This study provides evidence that two laccases, LAC4 and LAC17, participate in the polymerization of lignins in Arabidopsis stems. These findings suggest that the genetic engineering of lignin-specific laccases is a potentially innovative and promising tool for the fine-tuning of lignin content and structure.
Abstract
Peroxidases have been shown to be involved in the polymerization of lignin precursors, but it remains unclear whether laccases (EC 1.10.3.2) participate in constitutive lignification. We addressed this issue by studying laccase T-DNA insertion mutants in Arabidopsis thaliana. We identified two genes, LAC4 and LAC17, which are strongly expressed in stems. LAC17 was mainly expressed in the interfascicular fibers, whereas LAC4 was expressed in vascular bundles and interfascicular fibers. We produced two double mutants by crossing the LAC17 (lac17) mutant with two LAC4 mutants (lac4-1 and lac4-2). The single and double mutants grew normally in greenhouse conditions. The single mutants had moderately low lignin levels, whereas the stems of lac4-1 lac17 and lac4-2 lac17 mutants had lignin contents that were 20 and 40% lower than those of the control, respectively. These lower lignin levels resulted in higher saccharification yields. Thioacidolysis revealed that disrupting LAC17 principally affected the deposition of G lignin units in the interfascicular fibers and that complementation of lac17 with LAC17 restored a normal lignin profile. This study provides evidence that both LAC4 and LAC17 contribute to the constitutive lignification of Arabidopsis stems and that LAC17 is involved in the deposition of G lignin units in fibers.
INTRODUCTION
Angiosperm lignins are complex phenolic polymers that consist mostly of guaiacyl (G) and syringyl (S) units, together with small or trace amounts of p-hydroxyphenyl (H) units. Monolignols are synthesized in the cytosol and transported to the cell wall, where their oxidation generates lignins (Vanholme et al., 2008). Laccases were first identified in the lacquer tree (Rhus vernicifera) secreted resin (Yoshida, 1883). It has been suggested repeatedly that these enzymes are involved in lignin biosynthesis, based on their capacity to oxidize lignin precursors in vitro (Higuchi and Ito, 1958; Sterjiades et al., 1992; Bao et al., 1993; Kärkönen et al., 2002; Liang et al., 2006) and their localization in zones of lignification in various plant species (Dean and Eriksson, 1994; Richardson and McDougall, 1997; Dean et al., 1998; Ranocha et al., 2002; Caparros-Ruiz et al., 2006). However, the use of an antisense strategy to decrease laccase expression in transgenic poplar (Populus tremula × Populus alba) lines affected the pool of soluble phenolic compounds but had no detectable impact on lignification (Ranocha et al., 2002). Arabidopsis thaliana transgenic lines overexpressing LAC1, which encodes a cotton (Gossypium hirsutum) root-secreted laccase, were found to be resistant to phenolic allelochemicals and 2,4,6-trichlorophenol (Wang et al., 2004). Expression of this laccase gene in transgenic poplars (Populus deltoides) increased the lignin content of stems (Wang et al., 2008). In the last decade, several studies have focused on Arabidopsis laccases (McCaig et al., 2005; Pourcel et al., 2005; Cai et al., 2006). Seventeen laccase genes have been identified in Arabidopsis and classified into six groups based on the alignment of their amino acid sequences with those of laccase-like multicopper oxidases (McCaig et al., 2005; Hoegger et al., 2006). Among T-DNA insertion mutants for 12 laccase genes, the mutants corresponding to LAC2, LAC8, and LAC15 displayed changes in root elongation, earlier flowering, and changes in seed color, respectively (Cai et al., 2006). The first biological function assigned to an Arabidopsis laccase was revealed by the study of tt10 mutants, which display changes in seed color (Pourcel et al., 2005). This work provided evidence for the involvement of LAC15 in the oxidative polymerization of flavonoids in the Arabidopsis seed coat. Another study suggested that the extractable lignin content of the seed coat was lower in the tt10 mutant (Liang et al., 2006).
In this study, we aimed to establish whether laccases were involved in the lignification of Arabidopsis stems. We addressed this issue by demonstrating that LAC4 and LAC17 were expressed in the lignified tissues of the inflorescence stems, making these genes good candidates for involvement in lignification. We produced two double mutants and studied the impact of single and double mutations on the lignification of inflorescence stems and on the corresponding lignified tissues recovered by laser capture microdissection. The various histological and chemical analyses, together with complementation of the lac17 mutant with LAC17 constructs, provided evidence that both the LAC4 and LAC17 genes are involved in the lignification of Arabidopsis stems.
RESULTS
Identification of Arabidopsis Laccases Strongly Expressed in Inflorescence Stems
According to the Web-based GeneCAT coexpression tool (http://genecat.mpg.de), eight of the 17 genes encoding laccases in Arabidopsis are expressed in the inflorescence stem to various extents (see Supplemental Figure 1 online) and might be involved in lignification (LAC2, LAC4, LAC5, LAC6, LAC10, LAC11, LAC12, and LAC17). We monitored the expression profiles of these genes in the stem during plant development by RT-PCR (Figure 1). The levels of most laccase transcripts increased from early developmental stages (stages 6.0 to 6.2; Boyes et al., 2001) to reach a plateau, decreasing thereafter when the plant reached maturity. This RT-PCR experiment revealed that LAC4 and LAC17 were strongly expressed in stems (Figure 1). In addition, the GeneCAT tool (http://genecat.mpg.de) indicated that LAC4 and LAC17 were coexpressed (see Supplemental Figure 1 online), suggesting functional redundancy between them. We therefore hypothesized that LAC4 and LAC17 were the candidate genes most likely to be involved in constitutive lignification.
Figure 1.
Laccase Gene Expression in Arabidopsis Floral Stems (Plants Grown under Long-Day Conditions).
RT-PCR expression profiles of eight laccase genes expressed in the inflorescence stem at eight different growth stages of Arabidopsis. Expression was normalized against the housekeeping β-TUBULIN (β-TUB) gene. Two signals were obtained for LAC6, corresponding to alternative splicing events. Growth stages, according to Boyes et al. (2001), are indicated at the top.
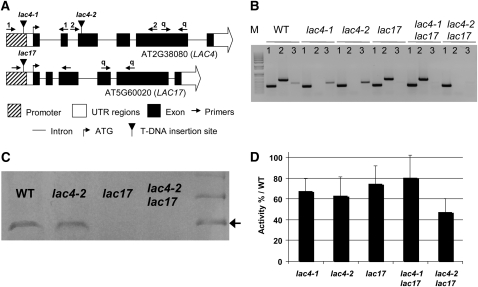
Molecular Characterization of LAC4 and LAC17 T-DNA Insertion Mutants and Production of the Corresponding Double Mutants
Two LAC4 (S_051892, referred to as lac4-1, corresponding to irx12 in Brown et al. [2005], and GabiKat-720G02, referred to as lac4-2) and one LAC17 (S_016748, referred to as lac17) mutants were identified in the SALK and GABI collections (Table 1). According to Brown et al. (2005), the irx12 mutant, identified on the basis of the coregulation of the corresponding gene, LAC4, with the CesA genes, has a weak irregular xylem (irx) phenotype. LAC4 homozygous lines were selected by PCR, with primers binding before and after the T-DNA insertion site (see Supplemental Table 1 online). The flanking regions of the T-DNA insertions were amplified by PCR with specific primers (see Supplemental Table 1 online) and sequenced. This analysis revealed that lac4-1 and lac17 had T-DNAs inserted in their promoter sequences, 125 and 145 bp upstream from the ATG, respectively, whereas lac4-2 had an insertion in the third exon of the coding sequence (Figure 2A). A single T-DNA insertion was found in the LAC4 mutants, whereas two contiguous T-DNAs were found to be inserted in tandem in the lac17 promoter. RT-PCR on total RNA extracted from the stem confirmed that these T-DNA insertion lines were null mutants for the corresponding genes (Figure 2B; see Supplemental Figure 2 online).
Table 1.
List of Single Mutants Used in This Study
| Gene Identification | Accession No. | Mutant Identification | Mutant Name | T-DNA Insertion Site | RT-PCR Confirmation |
| LAC4 | At2g38080 | Salk_051892 GabiKat-720G02 | lac4-1 lac4-2 | Promoter Exon | Knockdown Knockout |
| LAC17 | At5g60020 | Salk_016748 | lac17 | Promoter | Knockdown |
Figure 2.
Characterization of Laccase T-DNA Insertion Mutants (Grown in Long-Day Conditions).
(A) Schematic diagram of the T-DNA insertion in lac4-1, lac4-2, and lac17 mutants. The positions of primers used for genotyping lac4-1 and lac4-2 and for quantitative PCR are indicated by 1, 2, and q, respectively.
