This study shows that geminivirus C2/L2 protein interferes with the derubylation of CUL1. Responses regulated by the CUL1-based SCF ubiquitin ligases, and particularly the response to jasmonates, are altered in transgenic Arabidopsis thaliana expressing C2/L2. The capability to selectively interfere with SCF complexes may define a novel and powerful strategy in viral infections.
Abstract
Viruses must create a suitable cell environment and elude defense mechanisms, which likely involves interactions with host proteins and subsequent interference with or usurpation of cellular machinery. Here, we describe a novel strategy used by plant DNA viruses (Geminiviruses) to redirect ubiquitination by interfering with the activity of the CSN (COP9 signalosome) complex. We show that geminiviral C2 protein interacts with CSN5, and its expression in transgenic plants compromises CSN activity on CUL1. Several responses regulated by the CUL1-based SCF ubiquitin E3 ligases (including responses to jasmonates, auxins, gibberellins, ethylene, and abscisic acid) are altered in these plants. Impairment of SCF function is confirmed by stabilization of yellow fluorescent protein–GAI, a substrate of the SCFSLY1. Transcriptomic analysis of these transgenic plants highlights the response to jasmonates as the main SCF-dependent process affected by C2. Exogenous jasmonate treatment of Arabidopsis thaliana plants disrupts geminivirus infection, suggesting that the suppression of the jasmonate response might be crucial for infection. Our findings suggest that C2 affects the activity of SCFs, most likely through interference with the CSN. As SCFs are key regulators of many cellular processes, the capability of viruses to selectively interfere with or hijack the activity of these complexes might define a novel and powerful strategy in viral infections.
INTRODUCTION
Members of the Geminivirus family are plant viruses with circular, single-stranded DNA genomes (Rojas et al., 2005) that infect a wide range of plant species and cause extensive losses in food and fiber crops. Geminiviruses have highly reduced genomes, encoding only six to eight proteins. Due to limiting coding capacity, to successfully accomplish infection, these viruses must rely on both their own multifunctional proteins and the host cell machinery to replicate, move within and between cells, and avoid plant defense mechanisms (Hanley-Bowdoin et al., 2004).
C2 (also known as L2, AC2, AL2, or TrAP, for transcriptional activator protein) is a multifunctional protein encoded by geminiviruses. In viruses belonging to the genus Begomovirus, C2 acts as a transcription factor required for the expression of viral genes needed late in infection (Sunter and Bisaro, 1992) and triggers transactivation of host genes (Trinks et al., 2005) through an indirect mechanism. C2 is also a pathogenicity factor that suppresses host defenses: constitutive expression of truncated C2 from the begomovirus Tomato golden mosaic virus or the related L2 protein from the curtovirus Beet curly top virus (BCTV) in transgenic plants conditions an enhanced susceptibility phenotype (Sunter et al., 2001) that correlates with their ability to interact with and inactivate SNF1-related kinase (Sunter et al., 2001; Hao et al., 2003). C2 and L2 are also gene silencing suppressors of both posttranscriptional gene silencing and transcriptional gene silencing (reviewed in Raja et al., 2010).
Plants are sessile organisms forced to face environmental variations and continuously challenged by potential pathogens. To mount a rapid response, plants extensively rely on proteomic plasticity, which is partially driven by ubiquitination, a highly dynamic posttranslational modification that controls most of the protein degradation events in eukaryotes. According to proteomic and genetic analyses, ubiquitination rivals transcription as the dominant regulatory mechanism in plants (Vierstra, 2009). Ubiquitination occurs through an enzymatic cascade comprising an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase that binds the substrate and thus confers specificity. In plants, the most abundant family of E3 ligases comprises the multisubunit Cullin RING Ligases (CRLs). Among these, the Cullin1-based group, also named SCF (for Skp1/Cullin1/F-box), is the largest and best characterized because of its unveiled roles in many cellular processes, such as hormonal responses (reviewed in Dreher and Callis, 2007; Santner and Estelle, 2009), light signaling (Dieterle et al., 2001; Harmon and Kay, 2003; Marrocco et al., 2006), or floral meristem and organ identity (Kuroda et al., 2002; Wang et al., 2003). SCF complexes are composed of four subunits: Cullin1 (CUL1), SKP1/ASK (S-phase kinase-associated protein), the RING subunit RBX1 (RING box 1), and an F-box substrate binding protein. The Arabidopsis thaliana genome encodes more than 700 predicted F-box proteins, which suggests a high targeting potential (Hua et al., 2011).
The activity of Cullin RING ligases is regulated by a cycle of covalent attachment and removal of a ubiquitin-like protein named RUB (for Related to Ubiquitin; known as Nedd8 in fission yeast and animals) (del Pozo and Estelle, 1999; reviewed in Hotton and Callis, 2008), which is needed for robust CRL activity (Lyapina et al., 2001). One of the regulators of this activity is a conserved protein complex named CSN (COP9 signalosome; reviewed in Wei et al., 2008). The CSN complex is comprised of eight subunits, named CSN1 to CSN8, where CSN5 is the only catalytic subunit described to date. The best-characterized biochemical activity assigned to the CSN is the isopeptidase activity that removes the RUB moiety from the cullin component of the CRL, which is essential for the function of CRLs in vivo. In addition to the CSN holocomplex, several other subcomplexes are formed by a subset of CSN subunits or by CSN5 and other proteins, but the composition and number of these small complexes still remain unclear (Mundt et al., 2002; Oron et al., 2002; Gusmaroli et al., 2004; Fukumoto et al., 2005; Tomoda et al., 2005). Ubiquitination has been shown to contribute to multiple levels of plant defense (reviewed in Dreher and Callis, 2007). Specifically, several lines of evidence suggest that SCF complexes function in plant–virus interactions: (1) SGT1, an essential SKP1-interacting eukaryotic protein, is required for host and nonhost resistance, virus-induced necrosis, and restrain of viral growth of Plantago asiatica mosaic virus and Potato virus X (Komatsu et al., 2010); (2) virus-induced gene silencing of SKP1, SGT1, or the CSN complex compromised N gene–mediated resistance to Tobacco mosaic virus (TMV) in Nicotiana benthamiana (Liu et al., 2002); (3) the F-box protein ACIF is needed for TMV-triggered hypersensitive response in Nicotiana tabacum and affects N gene–mediated responses to TMV (van den Burg et al., 2008).
A large number of both animal and plant viruses have been described to interfere, inhibit, or usurp the ubiquitination machinery in the cell (reviewed in Isaacson and Ploegh, 2009) by encoding their own ubiquitination components (ubiquitin-like proteins, E3 ligases, adaptors, or deubiquitinating enzymes) or redirecting host ubiquitination. In plants, the Polerovirus P0 protein carries an F-box domain that allows its incorporation into an SCF complex to mediate degradation of AGO1, modulating gene silencing (Baumberger et al., 2007; Bortolamiol et al., 2007). The nanovirus Clink protein is also an F-box protein and can bind to both SKP1 and the cell cycle protein pRBR, affecting cell cycle regulation (Aronson et al., 2000).
Ubiquitination controls most of the hormonal responses in plants (reviewed in Dreher and Callis, 2007; Santner and Estelle, 2009). Among them, the jasmonate response, dependent on the SCFCOI1 complex, plays a crucial role in pathogen defenses. Not much information about the role of jasmonates on viral infection is currently available, but recent works revealed jasmonate signaling as an emerging topic in plant–virus interaction research (Vigliocco et al., 2002; Liu et al., 2004; Agudelo-Romero et al., 2008; Ascencio-Ibáñez et al., 2008; Yang et al., 2008; Kovac et al., 2009).
In this article, we demonstrate that geminiviruses, through their C2 protein, interact and interfere with the derubylation activity of the CSN complex. The activity of the CSN over CUL1 seems to be compromised when C2 is present; consequently, processes regulated by SCF complexes are altered. Since SCFs are key regulators of many cellular processes, the capability of geminiviruses to selectively interfere with or hijack the activity of these complexes might represent a powerful strategy in the viral infection. According to our results, one of the main targets of geminiviral inhibition of SCFs might be the suppression of the jasmonate response. This work demonstrates that geminiviruses are capable of interfering with the ubiquitination pathway and jasmonate signaling through a novel mechanism.
RESULTS
TYLCSV C2 Is Required for Full Infection
Several geminiviral proteins are required to accomplish full infection. Among them, C2 from several begomoviruses has been shown to be needed for viral propagation (Etessami et al., 1988; Wartig et al., 1997). To confirm if this is also applicable to Tomato yellow leaf curl Sardinia virus Spain isolate (TYLCSV; accession number L27708), one of the begomoviruses responsible for the Tomato yellow leaf curl disease (TYLCD) in Spain, we constructed a null mutant virus for the C2 gene with a T-C transition in the start codon, hereafter called TYLCSV C2T2C. This mutation also affects the nucleotide sequence of Rep (for Replication-associated protein) viral gene but does not result in an amino acid change. TYLCSV C2T2C was unable to infect tomato, while N. benthamiana plants infected with the mutant developed very mild or no symptoms and the level of viral DNA was severely reduced (see Supplemental Figure 1A online). To confirm that the viral DNA accumulated in plants infected with the mutant does not result from replication of revertants, we extracted DNA from young leaves collected at 28 d after inoculation (DAI) from plants inoculated with the mutant virus (three infected plants). A 625-bp fragment containing the mutation site was PCR amplified and fully sequenced. All analyzed fragments contained the mutation, confirming that the T-C transition is stable in infected plants.
To determine if the mutant is affected in replication, we evaluated the level of viral DNA accumulated in agroinfiltrated leaf patches of N. benthamiana. Total DNA was extracted 7 d after infiltration and hybridized with a TYLCSV probe. TYLCSV C2T2C DNA accumulates to levels comparable to those of the wild-type virus, indicating that the mutant is not impaired in replication (see Supplemental Figure 1B online). These results indicate that C2 from the Spanish isolate of TYLCSV is required for the establishment of a systemic infection but not for viral replication.
C2 Interacts with the Plant CSN5
Protein–protein interactions between viral and host proteins are one of the main mechanisms used by viruses to create a proper environment for the infection. To identify plant proteins interacting with C2, we performed a wide yeast two-hybrid screen using an Arabidopsis cDNA library (F. Hericourt, C. Grevesse, D. Colinet, S. Déret, E.R. Bejarano, and A. Khashoggi, unpublished data). For the screening, a partial clone of C2, named C2-TS1-78, lacking 59 amino acids of the C terminus, was expressed fused to GAL4 DBD. This truncated C2 protein lacks the transcriptional activation domain, since this domain has been previously shown to activate the expression of yeast two-hybrid GAL4 system reporter genes by itself (Hartitz et al., 1999). One of five clones identified in the screening (from 2 × 107 transformants) corresponds to a truncated version of the Arabidopsis CSN5A lacking the 44 N-terminal amino acids (CSN5A44-357).
CSN5A is the only catalytic subunit of the conserved eukaryotic multiprotein complex named CSN described to date. CSN, originally identified through genetic screening as a negative regulator of photomorphogenesis in Arabidopsis, has been subsequently involved in the regulation of a wide variety of signaling and developmental processes in multiple organisms across all eukaryotic kingdoms, and its activity has proven to be essential. In Arabidopsis, transgenic lines expressing dominant-negative versions of CSN5A (Gusmaroli et al., 2004) or mutants partially defective in CSN5 activity (Gusmaroli et al., 2007) display severe pleiotropic developmental defects. On the other hand, complete loss of function of any of the eight CSN subunits results in a lethal phenotype characterized by postembryonic arrest at seedling stage (Gusmaroli et al., 2007). Despite its involvement in the regulation of a plethora of developmental and environmental responses, the major biochemical activity ascribed to date to the CSN is the removal of RUB1 from cullins.
