This study identified WRKY8 as a downstream target of three mitogen-activated protein kinases in Nicotiana benthamiana. Phosphorylation of WRKY8 increased its DNA binding activity, and ectopic expression of WRKY8 induced the expression of various defense-related genes.
Abstract
Mitogen-activated protein kinase (MAPK) cascades have pivotal roles in plant innate immunity. However, downstream signaling of plant defense-related MAPKs is not well understood. Here, we provide evidence that the Nicotiana benthamiana WRKY8 transcription factor is a physiological substrate of SIPK, NTF4, and WIPK. Clustered Pro-directed Ser residues (SP cluster), which are conserved in group I WRKY proteins, in the N-terminal region of WRKY8 were phosphorylated by these MAPKs in vitro. Antiphosphopeptide antibodies indicated that Ser residues in the SP cluster of WRKY8 are phosphorylated by SIPK, NTF4, and WIPK in vivo. The interaction of WRKY8 with MAPKs depended on its D domain, which is a MAPK-interacting motif, and this interaction was required for effective phosphorylation of WRKY8 in plants. Phosphorylation of WRKY8 increased its DNA binding activity to the cognate W-box sequence. The phospho-mimicking mutant of WRKY8 showed higher transactivation activity, and its ectopic expression induced defense-related genes, such as 3-hydroxy-3-methylglutaryl CoA reductase 2 and NADP-malic enzyme. By contrast, silencing of WRKY8 decreased the expression of defense-related genes and increased disease susceptibility to the pathogens Phytophthora infestans and Colletotrichum orbiculare. Thus, MAPK-mediated phosphorylation of WRKY8 has an important role in the defense response through activation of downstream genes.
INTRODUCTION
Plants have evolved a system of defense responses to protect themselves against colonization by pathogens. The plant innate immunity system consists of two primary layers. One layer relies on the perception of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (Zipfel, 2008). This recognition activates basal defense to prevent penetration and to restrict growth of pathogens (Jones and Dangl, 2006). The second layer is the recognition of pathogen effector molecules through host resistance (R) proteins. R gene–mediated resistance triggers strong gene-for-gene resistance that often includes hypersensitive response (HR) cell death.
Mitogen-activated protein kinase (MAPK) signaling pathways have important roles in both basal defense and R gene–mediated resistance. Many studies have characterized the tobacco (Nicotiana tabacum) MAPKs, wound-induced protein kinase (WIPK; Seo et al., 1995), and salicylic acid–induced protein kinase (SIPK; Zhang and Klessig, 1997) and their orthologs in other plant species (Katou et al., 2005; Nakagami et al., 2005). WIPK and SIPK participate either in N gene–dependent resistance against Tobacco mosaic virus (Zhang and Klessig, 1998; Jin et al., 2003), Cf-9–dependent responses to Cladosporium fulvum–derived elicitor Avr9 (Romeis et al., 1999), or in PAMP-mediated basal defense against the fungal pathogen Colletotrichum orbiculare (Tanaka et al., 2009). WIPK and SIPK share MEK2, a common upstream mitogen-activated protein kinase kinase (MAPKK; Yang et al., 2001). Expression of MEK2DD, a constitutively active mutant of MEK2, induces HR-like cell death, defense gene expression, and generation of nitric oxide and reactive oxygen species, all preceded by activation of endogenous WIPK and SIPK (Yang et al., 2001; Ren et al., 2002; Yoshioka et al., 2003; Asai et al., 2008). Transgenic potato (Solanum tuberosum) plants carrying MEK2DD driven by a pathogen-inducible promoter show resistance to Phytophthora infestans and Alternaria solani (Yamamizo et al., 2006). MPK3 and MPK6 (WIPK and SIPK orthologs, respectively, in Arabidopsis thaliana) cooperatively regulate the production of the antifungal compound camalexin, an indolic phytoalexin of Arabidopsis (Ren et al., 2008). NTF4, which shares 93.6% identity with SIPK, is thought to be functionally redundant with SIPK in the defense-related signaling pathway (Ren et al., 2006; Asai et al., 2008). The cytokinesis-related MAPK cascade NPK1-MEK1-NTF6 is also essential for N- and Pto-dependent resistance (Jin et al., 2002; del Pozo et al., 2004; Liu et al., 2004), as well as for reactive oxygen species production (Asai et al., 2008). Despite extensive accumulation of evidence that shows the importance of MAPKs in basal defense and R gene–mediated resistance, the mechanisms through which MAPKs transduce the signals are largely unknown.
