This study demonstrates that APO1, originally proposed to be involved in the maturation of proteins with [4Fe-4S] ligands, is instead required for the splicing of several chloroplast introns. It shows further that DUF794, the plant-specific domain of unknown function that makes up the bulk of APO1, is an RNA binding domain harboring zinc binding motifs.
Abstract
Arabidopsis thaliana APO1 is required for the accumulation of the chloroplast photosystem I and NADH dehydrogenase complexes and had been proposed to facilitate the incorporation of [4Fe-4S] clusters into these complexes. The identification of maize (Zea mays) APO1 in coimmunoprecipitates with a protein involved in chloroplast RNA splicing prompted us to investigate a role for APO1 in splicing. We show here that APO1 promotes the splicing of several chloroplast group II introns: in Arabidopsis apo1 mutants, ycf3-intron 2 remains completely unspliced, petD intron splicing is strongly reduced, and the splicing of several other introns is compromised. These splicing defects can account for the loss of photosynthetic complexes in apo1 mutants. Recombinant APO1 from both maize and Arabidopsis binds RNA with high affinity in vitro, demonstrating that DUF794, the domain of unknown function that makes up almost the entirety of APO1, is an RNA binding domain. We provide evidence that DUF794 harbors two motifs that resemble zinc fingers, that these bind zinc, and that they are essential for APO1 function. DUF794 is found in a plant-specific protein family whose members are all predicted to localize to mitochondria or chloroplasts. Thus, DUF794 adds a new example to the repertoire of plant-specific RNA binding domains that emerged as a product of nuclear-organellar coevolution.
INTRODUCTION
The biogenesis of the photosynthetic apparatus in chloroplasts requires a complex interplay between nuclear and chloroplast genes. The multisubunit complexes that mediate photosynthetic electron transport, ATP synthesis, and carbon fixation consist of proteins of cyanobacterial ancestry whose genes are distributed between the nuclear and chloroplast genomes. Additional nuclear genes promote the expression of genes in the chloroplast, coordinate the expression of a nuclear photosynthetic transcriptome, encode enzymes involved in prosthetic group synthesis and attachment, and mediate protein targeting and assembly (reviewed in Cline and Dabney-Smith, 2008; Kleine et al., 2009; Waters and Langdale, 2009; Stern et al., 2010). It can be anticipated that a substantial fraction of the ~3000 nuclear-encoded proteins that localize to chloroplasts contribute to the assembly and regulation of the photosynthetic apparatus. The identification and functional characterization of such proteins is an active area of research.
Phenotype-driven genetic screens have been an important means for identifying nuclear genes that are required for chloroplast biogenesis. This approach has revealed that many such genes were not derived from the chloroplast’s cyanobacterial ancestor, but rather emerged after endosymbiosis. For example, genetic analyses identified 12 nucleus-encoded proteins that are required for the splicing of one or more of the ~18 group II introns in angiosperm chloroplasts, and most of these belong to RNA binding protein families that evolved in eukaryotes (summarized in Kroeger et al., 2009). These proteins function combinatorially to promote the splicing of different intron subsets and are found in large ribonucleoprotein particles that include their cognate intron RNAs (reviewed in Kroeger et al., 2009; Stern et al., 2010).
In this study, we identify another chloroplast splicing factor, APO1, and we show that the plant-specific domain of unknown function that makes up the bulk of APO1 is a previously unrecognized RNA binding domain. APO1 was originally identified in a genetic screen for nonphotosynthetic mutants in Arabidopsis thaliana (Amann et al., 2004). APO1 was proposed to be involved in the maturation of proteins with [4Fe-4S] ligands because apo1 mutants have reduced levels of two complexes harboring [4Fe-4S] clusters: photosystem I (PSI) and the thylakoid NADH dehydrogenase (NDH). APO1 came to our attention when we identified its maize (Zea mays) ortholog in a coimmunoprecipitate with the chloroplast splicing factor CAF1. We show here that Arabidopsis apo1 mutants fail to splice the second intron in the ycf3 pre-mRNA. In addition, apo1 mutants exhibit reduced petD and clpP-int1 splicing and have mild defects in the splicing of the ndhA and ndhB introns. These RNA splicing defects can account for the thylakoid protein deficiencies observed in apo1 mutants, so a direct role for APO1 in [Fe-S] cluster metabolism now seems very unlikely. We show further that recombinant APO1 binds RNA with high affinity in vitro, that DUF794, the domain of unknown function that makes up the entirety of mature APO1, harbors motifs resembling zinc fingers, and that DUF794 binds zinc. Mutagenesis of the putative zinc binding motifs disrupts zinc binding and APO1 function. The rice (Oryza sativa) and Arabidopsis genomes encode four proteins harboring DUF794, all of which are predicted to localize to mitochondria or chloroplasts. Thus, DUF794 represents a new example of a plant-specific RNA binding domain that functions specifically in organellar gene expression.
RESULTS
Previously, we discovered three chloroplast splicing factors (RNC1, WTF1, and WHY1) via a coimmunoprecipitation approach in which antibodies to known chloroplast splicing factors were used for immunoprecipitations from a size-enriched fraction of maize chloroplast stroma, and coprecipitating proteins were identified by mass spectrometry. Reverse-genetic and RNA coimmunoprecipitation analyses then confirmed the roles of these proteins in splicing (Watkins et al., 2007; Prikryl et al., 2008; Kroeger et al., 2009). The maize ortholog of APO1, Zm-APO1, was detected in a coimmunoprecipitate with the splicing factor CAF1 (see Supplemental Figure 1 online), prompting us to consider a role for APO1 in chloroplast group II intron splicing.
CAF1 is required for the splicing of several chloroplast introns and is found in stable complexes with those introns in vivo (Ostheimer et al., 2003; Asakura and Barkan, 2006). Among CAF1’s intron substrates is an intron in the ycf3 gene, which is required for PSI assembly (Boudreau et al., 1997; Ruf et al., 1997). Thus, the coimmunoprecipitation of Zm-APO1 with CAF1 suggested an alternative hypothesis for the role of APO1 in PSI biogenesis: that APO1 promotes ycf3 intron splicing and that it is a defect in Ycf3 expression that underlies the PSI deficiency in apo1 mutants. The results presented below support this view.