(B) Confirmation of the downregulation of laccase transcripts by RT-PCR. Lane 1, expression of the β-tubulin gene (housekeeping gene); lane 2, LAC4 expression; lane 3, LAC17 expression; lane M, 1 kb + ladder (Invitrogen).
(C) Immunoblot analysis with an antibody against Arabidopsis LAC17. The molecular mass of Arabidopsis LAC17 is indicated by an arrow at 75 kD.
(D) Quantification of laccase activity in partially purified protein extracts, with ABTS as the substrate. Data represent means ± sd (n = 3).
Homozygous lac4-1 and lac17 mutants were crossed to generate a double mutant. We checked the double mutant status of the offspring using primers specific for each mutation in the T2 progeny (see Supplemental Table 1 online). Surprisingly, lac4-1 lac17 was found to display effective downregulation of LAC17 but with the partial restoration of LAC4 expression (Figure 2B). We therefore generated another double mutant by crossing lac4-2 and lac17 and established that both LAC4 and LAC17 were knocked out in this mutant (Figure 2B). Immunoblot analyses were performed with partially purified protein extracts from stem samples and an antibody specific for LAC17 (Figure 2C). Consistent with the findings of the transcriptomic studies, no LAC17 was detected in lines in which the LAC17 gene was disrupted (Table 2).
Table 2.
A Selection of the Differentially Expressed Genes in the Five Laccase Mutants
| Selected Gene Chip Hybridization Data | ||||||
| Putative Gene Function | Accession No. | log2 Ratio lac4-1/WT | log2 Ratio lac4-2/WT | log2 Ratio lac17/WT | log2 Ratio lac4-1 lac17/WT | log2 Ratio lac4-2 lac17/WT |
| Monolignol Genes | ||||||
| Phenylalanine ammonia-lyase 1 (PAL1) | At2g37040 | −0.25 | −0.57 | −0.24 | −0.92 | −1.09 |
| Phenylalanine ammonia-lyase 3 (PAL3) | At5g04230 | −0.47 | −0.76 | −0.57 | −1.06 | −1.59 |
| Phenylalanine ammonia-lyase 4 (PAL4) | At3g10340 | 0.01 | −0.40 | −0.01 | −1.43 | −2.03 |
| Cinnamate 4-hydroxylase (C4H) | At2g30490 | −0.28 | −0.53 | −0.32 | −1.25 | −1.24 |
| 4-Coumarate:CoA ligase 2 (4CL2) | At3g21240 | −0.32 | −0.75 | −0.19 | −0.76 | −0.78 |
| Coumarate 3-hydroxylase (C3H) | At2g40890 | −0.07 | −0.31 | −0.14 | −0.76 | −1.10 |
| Ferulate 5-hydroxylase (F5H) | At4g36220 | 0.04 | −0.30 | −0.07 | −0.52 | −1.04 |
| O-methyltransferase 1 (OMT1) | At5g54160 | 0.61 | 0.55 | 0.31 | −0.25 | −0.22 |
| Caffeoyl-CoA 3-O-methyltransferase (CCoAOMT1) | At4g34050 | −0.16 | −0.17 | 0.13 | −0.99 | −0.63 |
| Cinnamyl alcohol dehydrogenase (CAD-C) | At3g19450 | −0.11 | −0.11 | −0.13 | −0.34 | −0.74 |
| Cinnamyl alcohol dehydrogenase (CAD-D) | At4g34230 | −0.18 | −0.45 | −0.22 | −0.81 | −0.51 |
| β-Glucosidase 46 (βGLU46) | At1g61820 | −0.13 | 0.01 | −0.10 | 0.03 | −0.62 |
| Laccase Genes | ||||||
| Laccase 2 (LAC2) | At2g29130 | −0.08 | −0.27 | −0.30 | −0.38 | −0.32 |
| Laccase 4 (LAC4) | At2g38080 | −4.53 | −4.21 | −0.05 | −1.03 | −3.76 |
| Laccase 5 (LAC5) | At2g40370 | 0.28 | 0.15 | −0.03 | 0.01 | −0.10 |
| Laccase 6 (LAC6) | At2g46570 | 0.01 | −0.07 | −0.04 | 0.15 | 0.13 |
| Laccase 10 (LAC10) | At5g01190 | −0.06 | −0.11 | 0.03 | −0.10 | 0.10 |
| Laccase 11 (LAC11) | At5g03260 | −0.12 | 0.02 | −0.41 | 0.00 | 0.19 |
| Laccase 12 (LAC12) | At5g05390 | 0.26 | 0.28 | 0.08 | −0.42 | −0.08 |
| Laccase 17 (LAC17) | At5g60020 | 0.49 | 0.43 | −3.93 | −4.36 | −4.24 |
The five mutant samples were analyzed with wild-type (WT) samples grown in the same conditions as references (in growth chamber and under long-day conditions). A negative ratio in bold is indicative of downregulation of the gene in mutants. A positive ratio in bold indicates that the gene is upregulated in mutants. Other ratios (not in bold) were not found to be statistically significant after Bonferroni correction (P < 0.05).
We evaluated laccase expression levels in the single and double mutants by comparing laccase activity in protein extracts recovered from stems harvested at stage 6.4 (Boyes et al., 2001; Figure 2D). Consistent with the levels of expression of the genes for several stem-specific laccases, laccase activity was ~30% below control levels in lac4-1, lac4-2, lac17, and lac4-1 lac17 stems. Moreover, in accordance with the simultaneous disruption of LAC4 and LAC17, the double mutant lac4-2 lac17 displayed a larger decrease (53%) in laccase activity (Figure 2D).
Phenotypes of the Lines Displaying LAC4 and/or LAC17 Downregulation
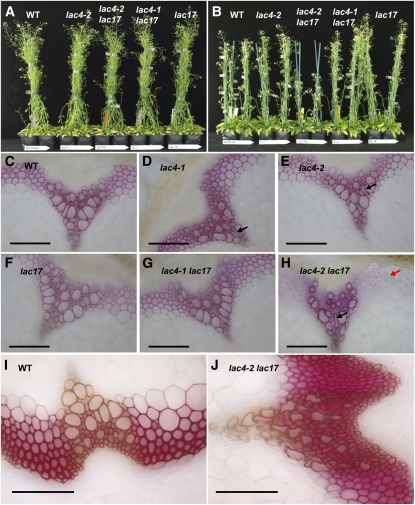
The lac4-1, lac4-2, lac17, and lac4-1 lac17 mutants had growth and development characteristics similar to those of the wild-type line grown under long-day or continuous light conditions (Figure 3). By contrast, the development of lac4-2 lac17 was dependent on the growth conditions. We repeatedly observed that this double mutant had a normal size (Figure 3A) when grown in long-day conditions (either in the growth chamber or in the greenhouse). By contrast, it displayed a semidwarf phenotype (Figure 3B) when grown under continuous light in the growth chamber.
Figure 3.
Phenotype of Laccase Mutants.
(A) Plants grown in the growth chamber (long-day conditions: no phenotype was observed).
(B) Plants grown in the growth chamber (continuous light: lac4-2 lac17 displayed a semidwarf phenotype).
(C) to (H) Wiesner staining of stem cross sections from plants grown under continuous light. The black arrows show collapsed xylem, and the red arrow shows hypolignified fibers. Bars = 100 μm.
(I) and (J) Maüle staining of stem cross sections from wild-type and lac4-2 lac17 plants grown under continuous light. Bars = 100 μm.
We further investigated the putative impact of the mutations at the tissue level by observing stem cross sections after Wiesner (Figures 3C to 3H) or Maüle (Figures 3I and 3J) staining. In the Wiesner test (phloroglucinol-HCl), lignins are stained a violet-red color, principally through reactions involving coniferaldehyde end groups (Nakano and Meshitsuka, 1992; Kim et al., 2002). Despite their low frequency in native lignins (<2% of the phenylpropane units) (Adler, 1977; Lapierre, 2010), these coniferaldehyde end groups efficiently detect lignins due to the high extinction coefficient of the coniferaldehyde-phloroglucinol adduct. Wiesner staining was weaker in the interfascicular fibers of lac4-2 lac17 mutants than in other lines, regardless of the growth conditions (Figure 3H for plants grown under continuous light and in the growth chamber; see Supplemental Figure 3 online for plants grown in long-day conditions). This suggests that this mutant had a lower lignin level and/or lower coniferaldehyde end group content. Maüle reagent specifically stains S lignin units bright red. Maüle staining revealed no major staining differences between the control and lac4-2 lac17 lines (Figures 3I and 3J). Accordingly, the deposition of S units did not seem affected in lac4-2 lac17 fibers.