To analyze if the interaction between C2 and CSN5 is conserved throughout the geminivirus family, we assayed the interaction of Arabidopsis CSN5A with C2 homologs from two other geminivirus species: C2 from the begomovirus Tomato yellow leaf curl virus (TYLCV), another causal agent of TYLCD, and L2 from the curtovirus BCTV, which is able to infect Arabidopsis. Hereafter, C2-TS stands for TYLCSV C2, C2-TM stands for TYLCV C2, and L2-BC stands for BCTV L2. Although C2 and L2 share similar roles, since both inhibit RNA silencing and act as pathogenicity factors, they show some functional divergence: C2 also functions as a transcription factor, while apparently L2 does not. Like for TYLCSV C2, partial clones encoding C-terminal truncated TYLCV C2 or BCTV L2, named C2-TM1-78 and L2-BC1-108, respectively, were used for the yeast two-hybrid assays. The C2 protein of both geminivirus species was shown to interact with CSN5A44-357 in a yeast two-hybrid assay (Table 1), indicating that this interaction is conserved among geminiviruses.
Table 1.
Interaction between Geminivirus C2 and the Plant CSN5 in Yeast
| Bait | Prey | Interaction |
| C21-78-TS | CSN5A44-357 | Yes |
| C21-78-TM | CSN5A44-357 | Yes |
| L21-108-BC | CSN5A44-357 | Yes |
| P53 | CSN5A44-357 | No |
| C21-78-TS | CSN5B44-358 | Yes |
| C21-78-TM | CSN5B44-358 | Yes |
| L21-108-BC | CSN5B44-358 | Yes |
| P53 | CSN5B44-358 | No |
| C21-78-TS | SlCSN557-367 | Yes |
| C21-78-TM | SlCSN557-367 | Yes |
| L21-108-BC | SlCSN557-367 | Yes |
| P53 | SlCSN557-367 | No |
| C21-78-TS | AgT | No |
| C21-78-TM | AgT | No |
| L21-108-BC | AgT | No |
C2-TS stands for TYLCSV C2; C2-TM stands for TYLCV C2; L2-BC stands for BCTV L2. CSN5A and CSN5B are from Arabidopsis; SlCSN5 is CSN5 from tomato cultivar Moneymaker. Interaction was indicated by the ability of cells to grow on medium lacking His and Ade and containing 50 mM 3-aminotriazole. P53 stands for the murine p53 protein fused to the GAL4 DBD (pGBKT7-53; Clontech), and AgT stands for the SV40 large T antigen fused to the GAL4 AD fused to the GAL4 (pGADT7-T; Clontech); both are used as negative controls.
Phylogenetic analysis shows that CSN5 is highly conserved among plants (see Supplemental Figure 2 online). In Arabidopsis, unlike any other plant species described so far, there are two different CSN5 subunits, named CSN5A and CSN5B. These subunits display very different abundance and incorporate into distinct CSN complexes (CSNCSN5A and CSNCSN5B) that play unequal roles in the regulation of plant development (Gusmaroli et al., 2004).
To determine if C2 also interacts with CSN5B, we cloned a partial CSN5B clone, equivalent to the CSN5A partial clone isolated in the screening, lacking the 44 N-terminal residues (CSN5B44-358), and found that all three tested C2/L2 proteins are able to interact with Arabidopsis CSN5B44-358 (Table 1). Given that TYLCSV and TYLCV are important pathogens for tomato (Solanum lycopersicum) crops and tomato is a host for BCTV, we also tested the interaction between C2/L2 and tomato CSN5. We cloned the CSN5 cDNA from S. lycopersicum cultivar Moneymaker (AC:FN820438) and generated a partial clone to express a truncated protein similar to the Arabidopsis CSN5A and CSN5B used in the binding assays (residues 57 to 367, SlCSN557-367). There are three amino acid differences between the cloned CSN5, obtained from Moneymaker, and a previously identified CSN5 obtained from the VFNT cultivar (AC:AF175964). Yeast two-hybrid assays demonstrate that all three tested C2 proteins are also able to interact with SlCSN557-367 (Table 1), suggesting that C2/L2-CSN5 interaction is also conserved in other plant species.
It has been previously reported that CSN5 interacts with the GAL4 DNA binding domain (Nordgård et al., 2001); thus, the isolation of CSN5 in a GAL4-based yeast two-hybrid screening should be considered cautiously. However, CSN5 homologs from different organisms have been isolated in this kind of screening in several independent studies (Kameda et al., 2006; Cho et al., 2008; Tanguy et al., 2008), and the interactions have been confirmed by other methods. Curiously, in two out of the three cited works, the isolated clone was partial, lacking at least the 44 N-terminal amino acids. We found that the complete CSN5 proteins from both Arabidopsis (CSN5A and CSN5B) and tomato, fused to the GAL4 AD, strongly interact with any protein fused to the GAL4 DBD or with the empty GAL4 DBD-containing vector. However, this unspecific interaction does not occur when the truncated version of CSN5 is used instead.
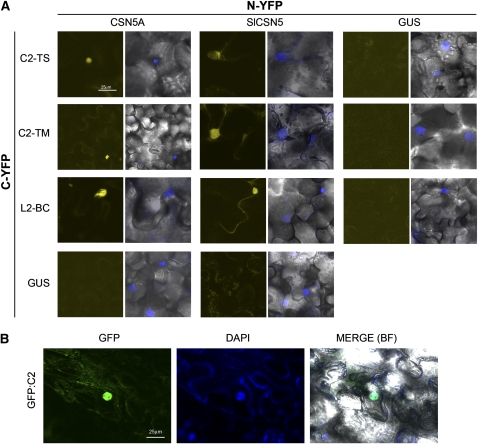
To confirm the C2/L2-CSN5 interaction in planta, we used a bimolecular fluorescence complementation (BiFC) assay. N. benthamiana leaves were coinfiltrated with Agrobacterium tumefaciens cells to express N-terminal fusions of C2 or L2 with N- or C-yellow fluorescent protein (YFP) and N-terminal fusions of Arabidopsis CSN5A or tomato CSN5 with C- or N-YFP. The infiltrated leaves were analyzed under the confocal microscope 3 d after infiltration. YFP fluorescence was observed in cells coinfiltrated with constructs corresponding to NYFP-CSN5A and any of the CYFP-C2/L2 constructs or vice versa. Similar results were obtained when leaves were coinfiltrated with tomato CSN5 and C2 constructs (Figure 1A). By contrast, expression of CSN5 or any of C2/L2 constructs alone (data not shown) or coexpression of any of those constructs with the β-glucuronidase protein (Kertbundit et al., 1991) fused to NYFP or CYFP did not restore the YFP fluorescence (Figure 1). Interactions between C2/L2 and CSN5 seem to occur mainly in the nuclei, as they colocalize with 4',6-diamidino-2-phenylindole (DAPI) staining (Figure 1A). The nuclear localization of C2 from some begomoviruses has been previously reported (van Wezel et al., 2001; Sharma et al., 2010). To confirm that this is also the case for C2-TS, subcellular localization was examined using a green fluorescent protein (GFP)-C2-TS fusion. Three days after agroinfiltration of the construct in N. benthamiana leaves, agroinfiltrated patches were observed under the confocal microscope. GFP-C2-TS localized mainly in the nucleus, as demonstrated by the colocalization with DAPI staining (Figure 1B).
Figure 1.
In Vivo Interaction between Geminivirus C2 and the Plant CSN5.
(A) BiFC analyses showing interaction between geminivirus C2/L2 (C2-TS stands for TYLCSV C2; C2-TM stands for TYLCV C2; L2-BC stands for BCTV L2) and the plant CSN5 (CSN5 stands for Arabidopsis CSN5A; SlCSN5 stands for S. lycopersicum cultivar Moneymaker CSN5). GUS stands for A. thaliana β-glucuronidase, used as a negative control. N. benthamiana leaves coinfiltrated with constructs expressing C2/L2, CSN5, or β-glucuronidase fused to the YFP C terminus (CYFP) or N terminus (NYFP) were observed under the confocal microscope 3 d after infiltration. Leaves were infiltrated with a 4 μg/mL DAPI solution 3 d after infiltration and observed under the confocal microscope 0.5 to 5 h later. No differences were observed between the two pair-wise combinations; only one of the combinations is shown.
(B) Subcellular localization of GFP-C2-TS fusion protein in epidermal cells of N. benthamiana. N. benthamiana leaves infiltrated with a construct expressing a GFP-C2-TS fusion protein were infiltrated with a 4 μg/mL DAPI solution 3 d after infiltration and observed under the confocal microscope 0.5 to 5 h later. GFP-C2-TS is mainly localized into the nucleus. GFP fluorescence, DAPI staining, and merge, including bright-field channel, are shown.
Taken together, these results demonstrate that C2/L2 from TYLCSV, TYLCV, and BCTV associate with CSN5 mainly in the nucleus of plant cells.
Expression of Viral C2/L2 Protein in Transgenic Arabidopsis Lines Specifically Interferes with CUL1 Derubylation without Affecting the Proper Assembly of CSN and SCF Complexes
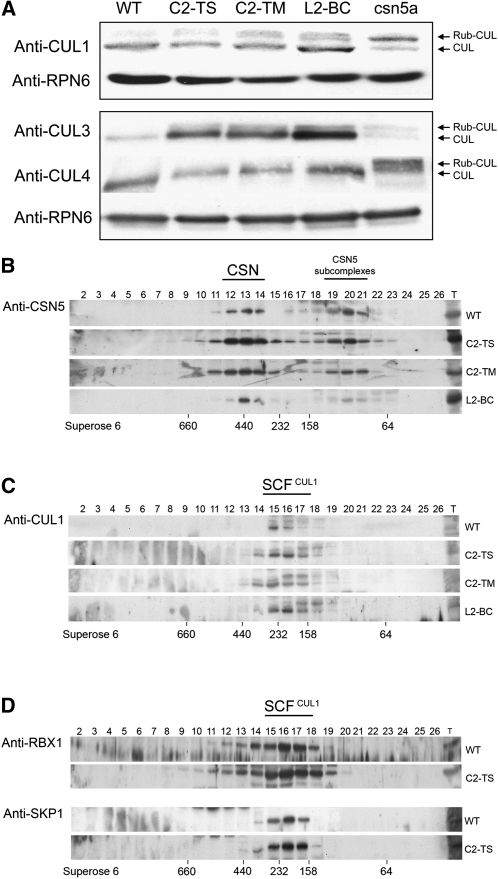
To determine if the interaction of C2/L2 with CSN5 might be affecting the derubylation activity of the CSN complex, we compared the relative levels of rubylated and derubylated cullins between the wild-type and transgenic Arabidopsis plants expressing C2/L2 from TYLCSV, TYLCV, and BCTV (details of these transgenic lines are shown in Supplemental Figure 3 online). None of these C2/L2-expressing transgenic lines displayed noticeable defects in development or morphology. Protein extracts from wild-type, transgenic C2/L2 lines and the csn5a-1 mutant (as a control) were subjected to immunoblot analysis using antibodies against Arabidopsis CUL1, CUL3, and CUL4, the three Arabidopsis cullins known to form CRLs. As shown in Figure 2A, the relative level of rubylated CUL1 observed in all transgenic lines expressing C2/L2 is higher than that of the wild-type plants, whereas we did not observe clear changes in the relative rubylation levels of CUL3 or CUL4 (see Supplemental Figure 4 online). This result suggests that C2/L2 may be hindering the derubylating activity of the CSN complex specifically over CUL1. It is noteworthy that the total cellular levels of CUL3 and CUL1 are slightly increased in the C2/L2 transgenic plants; however, no changes in CUL4 accumulation are detected.
Figure 2.
Inmmunoblot Analysis of 2-Week-Old Wild-Type, csn5a, and Kanamycin-Resistant Transgenic C2-TS, C2-TM, and L2-BC Arabidopsis Seedlings.
(A) Detection of Arabidopsis CUL1, CUL3, and CUL4. Total proteins were subjected to SDS-PAGE and immunoblot analysis with α-CUL1, α-CUL3, and α-CUL4. Equal protein loads were confirmed using α-RPN6 (RPN6 is a non-ATPase regulatory subunit of the 26S proteasome). Rubylated (Rub-CUL) and derubylated Cullins (CUL) are indicated by arrows. WT, wild type.
(B) to (D) Immunoblot analyses of Superose 6 gel filtration fractions. Column fractions were subjected to SDS-PAGE and immunobloted with α-CSN5 (B), α-CUL1 (C), α-RBX1, or α-ASK1 (D). Fraction numbers are indicated. Lane T contains the total unfractionated extracts.