APO1 Is Required for ycf3-int2 Splicing
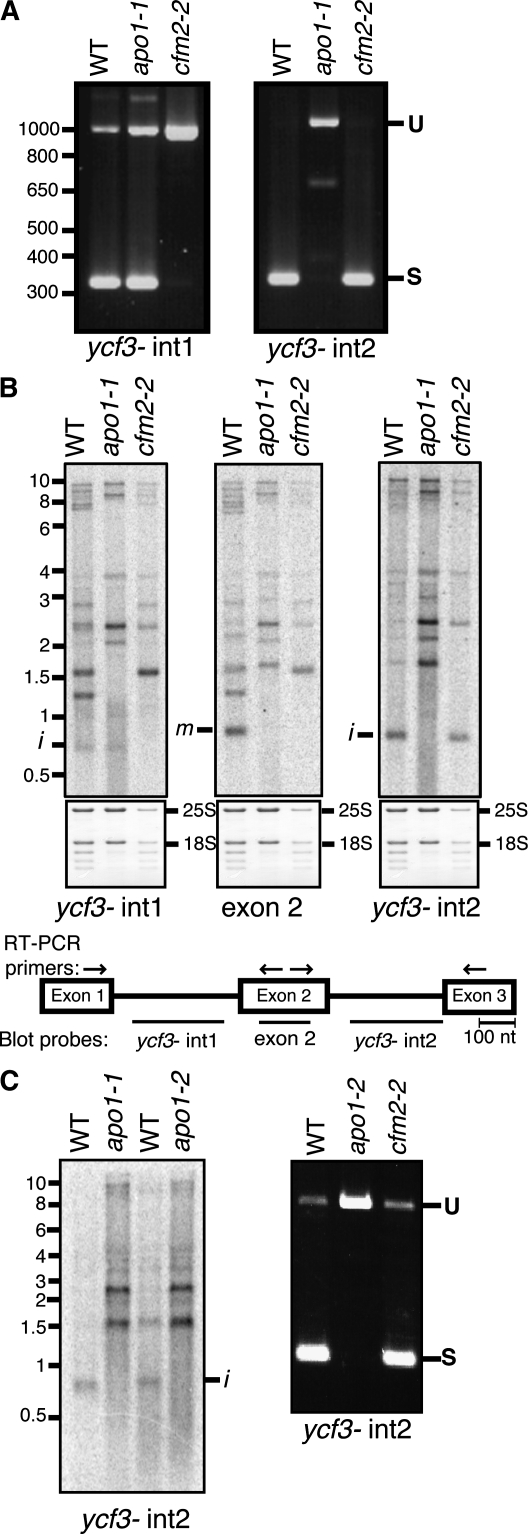
It had been reported previously that APO1 is required for the accumulation of PSI and that it does not influence the abundance or processing of chloroplast mRNAs encoding PSI subunits (Amann et al., 2004). However, the expression of the chloroplast ycf3 gene, which is required for the assembly and accumulation of PSI (Boudreau et al., 1997; Ruf et al., 1997), had not been investigated in apo1 mutants. The ycf3 gene in Arabidopsis, maize, and most other angiosperms includes two group II introns (ycf3-int1 and ycf3-int2); CAF1, with which Zm-APO1 coimmunoprecipitated, is required for the splicing of one of these introns (ycf3-int1). Thus, we hypothesized that APO1 might promote PSI accumulation via a role in ycf3 RNA splicing. To address this possibility, the splicing of both ycf3 introns was examined in apo1 mutants. An Arabidopsis cfm2 mutant, which fails to splice ycf3-int1 (Asakura and Barkan, 2007), was analyzed in parallel. RT-PCR using primers flanking each intron showed that APO1 is indeed required for ycf3 splicing: ycf3-int2 remained completely unspliced in the apo1 mutant, whereas ycf3-int1 appeared to be spliced normally (Figure 1A). The complete absence of the RT-PCR product representing spliced ycf3-int2 indicates a profound defect in splicing.
Figure 1.
APO1 Is Required for ycf3-int2 Splicing.
(A) RT-PCR assay of ycf3 splicing. RT-PCR was performed with total leaf RNA. Primers flanked either ycf3-int1 or ycf3-int2, as shown by the arrows in the map in (B). The predicted sizes of the RT-PCR products representing spliced (S) and unspliced (U) ycf3-int1 are 357 and 1071 bp, respectively. The predicted sizes of the RT-PCR products representing spliced and unspliced ycf3-int2 are 383 and 1170 bp, respectively. WT, wild type.
(B) RNA gel blot assay of ycf3 splicing. The sequences represented in each probe are diagrammed on the map below. Bands corresponding to excised intron (i) and mature spliced RNAs (m) are marked. The same blots were stained with methylene blue to image the rRNAs as a loading control and are shown below.
(C) Analysis of ycf3-int2 splicing in a second mutant APO1 allele. apo1-1 is the allele described by Amann et al. (2004). The apo1-1 and apo1-2 alleles arose in different ecotypes and harbor T-DNA insertions mapping ~20 bp apart near the beginning of the open reading frame. RT-PCR (right) and RNA gel blot (left) assays were performed as in (A) and (B), respectively.
These findings were validated by RNA gel blot hybridizations using intron- and exon-specific probes (Figure 1B). Fully spliced ycf3 mRNA (band m) is absent in the apo1-1 mutant as in the cfm2 mutant. However, the populations of precursor RNAs differ in these mutants and are consistent with the loss of intron 1 and intron 2 splicing in the cfm2 and apo1 mutant, respectively. Furthermore, excised ycf3-int2 accumulates to reduced levels in apo1 mutants (Figure 1B, transcript i detected with the ycf3-int2 probe), demonstrating that the loss of spliced RNA is due to a defect in splicing rather than to instability of the spliced product. The ycf3-int2 splicing defect was observed also in a second mutant allele of APO1 (apo1-2 in Figure 1C), confirming that the splicing defect is due to the disruption of APO1. Given that deletion of ycf3 results in the loss of PSI (Boudreau et al., 1997; Ruf et al., 1997; Landau et al., 2009), this splicing defect is sufficient to account for the absence of PSI in apo1 mutants.