The weak irx phenotype reported by Brown et al. (2005) for the irx12 mutant could not be observed in the stem sections of lac4-1, lac4-2, lac17, and lac4-1 lac17 when grown in long-day conditions (see Supplemental Figure 3 online). In conditions that generate the semidwarf phenotype for lac4-2 lac17 (i.e., continuous light in the growth chamber), some collapsed vessels were observed in the stem cross sections of lac4-1 and lac4-2 (Figures 3D and 3E). When its growth was not affected, the double mutant lac4-2 lac17 still had vessels with a slight irx phenotype (see Supplemental Figure 3 online). By contrast, the semidwarf stems of lac4-2 lac17 had more severely collapsed vessels (Figures 3H and 3J).
Effect of the LAC4 and/or LAC17 Mutations on the Expression of Other Genes
Complete Arabidopsis transcriptome microarray experiments were performed on RNA extracted from stems grown in a growth chamber under continuous light and collected at stage 6.2 (Boyes et al., 2001). LAC4 and/or LAC17 repression had no significant impact on the expression of other stem-specific laccases (Table 2). We also investigated the expression of some of the genes involved in the monolignol biosynthesis pathway. The lac4-1, lac4-2, and lac17 single mutants displayed no significant modification of the level of expression of these genes (Table 2). In addition, the two double mutants, lac4-1 lac17 and lac4-2 lac17, displayed decreases in the expression of 8 and 10 genes involved in lignin biosynthesis, respectively. Thus, disruption of both the LAC4 and LAC17 genes induced some feedback control on the genes of the phenylpropanoid (C4H, PAL1, PAL3, and PAL4) or lignin-specific (CAD-C, CAD-D, CCoAOMT1, OMT1, F5H, C3H, 4CL2, and β-GLU46) pathways. Only a few other genes were affected in the single and/or double mutants. The functions of these genes were not known, with the exception of a small number of genes involved in tolerance to water stress or in copper metabolism (see Supplemental Data Set 1 online).
Impact of LAC4 and/or LAC17 Silencing on Lignification, Enzymatic Hydrolysis, and Soluble Phenolic Compounds in Mature Arabidopsis Stems
Lignin content was determined by measuring both the Klason lignin and acid-soluble lignin concentrations of the extract-free stems. Regardless of the growth conditions (long-day or continuous light), the single mutants lac4-1, lac4-2, and lac17 consistently had lower Klason lignin levels (lignin levels 8 to 14% lower; Table 3) than the corresponding control. These levels were even lower in the double mutant lac4-1 lac17 (~20% lower than the wild-type level). The lac4-2 lac17 double mutant had much lower lignin levels (40% less than the control sample) regardless of its phenotype (normal or semidwarf size). Acid-soluble lignin contents were similarly low, whatever the sample (in the 1 to 2% range; Table 3) and did not compensate for the lower Klason lignin levels of the single and double mutants.
Table 3.
Lignin Content and Enzymatic Hydrolysis of Mature Stems from Laccase Mutants
| Lignin Content |
Enzymatic Hydrolysis |
||||
| Growth Conditions | Line | KL% | ASL% | Weight Loss% | Released Glc% |
| Long-day (no phenotype) | Wild type | 20.05 ± 0.18 (100) | 1.37 ± 0.07 | 28.2 ± 1.0 (100) | 9.0 ± 0.3 |
| lac4-2 | 17.35 ± 0.13 (87) | 1.70 ± 0.01 | 31.4 ± 0.1 (111) | 9.3 ± 0.9 | |
| lac17 | 17.26 ± 0.12 (86) | 1.63 ± 0.05 | 28.2 ± 0.5 (100) | 9.7 ± 0.4 | |
| lac4-1 lac17 | 15.37 ± 0.02 (77) | 1.95 ± 0.02 | 36.9 ± 0.1 (131) | 11.0 ± 0.0 | |
| lac4-2 lac17 | 12.66 ± 0.16 (63) | 1.70 ± 0.01 | 52.6 ± 0.4 (187) | 23.4 ± 0.4 | |
| Continuous light (semidwarf phenotype for lac4-2 lac17) | Wild type | 20.13 ± 0.09 (100) | 1.04 ± 0.02 | 29.4 ± 0.4 (100) | 9.9 ± 0.5 |
| lac4-1 | 18.42 ± 0.01 (92) | 1.10 ± 0.03 | 35.8 ± 0.6 (122) | 13.6 ± 0.5 | |
| lac4-2 | 18.27 ± 0.10 (91) | 0.96 ± 0.01 | 35.9 ± 1.1 (122) | 13.1 ± 0.7 | |
| lac17 | 17.97 ± 0.13 (89) | 1.20 ± 0.01 | 29.0 ± 0.4 (99) | 9.4 ± 0.3 | |
| lac4-1 lac17 | 16.22 ± 0.00 (81) | 1.10 ± 0.01 | 30.6 ± 0.5 (104) | 11.0 ± 0.2 | |
| lac4-2 lac17 | 12.24 ± 0.01 (61) | 1.56 ± 0.01 | 61.9 ± 0.5 (210) | 30.5 ± 0.6 | |
The Klason lignin (KL) and the acid-soluble lignin (ASL) contents are expressed as a percentage of the extract-free samples, in terms of weight. Data are mean values and standard errors from duplicate experiments. The values in parentheses are the percentages relative to the corresponding control.
Lignin content is known to be a factor in decreasing cell wall susceptibility to enzymatic hydrolysis. We therefore subjected the various extract-free samples to treatment with a commercial cellulase preparation to evaluate the saccharification potential of the various single and double mutants. Consistent with their low Klason lignin levels, commercial cellulase treatment (cellulase and hemicellulase activities) resulted in weight losses of 53 to 62% for lac4-2 lac17 samples, much greater than those observed for the control samples (Table 3). About half of this loss could be accounted for by Glc (Table 3). Both the lac4-1 and lac4-2 stems displayed moderately higher levels of saccharification than the wild type, as shown by weight loss after cellulase treatment. By contrast, lac17 was no more easily hydrolyzed than control samples despite having 11 to 14% lower lignin levels.
We evaluated the impact of laccase deficiency on lignin structure by thioacidolysis (Table 4). This analytical degradation method generates H, G, and S monomers from H, G, and S lignin units involved in β-O-4 bonds only (Lapierre et al., 1995). Thus, total monomer yield closely reflects the amounts of these lignin structures in the sample. In addition, when calculated on the basis of Klason lignin content, this yield is inversely correlated with the frequency of lignin units involved in resistant interunit linkages (condensed linkages). Whatever the sample, only small amounts of H thioacidolysis monomers were recovered (0.6 to 2 μmol per gram of extract-free sample); therefore, these monomers will not be discussed further below. Thioacidolysis yields calculated on the basis of lignin content did not significantly differ from control levels (arbitrarily set at 100) other than for the semidwarf lac4-2 lac17 sample (obtained under continuous light). Thus, laccase deficiency did not affect the frequency of condensed bonds in the lignins of the single and double mutants, except for the semidwarf lac4-2 lac17 sample, which contained lignins enriched in condensed bonds.
Table 4.
Determination of the Main H, G, and S Thioacidolysis Monomers Released by the Lignins of Extract-Free Mature Stems of Laccase Mutants
| Thioacidolysis Yield in μmole per Gram of Extract-Free Sample |
S/G Molar Ratio | Relative Yield Based on Lignin Contenta | |||||
| Growth Conditions | Line | H | G | S | Total | ||
| Long-day (no phenotype) | Wild type | 2.0 ± 0.3 | 179 ± 7 | 74 ± 4 | 254 ± 11 | 0.41 ± 0.02 | 100 |
| lac4-2 | 1.4 ± 0.3 | 153 ± 17 | 62 ± 7 | 216 ± 24 | 0.41 ± 0.01 | 91 ± 10 | |
| lac17 | 1.0 ± 0.2 | 143 ± 10 | 72 ± 2 | 215 ± 13 | 0.50 ± 0.02 | 93 ± 6 | |
| lac4-1 lac17 | 1.1 ± 0.1 | 146 ± 9 | 87 ± 7 | 234 ± 16 | 0.59 ± 0.01 | 107 ± 7 | |
| lac4-2 lac17 | 0.6 ± 0.2 | 114 ± 2 | 83 ± 6 | 198 ± 8 | 0.71 ± 0.04 | 107 ± 5 | |
| Continuous light (semidwarf phenotype for lac4-2 lac17) | Wild type | 1.4 ± 0.8 | 213 ± 1 | 93 ± 2 | 308 ± 4 | 0.44 ± 0.01 | 100 |
| lac4-1 | 1.0 ± 0.1 | 177 ± 1 | 83 ± 2 | 261 ± 3 | 0.47 ± 0.01 | 93 ± 1 | |
| lac4-2 | 1.1 ± 0.0 | 178 ± 3 | 79 ± 1 | 258 ± 2 | 0.45 ± 0.01 | 93 ± 2 | |
| lac17 | 1.2 ± 0.1 | 171 ± 9 | 91 ± 4 | 263 ± 13 | 0.53 ± 0.01 | 96 ± 5 | |
| lac4-1 lac17 | 1.1 ± 0.0 | 161 ± 1 | 101 ± 2 | 263 ± 3 | 0.63 ± 0.01 | 106 ± 3 | |
| lac4-2 lac17 | 1.4 ± 0.0 | 78 ± 3 | 64 ± 1 | 143 ± 4 | 0.82 ± 0.02 | 76 ± 1 | |
Data are mean values and standard errors from duplicate analyses.
