1 cells are µ1B negative (Ohno et al., 1999), these two cell lines can also be used to determine differences in localization between µ1B-expressing cells (LLC-PK1::µ1B-HA) and those that do not express µ1B (LLC-PK1::µ1A-HA).
LLC-PK1 cells were grown on coverslips and infected with defective adenoviruses encoding EGFP-tagged ARH (ARH-GFP), resulting in low levels of expression. 24 h after infection, specimens were labeled for AP-1A-HA or AP-1B-HA (Fig. 2 A), or for the RE marker transferrin receptor (TfnR; Fig. 2 B). Specimens were analyzed by confocal microscopy, and the percent overlap between ARH-GFP and AP-1A-HA, AP-1B-HA, or TfnR in the perinuclear region was determined according to Manders et al. (1993) using Volocity software as described in the Material and methods section. First, we analyzed ARH-GFP localization with respect to AP-1A or AP-1B. In addition to the plasma membrane, we detected ARH-GFP on intracellular structures where it partially colocalized with AP-1B-HA (∼50%). In contrast, the partial colocalization of AP-1A-HA with ARH-GFP was much less pronounced (∼25%; Fig. 2, A, A′, and C). Next, we tested ARH-GFP localization in REs by co-staining for TfnR. ARH-GFP and TfnR colocalized to ∼55% in REs of LLC-PK1::µ1B-HA cells. Remarkably, this partial colocalization dropped to ∼30% in LLC-PK1::µ1A-HA cells (Fig. 2, B, B′, and C). This data agrees with our hypothesis that ARH may localize in PI(3,4,5)P3-positive REs, which are exclusively present in AP-1B–expressing epithelial cells (Fields et al., 2010), but not in PI(3,4,5)P3-negative REs. Our data complement and extend previous studies that found ARH localizing in clathrin-coated pits at the plasma membrane of HeLa cells (Mishra et al., 2005), or together with internalized megalin and TfnR in rat embryonic yolk sac L2 cells (Nagai et al., 2003).
ARH localization in REs was confirmed in polarized MDCK cells. We expressed ARH-GFP in filter-grown cells and stained for TfnR. Specimens were analyzed by confocal microscopy and galleries were assembled using Volocity software to build 3D reconstructions of imaged cells. Like in coverslip-grown LLC-PK1::µ1B-HA cells, ARH-GFP localized to the plasma membrane and TfnR-positive REs in polarized MDCK cells. Fig. 2 D shows corresponding xx, xz, and yz slices, the JCB DataViewer shows the individual xy sections, and Video 1 shows a 3D reconstruction of the same cell.
Collectively, our data show that ARH and AP-1B colocalize in TfnR-positive REs, which indicates that they may cooperate in basolateral sorting from this compartment. This preference of ARH for AP-1B in vivo is perhaps a reflection of the different lipid environments of the TGN (phosphatidylinositol 4-phosphate) and REs (PI[3,4,5]P3; Wang et al., 2003; Fields et al., 2010).
ARH knockdown leads to specific apical missorting of LDLR-CT27
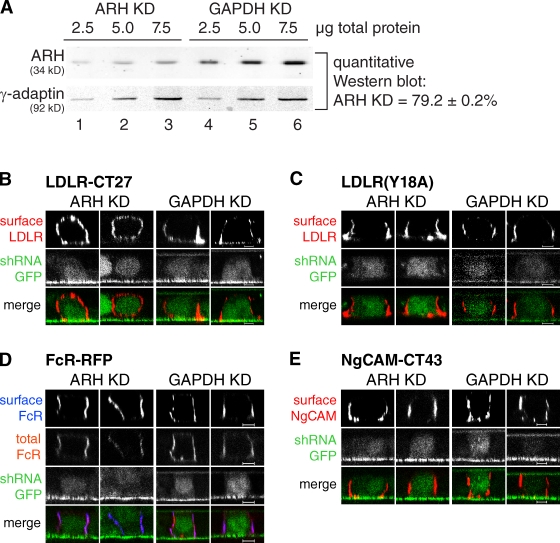
To test whether ARH may function in exocytosis, we next depleted human ARH in HBE cells, which resemble MDCK cells with respect to AP-1B expression and other polarity features (Nokes et al., 2008). To this aim, we used a lentiviral vector that encodes GFP-tagged short hairpin RNA (shRNA) targeting the 3′ untranslated region (UTR) of human ARH to generate HBE cells stably depleted of ARH. Vectors targeting GAPDH were used as controls. ARH knockdown was measured using two different methods. First, we performed quantitative Western blot analysis using the LI-COR Odyssey system. On average, ARH protein levels were reduced by ∼80% in knockdown cells (Fig. 3 A). This was confirmed in individual cells using a confocal microscopy–based assay as detailed in Materials and methods (Fig. S1, A and B).
Figure 3.