APO1 Enhances the Splicing of Several Additional Chloroplast Introns
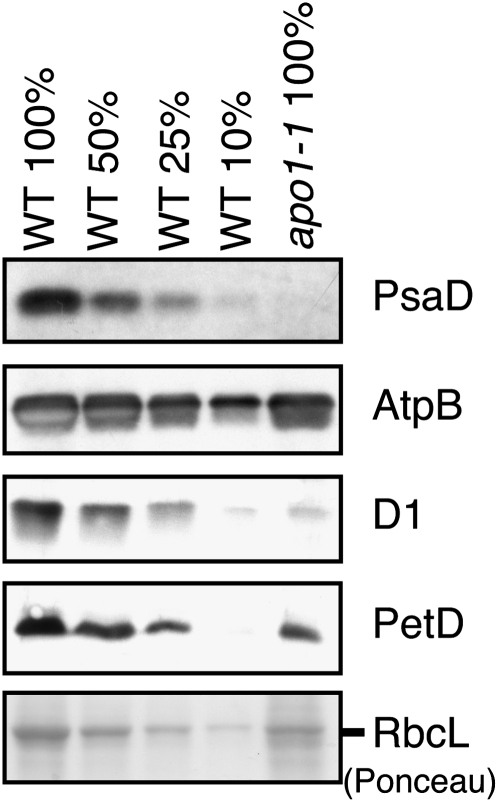
Group II introns are found in several chloroplast genes encoding components of the photosynthetic apparatus: NDH subunits NdhA and NdhB, the ATP synthase subunit AtpF, and cytochrome b6f subunits PetB and PetD. Defective splicing of these introns is expected to decrease the levels of the corresponding protein and of other closely associated subunits in the same complex. Previously, apo1 mutants were reported to lack PSI, to have reduced levels of several NDH subunits, and to accumulate ATP synthase, photosystem II (PSII), and cytochrome b6f subunits to near normal levels (Amann et al., 2004). However, the cytochrome b6f data were ambiguous: the PetA subunit appeared to accumulate normally, whereas PetB appeared to be reduced approximately fourfold. Clarification of this issue was relevant to understanding APO1 function, so we examined the abundance of several thylakoid proteins by immunoblot analysis (Figure 2). Our results revealed a >10-fold loss of the PsaD subunit of PSI and normal levels of the AtpB subunit of the ATPase in apo1-1 mutants, consistent with the prior report. In addition, we found that the PetD subunit of the cytochrome b6f complex was reduced to ~25% of its normal level, consistent with the prior data for PetB and indicating a defect in the biogenesis of the cytochrome b6f complex. In contrast with the prior report that PSII core subunits are reduced approximately twofold in apo1 mutants, we observed a reduction of approximately fivefold in the D1 reaction center protein of PSII. This difference may result from the use of different growth conditions in the two studies, as D1 is highly sensitive to photooxidative damage when photosynthetic electron transport is compromised (Voelker and Barkan, 1995).
Figure 2.
Abundance of Selected Thylakoid Membrane Proteins in apo1 Mutants.
Duplicate immunoblots of seedling leaf protein extracts were probed with antibodies for the indicated proteins. A portion of the Ponceau S–stained filter is shown below to demonstrate equal sample loading and the normal accumulation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RbcL) in apo1 mutants. WT, wild type.
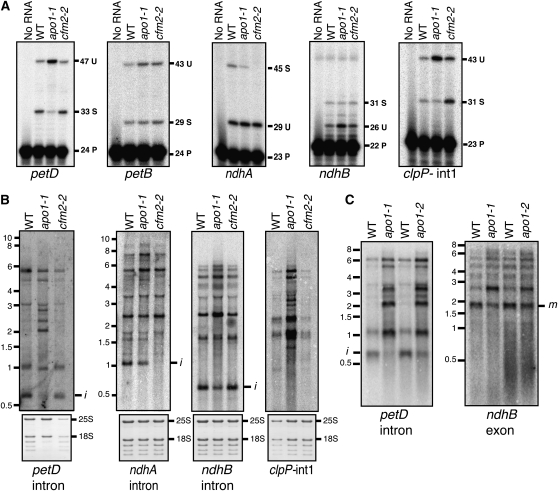
To determine whether splicing defects underlie the reductions in NDH and cytochrome b6f proteins in apo1 mutants, we examined the splicing of the petB, petD, ndhA, and ndhB introns. Poisoned primer extension assays (Figure 3A) showed petB splicing to be unaffected but revealed a substantial decrease in the ratio of spliced to unspliced petD RNA and small decreases in the ratios of spliced to unspliced ndhA and ndhB RNAs. RNA gel blot hybridizations (Figure 3B) using intron-specific probes support these findings, as they detected a reduction in excised intron and an increase in unspliced precursors in each case. The petD splicing defect was observed for both mutant alleles (Figure 3C) and is likely to account for the loss of cytochrome b6f subunits in apo1 mutants. The ndhB and ndhA splicing defects were modest, so it is unclear whether they account fully for the NDH deficiency in apo1 mutants. It is possible, for example, that YCF3 itself is required for normal NDH accumulation, a hypothesis that has not been addressed in published studies of ycf3 mutants.
Figure 3.
APO1 Enhances the Splicing of Several Chloroplast Introns.
(A) Poisoned primer extension assays. DNA primers complementary to exon sequences near the indicated 3′ splice junction were used to prime reverse transcription in the presence of a dideoxynucleotide that terminates chain extension after different distances on spliced (S) and unspliced (U) RNA templates. The length in nucleotides of the primer (P) and the extension products is indicated. WT, wild type.
(B) RNA gel blot hybridizations using the indicated intron-specific probes. Bands marked i represent excised intron. The size (in kilobases) and positions of RNA markers are shown to the left. Markers for the ndhB and clpP-int1 blots were at the same positions as those on the ndhA blot. The same blots were stained with methylene blue and are shown below to illustrate the abundance of rRNA in each sample.
(C) RNA gel blot hybridization assays illustrating petD and ndhB splicing defects in apo1-2 mutants. Hybridization probes were specific for the indicated sequences. Bands corresponding to excised intron (i) and mature spliced RNAs (m) are marked.
A survey of other chloroplast introns in apo1-1 mutants revealed a reduction in the splicing of clpP-int1 (Figure 3A), normal splicing of the atpF, trnV, trnK, trnI, trnA, trnG, rpl16, and clpP-int2 group II introns, and normal splicing of the trnL group I intron (see Supplemental Figure 2 online). Introns in three genes involved in basal chloroplast gene expression (rps12, rpl2, and rpoC) were not assayed; however, a substantive defect in the splicing of any of these introns is predicted to cause a global loss of chloroplast-encoded proteins and chloroplast rRNAs, neither of which is observed in apo1 mutants (Figure 2; see Supplemental Figure 2C online).
The spectrum of splicing defects in apo1 mutants is consistent with the recovery of Zm-APO1 in a CAF1 coimmunoprecipitate. Both APO1 and CAF1 promote the splicing of the ycf3 and petD pre-mRNAs, although CAF1 and APO1 target different ycf3 introns (intron1 and intron 2, respectively) (Ostheimer et al., 2003; Asakura and Barkan, 2006). CAF1 is an RNA binding protein that is found in stable complexes with its intron targets in vivo (Ostheimer et al., 2003). Results described below show that APO1 is, likewise, an RNA binding protein. Therefore, the coimmunoprecipitation of Zm-APO1 and CAF1 may be due either to their association with a shared RNA tether or to a direct protein–protein interaction.