The relative thioacidolysis yield corresponds to the total yield of (H+G+S) monomers calculated per gram of Klason lignin content and compared to the corresponding control value set at 100.
When calculated on the basis of extract-free stems, the total yields of monomers released by the laccase mutants were lower than the control value, due directly to the lower lignin levels of these mutants. The extract-free stems of the mutant lines with a LAC17 gene knockout released moderately (lac17 and lac4-1 lac17) or substantially (lac4-2 lac17) smaller amounts of G thioacidolysis monomers than the other lines (Table 4). By contrast, they released S monomers in similar (lac17 and lac4-1 lac17) or slightly smaller amounts (lac4-2 lac17, only when semidwarf) than the control. These variations made the S/G ratios of these mutants higher than those of the wild type or LAC4 single mutants.
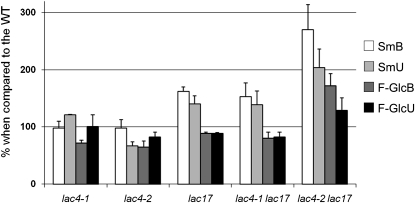
Several recent studies have shown soluble phenolic content to be higher in the hypolignified stems of various Arabidopsis mutants with impaired lignin biosynthesis than in the wild type (Abdulrazzak et al., 2006; Besseau et al., 2007; Mir Derikvand et al., 2008). We further evaluated the impact of laccase mutations on phenylpropanoid metabolism by studying the soluble phenolic compounds extracted from the basal and the upper parts of stems (Figure 4). The lac17, lac4-1 lac17, and lac4-2 lac17 mutants released substantially more sinapoyl malate (162, 153, and 270 in the basal part of stems; 140, 139, and 204 in the upper part of stems, respectively) than the wild type (level arbitrarily set at 100). We also investigated the flavonol glycoside pool of the samples, considering three kaempferol glycosides as the major representatives of this pool (Mir Derikvand et al., 2008). These flavonol glycosides were recovered in larger amounts from the basal and upper parts of lac4-2 lac17 stems (172 and 129, respectively) than from the wild type (level arbitrarily set at 100). By contrast, the amount of soluble phenolic compounds released from lac4-1 or lac4-2 samples was similar to that from the wild type (Figure 4). Thus, disruption of the LAC17 gene induced redirection of the phenylpropanoid pathway. Greater redirection was observed in lac4-2 lac17 stems, which had the lowest lignin content, than in other mutant stems.
Figure 4.
Analyses of Soluble Phenolic Compounds Extracted from the Stems of lac4-1, lac4-2, lac17, lac4-1 lac17, and lac4-2 lac17 Mutants (Grown in Long-Day Conditions).
Data are mean values from three to nine biological replicates. Error bars indicate sd (n = 3 to 9). All results are expressed as percentages of wild-type levels (arbitrarily set at 100). SmB, sinapoyl malate in the basal part of stems; SmU, sinapoyl malate in the upper part of stems; F-Glc B, flavonol glycosides in the basal part of stems; F-Glc U, flavonol glycosides in the upper part of stems.
Evaluation of the Tissue Specificity of LAC4 and LAC17
According to in silico expression data, LAC4 and LAC17 are strongly expressed in the developing stems of Arabidopsis. We tried to localize the expression of these genes more precisely at the tissue level by inserting a 2.0-kb fragment corresponding to the promoters of LAC4 and LAC17 into pBI101GUS-GTW, a vector containing the β-glucuronidase (GUS) reporter gene (Baudry et al., 2006; Dubos et al., 2008). The resulting constructs were introduced into Arabidopsis via Agrobacterium tumefaciens transformation, and several transgenic lines were obtained for each construct. GUS expression was monitored in T1 lines and evaluated more precisely in selected T2 plants. LAC4 was strongly expressed in the differentiating xylem and stem fibers, both highly lignified tissues (Figure 5A). By contrast, LAC17 seemed to be more specifically expressed in the interfascicular fibers (Figure 5B). The localization of the LAC4 and LAC17 proteins was further studied by confocal microscopy with fluorophore-conjugated specific antibodies (see Supplemental Figure 4 online). LAC4 could be detected in the cambium of vascular bundles (see Supplemental Figure 5 online). It was also observed in the secondary wall and in the middle lamella of interfascicular fibers (see Supplemental Figure 6 online). By contrast, LAC17 could be detected only in fibers and not in vascular bundles (see Supplemental Figure 5 online).
Figure 5.
Localization of Laccase Transcripts in Inflorescence Stem Cross Sections of GUS Promoter Fusion Lines (from Plants Grown under Long-Day Conditions).
(A) Expression profile of the 2-kb ProLAC4:GUS.
(B) Expression profile of the 2-kb ProLAC17:GUS.
fi, fibers; vb, vascular bundle; c, cambium. Bars = 100 μm.
As these experiments revealed the tissue specificities of LAC4 (vascular bundles and fibers) and LAC17 (fibers), we then investigated the impact of the downregulation of these genes on the lignification of specific tissues. We performed thioacidolysis on stem tissues recovered by laser capture microdissection. Vascular bundles and interfascicular fibers were recovered from stems collected at stage 6.4 (Boyes et al., 2001) for the wild-type, lac4-2, and lac17 lines. The reproducibility of thioacidolysis yields between biological duplicates was poor. By contrast, measurements of S/G ratio were reproducible (standard errors in the 2 to 10% range). Consistent with previous findings (Ruel et al., 2009), analyses of microdissected samples confirmed that interfascicular fibers were richer in S units than vascular bundles (Table 5). The S/G ratios of vascular bundles did not clearly distinguish the mutant lines from the wild type. By contrast, the S/G ratio was higher in lac4-2 fibers (S/G = 0.75) and much higher in lac17 fibers (up to 0.91) than in wild-type fibers (S/G = 0.57). This finding supports the hypothesis that a LAC17 deficiency affects the deposition of G lignin units in interfascicular fibers.
Table 5.
Lignin Composition in the Vascular Bundles and in the Interfascicular Fibers of Wild-Type, lac4-2, and lac17 Inflorescence Stems (of Plants Grown under Long-Day Conditions)
| S/G Molar Ratio |
||
| Lines | Vascular Bundles | Fibers |
| Wild type | 0.36 ± 0.05 | 0.57 ± 0.04 |
| lac4-2 | 0.30 ± 0.01 | 0.75 ± 0.01 |
| lac17 | 0.28 ± 0.02 | 0.91 ± 0.04 |
The S/G ratio was measured by thioacidolysis of selected tissues collected by laser capture microdissection. Data are mean values and standard errors from two samples (50 microdissections for each one) per tissue type.
Complementation of lac17 with Various Constructs
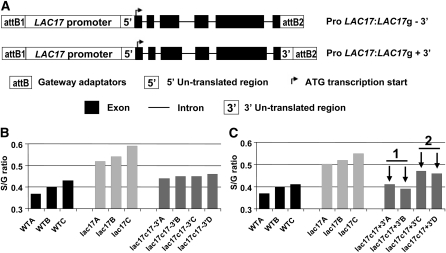
For confirmation of the role of LAC17 in lignification, lac17 was complemented with various constructs (Figure 6A). As a higher thioacidolysis S/G ratio was systematically obtained in LAC17-silenced mutants, the effectiveness of complementation was checked by subjecting the stems of the various transformants to thioacidolysis based on the rationale that effective complementation should restore the S/G ratio to control levels. Complementation was first assayed with the coding sequences of LAC17 (from the ATG to the stop codon) under the control of the cauliflower mosaic virus 35S promoter. This complementation was unsuccessful because the S/G ratio of the mutant was not restored to control levels (see Supplemental Figure 7 online).
Figure 6.
Complementation of lac17 with Various Constructs.
The effectiveness of the complementation was tested by thioacidolysis of mature stems (from plants grown under long-day conditions), assuming that lac17 mutants had stem lignins with a higher S/G thioacidolysis ratio.
(A) Schematic diagram of the Gateway constructs used for lac17 complementation.