WRKY proteins comprise a large family of plant transcription factors (Eulgem et al., 2000). WRKY family members are divided into three groups based on the number of WRKY domains and certain features of the zinc finger–like motifs (Eulgem et al., 2000). WRKY proteins with two WRKY domains belong to group I, and proteins with one WRKY domain belong to group II or III based on features of the zinc finger motif. WRKY proteins bind W-box sequences (TTGACC/T) in the promoter region of target genes (Eulgem et al., 2000; Ciolkowski et al., 2008). Loss-of-function and gain-of-function studies showed that WRKY transcription factors participate in defense responses either as positive or as negative regulators (Eulgem and Somssich, 2007; Shen et al., 2007). Some WRKYs are regulated by MAPKs at the transcriptional and posttranscriptional levels in defense-related signaling pathways. Arabidopsis WRKY22 and WRKY29 are transcriptionally induced by MPK3 and MPK6 and confer resistance to both bacterial and fungal pathogens (Asai et al., 2002). WRKYs are phosphorylated by MAPKs in vitro (Andreasson et al., 2005; Katou et al., 2005; Menke et al., 2005; Popescu et al., 2009). However, whether these WRKYs are directly phosphorylated by MAPKs in vivo is unknown. Likewise, the effect of MAPK-mediated phosphorylation on the function of WRKY remains to be defined.
Here, we report that Nicotiana benthamiana WRKY8 is a physiological substrate of SIPK, NTF4, and WIPK. SIPK phosphorylates the SP cluster of WRKY8, which is highly conserved in the N-terminal region of group I WRKY proteins. SIPK-mediated phosphorylation of WRKY8 increased its binding activity to the W-box sequence and transactivation activity. Phospho-mimicking mutant and virus-induced gene silencing (VIGS) of WRKY8 showed that MAPK-mediated phosphorylation of WRKY8 has an important role in plant immunity through activation of downstream genes.
RESULTS
Multisite Phosphorylation of WRKY8 by SIPK
We previously identified proteins phosphorylated in vitro by St MPK1, an SIPK ortholog in potato, using an in vitro expression cloning method (Katou et al., 2005). In this study, we designated a clone that contains two putative WRKY domains as St WRKY8. N. benthamiana, like potato, belongs to Solanaceae and is useful to study biological functions by Agrobacterium tumefaciens–mediated transient expression and VIGS. We isolated a cDNA fragment of Nb WRKY8 from N. benthamiana by PCR using primers designed based on the St WRKY8 sequence. The deduced amino acid sequence of Nb WRKY8 contained two WRKY domains and belonged to group I WRKY proteins (see Supplemental Figure 1A online). Ser or Thr followed by Pro (SP or TP) is a minimal consensus motif for MAPK phosphorylation (Sharrocks et al., 2000). WRKY8 contains seven potential MAPK phosphorylation sites, five of which are concentrated in the N-terminal region (SP cluster). Interestingly, WRKY8 also contained a deduced D domain adjacent to the N-terminal side of the SP cluster (see Supplemental Figure 1A online). The ability of MAPKs to effectively and specifically recognize their substrates is increased by their capacity to bind to those substrates with relatively high affinity. Often, this direct interaction occurs by way of MAPK-docking sites on substrate proteins (Sharrocks et al., 2000). The D domain is the best described docking motif whose general features include a cluster of basic residues upstream of the LxL motif ([K/R]1-2-x2-6-[L/I]-x-[L/I]) (Sharrocks et al., 2000). A closer examination of Arabidopsis, rice (Oryza sativa), and Solanaceae plant group I WRKY amino acid sequences showed that the SP cluster and deduced D domain are conserved in the N-terminal regions of several group I WRKYs, inferring a functional importance (see Supplemental Figure 1B and Supplemental Data Set 1 online). The closest homolog of Nb WRKY8 in Arabidopsis is WRKY33 (48.2% amino acid identity).