Viral proteins have often been shown to sequester host proteins to co-opt or redirect pivotal cellular machineries to viral function. In this context, it is possible to speculate that C2/L2 might sequester CSN5, preventing its assembly into the complex, or that C2/L2-CSN5 interaction might affect the distribution of CSN5 between the CSN holocomplex and the CSN5-containing subcomplex forms. Based on this idea, a gel filtration experiment was performed in which the fractionation pattern of CSN5 was analyzed (Figure 2B). The comparison of the gel filtration profiles of wild-type plants and transgenic lines expressing C2/L2 demonstrates that CSN5 is normally assembled into both the CSN holocomplex, where it exercises its derubylation activity, and into the subcomplex forms. Keeping in mind that the expression of C2/L2 results in the accumulation of preferentially rubylated CUL1, it is also possible that these viral proteins could alter CUL1 assembly into the SCF complex. However, the analyses of CUL1 fractionation patterns did not reveal any significant changes in the presence of C2/L2 (Figure 2C). These results imply that C2/L2 does not interfere with the proper assembly of CUL1 into the SCF complex. This is in agreement with the observation that RBX1 and SKP1, two other components of the SCF complex, accumulate in the same fractions in one representative C2 transgenic line (Figure 2D). Taken together, these data indicate that C2/L2 does not interfere with the assembly of either the CSN or the SCF complexes and that the observed accumulation of rubylated CUL1 is therefore consistent with a specific interference of C2/L2 with the CSN-mediated CUL1 derubylation.
C2/L2 Arabidopsis Transgenic Plants Share Phenotypes with cul1 Mutants, Including Altered SCF-Dependent Hormonal Responses
Given that C2/L2 transgenic plants display an altered CUL1 rubylated/derubylated ratio, CUL1 function could be impaired in these plants. cul1 mutants are altered in a plethora of developmental processes, such as root growth, skotomorphogenesis, and hormonal responses (Moon et al., 2007; Gilkerson et al., 2009); consequently, it is conceivable that the C2/L2 transgenic plants could also display these defects.
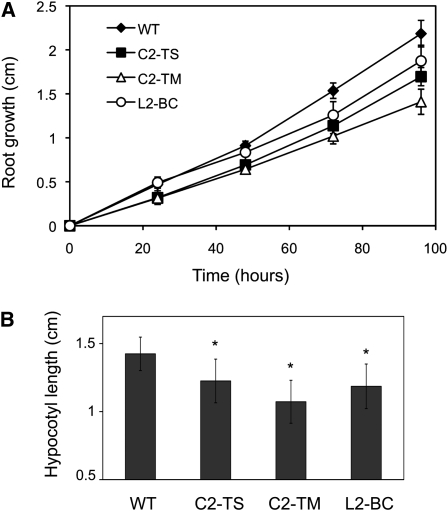
To evaluate root growth rate in the C2/L2 transgenic plants, the root length of 4-d-old seedlings was measured every 24 h for 4 d. Data show that C2/L2 transgenic roots are smaller and grow more slowly than wild-type roots (Figure 3A).
Figure 3.
Root and Hypocotyl Length Analysis of C2/L2 Transgenic Plants.
(A) Total root length of transgenic C2-TS, C2-TM, and L2-BC or wild-type Arabidopsis seedlings (WT) was measured every 24 h beginning 4 d after germination (n ≥ 14). Bars represent se.
(B) Hypocotyl length of 9-d-old dark-grown transgenic C2-TS, C2-TM, and L2-BC or wild-type Arabidopsis seedlings. Bars represent sd. Asterisks indicate a statistically significant difference when compared with the wild-type value according to Mann-Whitney rank sum test. n ≥ 30; the experiment was repeated three times.
Skotomorphogenesis is also altered in transgenic C2/L2 plants: etiolated transgenic seedlings differ from the wild type in hypocotyl length. All three transgenic lines display significantly shorter hypocotyls than the wild type as determined by Mann-Whitney rank sum test (Figure 3B). The reduction in hypocotyl size correlates with the RNA expression level of the transgenes (see Supplemental Figure 5 online).
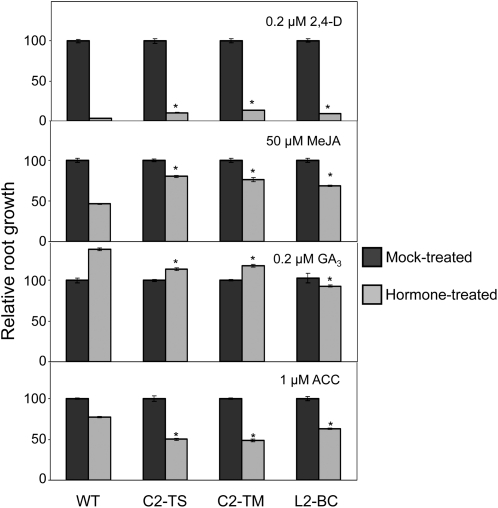
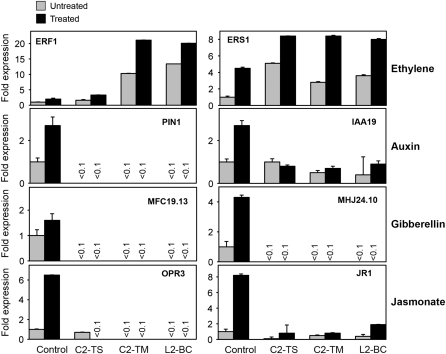
Because SCF complexes play a role in the signaling pathways of several hormones, and most of these responses have been shown to be altered in cul1 mutants (Moon et al., 2007; Gilkerson et al., 2009), we investigated how C2/L2 transgenic lines respond to ethylene, auxins, gibberellins, and jasmonates. In all cases, we measured inhibition of primary root elongation caused by treatment with the exogenous compound (1-aminocyclopropane-1-carboxylic acid [ACC], 2,4-D, gibberellin A3 [GA3], or methyl jasmonate [MeJA]) as a measure of the response to the hormone. The results show that Arabidopsis transgenic plants expressing C2 or L2 were less sensitive to 2,4-D, GA3, and MeJA and more sensitive to ACC (Figure 4). The differential sensitivity of C2/L2 transgenic plants is thus consistent with a malfunction of the corresponding SCF complex in all cases. We tested the differential sensitivity to MeJA in independent transgenic lines expressing different levels of C2/L2 mRNA and found a correlation between lower sensitivity and higher mRNA expression (see Supplemental Figure 6A online). To confirm these results, quantitative real-time PCR was used to quantify the mRNA expression level of marker genes for each of the assayed hormones. We selected ERF1 and ERS1 as marker genes for the ethylene response, PIN1 and IAA19 for the auxin response, MFC19.13 and MHJ24.10 for the gibberellin response, and OPR3 and JR1 for the jasmonate response. In all cases, the expression level of the marker genes correlated with the observed differential sensitivity phenotype (Figure 5). We also tested if the expression of C2-TS alters the sensitivity to hormones in a different plant species: transgenic N. benthamiana plants containing a TYLCSV C2 expression cassette were tested for their sensitivity to auxins and jasmonates. As shown in Supplemental Figure 6B online, transgenic N. benthamiana plants expressing C2-TS are also less sensitive to 2,4-D and MeJA, which demonstrates that C2-mediated lower sensitivity to these hormones is not host specific.
Figure 4.
Reduced Auxin, Jasmonate, and Gibberellin Response and Enhanced Ethylene Response in Transgenic C2/L2 Arabidopsis Lines.
Hormone sensitivity was measured as root growth inhibition. Experiments were repeated at least three times independently; results from one of the replicates are represented (n ≥ 15). Five-day-old seedlings were grown on exogenous hormone for an additional 5 d. Bars represent se. Asterisks indicate a statistically significant difference when compared with the wild-type value according to Mann-Whitney rank sum test. WT, wild type.
Figure 5.
Expression Level of Hormone-Responsive Genes in C2/L2 Transgenic Plants.
Relative expression level of marker genes of the ethylene (ERF1 and ERS2), auxin (PIN1 and IAA19), gibberellin (MFC19.13 and MHJ24.10), and jasmonate (OPR3 and JR1) response in mock- or hormone-treated transgenic C2-TS, C2-TM, and L2-BC and control Arabidopsis seedlings determined by quantitative real-time PCR. C2/L2-expressing lines are compared with the control in each condition. Actin was used as the internal control.
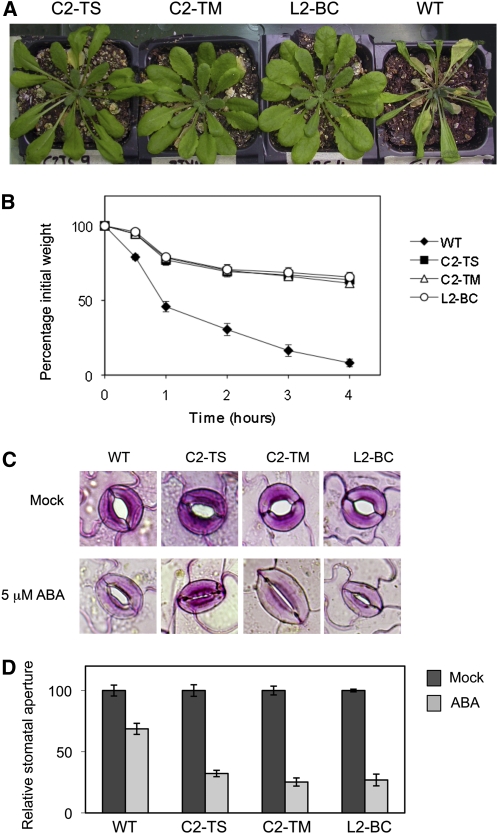
Besides the previously described phenotypes, Arabidopsis C2/L2 transgenic plants are more resistant to drought (Figure 6A). This enhanced tolerance correlated with a slower weight loss in detached leaves (Figure 6B), suggesting that the stomata are more efficiently closed in these plants. Recently, an F-box protein named DOR was described to function as an inhibitor for abscisic acid (ABA)-induced stomatal closure under drought stress, most probably through its activity in a SCFDOR complex (Zhang et al., 2008). The DOR gene is preferentially expressed in the guard cells and affects the stomatal response to ABA: guard cells of the dor mutant are hypersensitive to this hormone. However, other well-characterized responses to ABA, such as the inhibition of seed germination or the reduction of vegetative growth, are not altered in this mutant (Zhang et al., 2008), consistently with the specific expression pattern. Based on our previous findings that C2/L2 seems to be interfering with the SCF complexes, it would be feasible to speculate that a defective SCFDOR activity could result in increased ABA sensitivity in the guard cells, which would in turn explain the observed drought tolerance phenotype. In line with this idea, when we tested the stomatal response to exogenously applied ABA, we found that the stomata in the C2/L2 transgenic plants are indeed more responsive to ABA (Figures 6C and 6D), even though the sensitivity to this hormone is not higher when measured as inhibition of either seed germination or root growth.
Figure 6.
Drought-Related Phenotypes of Transgenic C2-TS, C2-TM, and L2-BC and Wild-Type Arabidopsis Plants.
(A) Phenotype of 7-week-old plants after 10 d without water supply. WT, wild type.
(B) Weight loss in detached leaves. Rosette leaves from 4-week-old plants were detached, placed on weighing dishes, and allowed to dry at room conditions. Weight of the samples was recorded at 0.5, 1, 2, 3, and 4 h. n = 5; bars represent se.
(C) Stomata in epidermal peels after inducing stomatal aperture and treating with 5 μM ABA or mock solution for 1 h.
(D) Stomatal aperture in epidermal peels after treatment with 5 μM ABA or mock solution. The experiments were repeated three times independently; at least 30 stomatal apertures were measured in each condition. Bars represent sd.
[See online article for color version of this figure.]