(B) Impact of lac17 complementation by the endogenous promoter and genomic sequence of LAC17 without the 3′ UTR (ProLAC17:LAC17g - 3′) on the S/G ratio.
(C) Impact of lac17 complementation by the endogenous promoter and genomic sequence of LAC17 with the 3′ UTR (ProLAC17:LAC17g + 3′) on the S/G ratio. The arrows indicate two lines selected for further lignin content determination on their pooled extract-free stems (lac17c17 +3′ 1; Table 6); the stems of the other two lines were pooled as lac17c17 +3′ 2 (lac17c17 +3′ 2; Table 6).
Another assay was then performed with the genomic sequence of LAC17 under the control of its endogenous promoter with and without the 3′ untranslated region (UTR). The degree of restoration of the S/G ratio indicated that complementation was partially effective in four transformants obtained without the 3′ UTR (Figure 6B), whereas it was complete in two transformants complemented with the construct containing the 3′ UTR (Figure 6C). The effectiveness of complementation with the construct containing the 3′ UTR was further confirmed by Klason analyses of the extract-free stems (Table 6).
Table 6.
Klason Lignin and Acid-Soluble Lignin Content in Extract-Free Mature Stems of lac17-Complemented Lines (Expressed as Weight Percentage)
| Line | KL% | ASL% |
| Wild type | 20.34 ± 0.05 | 0.89 ± 0.01 |
| lac17 | 16.75 ± 0.03 | 0.85 ± 0.01 |
| lac17c17+3′ 1 | 22.66 ± 0.13 | 0.81 ± 0.01 |
| lac17c17+3′ 2 | 20.39 ± 0.02 | 0.87 ± 0.02 |
The analyses were performed for two sets of transgenic lines obtained with the endogenous promoter and genomic sequence of LAC17 with the 3′ UTR. The two complemented samples were obtained by pooling the stems from transgenic lines in which the S/G ratio was completely (lac17c17 +3′ 1) or partially (lac17c17 +3′ 2) restored to wild-type levels. Data are mean values and standard errors from duplicate analyses. KL, Klason lignin; ASL, acid-soluble lignin.
DISCUSSION
The peroxidases and laccases potentially involved in the oxidation of lignin precursors in plant cell walls are encoded by multigene families. In Arabidopsis, 73 peroxidases and 17 laccases have been identified (Tognolli et al., 2002; Pourcel et al., 2005). Despite the high degree of redundancy of peroxidase genes, making it difficult to identify lignin-specific enzymes, peroxidase activity has clearly been implicated in the polymerization of lignins. The most compelling evidence of lignin-specific peroxidases has been provided by a small number of studies of transgenic or mutant plants in which the down- or upregulation of genes encoding particular peroxidases affected lignification (reviewed in Fagerstedt et al., 2010). By contrast, despite the smaller number of laccase genes, the role of laccases in lignification remains less clear. No one has yet demonstrated conclusively that laccases are involved in the lignification of stems. Only two studies have reported alterations to lignification in mutants or transgenic plants with impaired laccase gene expression. Levels of lignin 30% lower than those of the wild type have been found in the seed coat of the LAC15-deficient tt10 Arabidopsis mutant, as determined in thioglycolic acid assays (Liang et al., 2006). Using the same method, Wang et al. (2008) reported 19.6% higher UV absorbance by the thioglycolic acid extract recovered from a poplar transgenic line overexpressing a cotton laccase gene than by the control.
Laccase Gene Expression Profiles in Arabidopsis Stems and the Generation of Two LAC4 LAC17 Double Mutants
Consistent with previous transcript profiling in Arabidopsis primary stems (Ehlting et al., 2005), we found that LAC4 and LAC17 were strongly expressed in stems. Other studies have confirmed that LAC4 and LAC17 are coexpressed with CesA genes (Brown et al., 2005) and with the cinnamyl alcohol dehydrogenase (CAD) genes CAD-C and CAD-D, which are specifically involved in monolignol biosynthesis (Sibout et al., 2005). We therefore selected two different LAC4 mutants, lac4-1 and lac4-2, from the SALK and GABI collections, respectively, and a lac17 mutant from the SALK collection. This choice was further supported by expression profiles from plants harboring the LAC4 or LAC17 promoter regions fused to the GUS reporter gene. LAC4 expression was observed in both vascular bundles and interfascicular fibers, whereas LAC17 expression seemed to be more specific to fibers. These results are consistent with the expression profile determined in another study using the same reporter gene (Koizumi et al., 2009).
We then generated two double mutants by crossing lac4-1 and lac17 and by crossing lac4-2 and lac17. Surprisingly, lac4-1 lac17 displayed a partial restoration of LAC4 transcript levels but efficient downregulation for LAC17. This restoration may be accounted for by the insertion of the lac4-1 T-DNA into the promoter of the LAC4 gene rather than its coding sequence. For unknown reasons, the promoter was not operational in the lac4-1 single mutant, but its function was partially restored in the lac4-1 lac17 double mutant. By contrast, the other double mutant, lac4-2 lac17, displayed effective silencing for both the LAC4 and LAC17 genes. In this double mutant lac4-2 lac17, laccase activity was reduced (to ~50% of control levels), further confirming that LAC4 and LAC17 make a major contribution to total laccase activity in stems.
Disruption of LAC4 and/or LAC17 Affects Plant Development, Vascular Tissues, Lignification, and Saccharification Efficiency to Various Extents
The finding that lac4-2 lac17 displayed either normal development or a semidwarf phenotype, depending on the growth conditions, suggested an effect of stress-inducing conditions. The LAC2-deficient mutant has also been reported to display developmental defects (altered root elongation) only when subjected to dehydration stress (Cai et al., 2006). The precise conditions inducing the semidwarf phenotype in lac4-2 lac17 should be investigated in a more systematic and comprehensive study.
The irx12 mutant (lac4-1 in this work) has been reported to display a weak irregular xylem phenotype that varies in severity between plants and even between the vascular bundles of the same plant (Brown et al., 2005). In our growth conditions, only lac4-2 lac17 consistently displayed a moderate to severe irx phenotype. In the other LAC4-deficient mutants (lac4-1, lac4-2, and lac4-1 lac17), some collapsed vessels were sporadically observed. These results suggest that xylem morphology is not greatly affected when mutants in which LAC4 is silenced are grown in optimum conditions. The lac17 mutant did not display any irx phenotype, consistent with the expression profile of LAC17, which tended to be specifically expressed in fibers.
The extract-free stems of the single mutants had consistently low lignin levels. Surprisingly, these lower lignin levels are reported here for the first time for the lac4-1 mutant, which has been studied before (Brown et al., 2005). Up to eight laccase genes are expressed in Arabidopsis stems. However, the silencing of a single laccase gene (LAC4 or LAC17) is sufficient to decrease lignin content slightly, but noticeably, as shown for two different LAC4 insertion mutants and one LAC17 insertion mutant. The larger decrease (by 40%) observed in the lignin level of the double lac4-2 lac17 knockout mutant was not necessarily associated with a smaller size. Indeed, the semidwarf phenotype of lac4-2 lac17 was obtained in continuous light, which could be related to growth in stress-inducing conditions. When lac4-2 lac17 was not subjected to stress during its growth, its size was similar to that of wild-type plants, despite its much lower lignin content. Several studies have reported the absence of a strict correlation between low lignin levels (30 to 40% lower than the wild type) and dwarfism. For instance, the cad c cad d double mutant has 40% lower Klason lignin levels than the wild type but displays no dwarfism, only a bending stem phenotype (Sibout et al., 2005). The stems of the CCoAOMT1 mutant are also normal in size, despite having lignin levels ~30% lower than those of the wild type (Do et al., 2007). By contrast, two CCR1-deficient mutants display a 40% decrease in lignin levels and a dwarf phenotype, whatever the growth conditions (Mir Derikvand et al., 2008). In this study, the hypolignified lac4-2 lac17 stems were either normal in size or semidwarf. The reason for the dwarfing of mutant stems does not seem to be directly related to their low lignin content. Instead, it seems to be more strongly correlated with the sites at which lignin levels are decreased and to the appearance of an irregular xylem phenotype.
It often has been suggested that lower levels of growth result from the accumulation of flavonol glycosides (Abdulrazzak et al., 2006; Besseau et al., 2007; Mir Derikvand et al., 2008). However, growth reduction in the hypolignified C3H Arabidopsis mutant has recently been shown to be independent of flavonoids (Li et al., 2010). The stems of LAC17 mutants (lac17, lac4-1 lac17, and lac4-2 lac17) contained high levels of sinapoyl malate. This accumulation seems to be correlated with the decrease in lignin levels, providing support for the hypothesis that phenolic intermediates not used for lignification in the LAC17-deficient mutants are redirected for sinapoyl malate biosynthesis. This redirection seems to occur even when the flux to lignin is decreased only slightly (by 10% in the lac17 single mutant). By contrast, the soluble phenolic pool is not affected in the lac4-1 and lac4-2 mutants, highlighting the complexity of crosstalk between lignin formation and the biosynthesis of other plant phenolics.