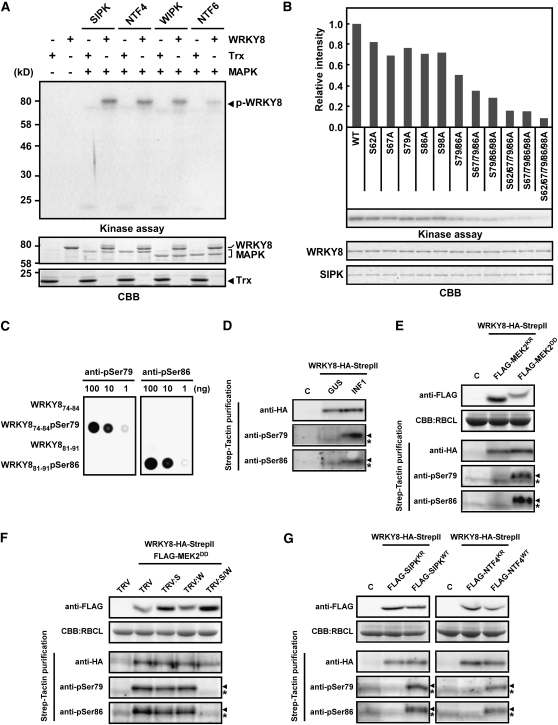
In tobacco, SIPK, NTF4, WIPK, and NTF6 are characterized as pathogen-responsive MAPKs (Nakagami et al., 2005; Ren et al., 2006). To determine which MAPK phosphorylates WRKY8, we performed in vitro assays using active SIPK, NTF4, WIPK, and NTF6 that had been phosphorylated by cognate recombinant constitutively active MAPKK in vitro. All the active MAPKs showed myelin basic protein (MBP) phosphorylation activity in vitro (see Supplemental Figure 2 online). Thioredoxin (Trx)-fused WRKY8 recombinant protein expressed in Escherichia coli was phosphorylated by SIPK, NTF4, and WIPK, but phosphorylation of Trx-WRKY8 by NTF6 was to a lesser degree (Figure 1A). To investigate the phosphorylation site, we prepared the Trx-fused N-terminal half (WRKY81-170) and C-terminal half (WRKY8171-538) of recombinant WRKY8, which then underwent an in vitro phosphorylation assay using SIPK. The WRKY81-170, but not WRKY8171-538, was phosphorylated, suggesting that the N-terminal region is phosphorylated by SIPK (see Supplemental Figure 3 online). Potential phosphorylation sites Ser-62, Ser-67, Ser-79, Ser-86, and Ser-98 were identified in the N-terminal region of WRKY8. We generated various recombinant full-length WRKY8 proteins with Ala substitutions at the potential phosphorylation sites. SIPK could hardly phosphorylate the S62A/S67A/S79A/S86A/S98A quintuple mutant, but all single mutations slightly reduced the level of phosphorylation (Figure 1B). The levels of phosphorylation were proportional to the number of phosphorylation sites (Figure 1B, top panel), indicating that Ser-62, Ser-67, Ser-79, Ser-86, and Ser-98 could be independently phosphorylated.
Figure 1.
Phosphorylation of WRKY8 in Vitro and in Vivo.
(A) In vitro phosphorylation of WRKY8 by MAPKs. Purified Trx and Trx-fused WRKY8 were used as substrates for active MAPKs. Proteins were separated by SDS-PAGE, were stained with Coomassie Brilliant Blue (CBB; bottom panel), and were exposed to x-ray film (kinase assay; top panel).
(B) Ala scanning analysis of SIPK-mediated phosphorylation sites in WRKY8. Phosphorylation of WRKY8 variants was detected by autoradiography (middle), and the intensities of each band were quantified (top). Protein loads were monitored by CBB staining (bottom).
(C) Specificity of anti-pSer79 and anti-pSer86 antibodies. Peptides of WRKY874-84, Ser-79–phosphorylated WRKY874-84 (WRKY874-84pSer79) and WRKY881-91, and Ser-86–phosphorylated WRKY881-91 (WRKY881-91pSer86) were spotted on nitrocellulose membranes. Immunoblot analyses were performed using anti-pSer79 antibody or anti-pSer86 antibody.
(D) Phosphorylation of Ser-79 and Ser-86 in WRKY8 by INF1 elicitin expression. WRKY8-HA-StrepII was expressed with INF1 or GUS in N. benthamiana leaves by agroinfiltration. Total proteins were prepared 30 h after agroinfiltration. After Strep-Tactin purification, immunoblot analyses were done using anti-HA, anti-pSer79, or anti-pSer86 antibody.