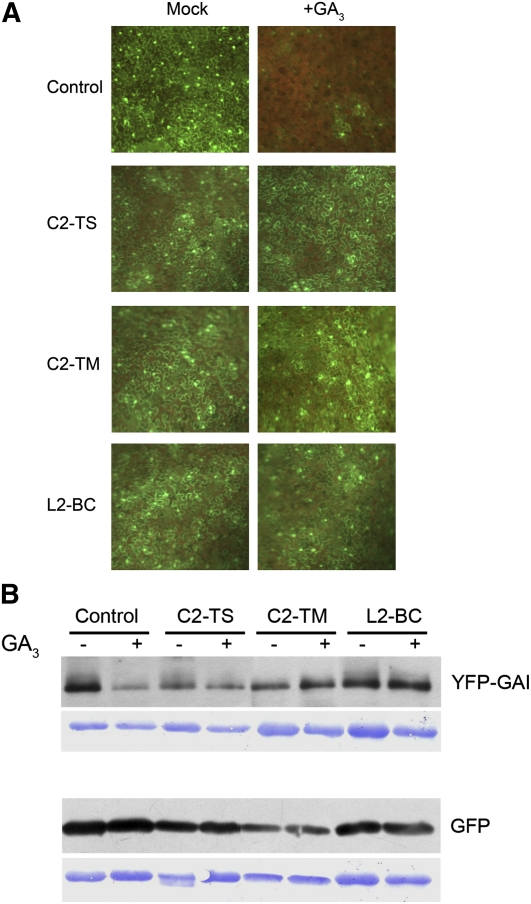
C2/L2 Hinders the Degradation of GAI, Target of the SCFSLY1 E3
If the differential sensitivity to hormones observed in the C2/L2-expressing lines is due to the inhibition of the SCF complexes, the substrates of these complexes must be accumulating in the presence of C2/L2. To check this possibility, we took advantage of a YFP-GAI expression construct (kindly provided by David Alabadí, Instituto de Biología Molecular y Celular de Plantas, Spain). GAI is a DELLA protein that is degraded by the SCFSLY1 in the presence of gibberellins. When the construct expressing YFP-GAI is agroinfiltrated in N. benthamiana leaves, yellow fluorescence can be observed in the nuclei 3 DAI (Figure 7A), indicating the expression and accumulation of the fusion protein. This fluorescence diminishes and eventually disappears when the leaves are treated with 100 μM GA3, since the hormone treatment triggers the ubiquitination of the fusion protein by the SCFSLY1 and its subsequent degradation by the 26S proteasome. As shown in Figure 7A, when we coinfiltrate YFP-GAI and C2/L2 expression construct, the decrease in fluorescence after GA3 treatment is less dramatic, indicating a stabilization of the DELLA protein caused by C2/L2 protein. These results were confirmed by immunoblot analyses using an anti-GFP antibody (Figure 7B). GA3 treatment clearly reduced the amount of YFP-GAI when this fusion protein is agroinfiltrated alone, but no significant differences were observed when it is coinfiltrated with any of the C2/L2 expression constructs. As an internal control, a GFP expression construct was agroinfiltrated alone or coinfiltrated with the C2/L2 expression construct. No differences in fluorescence (data not shown) or GFP protein accumulation were detected between treated and untreated plants (Figure 7B).
Figure 7.
In Vivo Degradation Assay of YFP-GAI Fusion Protein.
(A) The construct expressing YFP-GAI was agroinfiltrated in N. benthamiana leaves alone (control) or coinfiltrated with constructs expressing C2-TS, C2-TM, or L2-BC. Three days after infiltration, agroinfiltrated leaves were sprayed with 100 μM GA3 or mock solution and visualized under the epifluorescence microscope 1 to 2 h later.
(B) Detection of YFP-GAI or GFP (as a control) in N. benthamiana leaves agroinfiltrated with the construct expressing YFP-GAI alone or together with constructs expressing C2-TS, C2-TM, and L2- BC and treated with 100 μM GA3 or mock solution. Total proteins were subjected to SDS-PAGE and immunoblot analysis with α-GFP, which also recognizes YFP. Coomassie blue staining of the protein blot is shown as loading control.
Transcriptomic Analysis Reveals a Clear Suppression of Jasmonate Responses in C2 Plants
To further characterize the global effects on gene expression induced by C2, we performed a transcriptomic analysis of the transgenic Arabidopsis plants expressing TYLCSV C2. Microarray examination reveals 606 genes that were upregulated and 644 that were downregulated in the transgenic plants with a P value below 0.05 compared with control plants. These microarray results were validated by quantitative real-time PCR (see Supplemental Figure 7 online). When we subjected the two groups of genes with altered expression to functional enrichment, we found several biological processes affected by C2-TS, including response to hormone stimulus and defense response (Table 2). As expected, the expression of genes involved in the hormonal responses previously tested is also altered: for example, the gibberellin-responsive GASA4 and GASA5 are downregulated, whereas the ethylene-responsive ATERF4, PDX1L4, and HRE1 are upregulated.
Table 2.
Gene Ontology Analysis of Differentially Expressed Genes
| Nonredundant GO Categories | Level | Differentially Expressed (%) | Expected Frequency | P Value |
| Downregulated Genes | ||||
| Response to stress | 3 | 16.7% (107) | 5.5% | 8.98e-20 |
| Secondary metabolic process | 3 | 6.7% (43) | 1.3% | 1.84e-15 |
| Immune response | 3 | 3.3% (21) | 0.6% | 5.27e-07 |
| Catabolic process | 3 | 6.2% (40) | 2.2% | 9.73e-06 |
| Defense response | 3 | 5.5% (35) | 1.9% | 8.21e-05 |
| Nitrogen compound metabolic process | 3 | 4.7% (30) | 1.6% | 0.00041 |
| Cellular biosynthetic process | 4 | 13.2% (85) | 4.8% | 9.45e-13 |
| Organic acid metabolic process | 4 | 7.3% (47) | 2.2% | 7.80e-09 |
| Aromatic compound metabolic process | 4 | 4.5% (29) | 1.0% | 1.32e-08 |
| Response to wounding | 4 | 2.8% (18) | 0.4% | 2.05e-07 |
| Innate immune response | 4 | 3.1% (20) | 0.6% | 6.74e-07 |
| Heterocycle metabolic process | 4 | 3.1% (20) | 0.7% | 1.89e-05 |
| Pigment metabolic process | 4 | 2.3% (15) | 0.4% | 2.04e-05 |
| Cellular catabolic process | 4 | 5.9% (38) | 2.1% | 2.59e-05 |
| Amine metabolic process | 4 | 4.7% (30) | 1.4% | 3.01e-05 |
| Response to water | 4 | 2.5% (16) | 0.5% | 0.00010 |
| Response to jasmonic acid stimulus | 4 | 2.5% (16) | 0.5% | 0.00031 |
| Sulfur metabolic process | 4 | 1.9% (12) | 0.3% | 0.00167 |
| Carboxylic acid metabolic process | 5 | 7.3% (47) | 2.2% | 7.27e-09 |
| Aromatic compound biosynthetic process | 5 | 3.3% (21) | 0.6% | 1.92e-07 |
| Response to heat | 5 | 2.5% (16) | 0.4% | 7.85e-06 |
| Nitrogen compound biosynthetic process | 5 | 3.0% (19) | 0.6% | 2.71e-05 |
| Response to water deprivation | 5 | 2.5% (16) | 0.5% | 4.14e-05 |
| Response to oxidative stress | 5 | 3.3% (21) | 0.8% | 5.57e-05 |
| Response to cold | 5 | 2.8% (18) | 0.7% | 0.00129 |
| Amino acid derivative biosynthetic process | 6 | 3.6% (23) | 0.6% | 5.91e-09 |
| Defense response, incompatible interaction | 6 | 2.6% (17) | 0.3% | 5.64e-08 |
| Cellular carbohydrate metabolic process | 6 | 4.2% (27) | 1.2% | 4.32e-05 |
| Response to desiccation | 6 | 1.1% (7) | 0.1% | 9.12e-05 |
| Amino acid metabolic process | 6 | 4.0% (26) | 1.2% | 9.23e-05 |
| Biogenic amine metabolic process | 6 | 1.4% (9) | 0.2% | 0.00025 |
| Toxin catabolic process | 6 | 1.1% (7) | 0.1% | 0.00398 |
| Porphyrin catabolic process | 6 | 0.8% (5) | 0.0% | 0.00412 |
| Jasmonic acid and ethylene-dependent systemic resistance | 7 | 1.2% (8) | 0.1% | 0.00010 |
| Indole derivative biosynthetic process | 7 | 1.2% (8) | 0.1% | 0.00010 |
| Phenylpropanoid biosynthetic process | 7 | 2.2% (14) | 0.4% | 0.00015 |
| Amino acid biosynthetic process | 7 | 2.5% (16) | 0.5% | 0.00017 |
| Biogenic amine biosynthetic process | 7 | 1.2% (8) | 0.1% | 0.00076 |
| Indolalkylamine metabolic process | 7 | 0.9% (6) | 0.1% | 0.00439 |
| Flavonoid biosynthetic process | 8 | 1.6% (10) | 0.2% | 0.00029 |
| Indoleacetic acid biosynthetic process | 8 | 0.8% (5) | 0.0% | 0.00054 |
| Tryptophan metabolic process | 8 | 0.9% (6) | 0.1% | 0.00439 |
| Glycosinolate biosynthetic process | 9 | 1.1% (7) | 0.1% | 2.89e-05 |
| Jasmonic acid metabolic process | 9 | 1.1% (7) | 0.1% | 5.27e-05 |
| Oxylipin biosynthetic process | 9 | 1.1% (7) | 0.1% | 5.27e-05 |
| Upregulated genes | ||||
| Response to hormone stimulus | 3 | 5.6% (34) | 2.4% | 0.00738 |
Nonredundant GO categories identified as enriched among down- or upregulated genes in C2-TS expressing Arabidopsis plants versus control plants. GO category levels are indicated. The percentages of genes belonging to each category are reported for the differentially expressed genes and for the genes present in the microarray. The absolute number of differentially expressed genes belonging to each category is reported in parentheses.
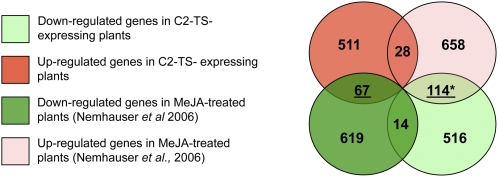
Although the number of up- and downregulated genes in the transgenic C2-TS plants is similar, the nonredundant analysis of GO categories did not reveal any specific hormonal response affected among the upregulated genes. Instead, in the subset of repressed genes, it is especially noticeable the presence of processes related to plant defense and response to jasmonates. Among these downregulated genes, some hallmarks of the jasmonates biosynthetic and perception pathways can be found (see Supplemental Table 3 online). When the list of downregulated genes is compared with that of the upregulated genes in a Columbia (Col-0) plant after MeJA treatment (jasmonate-responsive genes) (Nemhauser et al., 2006), the intersection contains 114 common genes (Figure 8). Gene ontology (GO) analysis of these 114 genes reveals that 32 out of 45 (71%) of the GO categories reported as overrepresented for the whole set of downregulated genes in C2-expressing plants are also overrepresented in this intersectional subset (Table 3). On the other hand, intersections between the set of upregulated genes in C2-expressing plants with either up- or downregulated genes in MeJA-treated plants or between the downregulated genes in C2-expressing plants and upregulated genes in MeJA-treated plants give no significant functional terms exceeding the P value cutoff of 0.01. Given that processes related to jasmonates biosynthesis and perception appear to be clearly repressed by C2, we can infer that interference with the jasmonate pathway might to some degree account for the suppression of the defense response, maybe linking this phenotype to the ability of C2 to hinder the activity of the SCFCOI1. Taking these results together, we conclude that the inhibition of the jasmonates response is the main process affecting downregulation of transcription in the C2-TS–expressing plants.
Figure 8.
Venn Diagrams Depicting the Intersection between Upregulated or Downregulated Genes in C2-TS Transgenic Plants or MeJA-Treated Col-0 Plants (Nemhauser et al., 2006).
The number of genes in each category is indicated. Numbers of genes higher than those expected in a random distribution, with α = 0.05, are underlined. The significance of matches being higher than explained by random sampling was tested by assuming a Poisson distribution from a mean value calculated as μ = G1x (G2/Gt), since any gene in G1 has a G2/Gt possibility to belong to G2, and being G1 and G2 the subsets of genes to be compared and Gt the total number of genes represented in the microarray. An asterisk by the number of genes in an intersection indicates that there are overrepresented GO terms in that subset of genes (Table 2).