Absence of ARH leads to apical missorting of LDLR-CT27. (A) Lysates of HBE cells stably depleted of ARH or GAPDH were analyzed by SDS-PAGE and quantitative Western blotting. Data represent mean values of three independent experiments, each with three different data points representing 2.5, 5, and 7.5 µg total protein. Errors are SD. (B–E) Knockdown cells were grown on filter supports and infected with defective adenoviruses expressing LDLR-CT27 (B), LDLR(Y18A) (C), FcR-RFP (D), or NgCAM-CT43 (E). Receptors at the surface were stained using cargo-specific antibodies. GFP expression indicates shRNA presence. Specimens were analyzed by confocal microscopy and representative images are shown. Data mentioned in the text were from three independent experiments scoring 30 Arf6 and 22 GAPDH knockdown cells for randomized apical and basolateral sorting of LDLR-CT27. Bars, 5 µm.
We infected filter-grown HBE cells stably depleted of ARH or GAPDH with defective adenoviruses encoding various cargo proteins, and analyzed their steady-state localization. First, we examined LDLR-CT27, which has only the proximal FxNPxY sorting motif and thus might be a cargo for ARH during exocytosis. Indeed, upon knockdown of ARH, LDLR-CT27 localization was randomized in 92 ± 8% of the cells analyzed (Fig. 3 B). Importantly, in GAPDH knockdown cells, LDLR-CT27 was sorted correctly to the basolateral membrane in 68.3 ± 3% of the cells analyzed (P < 0.0001). This indicated that the expression of shRNAs in the cells did not lead to nonspecific missorting LDLR-CT27. Next we tested additional cargos with either YxxØ- or LL-based motifs. We found that LDLR(Y18A), which contains only the distal YxxØ sorting signal, was sorted to the basolateral membrane even when ARH levels were reduced (Fig. 3 C). Moreover, Fc receptors (FcRs), which contain LL-based sorting information and are sorted to the basolateral membrane independent of AP-1B (Matter et al., 1994; Roush et al., 1998), were also correctly sorted in the absence of ARH (Fig. 3 D). Finally, we analyzed the truncation mutant neuron–glia cell adhesion molecule (NgCAM)-CT43. NgCAM-CT43 contains a YxxØ sorting motif and requires AP-1B expression for basolateral sorting (Anderson et al., 2005). NgCAM-CT43 was correctly sorted in ARH knockdown cells (Fig. 3 E).
In summary, knockdown of ARH led to specific apical missorting of an AP-1B–dependent cargo with an FxNPxY sorting motif, but not of cargos that contained YxxØ- or LL-based sorting signals. These data support the hypothesis that ARH and AP-1B may cooperate in basolateral sorting of cargos with FxNPxY motifs.
Dominant-negative ARH mutants abolish exocytosis of LDLR-CT27
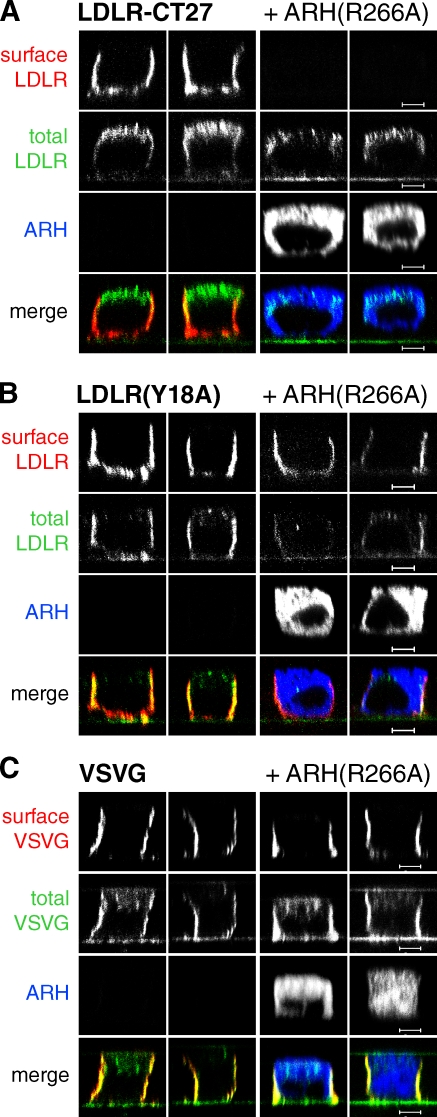
To directly test whether ARH and AP-1B cooperate in basolateral sorting of LDLR-CT27, we generated ARH with a point mutation in the β1/β2-binding site (ARH[R266A]; He et al., 2002). This mutant protein still binds FxNPxY motifs and clathrin; however, the binding to β1/β2 adaptins is abolished (He et al., 2002). Thus, ARH(R266A) can no longer facilitate cargo selection into nascent clathrin-coated vesicles (Mishra et al., 2005). If our model is correct, we would expect that overexpression of ARH(R266A) using a microinjection-based assay should impair surface delivery of LDLR-CT27. We co-injected plasmids expressing ARH(R266A) with plasmids encoding LDLR-CT27 into polarized MDCK cells. Subsequently, cells were incubated at 20°C to block LDLR-CT27 exit from the TGN while ARH(R266A) accumulated in the cytosol followed by a 2-h chase at 37°C in the presence of cycloheximide to inhibit further protein synthesis. During this chase period, LDLR-CT27 will move from the TGN into REs for basolateral sorting along the AP-1B pathway (Fields et al., 2007; Nokes et al., 2008). Receptors at the surface were then stained with antibodies recognizing their ectodomains before fixation and total staining of LDLR-CT27 and ARH(R266A). Under control conditions, LDLR-CT27 was correctly sorted to the basolateral membrane. Correct sorting of LDLR-CT27 was further observed upon coexpression of wild-type ARH-GFP (Fig. S1 