(E) Phosphorylation of Ser-79 and Ser-86 in WRKY8 by MEK2DD expression. WRKY8-HA-StrepII was expressed with FLAG-MEK2KR or FLAG-MEK2DD in N. benthamiana leaves by agroinfiltration. Total proteins were prepared 36 h after agroinfiltration. Anti-FLAG antibody was used to detect accumulation of FLAG-MEK2KR and FLAG-MEK2DD. Protein loads were monitored by CBB staining of the bands corresponding to the ribulose-1,5-bisphosphate carboxylase large subunit (RBCL). Phosphorylation of Ser-79 and Ser-86 in WRKY8 was detected as described in (D).
(F) Effects of infection of TRV:SIPK (S), TRV:WIPK (W), or TRV:SIPK/WIPK (S/W) on phosphorylation of Ser-79 and Ser-86 in WRKY8 in response to MEK2DD expression. Ten micromolars of 17-β-estradiol was injected into the leaves 18 h after coinoculation with Agrobacterium strains containing pER8:FLAG-MEK2DD and pER8:WRKY8-HA-StrepII. Total proteins were prepared 12 h after estradiol injection, and phosphorylation of Ser-79 and Ser-86 in WRKY8 was detected as described in (D).
(G) Phosphorylation of Ser-79 and Ser-86 in WRKY8 by SIPKWT or NTF4WT expression. WRKY8-HA-StrepII was expressed with FLAG-tagged SIPKWT, SIPKKR, NTF4WT, or NTF4KR in N. benthamiana leaves by agroinfiltration. Total proteins were prepared at 48 h after agroinfiltration, and phosphorylation of Ser-79 and Ser-86 in WRKY8 was detected as described in (D).
Arrowheads refer to the pertinent bands. Asterisks indicate nonspecific bands.
To determine if WRKY8 is phosphorylated in vivo, we prepared antiphosphopeptide antibodies against peptides, including phospho-Ser-79 (pSer79) or phospho-Ser-86 (pSer86) that are highly conserved among group I WRKY proteins (see Supplemental Figure 1B online). Immunoblot analyses showed that antibodies specifically recognized each phosphopeptide used as an antigen (Figure 1C). We prepared total proteins from N. benthamiana leaves coexpressing WRKY8-HA-StrepII with β-glucuronidase (GUS) or INF1 (a protein elicitor produced by P. infestans), which activates SIPK, NTF4, and WIPK (Asai et al., 2008). After purification of StrepII-tagged proteins with Strep-Tactin chromatography, we conducted immunoblot analyses using anti-HA, anti-pSer79, and anti-pSer86 antibodies. Phosphorylation of Ser-79 and Ser-86 was only detected in leaves expressing WRKY8-HA-StrepII with INF1, but not with GUS (Figure 1D). Immunoblot analysis using anti-HA antibody confirmed similar amounts of WRKY8-HA-StrepII accumulation irrespective of coexpression with INF1 or GUS (Figure 1D). To confirm that phosphorylation of Ser-79 and Ser-86 of WRKY8 is dependent on SIPK, NTF4, and WIPK activities, we analyzed the phosphorylation state of WRKY8-HA-StrepII in response to coexpression of FLAG-MEK2DD or FLAG-MEK2KR (inactive forms of MEK2). Phosphorylation of Ser-79 and Ser-86 was also induced by expression of FLAG-MEK2DD, but not by MEK2KR (Figure 1E). MEK2DD did not directly phosphorylate WRKY8 in vitro, suggesting that WRKY8 is phosphorylated by downstream MAPKs of MEK2 (see Supplemental Figure 4 online).