[See online article for color version of this figure.]
Table 3.
GO Analysis of the Intersection between C2 Downregulated or Upregulated Genes and the Upregulated Genes in the MeJA Microarray from Nemhauser et al. (2006)
| Downregulated Genes | Level | Differentially Expressed (%) | Expected Frequency | P Value |
| Nonredundant GO Categories | ||||
| Response to stress | 3 | 22.8% (26) | 5.5% | 5.04e-08 |
| Secondary metabolic process | 3 | 8.8% (10) | 1.3% | 0.00024 |
| Immune response | 3 | 6.1% (7) | 0.7% | 0.00140 |
| Catabolic process | 3 | 3.5% (4) | 0.2% | 0.00383 |
| Defense response | 3 | 5.3% (6) | 0.5% | 0.00160 |
| Nitrogen compound metabolic process | 3 | 9.6% (11) | 1.5% | 0.00011 |
| Cellular biosynthetic process | 4 | 19.3% (22) | 7.3% | 0.00263 |
| Organic acid metabolic process | 4 | 18.4% (21) | 2.1% | 4.07e-12 |
| Aromatic compound metabolic process | 4 | 9.6% (11) | 1.0% | 1.36e-06 |
| Response to wounding | 4 | 9.6% (11) | 0.4% | 6.46e-11 |
| Innate immune response | 4 | 6.1% (7) | 0.7% | 0.00140 |
| Heterocycle metabolic process | 4 | 7.9% (9) | 0.6% | 4.53e-06 |
| Amine metabolic process | 4 | 5.3% (6) | 0.1% | 1.64e-06 |
| Response to jasmonic acid stimulus | 4 | 10.5% (12) | 0.5% | 3.64e-11 |
| Carboxylic acid metabolic process | 5 | 18.4% (21) | 2.1% | 3.94e-12 |
| Aromatic compound biosynthetic process | 5 | 5.3% (6) | 0.6% | 0.00577 |
| Nitrogen compound Biosynthetic process | 5 | 7.9% (9) | 0.6% | 2.49e-06 |
| Amino acid derivative biosynthetic process | 6 | 5.3% (6) | 0.6% | 0.00702 |
| Defense response, incompatible interaction | 6 | 5.3% (6) | 0.5% | 0.00160 |
| Amino acid metabolic process | 6 | 8.8% (10) | 1.1% | 5.29e-05 |
| Biogenic amine metabolic process | 6 | 5.3% (6) | 0.1% | 1.64e-06 |
| Toxin catabolic process | 6 | 3.5% (4) | 0.2% | 0.00383 |
| Jasmonic acid and ethylene-dependent systemic resistance | 7 | 5.3% (6) | 0.1% | 2.38e-07 |
| Indole derivative biosynthetic process | 7 | 5.3% (6) | 0.1% | 3.56e-07 |
| Amino acid biosynthetic process | 7 | 7.0% (8) | 0.5% | 7.10e-06 |
| Biogenic amine biosynthetic process | 7 | 4.4% (5) | 0.1% | 3.67e-05 |
| Indolalkylamine metabolic process | 7 | 4.4% (5) | 0.1% | 3.72e-06 |
| Indoleacetic acid biosynthetic process | 8 | 2.6% (3) | 0.0% | 0.00035 |
| Trp metabolic process | 8 | 4.4% (5) | 0.1% | 3.72e-06 |
| Glycosinolate biosynthetic process | 9 | 2.6% (3) | 0.1% | 0.00930 |
| Jasmonic acid metabolic process | 9 | 5.6% (3) | 0.1% | 2.48e-08 |
| Oxylipin biosynthetic process | 9 | 5.6% (3) | 0.1% | 3.35e-08 |
GO category levels are indicated. The percentages of genes belonging to each category are reported for the differentially expressed genes and for the genes present in the microarray. The absolute number of differentially expressed genes belonging to each category is reported in parentheses.
Jasmonate Treatment Reduces the Susceptibility to Geminivirus Infection
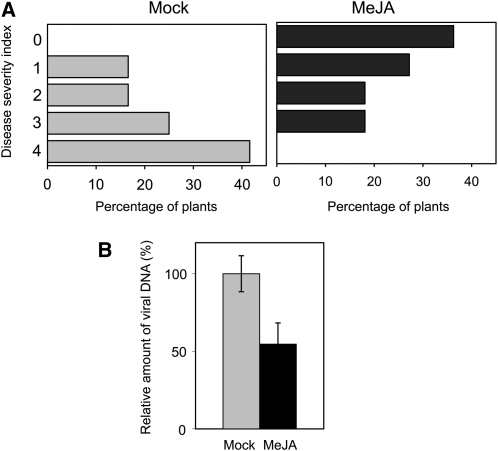
Previous results demonstrated that expression of Tomato golden mosaic virus C2 or BCTV L2 in N. benthamiana plants produced an enhanced susceptibility to DNA and RNA viruses (Sunter et al., 2001), suggesting that C2/L2 proteins have the ability to suppress host stress or defense responses. Since jasmonate signaling has been extensively implicated in defense responses (reviewed in Bari and Jones, 2009), the changes in hormonal sensitivity observed in the C2/L2 transgenic plants could be responsible for this enhanced susceptibility phenotype, suggesting that repression of the jasmonate response could favor viral infection. To determine whether jasmonate response affects geminivirus infection, we inoculated MeJA and mock-treated Arabidopsis plants with BCTV. Total DNA was extracted from these samples and subjected to nucleic acid hybridization with a viral probe. Results from symptom evaluation and viral DNA accumulation are presented in Figure 9. The application of exogenous MeJA results in milder symptoms and lower viral DNA accumulation, indicating a disruption of the geminivirus infection by this compound.
Figure 9.
BCTV Infection of Arabidopsis Plants Treated with MeJA or Mock Solution.
Four- to five-week-old Arabidopsis plants were agroinoculated with BCTV, treated every other day with MeJA or mock solution (12 plants per treatment), and scored for the appearance of symptoms at 28 DAI. Total DNA was extracted from each plant independently and subjected to DNA gel blot to quantify viral DNA accumulation. The experiment was repeated twice; no differences in symptom development or viral DNA accumulation were observed between the replicates. The results of one of the experiments are shown.
(A) Symptom severity at 28 DAI according to the severity index described in Baliji et al. (2007), where 0 represent symptomless plants and 1 to 4 represent plants showing increasing symptom severity.
(B) Relative viral DNA accumulation. Bars represent se.
DISCUSSION
C2, a Protein Required for Virus Infectivity, Interacts with CSN5 and Exerts a Specific Effect on CUL1 Rubylation State
Using TYLCSV C2 as bait protein in a yeast two-hybrid screening, we isolated the Arabidopsis protein CSN5A. Binding assays in yeast and plant confirm C2-CSN5A interaction and demonstrate that this viral protein also binds the tomato ortholog and the Arabidopsis paralog (CSN5B). Interaction experiments using these three CSN5 proteins further demonstrate that they interact with the C2 protein from another begomovirus, TYLCV, and with the homologous protein in the curtovirus BCTV. Taken together, these results suggest that binding to CSN5 is a conserved function of geminivirus C2/L2 protein.
Although interactions with components of the CRLs have been described previously for DNA and RNA viruses, only few examples of interactions with the CSN complex have been reported, all limited to animal viruses (Mahalingam et al., 1998; Oh et al., 2006; Tanaka et al., 2006; Hsieh et al., 2007). Even though, as for C2-CSN5, those interactions seem to play a role during virus infection (Oh et al., 2006; Tanaka et al., 2006; Hsieh et al., 2007), the mechanisms proposed point to a redirection of proteosomal degradation rather than to an effect on the derubylating activity of the CSN complex itself.
Given that the main biochemical activity of the CSN complex is the derubylation of cullins, we checked the rubylated/derubylated ratio of CUL1, CUL3, and CUL4 in transgenic Arabidopsis lines expressing geminivirus C2/L2 and found that these plants contain a higher proportion of rubylated CUL1; nevertheless, CUL3 and CUL4 rubylation ratio is not altered. Although we cannot rule out the possibility that subtle changes in CUL3 to CUL4 ratio cannot be detected by immunoblots, this result suggests that C2/L2-CSN5 interaction specifically inhibits the derubylation activity of CSN over CUL1. In spite of the fact that the precise mechanism conferring specificity to the action of C2 remains elusive, the interaction between several CSN subunits and SCF components in plants (Schwechheimer et al., 2001) raises the possibility that the reduction in CUL1 derubylation could be the result of the specific interference of C2 with the CSN-SCF binding.
Strikingly, even though the CUL3 rubylated/derubylated ratio is not affected by the presence of C2/L2, the transgenic plants expressing these viral genes show a slight increase in the accumulation of CUL3, which seems to be the result of posttranslational regulation, since the level of CUL3 mRNA is reduced in these plants (see Supplemental Figure 8 online). Interestingly, a genetic interaction has been described for CSN5 and CUL3, and CSN5 and CUL3 have been proposed to regulate each other’s abundance in an opposite manner (Gusmaroli et al., 2007). In this context, one would expect that C2/L2 interference with CSN function would trigger the same reduction in CUL3 accumulation. However, C2/L2 expression is instead accompanied by an increase in CUL3 abundance. The fact that CUL1 derubylation is affected in the same transgenic lines allows the intriguing possibility that a defective SCF activity could be responsible for CUL3 accumulation. Alternatively, a tantalizing, nonexclusive hypothesis could be that C2-mediated blocking of the derubylation of CUL1 could increase the number of CSN complexes available to remove RUB from other cullins, thus reducing CUL3 autodegradation. It has been previously proposed that CSN5A reduced activity could trigger the autoubiquitination and degradation of rubylated CUL1 (Stuttmann et al., 2009). However, in spite of a clearly higher rubylated/derubylated CUL1 ratio, we did not observe any destabilization of rubylated CUL1 in either transgenic lines expressing C2/L2 or in csn5a mutants, in agreement with previous data indicating that CUL1 is not destabilized in the same csn5a background (Gusmaroli et al., 2007).
C2 Affects the Activity of Several SCF Complexes
Since C2/L2 expression increases the CUL1 rubylated/derubylated ratio, it is reasonable to expect that CUL1-based SCF functions would be compromised in the presence of C2/L2. Transgenic C2/L2 plants share phenotypes with csn and cul1 mutants, including decreased sensitivity to auxins, gibberellins, and jasmonates and enhanced sensitivity to ethylene and ABA in the guard cells, results that are consistent with a general impairment in the activity of several known SCF complexes (SCFTIR1, SCFSLY1, SCFCOI1, SCFEBF1/2, and SCFDOR) and the subsequent accumulation of their substrates. The stabilization of YFP-GAI, target of the SCFSLY1 complex, produced when C2/L2 is expressed supports this hypothesis.
The observation that the pleiotropic defects of transgenic C2/L2-expressing plants are not as severe as the multifaceted developmental phenotype of cul1 or csn5 mutants could be explained by the fact that C2/L2 does not completely impair derubylation but rather hinders it, so that the downstream changes are more subtle. On the other hand, and given that the CSN and the SCF complexes are essential for cell viability, it might also be feasible that the expression of C2/L2 could be counterselected, and consequently the expression level of the selected transgenic lines would be low. The fact that overexpression of some C2 proteins from a potato virus X–based vector induces severe developmental changes and the subsequent collapse of the plant (A.P. Luna and E.R. Bejarano, unpublished data) is in agreement with this idea. Another possibility would be that the effect of C2 on the SCF complexes could be specific rather than generalized, as suggested by the transcriptomic data. Curiously, Stuttmann et al. (2009) propose that defects in cullin derubylation may be tolerated without causing obvious physiological defects.