C). However, coexpression of ARH(R266A) completely abolished surface delivery in virtually every cell analyzed, and LDLR-CT27 accumulated within the cells (Fig. 4 A and Fig S1 C). In contrast, sorting of LDLR(Y18A), which does not involve movement through REs during biosynthetic delivery (Nokes et al., 2008), was not affected by ARH(R266A) coexpression (Fig. 4 B). Furthermore, basolateral sorting of vesicular stomatitis virus glycoprotein (VSVG) was also not influenced by the presence of ARH(R266A) (Fig. 4 C). Like LDLR-CT27, VSVG moves from the TGN into REs during biosynthetic delivery (i.e., during the chase period of our protocol) to be sorted to the basolateral membrane, dependent on AP-1B (Ang et al., 2004; Nokes et al., 2008). However, unlike LDLR-CT27, VSVG has a YxxØ sorting motif that interacts with µ1B/AP-1B (Fields et al., 2007). Thus, the dominant-negative action of ARH(R266A) did not disrupt global AP-1B–dependent sorting, but was specific for LDLR-CT27.
Figure 4.
Expression of ARH(R266A) specifically interrupts exocytosis of LDLR-CT27. Filter-grown MDCK cells were co-injected with cDNAs encoding V5-tagged ARH(R266A) and LDLR-CT27 (A), LDLR(Y18A) (B), or VSVG (C). Cells were subjected to cell surface staining using cargo-specific antibodies, fixed, and permeabilized to stain for total cargo (A and B) and ARH(R266A)-V5. Specimens were analyzed by confocal microscopy and representative xz sections are shown. Bars, 5 µm.
Conclusions
We propose that during biosynthetic delivery or endocytic recycling, the FxNPxY sorting determinant of LDLR-CT27 is recognized by ARH, which subsequently bridges LDLR-CT27 to AP-1B by interacting with AP-1B’s β1 subunit, thereby facilitating incorporation of LDLR-CT27 into nascent clathrin-coated vesicles (Fig. 5 B). Disruption of ARH binding to β1 may result in the capture of LDLR-CT27; however, incorporation of LDLR-CT27 into clathrin-coated vesicles may be stalled, leading to impaired surface delivery (Fig. 5 C). Successfully formed AP-1B vesicles with or without LDLR cargo are then thought to tether to the basolateral membrane via the exocyst complex followed by membrane fusion facilitated by the SNARE proteins cellubrevin and syntaxin 4 (Fölsch, 2005; Fields et al., 2007).
LDLR’s FxNPxY motif is well conserved in different genes of the LDLR gene family, including the basolaterally localized LDLR-related protein (LRP) and the apically targeted megalin (Chen et al., 1990). LRP contains an FxNPxY motif with basolateral sorting information (Takeda et al., 2003). We propose that LRP is also sorted in REs via ARH and AP-1B. The situation for megalin is more complex. Megalin contains two FxNPxY motifs and an NxxY apical sorting motif (Takeda et al., 2003). Of the two FxNPxY motifs, only the first one interacts with ARH (Nagai et al., 2003), whereas the second one is recognized by Dab2 (Oleinikov et al., 2000). Perhaps the first FxNPxY motif is inactivated in REs, possibly through phosphorylation (Yuseff et al., 2007), or the apical NxxY motif is the dominant signal in REs. Regardless of the exact nature of megalin trafficking, our data suggest that basolateral sorting of cargos with a functional, ARH-interacting FxNPxY motif may require the presence of both ARH and AP-1B.
Materials and methods
DNA constructs and plasmids
To generate ARH with a C-terminal EGFP tag, we first PCR amplified ARH using full-length human ARH (clone ID 5197824; Thermo Fisher Scientific) as template and ARH-FL-N and ARH-FL-C as N- and C-terminal primers (Table S1). The resulting PCR products were cloned as EcoRI–XbaI fragments into the microinjection vector pRKV. In addition, DNA encoding EGFP was inserted as XbaI–HindIII PCR products between the corresponding restriction sites of pRKV, creating DNA encoding EGFP in frame behind ARH. ARH-GFP was subsequently subcloned into the adenovirus shuttle vector pShuttle-CMV. For cloning of His-ARH, we amplified ARH using the primers ARH-FL-N and ARH-FL-C2 (Table S1), and PCR products were cloned as EcoRI–HindIII fragments in frame into the pET28 expression vector, thereby introducing an N-terminal His6 tag.