We evaluated whether phosphorylation of Ser-79 and Ser-86 induced by MEK2DD depends on SIPK, NTF4, or WIPK. We silenced these MAPKs in N. benthamiana using Tobacco rattle virus (TRV)-VIGS constructs (Asai et al., 2008), and WRKY8 phosphorylation was monitored. NTF4 showed high sequence identity to SIPK (Ren et al., 2006), and the VIGS construct of SIPK also silenced NTF4, as also reported by Asai et al. (2008). Because we detected no protein accumulation of WRKY8-HA-StrepII under the control of Cauliflower mosaic virus (CaMV) 35S promoter without a p19 silencing suppressor, we used an estradiol-inducible expression system to express WRKY8-HA-StrepII and MEK2DD only in the silenced plants. The estradiol-inducible expression system can control the timing and expression level of the transgene in a dose-dependent manner, and 5 μM estradiol treatment induces the expression of a transgene approximately fourfold higher than a transgene controlled by the CaMV 35S promoter (Zuo et al., 2000). In this study, phosphorylation of Ser-79 and Ser-86 was detected in SIPK/NTF4- or WIPK-silenced leaves in response to FLAG-MEK2DD, but not in SIPK/NTF4/WIPK-silenced leaves (Figure 1F). This result suggests that SIPK/NTF4 and WIPK contribute redundantly to Ser-79 and Ser-86 phosphorylation of WRKY8 in response to MEK2DD expression. We further evaluated the individual contribution of SIPK and NTF4 in WRKY8 phosphorylation. Overexpression of SIPK and NTF4, but not WIPK, show activity by its endogenous upstream kinase-mediated phosphorylation (Zhang and Liu, 2001; Ren et al., 2006). We analyzed the phosphorylation state of WRKY8-HA-StrepII in response to coexpression of FLAG-SIPKWT or FLAG-NTF4WT. Phosphorylation of Ser-79 and Ser-86 was induced by expression of FLAG-SIPKWT and FLAG-NTF4WT, but not by FLAG-SIPKKR and FLAG-NTF4KR (inactive mutants of SIPK and NTF4, respectively) (Figure 1G). These results indicate that overexpression of either SIPK or NTF4 is sufficient to phosphorylate Ser-79 and Ser-86 in plants. The results from these gain-of-function and loss-of-function analyses suggest that SIPK, NTF4, and WIPK are kinases that function upstream of WRKY8.
D Domain–Dependent Interactions Are Required for Phosphorylation of WRKY8 by SIPK
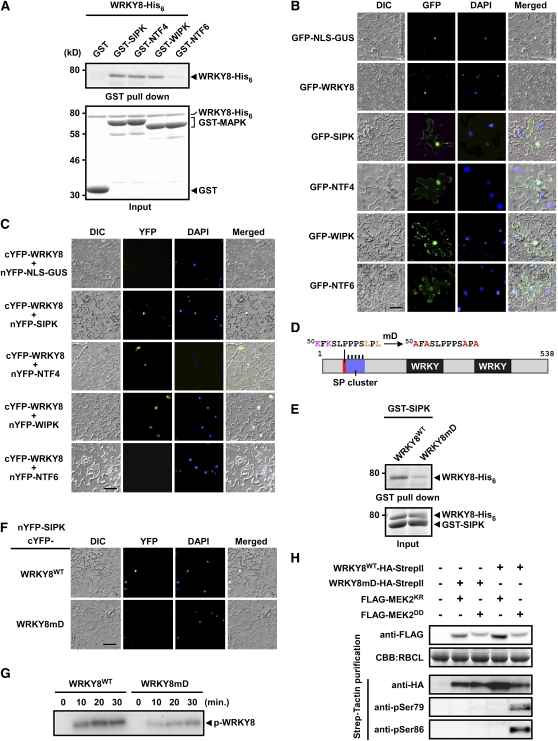
We examined the interaction between WRKY8 and MAPKs by in vitro glutathione S-transferase (GST) pull-down assays. WRKY8-His6 was incubated with glutathione–agarose-bound GST, GST-SIPK, GST-NTF4, GST-WIPK, or GST-NTF6, and the pull-down products were analyzed by immunoblot using anti-His6 antibody. WRKY8-His6 was pulled down with GST-SIPK, GST-NTF4, and GST-WIPK, but not GST and GST-NTF6, suggesting that WRKY8 could interact with SIPK, NTF4, and WIPK (Figure 2A). These results indicate that no other factor is required for interactions between WRKY8 and these MAPKs. Interaction of WRKY8 with SIPK was also examined using a yeast two-hybrid (Y2H) assay. The yeast transformed with full-length WRKY8 that was fused to an activation domain grew on selective plates without specific bait protein. Therefore, the Y2H assay was performed using the N-terminal region of WRKY8 (WRKY81-170), which is sufficient for in vitro phosphorylation by SIPK (see Supplemental Figure 3 online). WRKY81-170 interacted with SIPK, but not with NTF6 (see Supplemental Figure 5 online).