APO1 Is an RNA Binding Protein
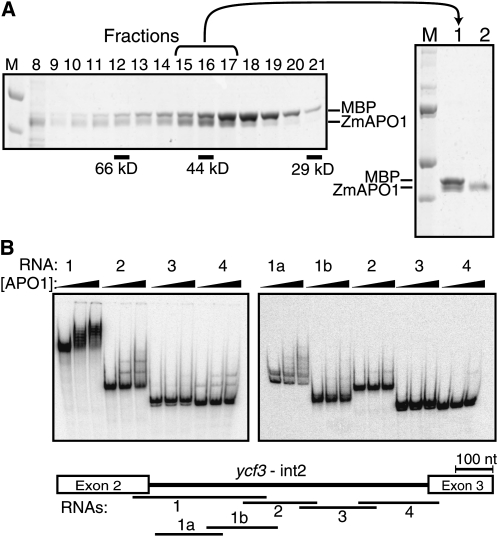
Mature APO1 (i.e., lacking its predicted chloroplast targeting sequence) consists entirely of DUF794, a plant-specific domain of unknown function (http://www.ebi.ac.uk/interpro/) (see Supplemental Figure 1C online). APO1’s role in group II intron splicing suggested that DUF794 might be an RNA binding domain. To address this possibility, recombinant APO1 was tested for RNA binding activity in vitro. We used the maize ortholog, Zm-APO1, for these experiments because the Arabidopsis ortholog, At-APO1, was more prone to aggregation when expressed in Escherichia coli. Zm-APO1 was expressed as a fusion with maltose binding protein (MBP), purified by amylose affinity chromatography, cleaved from the MBP moiety, and purified further by gel filtration chromatography (Figure 4A). Zm-APO1 eluted at the same position as did the ovalbumin marker (44 kD), consistent with Zm-APO1’s monomeric molecular mass (43 kD). Contaminating MBP was removed from the final preparations via a second amylose-affinity step (Figure 4A).
Figure 4.
APO1 Is an RNA Binding Protein.
(A) Purification of recombinant Zm-APO1. MBP-ZmAPO1 was expressed in E. coli, enriched by amylose affinity chromatography, and cleaved with TEV protease to separate the MBP and Zm-APO1 moieties. This material was fractionated on a Superdex 200 gel filtration column, and equal proportions of the indicated fractions were analyzed by SDS-PAGE and staining with Coomassie blue (shown on the left). The bracketed fractions were pooled (lane 1 in the panel to the right), and the MBP was removed by application to amylose-affinity resin. The material used for RNA binding assays is shown in lane 2 in the panel to the right. MBP expressed from the empty vector and purified by successive amylose-affinity and gel filtration chromatography has been shown previously not to bind RNA (Kroeger et al., 2009). M, marker.
(B) Gel mobility shift assays using recombinant Zm-APO1 and RNAs derived from ycf3-int2. Sequences represented by each RNA are diagrammed on the map below. The first lane in each series shows a reaction lacking Zm-APO1. The concentrations of protein were 0, 31, and 62 nM (left panel) or 0, 63, and 125 nM (right panel). The reactions within each panel were performed in parallel and can be compared with one another to infer relative affinities. However, the reactions in the two panels were performed on separate occasions and under slightly different conditions: the left-hand panel used tRNA at 0.4 ng/μL and freshly prepared Zm-APO1, whereas the right-hand panel used tRNA at 0.6 ng/μL and Zm-APO1 that had been stored for several weeks at −20°C. Preferential binding to fragment 1a was detected in two independent experiments (M. Rojas, unpublished data). nt, nucleotide.
To determine whether Zm-APO1 has RNA binding activity, gel mobility shift assays were performed with radiolabeled RNAs representing different regions of maize ycf3-int2 (see map in Figure 4B). Under low stringency binding conditions (150 mM NaCl and no nonspecific competitor), Zm-APO1 bound similarly to RNAs 1 though 4 (M. Rojas, unpublished data). However, increasing the NaCl concentration to 260 mM and adding tRNA to the binding reactions revealed preferential binding to the 5′ half of the intron (compare RNA1 to RNAs 2, 3, and 4 in Figure 4B). Results obtained with two smaller RNAs spanning this region (RNAs 1a and 1b) suggested that Zm-APO1 may bind preferentially to RNA1a, representing nucleotides 40 to 240 in intron domain 1. However, quantification of the binding data was challenging for two reasons. First, a doublet of bands was reproducibly observed with RNA1a in the absence of protein and presumably reflects the propensity of this RNA to adopt multiple conformations. Second, multiple shifted bands were observed in the presence of Zm-APO1, suggesting that multiple copies of APO1 can bind to the RNA. More detailed experiments will be required to understand these interactions and to pinpoint specific binding sites for APO1. Nonetheless, these results show that Zm-APO1 binds RNA with high affinity. As discussed below, Arabidopsis APO1 likewise binds RNA in vitro (Figure 5E). Therefore, DUF794, which makes up essentially all of mature APO1, is an RNA binding domain.
Figure 5.
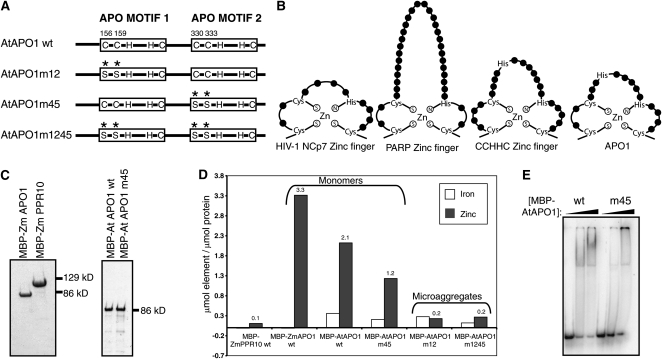
Evidence that APO1 and DUF794 Harbor Variant Zinc Fingers.
(A) Diagrammatic representation of the putative metal binding motifs and the Cys-to-Ser substitutions made in MBP-AtAPO1.
(B) Comparison of characterized zinc fingers with the proposed zinc fingers of APO1. This representation is modeled after that by Huang et al. (1998). The CCHHC NZF-1 model is based on information by Berkovits-Cymet et al. (2004). The consensus sequence of CCHC zinc fingers at Pfam and the sequences of the corresponding motifs in APO1 are shown in Supplemental Figure 1C online.
(C) Purified MBP-ZmAPO1, MBP-PPR10, and MBP-AtAPO1 fusion proteins used for ICAP spectroscopy. The APO1 preparations shown here were monomeric, as indicated by their elution from the gel filtration column. The gel filtration elution profiles for the other MBP-AtAPO1 mutants (which were found primarily in microaggregates) and the fractions used for ICAP spectroscopy are shown in Supplemental Figure 3 online.
(D) Summary of ICAP spectroscopy data for zinc and iron. Results for other metals are shown in Supplemental Tables 1 and 2 online. The molar ratio of metal-to-protein was calculated after subtracting values in the buffer control. wt, wild type.
(E) RNA binding activity of MBP-AtAPO1 and MBP-AtAPO1m45. The monomeric protein preparations used for ICAP spectroscopy were used for gel mobility shift assays with a nonspecific radiolabeled 50-mer RNA. Binding reactions contained 0.4 ng/μL tRNA and proteins at 0, 315, and 630 nM. The wild-type protein bound with higher affinity to RNA than did the mutant, as indicated by the loss of unbound RNA at lower protein concentrations. MBP-AtAPO1m12 and MBP-AtAPO1m1245 were not assayed because they were found solely in microaggregates, and microaggregates of wild-type MBP-AtAPO1 did not bind RNA (M. Rojas, unpublished data).