Full-length T7-tagged human ARH including either short (UTR1) or long (UTR2) sections of its 3′ UTR were cloned from mRNA extracted from HBE cells using TRIzol (Invitrogen) followed by coupled RT-PCR using ARH-T7-N, ARH-UTR1-C, and ARH-UTR2-C primers (Table S1). The T7-ARH PCR products were cloned as SalI–XbaI fragments behind the CMV promoter in pBUDCE4. Subsequently, DNA encoding monomeric RFP was inserted as KpnI–BamHI PCR fragments behind the EF-1α promoter of pBUDCE4.
ARH(R266A) was generated by QuikChange site-directed mutagenesis (Agilent Technologies) using ARH-GFP as a template and corresponding sense and anti-sense primers (see Table S1 for sense primer ARH[R266A]). Subsequently, ARH(R266A) with a C-terminal V5-tag was generated by PCR using ARH(R266A)-GFP as template and ARH-FL-N and ARH-FL-V5-C as primers (Table S1). PCR products were cloned as EcoRI–HindIII fragments into pRKV. All constructs have been sequenced, no errors were found.
Plasmids encoding VSVGts045-GFP, LDLR-CT27, and LDLR(Y18A) have been described previously (Nokes et al., 2008).
RT-PCR and qRT-PCR
To perform RT-PCR and qRT-PCR, MDCK cells were seeded at a 1:1 dilution onto 10-cm plates. 1 d after seeding, total RNA was extracted from 6 × 106 cells from confluent cultures using Nucleospin RNA II (Macherey-Nagel). Subsequently, 1 µg of purified RNA was used for reverse transcription using reverse transcription Superscript III (Invitrogen). Reversed transcribed RNA was then used in conventional RT-PCR or qRT-PCR. Primers for RT-PCR were directed against the N- and C-termini of canine ARH, Rab8, and Rab10 (Table S1, RT primers). qRT-PCR was performed using the MX3000P real-time PCR system (Agilent Technologies) and SYBR green detection. The primer sequences used to amplify same length fragments of canine GAPDH, ARH, Rab8, and μ1B are shown in Table S1 (qRT primers).
Adenoviruses and lentiviruses for gene knockdown
Defective adenoviruses encoding human ARH-GFP were generated as described previously (Fölsch et al., 1999). Adenoviruses encoding FcR-RFP, LDLR-CT27, LDLR(Y18A), and NgCAM-CT43 were as described previously (Anderson et al., 2005; Fields et al., 2007).
The shRNAmir construct in the lentiviral pGIPZ vector targeting human ARH (LDLRAP1, oligo ID V2LHS_114399, short hairpin sequence: 5′-CGCTTGGCACTTTAAAGCATTATAGTGAAGCCACAGATGTATAATGCTTTAAAGTGCCAAGCT-3′) was obtained from Thermo Fisher Scientific. Constructs targeting human GAPDH have been described previously (Nokes et al., 2008). Replication-defective lentiviruses were generated by transfecting HEK293T cells with 30 µg of pGIPZ lentiviral vector encoding each shRNA, 9 µg pMD2G, and 24 µg psPAX2 by calcium phosphate precipitation using standard techniques (Hunziker et al., 1991). Replication-defective lentiviruses were then harvested from transfected HEK293T cells essentially as described previously (Salmon and Trono, 2006).
Antibodies
Polyclonal antibodies recognizing ARH were a gift from L. Traub (University of Pittsburgh, Pittsburgh, PA), and polyclonal anti-His6 antibodies (sc-803) were obtained from Santa Cruz Biotechnology, Inc. Monoclonal anti-HA antibodies (16B12) were from Covance, anti–γ-adaptin antibodies (100/3) were from Sigma-Aldrich, anti-T7 antibodies (69522–3) were from EMD, and anti-V5 antibodies were produced by R. Randall (St. Andrews University, St. Andrews, Scotland, UK) and obtained from R. Lamb (Northwestern University, Evanston, IL).
Hybridomas producing antibodies against TfnR (H68.4), LDLR (C7), FcR (2.4G2), or VSVG (TK1) were as described previously (Fields et al., 2007). Hybridomas developed by V. Lemmon (University of Miami School of Medicine, Miami, FL) to produce antibodies recognizing NgCAM (8D9) were obtained from the Developmental Studies Hybridoma Bank.
Secondary antibodies labeled with Alexa Fluor dyes were obtained from Invitrogen. HRP-conjugated secondary antibodies, Cy5-labeled goat anti–mouse antibodies, and goat anti–rat antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. Cy5-labeled goat anti–mouse IgG2a antibodies were obtained from SouthernBiotech. IRDye 680– and IRDye 800–conjugated secondary antibodies were from LI-COR Biosciences.