We evaluated lignin structure by thioacidolysis. As the lignin structures generating the H, G, and S thioacidolysis monomers are restricted to those involved in β-O-4 bonds, a higher frequency of resistant bonds (referred to as condensed bonds) in lignins has been demonstrated by low thioacidolysis yields (expressed in μmoles per gram of lignin) in various Arabidopsis mutants, such as COMT-, CAD-, C3H-, and CCR-deficient mutants (Goujon et al., 2003; Sibout et al., 2005; Abdulrazzak et al., 2006; Mir Derikvand et al., 2008). By contrast, the silencing of LAC4 or LAC17 had no effect on the frequency of condensed bonds in stem lignins, except for the semidwarf lac4-2 lac17 samples (thioacidolysis yield 25% lower than the control level when calculated in μmoles per gram of lignin). This result provides further support for the hypothesis that the semidwarf phenotype of lac4-2 lac17 is induced by stress, as lignins formed in response to stress are enriched in condensed bonds (Cabané et al., 2004; Betz et al., 2009).
Lignin composition, as reflected by the S/G thioacidolysis ratio, was affected to various extents. No clear change was observed in the lac4-1 and lac4-2 mutants. By contrast, the S/G ratio of stem lignins was altered in LAC17-deficient mutants to an extent that seemed to be correlated with the decrease in lignin levels. The higher S/G ratio of LAC17-silenced mutants resulted principally from a lower recovery of G monomers, whereas the recovery of S monomers from the extract-free cell walls was not affected. In other words, the lower lignin levels of LAC17-deficient mutants resulted essentially from lower levels of G lignin unit deposition.
Thioacidolysis of laser capture microdissected vascular bundles and of interfascicular fibers provided conclusive evidence about the tissue specificity of the LAC4 and LAC17 enzymes. Consistent with previous findings (Chapple et al., 1992; Ruel et al., 2009), the interfascicular fibers of the wild type had a higher S/G ratio than the corresponding vascular bundles. This result is consistent with many published data for angiosperm lignins, reporting a higher abundance of S units in supportive tissues, with G-rich lignins found in the xylem. The silencing of LAC4 and LAC17 induced a weak or more pronounced increase in fiber S/G ratio, respectively, whereas the analyses of lac4-1 or lac4-2 stems did not reveal any effect on S/G. This highlights the importance of using analytical methods capable of providing information about lignin variability in individual tissues.
Unsurprisingly, the stems of lac4-2 lac17 with 40% lower lignin levels displayed markedly higher levels of saccharification when treated with a commercial cellulase preparation in the absence of pretreatment. Accordingly, laccase silencing may be a promising strategy for increasing the saccharification of plant cell walls when used for lignocellulose-to-ethanol biological conversion. Consistent with the small decrease in lignin content observed in the lac4-1 and lac4-2 mutants, only a slight increase in saccharification was observed in these mutants. By contrast, despite having a substantially lower lignin content, lac17 had a level of saccharification similar to that of the control sample. The reasons for this unexpected result remain unclear, but it shows that factors other than lignin content may affect the susceptibility of cell walls to enzymatic hydrolysis.
The Large Decrease in Lignin Biosynthesis in the Double Mutant Silenced for LAC4 and LAC17 May Result from Broader Changes to the Phenylpropanoid Pathway
Disrupting both LAC4 and LAC17 had no significant effect on the expression of the other stem-specific laccase genes. By contrast, in lac4-2 lac17 and, to a lesser extent, in lac4-1 lac17, several genes involved in the phenylpropanoid pathway were found to be downregulated. This coordinated downregulation may partly account for the large decrease in metabolic flux to lignins in this mutant. Microarray analyses revealed no other major changes in the expression of genes for other metabolic pathways, by contrast with the pal1 pal2 Arabidopsis mutant (Rohde et al., 2004), the cad c cad d Arabidopsis mutant (Sibout et al., 2005), and the poplar (Leplé et al., 2007) or tobacco (Nicotiana tabacum; Dauwe et al., 2007) lines presenting CCR downregulation.
The Final Piece in the Puzzle, Demonstrating the Existence of Lignin-Specific Laccases, Is Provided by Complementation Experiments with a Laccase-Deficient Mutant
For a conclusive demonstration of the involvement of LAC4 or LAC17 in the constitutive lignification of Arabidopsis stems, it was of the utmost importance to restore the control profile from the altered lignin profile through appropriate complementation experiments. Therefore, we chose to complement the lac17 mutant, based on the rationale that there are two lignin parameters to be restored: S/G ratio and lignin content. Complete restoration could be obtained for transgenic lines only when the LAC17 genomic sequence was used under the control of its own 2-kb promoter, including the 3′ UTR region. Complementation was partial in the absence of the 3′ UTR, as already reported for complementation studies of the Arabidopsis tt10 mutant (Pourcel et al., 2005). This 3′ UTR may help to stabilize the transcript, as proposed by Gutiérrez et al. (1999). The effective complementation of lac17 by the genomic sequence of LAC17 under the control of its full-length promoter involved full restoration not only of S/G ratios but also of Klason lignin levels in stems to control values. This provides definitive proof of the involvement of LAC17 in the constitutive lignification of Arabidopsis inflorescence stems.
This study demonstrates that both LAC4 and LAC17 are involved in the constitutive lignification of Arabidopsis floral stems. Based on all the results obtained in expression profile studies, histochemical analyses, and wet chemistry analyses, we can suggest the following. Both the LAC4 and LAC17 laccases play major roles in the lignification of Arabidopsis stems, with LAC17 being more specific to interfascicular fibers. The silencing of these lignin-specific laccase genes markedly decreases lignin biosynthesis, with some redirection to sinapoyl malate synthesis. The silencing of LAC17 specifically affects the deposition of G lignin units in fibers, whereas the unit specificity of LAC4 is less clear. Previous studies (Sterjiades et al., 1992; Donaldson, 2001) suggested a role for laccases in the early steps of lignin polymerization. It is now well established that lignification begins with the deposition of G lignin units (Terashima and Fukushima, 1993). Therefore, we can hypothesize that LAC17 participates in the early stages of lignification accompanying the deposition of G lignin units. These findings suggest that the genetic engineering of lignin-specific laccases is a potentially innovative and promising tool for the fine-tuning of lignin content and structure.
METHODS
Plant Materials and Growth Conditions
All the plants used were from the Columbia background (Col-0). The lac4-1 (S_051892) and lac17 (S_016748) mutants were obtained from the Salk Institute T-DNA insertion collection. The lac4-2 (GK-720G02) mutant was obtained from the GabiKat collection. The lac4-1 lac17 and lac4-2 lac17 double mutants were isolated from the F2 population of the lac4-1 lac17 and lac4-2 lac17 crosses, respectively. Homozygous plants were obtained by genomic PCR. The list of primers is reported in Supplemental Table 1 online. Most cultures were performed in long-day conditions (16/8 h light/dark) in a greenhouse or growth chamber (20°C, 60% relative humidity). Some cultures were performed in a growth chamber under continuous light (21°C, 60% relative humidity). For RT-PCR and microarray analyses, plants were arranged at random in the same growth chamber and grown under long-day conditions.
Histology
Cross sections were cut from stems collected at development stage 6.4 (Boyes et al., 2001), corresponding to a plant height of 30 cm. Wiesner staining was performed with phloroglucinol-HCl (Prolabo), and Maüle staining was performed by incubating sections in 1% KMnO4. After incubation in the staining solution for 7 min, sections were washed and acidified by incubation with 30% HCl for 1 min. They were then washed again and incubated with 5% NaHCO3.
GUS Fusion Constructs for the Analysis of Promoter Activity and Histochemical Detection of GUS Activity
We used a 1.95-kb promoter fragment for LAC4 and a 2-kb promoter fragment for LAC17. These promoter fragments were amplified by PCR with primers containing attB1 and attB2 GATEWAY recombination sites (see Supplemental Table 1 online) and introduced into the pDONR207 vector by BP recombination. The promoter fragments were transferred into the binary vector pBI101GUS-GTW by LR recombination (Baudry et al., 2006; Dubos et al., 2008). The integrity of the constructs was checked by sequencing and plants were transformed as described by Clough and Bent (1998).
The promoter expression profile was characterized by measuring GUS activity in at least five independent lines of transformants. All the plants were genotyped for the presence of the corresponding constructs. The same pattern of promoter activity was observed in T2 plants.