APO1 Is a Zinc Binding Protein and Harbors Motifs That Resemble Zinc Fingers
DUF794 includes two related motifs that have been named APO motifs (Amann et al., 2004). Each APO motif has a conserved arrangement of Cys and His residues suggestive of a metal binding motif (Figure 5A; see Supplemental Figure 1C online), but they were not recognized as such by the algorithms employed at InterPro. Previously, it was suggested that these motifs bind iron-sulfur clusters, in concordance with the view at that time that APO1 functioned in the metabolism of iron-sulfur clusters (Amann et al., 2004). However, our finding that DUF794 binds RNA prompted us to consider whether these motifs might instead bind zinc, as zinc fingers are commonly observed in RNA binding proteins. In fact, the spacing of the conserved Cys and His residues in the APO motifs is similar to that in CCHC and CCHHC zinc fingers (Figure 5B). To address the possibility that APO1 binds zinc, Zm-APO1 and At-APO1 were analyzed by inductively coupled argon plasma (ICAP) spectroscopy to determine their metal content. As a control, we analyzed the chloroplast RNA binding protein PPR10 (Pfalz et al., 2009), which consists almost entirely of PPR motifs and is not expected to bind metals. All three proteins were expressed as fusions to MBP in E. coli grown in standard medium without metal supplementation. They were purified by successive amylose affinity and gel filtration chromatography (see Supplemental Figure 3 online), yielding highly purified preparations (Figure 5C).
The ICAP spectroscopy data (Figure 5D; see Supplemental Tables 1 and 2 online) showed similar low levels of iron in all three preparations. However, zinc levels were dramatically elevated in both the MBP-ZmAPO1 and MBP-AtAPO1 samples, in comparison with MBP-ZmPPR10. These results argue against the previous hypothesis that APO1 binds iron and provide strong evidence that it does bind zinc. Comparison of the protein and zinc concentrations leads to an estimate of two or three zinc atoms bound per APO1 monomer (Figure 5D). The variation is likely to arise from imprecision in the determination of protein concentration, as different assay methods gave slightly different results (see Methods). Nonetheless, the ICAP spectroscopy data are consistent with the view that each of the two APO motifs in APO1 coordinates one zinc atom. All other metals assayed except for phosphorous were found at similar levels in the three protein samples (see Supplemental Tables 1 and 2 online). MBP-ZmAPO1 had elevated levels of phosphorous but MBP-AtAPO1 did not. This correlates with the differing levels of nucleic acid contamination in the two samples, as indicated by their ratios of absorbance at 260 and 280 nm (see Supplemental Figure 3 online). Therefore, it is likely that the phosphorous arose from residual E. coli RNA in the MBP-ZmAPO1 sample.
To determine whether the zinc finger-like motifs in APO1 are responsible for zinc binding, mutant isoforms of MBP-AtAPO1 were generated in which Cys pairs were changed to Ser in the first (AtAPO1m12), the second (AtAPO1m45), or both (AtAPO1m1245) motifs (Figure 5A). As for the wild-type proteins, these were purified by amylose affinity chromatography followed by gel filtration chromatography. The mutant proteins were more prone to aggregation than were the wild-type proteins, as shown by an increase in their recovery as microaggregates that elute near the void volume during gel filtration and a decreased recovery as monomers (see Supplemental Figure 3 online). The mutant isoforms were also more prone to proteolysis than were the wild-type proteins (see Supplemental Figure 3 online). These results indicate that the Cys pair in each APO motif makes an important contribution to protein folding. This is typical of zinc binding motifs (Witkowski et al., 1995; Liu et al., 2004).
A sufficient quantity of monomeric MBP-AtAPO1m45 was recovered for ICAP analysis. The zinc content of this preparation was approximately half that in MBP-AtAPO1 (Figure 5D), supporting the view that the second proposed zinc binding motif binds one zinc atom. These Cys residues are important for RNA binding activity, as monomeric MBP-AtAPO1m45 bound RNA less avidly than did monomeric MBP-AtAPO1 (Figure 5E).
No monomeric protein was recovered for MBP-AtAPO1m12 or MBP-AtAPO1m1245, so ICAP spectroscopy was performed with column fractions harboring microaggregates (see Supplemental Figure 3 online). The zinc content of this material was considerably lower than that of the monomeric wild-type and MBP-AtAPO1m45 preparations (Figure 5D). Further evidence for the critical nature of these Cys residues came from attempts to complement the apo1 phenotype by expression of the AtAPO1m1245 mutant isoform: transformation experiments using the same vector that supported complementation by the wild-type gene (Amann et al., 2004) failed to rescue the mutant phenotype (J. Meurer, unpublished data). Together, these findings support the conclusion that each APO motif coordinates zinc and that zinc binding is critical for the folding and function of DUF794 and APO1.
DISCUSSION
Results presented here show that the plant-specific domain of unknown function DUF794 is an RNA binding domain harboring zinc binding motifs and demonstrate that APO1, the founding member of the DUF794 family, is required for the splicing of several group II introns in chloroplasts. These findings add a new player to the set of proteins known to promote the splicing of chloroplast introns, and they clarify the role of APO1 in PSI biogenesis. It had been proposed previously that APO1 promotes PSI accumulation by facilitating the incorporation of [4Fe-4S] clusters into PSI subunits (Amann et al., 2004). However, our results show that APO1 is required for PSI accumulation due to its role in promoting the splicing of the ycf3 mRNA, which encodes a PSI assembly factor.
This revised view of APO1’s function is consistent with the observation that APO1 is associated with chloroplast nucleoids (Amann et al., 2004). Several other chloroplast splicing factors are likewise associated with nucleoids (G. Friso and K.J. van Wijk, unpublished data), implying that splicing factors load cotranscriptionally on to pre-mRNAs. Our findings warrant reconsideration of the underlying biochemical basis for several aspects of the apo1 mutant phenotype. For example, apo1 mutants exhibit a reduced rate of translation elongation on the chloroplast psaA/B mRNA, which encodes PSI reaction center proteins (Amann et al., 2004). Given APO1’s RNA binding activity, it now seems plausible that this reflects a direct interaction between APO1 and the psaA/B mRNA. Alternatively, Ycf3 may influence psaA/B translation by promoting the cotranslational assembly of nascent PsaA and/or PsaB proteins. Another notable aspect of the apo1 mutant phenotype is the absence of NDH and LHC1 proteins (Amann et al., 2004). LHC1 subunits mediate the association of the PSI and NDH complexes into a supercomplex (Peng et al., 2009). Our findings suggest a new hypothesis to explain the LHC1/NDH defects in apo1 mutants: perhaps Ycf3 is necessary to assemble the PSI/NDH supercomplex, and in the absence of Ycf3, the LHC1 and NDH proteins become unstable. Analysis of psaA/B mRNA translation and NDH and LHC1 protein levels in ycf3 mutants (Ruf et al., 1997; Landau et al., 2009) can be used to address whether Ycf3 indeed plays these unanticipated roles. The reduced splicing of the clpP mRNA may also contribute to the protein defects in apo1 mutants, as the Clp protease complex plays wide-ranging roles in chloroplast biogenesis and homeostasis (Kim et al., 2009).