Cell culture, preparation of cell lysates, and Western blot analysis
All cells were grown at 37°C in the presence of 5% CO2, and respective media were supplemented with 2 mM l-glutamine and 0.1 mg/ml penicillin/streptomycin (except where otherwise stated). MDCK and HBE cells were maintained in MEM (7% fetal bovine serum). LLC-PK1::μ1B, LLC-PK1::μ1B-HA, and LLC-PK1::μ1A-HA cells were grown in α-MEM (7% fetal bovine serum) containing 1 mg/ml geneticin. EFA47 fibroblasts were grown in DME (10% fetal bovine serum) containing 200 µg/ml hygromycin, HEK293T cells were grown in DME (10% fetal bovine serum), and 3T3 cells were grown in DME (10% fetal bovine serum) containing 4 mM l-glutamine. HBE, HEK293T, and 3T3 cells were grown on coated surfaces. The coating solution contained LHC basal medium with 10 mg/100 ml bovine serum albumin, 3 mg/100 ml bovine collagen I, and 1 mg/100 ml fibronectin.
HBE cell lines stably knocking down either ARH or GAPDH were generated using lentiviruses encoding ARH or GAPDH shRNA constructs and selected with 8 µg/ml puromycin exactly as described previously (Anderson et al., 2005). They were maintained in 8–12 µg/ml puromycin added to the growth medium.
For Western blot analysis of total cell lysates, cells were seeded at a 1:1 dilution onto 10-cm plates and grown for 1 d. Cells were then lysed in 600 µl RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1.0% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, and 1× protease inhibitors [Boehringer Ingelheim]), incubated for 15 min on ice, and subsequently passed five times through a 22.5-gauge needle and 1-ml syringe. Samples were then centrifuged at 15,000 rpm for 25 min at 4°C (microcentrifuge; Eppendorf). Supernatants were transferred to a new tube, and total protein concentrations were quantified using the BCA protein assay (Thermo Fisher Scientific). Protein lysates were analyzed by SDS-PAGE and Western blot analysis using HRP-labeled secondary antibodies and SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
For quantitative Western blot analysis, we used an Odyssey infrared imaging system (Odyssey application software v2.0; LI-COR). Specifically, to determine ARH knockdown, we analyzed cell lysates of HBE cells stably depleted of GAPDH or ARH by SDS-PAGE and Western blotting using anti-ARH primary antibodies and IRDye 680–labeled secondary antibodies. For quantitation of ARH knockdown, ARH expression in cells depleted of GAPDH was set as 100%. Experiments were repeated three times. Each experiment had three data points: 2.5, 5, and 7.5 µg of total protein. First, we determined the percent knockdown of ARH for each data point. We then calculated the mean knockdown for each experiment. Finally, we calculated the mean values of the independent experiments. Errors are SD. Note that the value for γ-adaptin expression in ARH knockdown cells in comparison to GAPDH knockdown cells was 88.8 ± 5%.
Pull-down assay
His-ARH proteins were expressed in BL21 bacteria that were subsequently lysed using a French press. His-ARH was then batch purified using the His•Bind protein purification kit (EMD), and desalted using a PD10 desalting column (GE).
For pull-down experiments, 24 µg of purified His-ARH was immobilized on HisLink protein purification resin (Promega). Subsequently, immobilized His-ARH was incubated with cell lysates (1 mg/ml) generated as follows: 3T3 and EFA47 cells were each grown in 2 × 15-cm plates and washed twice with PBS2+ (PBS [0.2 g/liter KCl, 0.2 g/liter KH2PO4, 8 g/liter NaCl, and 2.17 g/liter Na2HPO4 × 7 H2O] plus 0.1 g/liter CaCl2 and 0.1 g/liter MgCl2 × 6 H2O) on ice, scraped in 1 ml of ice-cold buffer A (20 mM Hepes-KOH, pH 7.6, 320 mM sucrose, 100 mM NaCl, 25 mM imidazole, 0.4% Triton X-100, and 1× protease inhibitor cocktail [Boehringer Ingelheim]), incubated for 10 min on ice, and subsequently passed five times through a 22.5-gauge needle and 1-ml syringe. Samples were then centrifuged at 15,000 rpm for 25 min at 4°C (microcentrifuge; Eppendorf). Supernatants were transferred to a new tube, and protein concentrations were quantified using the BCA protein assay (Thermo Fisher Scientific). Samples were adjusted to a final concentration of 1 mg/ml, and bovine serum albumin was added to a final concentration of 0.1% [wt/vol]. Pull-down reactions were incubated for 4 h at 4°C with gentle rotation. Subsequently, samples were washed three times in buffer A plus 0.1% Triton X-100. Bound proteins were eluted by vigorously shaking for 20 min and boiling in SDS sample buffer. Samples were analyzed by SDS-PAGE and Western blot analysis.
Immunofluorescence assays
For immunofluorescence experiments with coverslip-grown cells, we seeded LLC-PK1 cell lines on Alcian blue–coated coverslips and HBE cells on collagen- and fibronectin-coated coverslips. Cells were grown for 3–4 d.