Histochemical analyses of GUS activity were performed as described by Debeaujon et al. (2003). Samples were incubated under a vacuum for 1 h and then for 12 h at 37°C in the dark. Chlorophyll was removed by incubating the samples in 70% (v/v) ethanol/water at room temperature. Tissues were stained by incubation with 5-bromo-4-chloro-3-indolyl glucuronide for 3 h at room temperature. The stained tissues were then cleared by incubation with a chloral hydrate solution (chloral hydrate:distilled water:glycerol [8:2:1,w/v/v]) and observed as whole mounts and stem cross sections. Tissues were observed under a binocular or Axioplan 2 microscope (Zeiss) equipped with bright-field optics and with Nomarski differential interference contrast optics.
Immunolocalization by Indirect Immunofluorescence Analysis
Immunolocalization was performed on stem cross sections from plants at stage 6.2 (Boyes et al., 2001) according to the method of Masclaux-Daubresse et al. (2006), modified as follows: (1) slides were incubated with the same rabbit antibodies as for immunoblots; (2) slides were then incubated in Evans Blue solution (0.001% in PBS: 10 min) to decrease the autofluorescence of lignified cells; and (3) after incubation with the secondary antibodies (goat anti-rabbit IgG labeled with Alexa 488; Molecular Probes), slides were treated with 4',6-diamidino-2-phenylindole. Immunofluorescence was observed with an epifluorescence microscope (DMRB DIC; Leica), and the specificity of the Alexa 488 signal was determined with a spectral confocal laser scanning microscope. No signal was observed when using the preimmune serum.
RNA Extraction and RT-PCR
Stems of plants grown in the same growth chamber were collected at stage 6.2 (Boyes et al., 2001). For each line, we pooled the entire stems of three plants to constitute one sample. Samples were immediately frozen in liquid nitrogen, ground to powder, and stored at −80°C until RNA extraction.
RNA was extracted with the Qiagen RNeasy Plant Minikit according to the manufacturer’s instructions. Qiagen DNase was applied to the column to eliminate contaminating DNA. RNA samples were quantified with a Nanodrop spectrophotometer (ND-1000; Labtech). Reverse transcription was performed with 1 μg of total RNA in a final volume of 20 μL, with Superscript Reverse Transcriptase II (Invitrogen), used according to the manufacturer’s instructions. PCR was performed in a thermocycler (GeneAmp PCR System 2700).
Transcriptome Studies
Microarray analysis was performed on complete Arabidopsis thaliana transcriptome microarrays containing 24,576 GSTs corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). Two independent biological replicates were performed. For each biological replicate and each point, we pooled the RNA from three plants to generate each sample. We collected 20-cm-long stems from three plants at growth stage 6.2 (Boyes et al., 2001). The plants concerned had been grown in a growth chamber under long-day conditions. Total RNA was extracted with the RNeasy Plant Kit (Qiagen) according to the manufacturer’s instructions. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e., four hybridizations per comparison). We labeled cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products) and performed hybridization and scanning of the slides as previously described (Lurin et al., 2004).
Statistical Analysis of Microarray Data
Experiments were designed with the statistics group of the Plant Genomics Research Unit. Statistical analysis was performed with normalization based on dye swapping (i.e., four arrays, each containing 24,576 GSTs and 384 controls) as previously described (Gagnot et al., 2008). For the identification of differentially expressed genes, we performed a paired t test on log ratios, assuming that the variance of the log ratios was similar for all genes. Spots with extreme variances (too small or too large) were excluded. The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate (with a type I error equal to 5%) to minimize the number of false positives in a multiple-comparison context (Ge et al., 2003). We considered genes with a Bonferroni P value ≤ 0.05 to be differentially expressed, as previously described (Gagnot et al., 2008).
Data Deposition
Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE22907) and at CATdb (http://urgv.evry.inra.fr/CATdb/; Project: RA10-01_Laccases) according to Minimum Information about a Microarray Experiment standards.
Purification of Arabidopsis Laccases
About 7 g of Arabidopsis stems was homogenized in 7 mL of extraction buffer (25 mM BisTris, pH 7, 200 mM CaCl2, 10% [v/v] glycerol, 4 μM sodium cacodylate, and 1/200 [v/v] protease inhibitor cocktail [P-9599; Sigma-Aldrich]) for 5 min in a blender. The homogenate was centrifuged twice at 8°C, for 5 min each, at 3000g and once at 4°C for 5 min at 13,000g. The supernatant was centrifuged at 8°C for 45 min at 15,000g. Proteins were purified by affinity chromatography on a 0.5 × 3-cm column filled with 1 mL of Concanavalin-A Sepharose (Sigma-Aldrich) and washed with 3 mL of 20 mM Tris-HCl and 0.5 M NaCl buffer, pH 7.4. The soluble protein extract was loaded and the column was washed with 10 mL of buffer. The proteins were eluted with 0.2 M methyl-α-glucopyranoside in the same buffer. The eluates were collected (1 mL per fraction), and 3 or 5 μL samples from each fraction were tested for laccase activity. Pooled fractions showing laccase activity were equilibrated in 25 mM Tris-HCl buffer, pH 7.4, supplemented with 5% glycerol (v/v) and 0.015% Triton X-100 (v/v). Glycerol was added to the buffer to prevent partial inactivation of the enzymes.
SDS-PAGE and Protein Gel Blot Analysis
Protein-denaturating SDS-PAGE was performed with 10% polyacryamide gels. Standard markers (molecular range 15 to 100 kD; Sigma-Aldrich) were used to determine the approximate molecular masses of purified proteins in Coomassie Brilliant Blue–stained gels. Proteins were transferred onto a 0.45-μm Hybond ECL membrane (Amersham Biosciences) by electroblotting. Proteins were detected with alkaline phosphatase–conjugated primary monoclonal antibodies against laccases and secondary antibodies against alkaline phosphatase conjugate.
In Vitro Laccase Activity Assays
An 11 mg mL−1 solution of the substrate, 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS), was prepared in DMSO and stored in aliquots at −20°C. Laccase activity was determined at 30°C by the oxidation of ABTS to generate a stable cationic radical assayed by spectrometry at 420 nm. The reaction mixture contained 100 mM acetate buffer, pH 5, 1 mM ABTS, and 20 μL of protein extract in a total volume of 200 μL.
Lignin Analysis
Dry mature stems from control and transgenic lines were harvested. Siliques and leaves were systematically removed. Extract-free samples were obtained by removing all soluble compounds with a Soxhlet apparatus by sequentially extracting the ground material with toluene:ethanol (2/1, v/v), ethanol, and water. The lignin content of dried mature stems was estimated from the extract-free samples by the standard procedures (Dence, 1992). The lignin composition of extract-free material was studied by thioacidolysis, as previously described (Lapierre et al., 1995). The lignin-derived thioacidolysis monomers were identified by gas chromatography–mass spectrometry as their trimethylsilylated derivatives.
Determination of Stem Soluble Phenolic Compounds
The soluble phenolic compounds present in stems collected at stage 6.4 (Boyes et al., 2001) were extracted in 80:20 methanol/water (v/v, HPLC quality) after the addition of the internal standard (morine, 10 μg) and identified by liquid chromatography–mass spectrometry analyses (electrospray ionization, negative mode) as previously described (Goujon et al., 2003; Mir Derikvand et al., 2008). These soluble phenolic compounds were extracted from 1-cm-long sections of 40-cm-high stems obtained from the basal part of the stem (1 cm from the base of the stem) or the upper part of the stem (3 cm below the inflorescence).
Cellulolysis
Cellulolysis assays were performed on extract-free cell walls. Twenty to thirty milligrams of sample was placed in 30 mL of 0.05 M sodium acetate buffer, pH 4.7, supplemented with 2 mg mL−1 commercial cellulase (cellulase Onozuka-R10; Serva). After 5 d of incubation at 37°C with magnetic stirring, samples were filtered and weighed again. The Glc released into the filtrate was quantified with the Biomerieux Glucose RTU kit (bioMérieux).
Procedure Used for Laser Capture Microdissection
For this protocol, no specific preparation of the tissues was required. Stem tissues harvested from plants at stage 6.4 (Boyes et al., 2001) were cut into pieces of ~1 to 2 cm each and immediately frozen in a cryotome (CM 1510S Leica) at −20°C, and 40-μm cross sections were cut. These sections were placed directly on slides for microdissection. A Palm Microbeam system (Zeiss microdissector) was used. This system is equipped for laser capture microdissection and pressure catapulting. Selected areas of the cross sections were subsequently cut with a UV A laser (337 nm). After the laser pressure catapulting pulse had been applied to the cut line, the samples were collected in the caps of Eppendorf tubes. Fifteen microliters of Versol water was added to each cap containing 50 microdissected samples. The samples were carefully transferred to a glass tube by repeated rinsing of the Eppendorf with small volumes of water. The samples in the glass tubes were then freeze-dried for thioacidolysis, which was performed as previously described (Ruel et al., 2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g29130 (LAC2), At2g38080 (LAC4), At2g40370 (LAC5), At2g46570 (LAC6), At5g01040 (LAC8), At5g01190 (LAC10), At5g03260 (LAC11), At5g05390 (LAC12), At5g48100 (LAC15), and At5g60020 (LAC17).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Laccase Gene Expression Profiles Obtained from the GeneCAT Database.