Group II Intron Splicing Factors in Chloroplasts
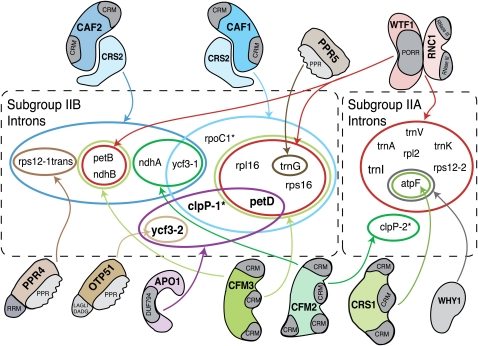
APO1 represents a new component of the complex network of RNA-protein interactions that contribute to chloroplast intron splicing (Figure 6). There is now strong evidence for the involvement of one chloroplast-encoded and 13 nucleus-encoded proteins in chloroplast intron splicing (reviewed in Kroeger et al., 2009; Stern et al., 2010). Unlike the nuclear spliceosome, whose core has a uniform composition, chloroplast splicing factors function in a combinatorial manner, with each intron requiring distinct but overlapping protein sets. Ycf3-int2 has been classified as a subgroup IIB intron (Michel et al., 1989), but it differs from all of the other chloroplast subgroup IIB introns in its splicing factor requirements (Figure 6). Ycf3-int2 is the only member of this group whose splicing does not require a heterodimer between the peptidyl-tRNA hydrolase homolog CRS2 and either of two paralogous proteins CAF1 or CAF2 (Jenkins et al., 1997; Jenkins and Barkan, 2001; Ostheimer et al., 2003). It is also the only cis-spliced member of this group that requires neither of the two paralogous proteins CFM2 or CFM3 (Asakura and Barkan, 2007; Asakura et al., 2008). In fact, only one protein had been shown previously to be required for ycf3-int2 splicing, the pentatricopeptide repeat protein OTP51 (de Longevialle et al., 2008). Like APO1, OTP51 is absolutely required for ycf3-int2 splicing and also stimulates the splicing of several other introns. However, the secondary targets of these proteins differ: whereas OTP51 enhances the splicing of the trnV, trnK, and atpF introns (de Longevialle et al., 2008), APO1 has a strong effect on petD and clpP-int1 splicing and a weak effect on ndhA and ndhB splicing. Detailed in vitro experiments will be required to understand how APO1 contributes to ycf3-int2 splicing and why the ycf3-int2 ribonucleoprotein particle has taken such a different evolutionary trajectory than the other group II intron ribonucleoproteins in chloroplasts.
Figure 6.
Summary of Nucleus-Encoded Proteins Involved in Group II Intron Splicing in Angiosperm Chloroplasts.
Introns are designated as Subgroup IIA or Subgroup IIB according to Michel et al. (1989). Introns found in Arabidopsis but not in maize are marked with an asterisk. Most, but not all, of the diagrammed interactions are supported by both genetic and coimmunoprecipitation data. Where analyzed, the functions of chloroplast splicing factors are conserved between maize and Arabidopsis (Asakura and Barkan, 2006). Experimental data are lacking for the Arabidopsis orthologs of CRS2, PPR4, WTF1, RNC1, and PPR5, so the set of splicing factors shown for introns found in Arabidopsis but not maize (clpP-1, clpP-2, and rpoC1) is likely to be incomplete. The chloroplast-encoded protein MatK associates in vivo with all of the subgroup IIA introns except for clpP-int2 (Zoschke et al., 2010) and is anticipated to be required for the splicing of its intron ligands. The ndhA and ndhB introns are not designated as APO1 ligands because the splicing defects in apo1 mutants were mild. Results are summarized from this work and from previously published work (Jenkins et al., 1997; Vogel et al., 1999; Jenkins and Barkan, 2001; Till et al., 2001; Ostheimer et al., 2003; Schmitz-Linneweber et al., 2006; Asakura and Barkan, 2007; Watkins et al., 2007; Asakura et al., 2008; Beick et al., 2008; de Longevialle et al., 2008; Prikryl et al., 2008; Kroeger et al., 2009).
DUF794 and the Plant APO Protein Family
APO1 belongs to a plant-specific gene family consisting of four members in Arabidopsis and rice (Amann et al., 2004) (see Supplemental Figure 1 online). All four APO proteins are organized similarly: a predicted mitochondrial or chloroplast targeting sequence at the N terminus is followed by DUF794. Our finding that DUF794 binds RNA suggests that other members of the DUF794 family function in RNA metabolism. Indeed, we observed chloroplast splicing defects in maize apo2 mutants (T. Kroeger and A. Barkan, unpublished data). Thus, it seems likely that APO3 and APO4, which are predicted to localize to mitochondria, function in mitochondrial RNA metabolism.
Organellar RNA metabolism in plants is characterized by a remarkable complexity that arose via the superposition of processes derived from the bacterial endosymbionts and processes that arose postsymbiosis. Examples of the latter include the acquisition of RNA editing and an abundance of group II introns in both organelles. It is becoming apparent that the emergence of these organelle-specific features of RNA metabolism was paralleled by the evolution of an RNA binding protein repertoire that is largely distinct from that in either bacteria or in the nuclear-cytosolic compartment. Previously described examples of RNA binding protein classes that are found specifically in organelles include the PPR, CRM, and PORR families, each of which functions in organelle-specific aspects of RNA metabolism (Small and Peeters, 2000; Barkan et al., 2007; Schmitz-Linneweber and Small, 2008; Kroeger et al., 2009). Results presented here strongly suggest that DUF794 is, likewise, the product of a coevolutionary process that spawned a diversity of noncanonical RNA binding motifs in conjunction with the emergence of the complex RNA metabolism characteristic of plant organelles.
METHODS
Identification of Zm-APO1 in CAF1 Coimmunoprecipitates
A peptide matching Zm-APO1 was identified by mass spectrometry in the same anti-CAF1 coimmunoprecipitate from which the splicing factors RNC1 and WTF1 were identified previously (Watkins et al., 2007; Kroeger et al., 2009). The methods are provided in those publications. and the mass spectrometry data are summarized in Supplemental Figure 1A online.