For experiments with polarized cells, we seeded 4 × 105 cells on 12-mm filter supports (0.4 µm pore size; Corning) and cultured for 3–4 d with changes of the medium in the basolateral chambers daily. For microinjection experiments, clear filter supports were used. The microinjection assay was performed exactly as described previously (Nokes et al., 2008) using a Femtojet (Injectman NI2; Eppendorf) mounted on a microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with a heated stage. Microinjected cells were processed for immunofluorescence microscopy essentially as described previously (Nokes et al., 2008) with the following alterations for LDLR proteins: LDLR proteins at the cell surface were stained for 1 h on ice with anti-LDLR antibodies (C7, IgG2b), fixed in 3% PFA for 15 min at room temperature followed by incubation in PBS2+ for 5 min. Subsequently, cells were incubated for 1 h in a blocking/permeabilization solution (BPS) (2% [wt/vol] bovine serum albumin, 0.4% [wt/vol] saponin in PBS2+) plus 2% (vol/vol) goat serum, then incubated for 1 h with goat anti–mouse IgG2b antibodies labeled with Alexa Fluor 594 in BPS. Cells were then fixed again in 3% PFA for 15 min at room temperature followed by incubation in PBS2+ for 5 min. Subsequently, cells were incubated in BPS plus 2% (vol/vol) goat serum for 1 h, followed by a 1-h incubation with C7 antibodies to stain for LDLR that was not delivered to the surface and anti-V5 antibodies (IgG2a) to detect ARH(R266A)-V5 proteins produced in the cytosol. Subsequently, cells were washed five times with BPS over 30 min. Finally, cells were incubated for 1 h with secondary antibodies (Alexa Fluor 488–labeled anti-IgG2b and Cy5-labeled anti-IgG2a) in BPS, followed by five washes in BPS over 30 min. Cells were mounted in a solution containing 10% (wt/vol) 1,4-diazabicyclo[2.2.2]octane (DABCO) and 50% (wt/vol) glycerol in water.
Infection of cells with defective adenoviruses was performed in growth media without fetal bovine serum by gently rocking cells with the viruses added to the apical surface in a 37°C incubator. After 2 h, media were exchanged with regular growth media and cells were incubated for 24 h before fixation and immunofluorescence staining as described in the previous paragraph. MDCK and HBE cells on filters were infected 2 d after seeding, and LLC-PK1 cells on coverslips were infected 3 d after seeding.
Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Images were acquired at room temperature using a confocal microscope (Microsystem LSM 510 with ConfoCor 3 software) equipped with a C-Apochromat 63×/1.2 NA water immersion objective (all from Carl Zeiss, Inc). The images were processed using Photoshop CS3 (Adobe), and combined using Illustrator (Adobe).
Statistical analysis and quantitative immunofluorescent assays
For the quantitation of shRNA knockdown in individual cells, HBE cells were grown on coverslips. 24 h after seeding, cells were cotransfected with shRNA constructs and pBUDCE4 encoding RFP and T7-ARH from different promoters. Because our knockdown construct targeted the 3′ UTR of ARH, we used two different constructs for this analysis: the first transcribed T7-ARH with the target sequence directly added to its 3′ end (ARHshort), and the second transcribed T7-ARH with its original 3′ UTR (ARHlong; Fig. S1 A). 24 h after transfection, cells were fixed, permeabilized, and stained for T7-ARH. Images of all samples were acquired using identical confocal settings. We then determined the mean pixel intensities using ConfoCor 3 software (Carl Zeiss, Inc.) and used these values to calculate the ratio between T7-ARH and RFP signals. The mean ratio of cells coexpressing a control shRNA construct targeting GAPDH was set as 100% (n = 82 cells for ARHshort and 50 cells for ARHlong), and the values of knockdown for the ARH constructs were determined as percent values of the control. We found that ARH expression was reduced by 90.7 ± 8.9% using ARHshort (n = 88 cells; Fig. S1B) and 77.8 ± 14.5% using ARHlong (n = 56 cells) as target sequences.
Quantitative image analysis determining the colocalization between ARH-GFP and AP-1A-HA, AP-1B-HA, or TfnR was performed on confocal raw data imported into Volocity 5.3 software (PerkinElmer). Backgrounds were corrected in the same manner for all images analyzed of the same set of markers. In cells with low-to-moderate expression levels of ARH-GFP, regions of interest defined by perinuclear AP-1A, AP-1B, or TfnR staining were selected for the calculation of the Mander’s overlap coefficients, which were directly converted into percent values (Manders et al., 1993).
To determine statistical significance, we first calculated the mean value and SD for each experimental condition. We then used the mean values, SD, and n values to calculate P-values in an unpaired student’s t test using GraphPad QuickCalcs (GraphPad Software). Experiments were repeated at least three times.
Online supplemental material
Fig. S1 shows a quantitation of ARH knockdown in HBE cells and correct LDLR-CT27 sorting in polarized MDCK cells in the presence of ARH. Video 1 is a Quicktime interactive movie of a 3D reconstruction of a fully polarized MDCK cell expressing ARH-GFP. ARH-GFP localizes to the plasma membrane and colocalizes with TfnR in the perinuclear region. The individual xy sections used to build the 3D model are shown in the JCB DataViewer. Table S1 shows the primer sequences used for cloning, RT-PCR, and qRT-PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012121/DC1.