C2 Expression Modulates Jasmonate Responses
Although transgenic C2/L2 Arabidopsis plants display multiple phenotypes derived from the interference with the function of the SCF complexes, microarray analysis of C2-TS transgenic plants highlighted the jasmonate response as the main SCF-dependent hormone signaling pathway impaired in these plants. Reduction in responsiveness to jasmonates has been reported for transgenic plants expressing antisense RNA of CSN5 (Schwechheimer et al., 2002), as well as for csn (Feng et al., 2003) and cul1 mutants (Ren et al., 2005; Moon et al., 2007). Jasmonates are important plant signaling molecules that mediate biotic and abiotic stress responses as well as several aspects of growth and development. Plants respond to jasmonates by degrading the JAZ family of transcriptional regulators in a SCFCOI1complex- and a proteasome-dependent manner (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). Therefore, it is likely that C2/L2 may alter the jasmonate response through its effect on CUL1 rubylation. However, and given that C2/L2 is a multifunctional protein, we cannot rule out that the observed phenotype might be driven, partially or completely, by a different mechanism, such as the transcriptional activation activity of this viral protein.
Several lines of evidence indicate that the suppression of the jasmonate response is required for geminivirus infectivity: (1) MeJA treatment of Arabidopsis plants reduces BCTV infection; (2) infection of Arabidopsis with CaLCuV induces repression of the jasmonate response (Ascencio-Ibáñez et al., 2008); (3) the pathogenesis factor βC1 from DNAβ of TYLCCNV can suppress expression of several jasmonate-responsive genes (Yang et al., 2008). Moreover, the fact that both localization of a large number of geminiviruses (e.g., TYLCSV, TYLCV, and BCTV) and jasmonate synthesis occur preferentially in the phloem cells (Stenzel et al., 2003) makes the suppression of the jasmonate response a feasible target during infection. This suppression could have a direct effect in virus movement or replication by leading to several changes in the plant advantageous for the virus, such as the inhibition of the synthesis of secondary metabolites deleterious for viral replication or movement (e.g., phenylpropanoids; (Kandan et al., 2002; Matros and Mock, 2004), or might be aimed at circumventing phloem cell wall in growth development (Amiard et al., 2007).
Additionally, the interference with the jasmonate signaling may have an impact on the viral insect vector. Jasmonates are the hormones mediating plant defense against insects and could therefore be indirectly affecting geminivirus spread. Both TYLCSV and TYLCV are transmitted by the whitefly Bemisia tabaci, and it has been described that whitefly nymphs trigger the expression of jasmonate-responsive genes, which are important in slowing nymphal development (Kempema et al., 2007; Valenzuela-Soto et al., 2010). Through the suppression of the jasmonate response, the virus might be accelerating its vector’s cycle, thus enhancing its own spread. On the other hand, this suppression could also prevent the synthesis of secondary metabolites that could interfere with the interaction between plant and insect (Bleeker et al., 2009).
C2/L2 Might Facilitate Co-Option of the SCF-Mediated Ubiquitination
According to our results, it seems that C2/L2 would be capable of hindering the activity of several SCF complexes in the plant cell, presumably conferring some biological advantage for the viral infection, such as suppression of hormone-mediated plant defense responses. It is an appealing hypothesis, however, that the virus might be not only impairing the function of the SCF complexes, but rather also redirecting them toward certain target proteins whose degradation would be advantageous for the viral infection. The fact that the overexpression of a given F-box protein can circumvent the general malfunction of the SCF complexes (Denti et al., 2006; Stuttmann et al., 2009) raises the idea that geminiviruses could be co-opting the SCF-mediated ubiquitination pathway for their own advantage through the promotion of the expression of selected adaptor subunits. It would be interesting to look for F-box proteins upregulated during geminivirus infection or in heterologous expression of geminiviral proteins to localize possible targets of this theoretical mechanism.
Some plant viruses have been shown to co-opt the SCF machinery by encoding their own viral F-box proteins, which are assembled into plant SCF complexes (Baumberger et al., 2007; Bortolamiol et al., 2007; Lageix et al., 2007), triggering the ubiquitination of plant proteins that interfere with viral infection. Thus, it would be a tempting hypothesis that, to maximize the effect of their encoded F-box proteins, promoting their efficient incorporation in the maximum possible number of SCF complexes, these plant viruses might have developed means to interfere with the assembly/disassembly cycle of SCF complexes, which is based in rubylation and derubylation.
In summary, viruses typically encode a few multifunctional proteins that enable them to redirect the host replication and transcriptional machineries to viral templates, reprogram host cells to provide an environment favorable for the viral infection, and counteract host defenses. The results obtained in this work unveil a powerful strategy used by geminiviruses, which involves the interaction with a hub regulator of protein ubiquitination, a mechanism that could allow the virus to trigger wide changes in the cellular homeostasis. Additional studies will be required to further dissect the molecular mechanisms underlying this strategy and to determine whether this is a generalized tactic for viruses.
METHODS
Microorganisms and General Methods
Manipulations of Escherichia coli and Saccharomyces cerevisiae strains and nucleic acids were performed according to standard methods (Ausubel et al., 1998; Sambrook and Russell, 2001). E. coli strain DH5-α was used for subcloning. All PCR-amplified fragments cloned in this work were fully sequenced. Agrobacterium tumefaciens GV3101 strain was used for the agroinfiltration assays, and LBA4404 was used for plant transformation. S. cerevisiae strain pJ696 (MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacZ), a derivative of PJ69-4A (James et al., 1996), was used for the two-hybrid experiments. Plant DNA gel blots were performed as described by Castillo et al. (2004).
Plant Materials and Growth Conditions
Wild-type Arabidopsis thaliana used in this study is the Col ecotype. Seeds were surface sterilized and sown on Murashige and Skoog (MS) agar plates with 30 g/liter sucrose. Plates were cold treated for 2 to 6 d at 4°C. Seedlings were grown at 20°C under fluorescent white light (fluence rate of 40 to 60 μmol m−2 s−1) with a 16-h-light/8-h-dark photoperiod. For far-red light treatments, seedlings were grown under continuous far-red light (fluence rate of 110 μmol m–2 s–1). For dark-grown seedlings, plates were wrapped in several layers of aluminum foil.
For root growth inhibition assays, MS plates were placed in a vertical orientation for 5 d, and seedlings were then transferred to MS plates containing the tested hormone. Root length was scanned 5 d later using ImageJ software (http://rsb.info.nih.gov/ij). The hormones and concentrations used in the root growth inhibition assays were the following: 2,4-D (Duchefa Biochemie; 0.1 μM), MeJA (Duchefa Biochemie; 50 or 100 μM), GA3 (Duchefa Biochemie; 0.2 μM), and ACC (Sigma-Aldrich A3903; 1 μM).
MeJA treatments for the infection experiments were as follows: a 50 μM MeJA solution or mock solution (containing 50 μM ethanol) were applied to 4-week-old Arabidopsis plants by spray every other day from 1 d before the inoculation to 28 DAI.
For the agroinfiltration experiments, N.benthamiana plants were grown in soil at 22°C in long-day conditions (16-h-light/8-h-dark photoperiod). For the root growth inhibition assays, wild-type and transgenic C2 N. benthamiana seeds were surface sterilized and sown on MS agar plates, and the seedlings were subjected to the corresponding treatments described for Arabidopsis.
The csn5a-1 mutant (SALK_063436 line) was previously described (Gusmaroli et al., 2007).
For the transcriptomic analysis, T2 seedlings were grown on MS with kanamycin for 7 d and then were treated with hormone-containing or mock solutions at the indicated concentrations for the indicated time: 1 μM 2,4D, 1 h; 50 μM MeJA, 10 h; 1 μM GA3, 1 h; 10 μM ACC, 1 h. Three independent replicates were performed. For these analyses, transgenic kanamycin-resistant plants containing with an expression cassette to express the firefly luciferase (LUC) reporter gene (Murray et al., 2002) were used as the control, and all seedlings were selected in kanamycin. Previously, the hormonal responses of the LUC plants were proved to be identical to those of the wild-type in the aforementioned assays.
Drought Tolerance Test
For the drought tolerance test, plants were initially grown on soil under a normal watering regime for 6 to 7 weeks. Watering was then halted and observations were taken after a further 10 d without water supply.
Weight Loss Measurements
For weight loss measurements, rosette leaves from 4-week-old plants were detached, placed on weighing dishes, and allowed to dry at room conditions. Weight of the samples was recorded at 0, 0.5, 1, 2, 3, and 4 h, and the percentage of initial weight was calculated for each point.
Stomatal Aperture Measurements
Rosette leaves from 4- to 5-week-old plants were exposed to white light for 2 h (fluence rate of 40 to 60 μmol m−2 s−1) while submerged in a solution containing 50 mM KCl, 10 μM CaCl2, 0.01% Tween 20, and 10 mM MES-KOH, pH 6.15, to induce stomatal aperture. Subsequently, 5 μM ABA (Sigma-Aldrich A4906) or mock solution was added to the buffer, and the samples were incubated under the same conditions for 1 h. Epidermal peels were stained with toluidine blue and observed under the microscope (TCS NT; Leica). Stomatal aperture was measured using ImageJ software.
Plasmids and Cloning
TYLCSV C2T2C mutant virus was generated by two-sided splicing by overlap extension (Ho et al., 1989). Primers pairs C2T2C-F/Fragment2C2T2C –R and C2T2C-R/ Fragment1C2T2C –F were used in the two initial PCR reactions; subsequent amplification used primers Fragment1C2T2C –F/ Fragment2C2T2C –R. The PCR product was cloned into the EcoRV site of pBluescript SKII+ (Stratagene) to yield pBSSK-TYA14NdeI/NcoI. An NdeI/NcoI fragment containing the wild-type C2 start codon in pGreenTYA14 was replaced by the NdeI/NcoI fragment in pBSSK-TYA14Nde/NcoI to generate pGreenTYA14C2T2C.
For the yeast two-hybrid constructs, cDNA from Arabidopsis and tomato (Solanum lycopersicum) were generated from total RNA extracted from seedlings and leaves respectively. One microgram of total RNA was used for first-strand cDNA synthesis using oligo(dT) primers and SuperScript II reverse transcriptase reagent (Invitrogen) following the manufacturer’s instructions. Arabidopsis CSN5A44-357 and CSN5B44-358 and tomato JAB (CSN5)57-357 were PCR amplified and cloned into pGADT7 vector (Clontech). Full C2/L2 open reading frame (ORF) from TYLCSV (accession number L27708), TYLCV (accession number AF071228), and BCTV (accession number AF379637) were amplified by PCR and cloned into pGBKT7 vector (Clontech).
For the BiFC experiments, cDNA from Arabidopsis and tomato were generated as indicated for the yeast two-hybrid constructs. Arabidopsis CSN5 and tomato JAB ORFs were PCR amplified and cloned into the pENTR/D-TOPO vector. Full C2/L2 ORF from TYLCSV, TYLCV, and BCTV was amplified by PCR and cloned into the pENTR/D-TOPO vector. Cloned ORFs were inserted by LR reaction (Invitrogen) into the binary pBiFP vectors pBiFP2 and pBiFP3 (de Lucas et al., 2008) containing the N- or C-terminal fragments of the eYFP fluorescent protein (NYFP and CYFP). The resulting constructs were used to transform A. tumefaciens GV3101.
For plant transformation and transient expression, PCR fragments containing the C2/L2 full ORF of TYLCSV, TYLCV, and BCTV were blunt-cloned in pBluescript SKII+ (Stratagene) (TYLCV C2 and BCTV L2) or cloned into the HpaI site of pSXSN1 (M.A. Sánchez-Durán, M.B. Dallas, J.T. Ascencio-Ibañez, M.I. Reyes, M. Arroyo-Mateos, J. Ruiz-Albert, L. Hanley-Bowdoin, and E.R. Bejarano, unpublished data) (TYLCSV C2) to yield pC2TM, pL2BC, and pSXC2TS, respectively. Fragments containing TYLCV C2 and BCTV L2 full ORFs were obtained from pC2TM and pL2BC by HpaI/KpnI digestion and subcloned in the HpaI/KpnI sites of pBINX (M.A. Sánchez-Durán, M.B. Dallas, J.T. Ascencio-Ibañez, M.I. Reyes, M. Arroyo-Mateos, J. Ruiz-Albert, L. Hanley-Bowdoin, and E.R. Bejarano, unpublished data) to yield pBINX-C2-TM and pBINX-L2-BC. A fragment comprising an expression cassette containing the TYLCSV C2 full ORF was obtained from pSXC2TS by XbaI digestion and subcloned into the XbaI site of the binary vector pBIN+ (van Engelen et al., 1995) to yield pBIN-C2-TS.