Phylogenetic Analysis
The maize (Zea mays) APO1 sequence was inferred from a full-length cDNA described in GenBank accession BT066015 and confirmed by the sequence of a cDNA recovered from our B73 cDNA library. This sequence was aligned with all members of the rice (Oryza sativa) and Arabidopsis thaliana APO family (as defined in Amann et al., 2004) using Muscle (default parameters). The alignment (available as Supplemental Data Set 1 online) was used to generate a maximum likelihood tree by PhyML at http://www.phylogeny.fr/ (Dereeper et al., 2008) using default parameters.
Plant Material
The apo1-1 allele was described originally by Amann et al. (2004) and has a T-DNA insertion at +32 bp with respect to the start codon. This allele is in a Wassilewskija ecotype background. The apo1-2 allele was obtained from the ABRC Stock Center (SALK_026463) and is in a Columbia-0 background. The position of the T-DNA insertion was determined by sequencing a PCR product obtained with a gene-specific and left border primer and found to map at +53 bp with respect to the start codon. The cfm2-2 mutant was used as a control and was described previously (Asakura and Barkan, 2007). Seeds were germinated on Murashige and Skoog agar medium under low light conditions (~10 μmol photons m−2 s−1) and harvested for RNA purification after ~4 weeks. Plant genotypes were confirmed by PCR using primers described in Supplemental Table 3 online.
Immunoblotting
Total leaf proteins were extracted, fractionated by SDS-PAGE, and analyzed by immunoblotting using methods and antibodies described previously (Watkins et al., 2007).
Splicing Assays
RNA was extracted from leaf tissue with TriZol reagent (Gibco BRL). Poisoned primer extension assays were performed as described by Asakura and Barkan (2006), with the exception that Transcriptor Reverse Transcriptase (Roche) was used for reverse transcription and the reactions contained 2 μg of total RNA.
RNA gel blot hybridizations were performed as described by Barkan (1998) using 2 μg of RNA per lane. The probes are summarized in Supplemental Table 3 online. Probes were either PCR products labeled by random-hexamer priming with [α-32P]dCTP or synthetic oligonucleotides that were 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase.
For RT-PCR, total leaf RNA was initially converted to cDNA by random hexamer priming: RNA (150 ng) was mixed with hexamers (500 ng) in a volume of 5 μL, heated to 95°C for 2 min, and placed on ice. Then, 5 μL of a mix containing 2× AMV buffer, 10 units of RNAsin, 1 mM each deoxynucleotide triphosphate, and 5 units of AMV Reverse Transcriptase (Promega) was added and incubated at 25°C for 10 min, and then at 48°C for an additional 50 min. The reactions were stopped by adding EDTA to 20 mM, and the nucleic acids were ethanol precipitated in the presence of Glycoblue (Ambion) and ammonium acetate. cDNAs were suspended in 25 μL of TE (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA), and 1 μL was used in a 25-μL PCR reaction with the RT-PCR primer pairs indicated in Supplemental Table 3 online. PCR was performed with Ex Taq (TaKaRa) using conditions recommended by the manufacturer and was performed for 25 to 30 cycles.
Expression and Purification of Recombinant Zm-APO1 and At-APO1
The coding sequence of mature Zm-APO1 (i.e., lacking its predicted chloroplast targeting sequence) was amplified from our maize leaf cDNA library (inbred line B73) (Fisk et al., 1999) using primers oAPO1for and oAPO1rev (see Supplemental Table 3 online) and Phusion DNA polymerase (New England Biolabs). The PCR product was digested with BamHI and SalI and cloned into the pMAL-TEV vector as previously described for RNC1 (Watkins et al., 2007). The sequence of the insert was confirmed to be identical to the corresponding region of a cDNA reported in GenBank under accession BT066015. The plasmid was transformed into Rosetta-2 cells, and Zm-APO1 was induced and purified as for RNC1 except that we did not include the polyethyleneamine precipitation step, and Buffer A included 450 mM NaCl. The protein was dialyzed against the same glycerol-containing buffer as used with RNC1 and stored at −20°C. The yield of purified protein was ~200 μg/L cell culture. Mature At-APO1 was cloned and expressed analogously, using oligonucleotides apo1E93F (5′-TATATAGATCTTTAGGAACGGTCTTTTGTTTTAATC-3′) and apo1E93R (5′-TATATGTCGACTTATGCGACCATGTCGGCTTCCTG-3′) for PCR from ABRC cDNA U82662 as template. Site-directed mutations were introduced into At-APO1 to exchange Cys residues Cys-156, Cys-159, Cys-330, and Cys-333 for Ser residues using a PCR-based approach (Kunkel et al., 1987) with the following primers: C156/159S, 5′-CATGTTGTTCCCGTCTTTGCTTCTAGTGAATCTGGAGCGGTTCATGTGGC-3′ and 5′-AGCAAAGACGGGAACAACATGAAGAAGCTGTGCTAAACCCTTG-3′; C330/333S, 5′-AGGAAATTCACAGTGAAAGCATCCGGATATTCTTCTGAAGTGCATGTAGG-3′ and 5′-TGCTTTCACTGTGAATTTCCTCATTAATTTCGTCAC-3′. These were subcloned into pMAL-TEV as described for the wild-type proteins. AtAPO1m12 has Cys-to-Ser substitutions at residues 156 and 159. AtAPO1m45 has Cys-to-Ser substitutions at residues 330 and 333. AtAPO1m1245 has Cys-to-Ser substitutions at all four positions. Protein concentrations were determined in two ways: by measuring A280 and calculations as described by Gill and von Hippel (1989) and by comparing Coomassie Brilliant Blue staining intensities of protein dilutions after SDS-PAGE. These methods gave slightly different results. We used the Gill and von Hippel method for proteins with negligible nucleic acid contamination (as indicated by A260/A280 ≤ 1), and we used the gel method for proteins whose A260/A280 was > 1.
Gel Mobility Shift Assays
Gel mobility shift assays were performed with trace amounts of radiolabeled RNA (40 pM) and protein concentrations as indicated in the figure legends using the gel conditions described previously (Watkins et al., 2007). Radiolabeled RNAs used in assays with Zm-APO1 were generated by in vitro transcription from PCR templates obtained with the primers shown in Supplemental Table 3 online. The MBP-AtAPO1 binding assay used a synthetic 50-mer RNA from the maize chloroplast atpB 5′ untranslated region as a nonspecific ligand; this RNA was labeled at its 5′ end with polynucleotide kinase and radiolabeled ATP. RNAs were purified by denaturing gel electrophoresis, followed by phenol-chloroform extraction and ethanol precipitation. Immediately prior to use in binding assays, they were heated in the absence of salts and snap-cooled on ice. Reactions included 40 pM RNA, 30 mM Tris-HCl, pH 7.5, 260 mM NaCl, 4 mM MgCl2, 10 μM ZnSO4, 0.08 units/μL RNAsin, 0.04 mg/mL BSA, and tRNA concentrations as indicated in the figure legends. Similar results were obtained when Mg2+ or Zn2+ were excluded from the reactions (M. Rojas, unpublished data).