Acknowledgments
We thank Dr. Linton Traub for anti-ARH antibodies and Dr. Bettina Winckler (University of Virginia) for comments on the manuscript.
This work was funded by a grant from the National Institutes of Health (GM070736) to H. Fölsch. R.S. Kang was supported by the Cellular and Molecular Basis of Disease Training Program (GM8061).
Footnotes
Abbreviations used in this paper:
- AP
- adaptor protein
- ARH
- autosomal recessive hypercholesterolemia protein
- BPS
- blocking/permeabilization solution
- FcR
- Fc receptor
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- HBE
- human bronchial epithelial
- LDLR
- low-density lipoprotein receptor
- LRP
- LDLR-related protein
- NgCAM
- neuron–glia cell adhesion molecule
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- qRT-PCR
- quantitative real-time RT-PCR
- RE
- recycling endosome
- shRNA
- short hairpin RNA
- TfnR
- transferrin receptor
- UTR
- untranslated region
- VSVG
- vesicular stomatitis virus glycoprotein
References
- Anderson E., Maday S., Sfakianos J., Hull M., Winckler B., Sheff D., Fölsch H., Mellman I. 2005. Transcytosis of NgCAM in epithelial cells reflects differential signal recognition on the endocytic and secretory pathways. J. Cell Biol. 170:595–605 10.1083/jcb.200506051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., Mellman I. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Bonifacino J.S. 2001. Adaptins: the final recount. Mol. Biol. Cell. 12:2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Brodsky F.M., Chen C.Y., Knuehl C., Towler M.C., Wakeham D.E. 2001. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17:517–568 10.1146/annurev.cellbio.17.1.517 [DOI] [PubMed] [Google Scholar]
- Chen W.J., Goldstein J.L., Brown M.S. 1990. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 265:3116–3123 [PubMed] [Google Scholar]
- Cui S., Guerriero C.J., Szalinski C.M., Kinlough C.L., Hughey R.P., Weisz O.A. 2010. OCRL1 function in renal epithelial membrane traffic. Am. J. Physiol. Renal Physiol. 298:F335–F345 10.1152/ajprenal.00453.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling M.A., Smith C., Owen D. 2006. Life of a clathrin coat: insights from clathrin and AP structures. Nat. Rev. Mol. Cell Biol. 7:32–44 10.1038/nrm1786 [DOI] [PubMed] [Google Scholar]
- Eskelinen E.L., Meyer C., Ohno H., von Figura K., Schu P. 2002. The polarized epithelia-specific mu 1B-adaptin complements mu 1A-deficiency in fibroblasts. EMBO Rep. 3:471–477 10.1093/embo-reports/kvf092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields I.C., Shteyn E., Pypaert M., Proux-Gillardeaux V., Kang R.S., Galli T., Fölsch H. 2007. v-SNARE cellubrevin is required for basolateral sorting of AP-1B-dependent cargo in polarized epithelial cells. J. Cell Biol. 177:477–488 10.1083/jcb.200610047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields I.C., King S.M., Shteyn E., Kang R.S., Fölsch H. 2010. Phosphatidylinositol 3,4,5-trisphosphate localization in recycling endosomes is necessary for AP-1B-dependent sorting in polarized epithelial cells. Mol. Biol. Cell. 21:95–105 10.1091/mbc.E09-01-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H. 2005. The building blocks for basolateral vesicles in polarized epithelial cells. Trends Cell Biol. 15:222–228 10.1016/j.tcb.2005.02.006 [DOI] [PubMed] [Google Scholar]
- Fölsch H. 2008. Regulation of membrane trafficking in polarized epithelial cells. Curr. Opin. Cell Biol. 20:208–213 10.1016/j.ceb.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H., Ohno H., Bonifacino J.S., Mellman I. 1999. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 99:189–198 10.1016/S0092-8674(00)81650-5 [DOI] [PubMed] [Google Scholar]
- Fölsch H., Pypaert M., Schu P., Mellman I. 2001. Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J. Cell Biol. 152:595–606 10.1083/jcb.152.3.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H., Pypaert M., Maday S., Pelletier L., Mellman I. 2003. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J. Cell Biol. 163:351–362 10.1083/jcb.200309020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H., Mattila P.E., Weisz O.A. 2009. Taking the scenic route: biosynthetic traffic to the plasma membrane in polarized epithelial cells. Traffic. 10:972–981 10.1111/j.1600-0854.2009.00927.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Gupta S., Yi M., Michaely P., Hobbs H.H., Cohen J.C. 2002. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277:44044–44049 10.1074/jbc.M208539200 [DOI] [PubMed] [Google Scholar]
- Hunziker W., Harter C., Matter K., Mellman I. 1991. Basolateral sorting in MDCK cells requires a distinct cytoplasmic domain determinant. Cell. 66:907–920 10.1016/0092-8674(91)90437-4 [DOI] [PubMed] [Google Scholar]