For the subcellular localization study, TYLCSV C2 ORF was fused to GFP at its N terminus, and the fusion protein or GFP alone was cloned under the control of the 35S promoter in a pBINX1 (M.A. Sánchez-Durán et al., unpublished data) to yield pBINX1-GFP-C2 and pBINX1-GFP. GFP ORF was PCR amplified from pSMGFP (Davis and Vierstra, 1998).
Supplemental Table 1 online contains all the oligonucleotides used in this study. Supplemental Table 2 online summarizes the engineering of the plasmids used in this work.
Phylogenetic Analysis
Amino acid plant CSN5 homolog proteins were aligned with ClustalW (http://www.ebi.ac.uk/clustalw/index.html). Afterwards, the alignment was checked and manually adjusted. The resultant aligned sequences (see Supplemental Data Set 1 online) were used to construct a phylogenetic tree (unrooted) using neighbor-joining clustering based on pairwise mean character differences conducted in SEAVIEW (SEAVIEW 4,2,12; http://pbil.univ-lyon1.fr/software/seaview.html; Gouy et al., 2010). Statistical support of branches was assessed by bootstrap analyses using 1000 resamples of the data matrix.
Yeast Two-Hybrid Assay
The yeast strain PJ696, which contains the reporter genes lacZ, HIS3, and ADE2, was used in the two-hybrid assays (Fields and Song, 1989). Assays were performed as described (Castillo et al., 2004). Yeast were cotransformed and selected for bait and prey plasmids, as described Yeast Protocols Handbook (Clontech Laboratories).
BiFC Assays
Different combination of the A. tumefaciens clones expressing the fusion proteins (NYFP-CSN5/CYFP-C2/L2 or NYFP-C2/L2/CYFP-CSN5) were coinfiltrated into the abaxial surface of 2- to 3-week-old N. benthamiana plants as described (Voinnet et al., 2003). The p19 protein of Tomato bushy stunt virus (pBIN61-p19, kindly provided by Olivier Voinnet, Strasbourg, France) was used to suppress gene silencing. A. tumefaciens strains containing the pBiFP constructs or the p19 plasmid were at a D600 ratio of 1:1:1 for infiltration. Fluorescence was visualized in epidermal cell layers of the leaves after 3 d of infiltration using a Leica DMR confocal microscope.
Subcellular Localization
A. tumefaciens GV3101 was transformed with pBIN1-GFP-C2 or pBIN1-GFP plasmids and coinfiltrated with p19 as described above. A. tumefaciens strains containing pBINX1-GFP-C2 or pBIN1-GFP (as a control) and the p19 silencing plasmid were at a D600 ratio of 1:1 for infiltration. Fluorescence was visualized in epidermal cell layers of the leaves after 3 d of infiltration using a Leica DMR fluorescence microscope.
Plant Transformation
Arabidopsis transformation was performed by floral dip (Clough and Bent, 1998) using A. tumefaciens GV3101 containing pBIN-C2-TS, pBINX-C2-TM, or pBINX-L2-BC. Transformants were selected with kanamycin (50 μg/mL). N. benthamiana plants were transformed with pBIN-C2-TS as described by Morilla et al. (2006). Ten independent lines per construct were selected and subjected to expression analysis by RNA gel blot. Unless otherwise indicated, T2 seeds from the lines C2-TS 9, C2-TM 1, and L2-BC 4 were used in this work. Further information about the transgenic lines is provided in Supplemental Figure 3 online.
Transient Expression Assays
For the in vivo degradation assay, the YFP-GAI construct (kindly provided by David Alabadí, Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain) and the C2/L2 expression constructs were used to transform A. tumefaciens GV3101. Three days after infiltration, the agroinfiltrated leaves were sprayed with a 100 μM GA3 solution or with mock solution containing the GA3 solvent (ethanol). Fluorescence was visualized 1 to 2 h later using an epifluorescence Leica microscope MZ FLIII.
Immunoblot and Gel Filtration Assays
Immunoblot and gel filtration analyses of plant extracts were performed as described by Gusmaroli et al. (2004). In all cases in which equal loading was required, the same samples were probed with α-RPN6 to confirm equal loading. For quantitative experiments, multiple exposures were obtained to assure that the film was not saturated. The antibodies used in this study are as follows: α-CUL1 (Wang et al., 2002), α-CUL3 (Figueroa et al., 2005), α-CUL4 (Chen et al., 2006), α-CSN5 (Kwok et al., 1998), α-RBX1 (Schwechheimer et al. 2002), α-SKP1 (Gray et al., 1999), and α-GFP (kindly provided by Olivier Voinnet, Strasbourg, France).
Quantitative Real-Time PCR
Primer pairs for real-time PCR were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3/; see Supplemental Table 1 online). Gene-specific primers were chosen so that the PCR products were 100 to 300 bp. Total RNA was extracted using RNAeasy Plant Mini Kit (Qiagen) and treated on column with Dnase (Qiagen). One microgram of total RNA was used for first-strand cDNA synthesis using oligo(dT) primers and SuperScript II reverse transcriptase reagent (Invitrogen) following the manufacturer’s instructions. For real-time PCR, the reaction mixture consisted of cDNA first-strand template, primer mix (5 μmol each) and SYBR Green Master Mix (Quanta Biosciences) in a total volume of 25 μL. The PCR conditions were as follows: 10 min at 95°C and 40 cycles of 30 s at 95°C and 30 s at 60°C. The reactions were performed using a Rotor-Gene real time cycler (Qiagen). A relative quantification real-time PCR method was used to compare expression of the genes in transgenic versus nontransgenic line (Panchuk et al., 2002). Relative quantification describes the change in expression of the target gene in a test sample relative to calibrator sample. Actin was used as the internal control. The sample of LUC transgenic plants was used as the calibrator, with the expression level of the sample set to 1. Each data point is the mean value from three experimental replicate determinations. Each cDNA sample used is a mixture from three biological replicates at a ratio of 1:1:1.
Transcriptomic Studies
Microarray analysis was performed at the Unité de Recherche en Génomique Végétale (Evry, France) using the CATMA arrays, containing 24,576 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). For each point, three independent biological replicates were produced. For each biological repetition, RNA samples were obtained by pooling RNAs from 8 to 10 plants. Samples were collected on plants at 1.10 to 1.12 developmental growth stages (Boyes et al., 2001), cultivated in MS plus kanamycin. Total RNA was isolated from three replicates of the 35S:LUC and transgenic C2-TS seedlings using TRIzol (Invitrogen) and subsequently cleaned using the RNeasy MinElute cleanup kit (Invitrogen). RNA quantity and quality were assessed with a Nanodrop ND-1000 spectrophotometer (Labtech) and an Agilent 2100 bioanalyzer (Agilent Technologies), respectively. For each comparison and each biological replicate, a dye-swap was performed (i.e., six hybridizations per comparison). The labeling of cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization to the slides, and the scanning were performed as described by Lurin et al. (2004).
For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green), and no background was subtracted. An array-by-array normalization was performed to remove systematic biases. First, spots considered badly formed features were excluded. Then, a global intensity-dependent normalization using the loess procedure (see Yang and Speed, 2002) was performed to correct the dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct effects on each block, as well as print tip, washing, and/or drying effects.
Differential analysis was based on the log ratios averaged on the dye-swap: the technical replicates were averaged to get one log-ratio per biological replicate, and these values were used to perform a paired t test. For each spot, the empirical variance was calculated and then a trimmed variance was calculated from spots, which did not display extreme variance. The spots that were excluded were those with a specific variance/common variance ratio smaller than the α-quantile of a χ2 distribution of two degree of liberty or greater than the 1-α-quantile of a χ2 distribution of two degree of liberty with α equal to 0.0001 (the same order of magnitude as the probe number). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate to keep a strong control of the false positives in a multiple-comparison context. We considered as being differentially expressed the spots with a Bonferroni P value ≤ 0.05. A detailed description of the normalization step and of the variance modeling used in the differential analysis is available in Gagnot et al. (2008).
Geminivirus Infection Assays
Twenty plants were agroinoculated with pGreenTYA14 (binary vector containing a partial dimer of TYLCSV; see Supplemental Table 2 online) or pGreenTYA14C2T2C (the same construct carrying a T-C transition in the start codon of the C2 ORF). For control, five plants were mock inoculated with A. tumefaciens harboring the empty binary vector pGreen-0229 (Hellens et al., 2000). Symptoms were evaluated every week until 42 DAI. Samples were taken at 21 DAI.
BCTV viral infections of Arabidopsis were performed by agroinoculation using wild-type virus (Briddon et al., 1989). Symptoms were evaluated every week until 28 DAI. Samples were taken at 28 DAI.
Viral DNA accumulation was quantified by DNA gel blot hybridization of total plant DNA. Two micrograms of total DNA were used. Membranes were hybridized with TYLCSV or BCTV radiolabeled probes. Viral DNA accumulation was quantified by phosphorimager analyses of DNA gel blots and normalized to genomic DNA.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes used in this article are as follows: AT1G22920 (CSN5A), AT1G71230 (CSN5B), AT4G02570 (CUL1), AT1G26830 (CUL3A), AT1G69670 (CUL3B), AT5G46210 (CUL4), AT3G23240 (ERF1), AT2G40940 (ERS1), AT1G73590 (PIN1), At3g15540 (IAA19), At5g45460 (MFC19.13), At5g64120 (MHJ24.10), At2G06050 (OPR3), and At3G16470 (JR1). Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24475 and at CATdb (http://urgv.evry.inra.fr/CATdb/; Project AU07-12_GeminiSelSup) according to the Minimum Information About a Microarray Experiment standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Infection of N. benthamiana Plants with TYLCSV Wild Type or C2T2C Mutant.
Supplemental Figure 2. Comparison of CSN5 from Several Plant Species.
Supplemental Figure 3. Characterization of the Transgenic Arabidopsis Lines Expressing C2/L2.
Supplemental Figure 4. CUL1 and CUL3 Protein Accumulation in Transgenic Arabidopsis Lines Expressing C2/L2.
Supplemental Figure 5. Hypocotyl Length of 9-d-Old Dark-Grown Transgenic C2-TS, C2-TM, and L2-BC or Wild-Type Arabidopsis Seedlings.
Supplemental Figure 6. Reduced Response to Hormones in Transgenic Arabidopsis and N. benthamiana Plants Expressing C2.
Supplemental Figure 7. Evaluation of the Expression of Two Differentially Expressed Genes in Transgenic C2-TS Arabidopsis Plants for Microarray Validation.
Supplemental Figure 8. Relative Expression of CUL3 in C2-TS, C2-TM, and L2-BC or Control Arabidopsis Seedlings Determined by Quantitative Real-Time PCR.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Table 2. Plasmids Generated in This Work.
Supplemental Table 3. Hallmark Genes of the Jasmonate Response Downregulated in the Microarray of C2-TS Transgenic Plants.
Supplemental Data Set 1. Text File of Alignment Corresponding to Phylogenetic Analysis in Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank David Alabadí for providing the YFP-GAI construct, Salome Prat for the BiFP vectors, and Olivier Voinnet for the anti-GFP antibody and pBIN61-p19. We thank Miguel A. Botella, Vicente Rubio, Jae Hoon Lee, and Ning Wei for helpful suggestions and discussions and Alberto Macho, Mayte Duarte, David Navas, and Marie-Laure Martin-Magniette for technical assistance. This research was supported by a grant from the Spanish Ministerio de Ciencia y Tecnología (AGL2007-66062-C02-02/AGR) and by National Science Foundation Grant MCB-0929100 to X.W.D. R.L.-D. was awarded a Predoctoral Fellowship from the Spanish Ministerio de Educación y Cultura and an EMBO Short-Term Fellowship (ASTF 234-2007). A.P.L was awarded a Predoctoral Fellowship from the Junta de Andalucia.