Supplemental Figure 2. Quantitative RT-PCR Validation of Microarray Data Performed in Triplicate on Stage 6.2 Primary Stems.
Supplemental Figure 3. Cross Sections of Arabidopsis Stems (Long-Day Conditions) Stained with Phloroglucinol.
Supplemental Figure 4. Peptide Sequences Used for the Generation of Antibodies against LAC4 and LAC17.
Supplemental Figure 5. Immunolocalization of LAC4 and LAC17.
Supplemental Figure 6. Subcellular Localization of LAC4 in the Fiber Cell Wall and Middle Lamella.
Supplemental Figure 7. Complementation of lac4-1 lac17 with Pro35S:LAC17c (ATG-Stop).
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Transcriptome Studies of lac4-1, lac4-2, lac17, lac4-1 lac17, and lac4-2 lac17 by Microarray Analysis.
Supplementary Material
Acknowledgments
We thank Frédéric Legée (Unité Mixte de Recherche 1318 Institut National de la Recherche Agronomique [INRA]-AgroParisTech) for running lignin determinations and the greenhouse management team (Unité Mixte de Recherche 1318 INRA-AgroParisTech) for growing the plants in greenhouses. This work was supported by a PhD grant from INRA to S.B. and was partly funded by the French Genoplante MAIZEWALL and the UE RENEWALL programs. The liquid chromatography–mass spectrometry analyses were performed with a mass spectrometer obtained with a Sesame Grant from INRA and Région Ile de France.
References
- Abdulrazzak N., et al. (2006). A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 140: 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E. (1977). Lignin chemistry. Past, present and future. Wood Sci. Technol. 11: 169–218 [Google Scholar]
- Bao W., O’malley D.M., Whetten R., Sederoff R.R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674 [DOI] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Besseau S., Hoffmann L., Geoffroy P., Lapierre C., Pollet B., Legrand M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz G.A., Knappe C., Lapierre C., Olbrich M., Welzl G., Langebartels C., Heller W., Sandermann H., Ernst D. (2009). Ozone affects shikimate pathway transcripts and monomeric lignin composition in European beech (Fagus sylvatica L.). Eur. J. For. Res. 128: 109–116 [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Zeef L.A.H., Ellis J., Goodacre R., Turner S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabané M., Pireaux J.-C., Léger E., Weber E., Dizengremel P., Pollet B., Lapierre C. (2004). Condensed lignins are synthesized in poplar leaves exposed to ozone. Plant Physiol. 134: 586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.N., Davis E.J., Ballif J., Liang M.X., Bushman E., Haroldsen V., Torabinejad J., Wu Y.J. (2006). Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 57: 2563–2569 [DOI] [PubMed] [Google Scholar]
- Caparros-Ruiz D., Fornale S., Civardi L., Puigdomenech P., Rigau J. (2006). Isolation and characterisation of a family of laccases in maize. Plant Sci. 171: 217–225 [Google Scholar]
- Chapple C.C., Vogt T., Ellis B.E., Somerville C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwe R., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Dean J.F., LaFayette P.R., Rugh C., Tristram A.H., Hoopes J.T., Eriksson K.E., Merckle S.A. (1998). Laccases associated with lignifying vascular tissue. Lignin and Lignan Biosynthesis, Lewis N.G., Sarkanen S., (Washington, DC: American Chemical Society; ), pp. 96–108 [Google Scholar]
- Dean J.F.D., Eriksson K.E.L. (1994). Laccase and the deposition of lignin in vascular plants. Holzforschung 48: 21–33 [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dence C. (1992). Lignin determination. Methods in Lignin Chemistry, Lin S.Y., Dence C.W., (Berlin: Springer-Verlag; ), pp. 33–61 [Google Scholar]
- Do C.T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Donaldson L.A. (2001). Lignification and lignin topochemistry - An ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Ehlting J., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Fagerstedt K., Kukkola E., Koistinen V.V.T., Takahashi J., Marjamaa K. (2010). Cell wall lignin is polymerized by class II secretable plant peroxidase in Norway spruce. J. Integr. Plant Biol. 52: 186–194 [DOI] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y.C., Dudoit S., Speed T.P. (2003). Resampling-based multiple testing for microarray data analysis. Test 12: 1–77 [Google Scholar]
- Goujon T., Sibout R., Pollet B., Maba B., Nussaume L., Bechtold N., Lu F., Ralph J., Mila I., Barrière Y., Lapierre C., Jouanin L. (2003). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Gutiérrez R.A., MacIntosh G.C., Green P.J. (1999). Current perspectives on mRNA stability in plants: Multiple levels and mechanisms of control. Trends Plant Sci. 4: 429–438 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegger P.J., Kilaru S., James T.Y., Thacker J.R., Kües U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273: 2308–2326 [DOI] [PubMed] [Google Scholar]
- Higuchi T., Ito Y. (1958). Dehydrogenation products of coniferyl alcohol formed by the action of mushroom phenol oxidase, rhus laccase and radish peroxidase. J. Biochem. 45: 575–579 [Google Scholar]
- Kärkönen A., Koutaniemi S., Mustonen M., Syrjänen K., Brunow G., Kilpeläinen I., Teeri T.H., Simola L.K. (2002). Lignification related enzymes in Picea abies suspension cultures. Physiol. Plant. 114: 343–353 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Lu F., Pilate G., Leplé J.-C., Pollet B., Lapierre C. (2002). Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J. Biol. Chem. 277: 47412–47419 [DOI] [PubMed] [Google Scholar]
- Koizumi K., Yokoyama R., Nishitani K. (2009). Mechanical load induces upregulation of transcripts for a set of genes implicated in secondary wall formation in the supporting tissue of Arabidopsis thaliana. J. Plant Res. 122: 651–659 [DOI] [PubMed] [Google Scholar]
- Lapierre C. (2010). Determining lignin structure by chemical degradations. Lignin and Lignans: Advances in Chemistry, Heitner C., Dimmel D., Schmidt J.A., (Boca Raton, FL: CRC Press, Taylor & Francis Group; ), pp. 11–48 [Google Scholar]
- Lapierre C., Pollet B., Rolando C. (1995). New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res. Chem. Intermediat. 21: 397–412 [Google Scholar]
- Leplé J.C., et al. (2007). Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bonawitz N.D., Weng J.-K., Chapple C. (2010). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M.X., Davis E., Gardner D., Cai X.N., Wu Y.J. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Pageau K., Lelandais M., Grandjean O., Kronenberger J., Valadier M.H., Feraud M., Jouglet T., Suzuki A. (2006). Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig B.C., Meagher R.B., Dean J.F.D. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221: 619–636 [DOI] [PubMed] [Google Scholar]
- Mir Derikvand M., Sierra J.B., Ruel K., Pollet B., Do C.-T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008). Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Nakano J., Meshitsuka G. (1992). The detection of lignin. Methods in Lignin Chemistry, Lin S.Y., Dence C.W., (Berlin: Springer-Verlag; ), pp 23–32 [Google Scholar]
- Pourcel L., Routaboul J.M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. (2005). TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P., Chabannes M., Chamayou S., Danoun S., Jauneau A., Boudet A.-M., Goffner D. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 129: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A., McDougall G.J. (1997). A laccase-type polyphenol oxidase from lignifying xylem of tobacco. Phytochemistry 44: 229–235 [Google Scholar]
- Rohde A., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K., Berrio-Sierra J., Derikvand M.M., Pollet B., Thévenin J., Lapierre C., Jouanin L., Joseleau J.P. (2009). Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjiades R., Dean J.F.D., Eriksson K.E.L. (1992). Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 99: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima N., Fukushima K. (1993). Comprehensive model of the lignified plant cell wall. Forage Cell Wall Structure and Digestibility, Jung H.G., Buxton D.R., Hatfield R.D., Ralph R., (Madison, WI: American Society of Agronomy; ), pp. 247–270 [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Wang G.D., Li Q.J., Luo B., Chen X.Y. (2004). Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat. Biotechnol. 22: 893–897 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang C.L., Zhu M.L., Yu Y., Zhang Y.B., Wei Z.M. (2008). Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult. 93: 303–310 [Google Scholar]
- Yoshida H. (1883). Chemistry of lacquer (urushi). J. Chem. Soc. 43: 472–486 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.