Analysis of Zinc in Zm-APO1, At-APO1, and At-APO1 Mutants
ICAP optical emission spectroscopy was performed at the University of Georgia Center for Applied Isotope Studies Chemical Analysis Lab (http://www.cais.uga.edu/analytical_services/chemical_analysis/). The MBP fusion constructs of Zm-APO1 and At-APO1 (and mutants thereof) are described above. The MBP-PPR10 construct was described previously (Pfalz et al., 2009). These were expressed and purified by amylose affinity chromatography followed by size exclusion chromatography on a Superdex 200 column. The column buffer used for MBP-PPR10 and MBP-ZmAPO1 contained 40 mM Tris-HCl, pH 7.5, and 400 mM NaCl. The column buffer used for MBP-AtAPO1 and its mutant variants contained 40 mM Tris-HCl, pH 7.5, and 200 mM KCl. Affinity-purified MBP-AtAPO1 and MBP-AtAPO1m45 eluted from Superdex 200 in two peaks: as microaggregates and as protein monomers. Column fractions corresponding to each peak were pooled separately for ICAP spectroscopy. Affinity-purified MBP-AtAPO1m12 and MBP-AtAPO1m1245 eluted from the Superdex 200 column entirely as microaggregates, and this material was used for ICAP spectroscopy (see Supplemental Figure 3 online). Nitric acid (TraceMetal grade; Fisher) was added to the buffer control and protein samples to a concentration of 2% (v/v). Samples were then analyzed on a Thermo Jarrell-Ash Enviro 36 instrument. EPA method 6010C was used with 10 ppm calibration standards purchased from Spex or Fisher Scientific. The analysis of MBP-AtAPO1 and its variants was performed on a separate occasion from that of MBP-PPR10 and MBP-ZmAPO1, so the results are presented in separate tables (see Supplemental Tables 1and 2 online). The following quantities of protein were submitted for analysis: MBP-ZmPPR10, 0.9 mL at 1.02 μg/μL; MBP-ZmAPO1, 0.9 mL at 0.35 μg/μL; MBP-AtAPO1(monomers), 0.85 mL at 0.27 μg/μL; MBP-AtAPO1 (microaggregates), 0.85 mL at 0.55 μg/μL; MBP-AtAPO1m12 (microaggregates), 0.85 mL at 0.37 μg/μL; MBP-AtAPO1m45 (microaggregates), 0.85 mL at 1.07 μg/μL; MBP-AtAPO1m45 (monomers), 0.85 mL at 0.42 μg/μL; MBP-AtAPO1m1245 (microaggregates), 0.85 mL at 0.45 μg/μL.
Accession Numbers
Arabidopsis APO1 is encoded by locus At1g64810. Zm-APO1 is encoded by a full-length cDNA deposited in GenBank under accession BT066015 and by locus GRMZM2G007453 in B73 RefGen_v2 (www.maizesequence.org).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of the Maize Ortholog of APO1 (Zm-APO1) in a CAF1 Coimmunoprecipitate.
Supplemental Figure 2. Assays of Introns Whose Splicing Is Not Affected in apo1 Mutants.
Supplemental Figure 3. Purification of MBP-AtAPO1 Variants Used for ICAP Spectroscopy.
Supplemental Table 1. Complete ICAP Data for MBP-ZmAPO1 and MBP-PPR10.
Supplemental Table 2.Complete ICAP Data for MBP-AtAPO1 and Its Mutant Variants.
Supplemental Table 3.Nucleic Acid Primers and Probes.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Supplemental Figure 1B.
Supplementary Material
Acknowledgments
We thank Kristina Djermanovic for performing the site-directed mutagenesis of the APO motifs in At-APO1. We thank Roz Williams-Carrier for help generating the CAF1 coimmunoprecipitates, for help with figures, and for useful discussions, Yukari Asakura for providing cfm2-2 tissue, Bobby Coalter and Sara Davidson for cloning the Zm-APO1 expression construct, and Rebecca Auxier for performing the ICAP analysis. This work was supported by National Science Foundation Grant IOS-0922560 to A.B. and K.J.v.W., National Science Foundation Grant MCB-0744960 to A.B., and Deutsche Forschungsgemeinschaft Grants SFB TR1 and ME 1794/4 to J.M.
References
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. (2006). Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. (2007). A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 19: 3864–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Bayraktar O.A., Barkan A. (2008). Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14: 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57 [Google Scholar]
- Barkan A., Klipcan L., Ostersetzer O., Kawamura T., Asakura Y., Watkins K.P. (2007). The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA 13: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits-Cymet H.J., Amann B.T., Berg J.M. (2004). Solution structure of a CCHHC domain of neural zinc finger factor-1 and its implications for DNA binding. Biochemistry 43: 898–903 [DOI] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Dabney-Smith C. (2008). Plastid protein import and sorting: Different paths to the same compartments. Curr. Opin. Plant Biol. 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle A.F., Hendrickson L., Taylor N.L., Delannoy E., Lurin C., Badger M., Millar A.H., Small I. (2008). The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue): W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.C., von Hippel P.H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Huang M., Maynard A., Turpin J.A., Graham L., Janini G.M., Covell D.G., Rice W.G. (1998). Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J. Med. Chem. 41: 1371–1381 [DOI] [PubMed] [Google Scholar]
- Jenkins B.D., Barkan A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek D.J., Barkan A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Maier U.G., Leister D. (2009). DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Kroeger T.S., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A., Roberts J.D., Zakour R.A. (1987). Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154: 367–382 [DOI] [PubMed] [Google Scholar]
- Landau A.M., Lokstein H., Scheller H.V., Lainez V., Maldonado S., Prina A.R. (2009). A cytoplasmically inherited barley mutant is defective in photosystem I assembly due to a temperature-sensitive defect in ycf3 splicing. Plant Physiol. 151: 1802–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Rigolet P., Dou S.X., Wang P.Y., Xi X.G. (2004). The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J. Biol. Chem. 279: 42794–42802 [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. (1989). Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Ostheimer G.J., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel H., Bock R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I.D., Peeters N. (2000). The PPR motif - A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R., Barkan A. (1995). Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol. Gen. Genet. 249: 507–514 [DOI] [PubMed] [Google Scholar]
- Vogel J., Börner T., Hess W.R. (1999). Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 27: 3866–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Langdale J.A. (2009). The making of a chloroplast. EMBO J. 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Kroeger T.S., Cooke A.M., Williams-Carrier R.E., Friso G., Belcher S.E., van Wijk K.J., Barkan A. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski R.T., Hattman S., Newman L., Clark K., Tierney D.L., Penner-Hahn J., McLendon G. (1995). The zinc coordination site of the bacteriophage Mu translational activator protein, Com. J. Mol. Biol. 247: 753–764 [DOI] [PubMed] [Google Scholar]
- Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. (2010). An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.