References
- Agudelo-Romero P., Carbonell P., de la Iglesia F., Carrera J., Rodrigo G., Jaramillo A., Pérez-Amador M.A., Elena S.F. (2008). Changes in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. Virol. J. 5: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard V., Demmig-Adams B., Mueh K.E., Turgeon R., Combs A.F., Adams W.W., III (2007). Role of light and jasmonic acid signaling in regulating foliar phloem cell wall ingrowth development. New Phytol. 173: 722–731 [DOI] [PubMed] [Google Scholar]
- Aronson M.N., Meyer A.D., Györgyey J., Katul L., Vetten H.J., Gronenborn B., Timchenko T. (2000). Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 74: 2967–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F., Brent R., Kingston R., Moore D., Seidman J., Smith J., Struhl K. (1998). Current Protocols in Molecular Biology. (New York: John Wiley & Sons; ). [Google Scholar]
- Baliji S., Sunter J., Sunter G. (2007). Transcriptional analysis of complementary sense genes in spinach curly top virus and functional role of C2 in pathogenesis. Mol. Plant Microbe Interact. 20: 194–206 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J.D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.H., Lie M., Havecker E., Baulcombe D.C. (2007). The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bleeker P.M., Diergaarde P.J., Ament K., Guerra J., Weidner M., Schütz S., de Both M.T., Haring M.A., Schuurink R.C. (2009). The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol. 151: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V. (2007). The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17: 1615–1621 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briddon R.W., Watts J., Markham P.G., Stanley J. (1989). The coat protein of beet curly top virus is essential for infectivity. Virology 172: 628–633 [DOI] [PubMed] [Google Scholar]
- Castillo A.G., Kong L.J., Hanley-Bowdoin L., Bejarano E.R. (2004). Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78: 2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., Zheng N., Deng X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cho J.H., Kim H.B., Kim H.S., Choi S.B. (2008). Identification and characterization of a rice MCM2 homologue required for DNA replication. BMB Rep. 41: 581–586 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Vierstra R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36: 521–528 [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Estelle M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96: 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Denti S., Fernandez-Sanchez M.E., Rogge L., Bianchi E. (2006). The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J. Biol. Chem. 281: 32188–32196 [DOI] [PubMed] [Google Scholar]
- Dieterle M., Zhou Y.C., Schäfer E., Funk M., Kretsch T. (2001). EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etessami P., Callis R., Ellwood S., Stanley J. (1988). Delimitation of essential genes of cassava latent virus DNA 2. Nucleic Acids Res. 16: 4811–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Ma L., Wang X., Xie D., Dinesh-Kumar S.P., Wei N., Deng X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., Feng S., Bostick M., Callis J., Hellmann H., Deng X.W. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto A., Tomoda K., Kubota M., Kato J.Y., Yoneda-Kato N. (2005). Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett. 579: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J., Hu J., Brown J., Jones A., Sun T.P., Callis J. (2009). Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 181: 945–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Gray W.M., del Pozo J.C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W.L., Yang M., Ma H., Estelle M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G., Feng S., Deng X.W. (2004). The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16: 2984–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G., Figueroa P., Serino G., Deng X.W. (2007). Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19: 564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Settlage S.B., Robertson D. (2004). Reprogramming plant gene expression: A prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5: 149–156 [DOI] [PubMed] [Google Scholar]
- Hao L., Wang H., Sunter G., Bisaro D.M. (2003). Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G., Kay S.A. (2003). The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr. Biol. 13: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Hartitz M.D., Sunter G., Bisaro D.M. (1999). The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263: 1–14 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Hotton S.K., Callis J. (2008). Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59: 467–489 [DOI] [PubMed] [Google Scholar]
- Hsieh Y.H., Su I.J., Wang H.C., Tsai J.H., Huang Y.J., Chang W.W., Lai M.D., Lei H.Y., Huang W. (2007). Hepatitis B virus pre-S2 mutant surface antigen induces degradation of cyclin-dependent kinase inhibitor p27Kip1 through c-Jun activation domain-binding protein 1. Mol. Cancer Res. 5: 1063–1072 [DOI] [PubMed] [Google Scholar]
- Hua Z., Zou C., Shiu S.H., Vierstra R.D. (2011). Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson M.K., Ploegh H.L. (2009). Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda K., Fukao M., Kobayashi T., Tsutsuura M., Nagashima M., Yamada Y., Yamashita T., Tohse N. (2006). CSN5/Jab1 inhibits cardiac L-type Ca2+ channel activity through protein-protein interactions. J. Mol. Cell. Cardiol. 40: 562–569 [DOI] [PubMed] [Google Scholar]
- Kandan A., Commare R.R., Nandakumar R., Ramiah M., Raguchander T., Samiyappan R. (2002). Induction of phenylpropanoid metabolism by Pseudomonas fluorescens against tomato spotted wilt virus in tomato. Folia Microbiol. (Praha) 47: 121–129 [DOI] [PubMed] [Google Scholar]
- Kempema L.A., Cui X., Holzer F.M., Walling L.L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143: 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertbundit S., De Greve H., Deboeck F., Van Montagu M., Hernalsteens J.P. (1991). In vivo random beta-glucuronidase gene fusions in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 88: 5212–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K., Hashimoto M., Ozeki J., Yamaji Y., Maejima K., Senshu H., Himeno M., Okano Y., Kagiwada S., Namba S. (2010). Viral-induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol. Plant Microbe Interact. 23: 283–293 [DOI] [PubMed] [Google Scholar]
- Kovac M., Müller A., Milanovic Jarh D., Milavec M., Düchting P., Ravnikar M. (2009). Multiple hormone analysis indicates involvement of jasmonate signalling in the early defence of potato to potato virus YNTN. Biol. Plant 53: 195–199 [Google Scholar]
- Kuroda H., Takahashi N., Shimada H., Seki M., Shinozaki K., Matsui M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Solano R., Tsuge T., Chamovitz D.A., Ecker J.R., Matsui M., Deng X.W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S., Catrice O., Deragon J.M., Gronenborn B., Pélissier T., Ramírez B.C. (2007). The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J. Virol. 81: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2004). Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38: 800–809 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Serino G., Deng X.W., Dinesh-Kumar S.P. (2002). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D.A., Wei N., Shevchenko A., Deshaies R.J. (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Mahalingam S., Ayyavoo V., Patel M., Kieber-Emmons T., Kao G.D., Muschel R.J., Weiner D.B. (1998). HIV-1 Vpr interacts with a human 34-kDa mov34 homologue, a cellular factor linked to the G2/M phase transition of the mammalian cell cycle. Proc. Natl. Acad. Sci. USA 95: 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K., Zhou Y., Bury E., Dieterle M., Funk M., Genschik P., Krenz M., Stolpe T., Kretsch T. (2006). Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45: 423–438 [DOI] [PubMed] [Google Scholar]
- Matros A., Mock H.P. (2004). Ectopic expression of a UDP-glucose:phenylpropanoid glucosyltransferase leads to increased resistance of transgenic tobacco plants against infection with Potato Virus Y. Plant Cell Physiol. 45: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Moon J., Zhao Y., Dai X., Zhang W., Gray W.M., Huq E., Estelle M. (2007). A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 143: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morilla G., Castillo A.G., Preiss W., Jeske H., Bejarano E.R. (2006). A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication. J. Virol. 80: 3624–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt K.E., Liu C., Carr A.M. (2002). Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell 13: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S.L., Thomson C., Chini A., Read N.D., Loake G.J. (2002). Characterization of a novel, defense-related Arabidopsis mutant, cir1, isolated by luciferase imaging. Mol. Plant Microbe Interact. 15: 557–566 [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nordgård O., Dahle O., Andersen T.O., Gabrielsen O.S. (2001). JAB1/CSN5 interacts with the GAL4 DNA binding domain: A note of caution about two-hybrid interactions. Biochimie 83: 969–971 [DOI] [PubMed] [Google Scholar]
- Oh W., Yang M.R., Lee E.W., Park K.M., Pyo S., Yang J.S., Lee H.W., Song J. (2006). Jab1 mediates cytoplasmic localization and degradation of West Nile virus capsid protein. J. Biol. Chem. 281: 30166–30174 [DOI] [PubMed] [Google Scholar]
- Oron E., Mannervik M., Rencus S., Harari-Steinberg O., Neuman-Silberberg S., Segal D., Chamovitz D.A. (2002). COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129: 4399–4409 [DOI] [PubMed] [Google Scholar]
- Panchuk I.I., Volkov R.A., Schöffl F. (2002). Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P., Wolf J.N., Bisaro D.M. (2010). RNA silencing directed against geminiviruses: Post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799: 337–351 [DOI] [PubMed] [Google Scholar]
- Ren C., Pan J., Peng W., Genschik P., Hobbie L., Hellmann H., Estelle M., Gao B., Peng J., Sun C., Xie D. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42: 514–524 [DOI] [PubMed] [Google Scholar]
- Rojas M.R., Hagen C., Lucas W.J., Gilbertson R.L. (2005). Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43: 361–394 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Callis J., Crosby W.L., Lyapina S., Deshaies R.J., Gray W.M., Estelle M., Deng X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292: 1379–1382 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Deng X.W. (2002). Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Ikegami M., Kon T. (2010). Identification of the virulence factors and suppressors of posttranscriptional gene silencing encoded by Ageratum yellow vein virus, a monopartite begomovirus. Virus Res. 149: 19–27 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I., Hause B., Maucher H., Pitzschke A., Miersch O., Ziegler J., Ryan C.A., Wasternack C. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato - Amplification in wound signalling. Plant J. 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stuttmann J., Lechner E., Guérois R., Parker J.E., Nussaume L., Genschik P., Noël L.D. (2009). COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284: 7920–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G., Bisaro D.M. (1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G., Sunter J.L., Bisaro D.M. (2001). Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285: 59–70 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., et al. (2006). The hepatitis B virus X protein enhances AP-1 activation through interaction with Jab1. Oncogene 25: 633–642 [DOI] [PubMed] [Google Scholar]
- Tanguy G., Drévillon L., Arous N., Hasnain A., Hinzpeter A., Fritsch J., Goossens M., Fanen P. (2008). CSN5 binds to misfolded CFTR and promotes its degradation. Biochim. Biophys. Acta 1783: 1189–1199 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tomoda K., Kato J.Y., Tatsumi E., Takahashi T., Matsuo Y., Yoneda-Kato N. (2005). The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood 105: 775–783 [DOI] [PubMed] [Google Scholar]
- Trinks D., Rajeswaran R., Shivaprasad P.V., Akbergenov R., Oakeley E.J., Veluthambi K., Hohn T., Pooggin M.M. (2005). Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79: 2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Soto J.H., Estrada-Hernández M.G., Ibarra-Laclette E., Délano-Frier J.P. (2010). Inoculation of tomato plants (Solanum lycopersicum) with growth-promoting Bacillus subtilis retards whitefly Bemisia tabaci development. Planta 231: 397–410 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Tsitsigiannis D.I., Rowland O., Lo J., Rallapalli G., Maclean D., Takken F.L., Jones J.D. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen F.A., Molthoff J.W., Conner A.J., Nap J.P., Pereira A., Stiekema W.J. (1995). pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4: 288–290 [DOI] [PubMed] [Google Scholar]
- van Wezel R., Liu H., Tien P., Stanley J., Hong Y. (2001). Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol. Plant Microbe Interact. 14: 1125–1128 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vigliocco A., Bonamico B., Alemano S., Miersch O., Abdala G. (2002). Stimulation of jasmonic acid production in Zea mays L. infected by the maize rough dwarf virus-Río Cuarto. Reversion of symptoms by salicylic acid. Biocell 26: 369–374 [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang X., Feng S., Nakayama N., Crosby W.L., Irish V., Deng X.W., Wei N. (2003). The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kang D., Feng S., Serino G., Schwechheimer C., Wei N. (2002). CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: A structure-function study of CSN1 subunit of COP9 signalosome. Mol. Biol. Cell 13: 646–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartig L., Kheyr-Pour A., Noris E., De Kouchkovsky F., Jouanneau F., Gronenborn B., Jupin I. (1997). Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology 228: 132–140 [DOI] [PubMed] [Google Scholar]
- Wei N., Serino G., Deng X.W. (2008). The COP9 signalosome: More than a protease. Trends Biochem. Sci. 33: 592–600 [DOI] [PubMed] [Google Scholar]
- Yang J.Y., Iwasaki M., Machida C., Machida Y., Zhou X., Chua N.H. (2008). betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Speed T. (2002). Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3: 579–588 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu W., Li Z., Deng X.W., Wu W., Xue Y. (2008). F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol. 148: 2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.