- Keyel P.A., Thieman J.R., Roth R., Erkan E., Everett E.T., Watkins S.C., Heuser J.E., Traub L.M. 2008. The AP-2 adaptor beta2 appendage scaffolds alternate cargo endocytosis. Mol. Biol. Cell. 19:5309–5326 10.1091/mbc.E08-07-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivisto U.M., Hubbard A.L., Mellman I. 2001. A novel cellular phenotype for familial hypercholesterolemia due to a defect in polarized targeting of LDL receptor. Cell. 105:575–585 10.1016/S0092-8674(01)00371-3 [DOI] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., Aten J.A. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375–382 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Mostov K. 2008. Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20:227–234 10.1016/j.ceb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Matter K., Mellman I. 1994. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr. Opin. Cell Biol. 6:545–554 10.1016/0955-0674(94)90075-2 [DOI] [PubMed] [Google Scholar]
- Matter K., Hunziker W., Mellman I. 1992. Basolateral sorting of LDL receptor in MDCK cells: the cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell. 71:741–753 10.1016/0092-8674(92)90551-M [DOI] [PubMed] [Google Scholar]
- Matter K., Yamamoto E.M., Mellman I. 1994. Structural requirements and sequence motifs for polarized sorting and endocytosis of LDL and Fc receptors in MDCK cells. J. Cell Biol. 126:991–1004 10.1083/jcb.126.4.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Nelson W.J. 2008. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9:833–845 10.1038/nrm2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S.K., Watkins S.C., Traub L.M. 2002. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl. Acad. Sci. USA. 99:16099–16104 10.1073/pnas.252630799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S.K., Keyel P.A., Edeling M.A., Dupin A.L., Owen D.J., Traub L.M. 2005. Functional dissection of an AP-2 beta2 appendage-binding sequence within the autosomal recessive hypercholesterolemia protein. J. Biol. Chem. 280:19270–19280 10.1074/jbc.M501029200 [DOI] [PubMed] [Google Scholar]
- Nagai M., Meerloo T., Takeda T., Farquhar M.G. 2003. The adaptor protein ARH escorts megalin to and through endosomes. Mol. Biol. Cell. 14:4984–4996 10.1091/mbc.E03-06-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Ohno H. 2003. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 28:419–429 10.1247/csf.28.419 [DOI] [PubMed] [Google Scholar]
- Nokes R.L., Fields I.C., Collins R.N., Fölsch H. 2008. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 182:845–853 10.1083/jcb.200802176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Tomemori T., Nakatsu F., Okazaki Y., Aguilar R.C., Foelsch H., Mellman I., Saito T., Shirasawa T., Bonifacino J.S. 1999. Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 449:215–220 10.1016/S0014-5793(99)00432-9 [DOI] [PubMed] [Google Scholar]
- Oleinikov A.V., Zhao J., Makker S.P. 2000. Cytosolic adaptor protein Dab2 is an intracellular ligand of endocytic receptor gp600/megalin. Biochem. J. 347:613–621 10.1042/0264-6021:3470613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Evans P.R. 1998. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 282:1327–1332 10.1126/science.282.5392.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Vallis Y., Pearse B.M., McMahon H.T., Evans P.R. 2000. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 19:4216–4227 10.1093/emboj/19.16.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Müsch A. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6:233–247 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- Roush D.L., Gottardi C.J., Naim H.Y., Roth M.G., Caplan M.J. 1998. Tyrosine-based membrane protein sorting signals are differentially interpreted by polarized Madin-Darby canine kidney and LLC-PK1 epithelial cells. J. Biol. Chem. 273:26862–26869 10.1074/jbc.273.41.26862 [DOI] [PubMed] [Google Scholar]
- Salmon, Trono D. 2006. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. Chapter 4:Unit 4.21 10.1002/0471142301.ns0421s37 [DOI] [PubMed] [Google Scholar]
- Simmen T., Höning S., Icking A., Tikkanen R., Hunziker W. 2002. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat. Cell Biol. 4:154–159 10.1038/ncb745 [DOI] [PubMed] [Google Scholar]
- Takeda T., Yamazaki H., Farquhar M.G. 2003. Identification of an apical sorting determinant in the cytoplasmic tail of megalin. Am. J. Physiol. Cell Physiol. 284:C1105–C1113 [DOI] [PubMed] [Google Scholar]
- Traub L.M. 2003. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J. Cell Biol. 163:203–208 10.1083/jcb.200309175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M. 2009. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10:583–596 10.1038/nrm2751 [DOI] [PubMed] [Google Scholar]
- Wang Y.J., Wang J., Sun H.Q., Martinez M., Sun Y.X., Macia E., Kirchhausen T., Albanesi J.P., Roth M.G., Yin H.L. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 114:299–310 10.1016/S0092-8674(03)00603-2 [DOI] [PubMed] [Google Scholar]
- Yuseff M.I., Farfan P., Bu G., Marzolo M.P. 2007. A cytoplasmic PPPSP motif determines megalin’s phosphorylation and regulates receptor’s recycling and surface expression. Traffic. 8:1215–1230 10.1111/j.1600-0854.2007.00601.x [DOI] [PubMed] [Google Scholar]