Figure 2.
D Domain–Dependent Interactions Required for Phosphorylation of WRKY8 by SIPK.
(A) In vitro interaction assays between WRKY8 and MAPKs. GST, GST-SIPK, GST-NTF4, GST-WIPK, and GST-NTF6 purified proteins were incubated with WRKY8-His6 as indicated. Pulled-down fractions were analyzed by immunoblotting using anti-His6 antibody (top). Input proteins were monitored by Coomassie blue staining (bottom).
(B) Subcellular localization of WRKY8, SIPK, NTF4, WIPK, and NTF6 in N. benthamiana epidermal cells. N. benthaniana leaves were transformed with GFP-NLS-GUS, GFP-WRKY8, GFP-SIPK, GFP-NTF4, GFP-WIPK, and GFP-NTF6 by agroinfiltration. DAPI, 4′,6–diamidino–2–phenylindole; DIC, differential interference contrast.
(C) BiFC visualization of WRKY8-SIPK, WRKY8-NTF4, and WRKY8-WIPK interactions. N. benthaniana leaves were cotransformed with the C-terminal part of YFP-fused WRKY8 (cYFP-WRKY8) and the N-terminal part of YFP-fused NLS-GUS or MAPKs (nYFP-SIPK, nYFP-NTF4, nYFP-WIPK, and nYFP-NTF6) by agroinfiltration.
(D) Deduced D domain found in WRKY8. Lys and Leu conserved in the D domain are indicated in pink and orange, respectively. The numbers indicate position of amino acids of WRKY8 protein. Key interacting residues were substituted with Ala (mD).
(E) In vitro interaction assays between D domain–mutated WRKY8 and SIPK.
(F) BiFC visualization of the interaction between D domain–mutated WRKY8 and SIPK.
(G) In vitro analysis of D domain–mutated WRKY8 phosphorylation by SIPK. Purified Trx-fused WRKY8WT and WRKY8mD were phosphorylated by active SIPK for the times indicated above each lane. Proteins were separated by SDS-PAGE and were exposed to x-ray film.
(H) In vivo phosphorylation analysis of D domain–mutated WRKY8 by MEK2DD or MEK2KR.
These experiments were repeated at least three times with similar results. Bars = 50 μm.
We investigated the subcellular localizations of WRKY8 and MAPKs. WRKY8 and MAPKs were fused to green fluorescent protein (GFP) and were expressed transiently in N. benthamiana leaves by Agrobacterium infiltration (agroinfiltration). Nuclear localization signal–fused GUS (NLS-GUS) was also expressed as a GFP fusion protein, as a positive control of nuclear localization showed by 4',6-diamidino-2-phenylindole staining (Figure 2B). GFP-NLS-GUS and GFP-WRKY8 were found in the nucleus (Figure 2B). GFP-SIPK, -NTF4, -WIPK, and -NTF6 were in both the cytosol and nucleus, but a larger proportion of GFP-NTF6 was in the cytosol compared with other GFP-MAPKs (Figure 2B). To study if WRKY8 interacts with MAPKs in plant cells, we used bimolecular fluorescence complementation (BiFC) (Hu et al., 2002). N- and C-terminal fragments of yellow fluorescent protein (YFP) were fused to either SIPK, NTF4, WIPK, or NTF6 and WRKY8 and were transiently coexpressed in N. benthamiana leaves. No fluorescence was detectable with combinations of cYFP-WRKY8/nYFP-NLS-GUS and cYFP-WRKY8/nYFP-NTF6 (Figure 2C). However, reconstituted YFP fluorescence in the nucleus was detected with combinations of cYFP-WRKY8/nYFP-SIPK, cYFP-WRKY8/nYFP-NTF4, and cYFP-WRKY8/nYFP-WIPK, indicating interactions between WRKY8 and these MAPKs in the nucleus (Figure 2C).
To investigate the contribution of the deduced D domain in the interaction between WRKY8 and SIPK, D domain–mutated WRKY8 (WRKY8mD) and wild-type WRKY8 (WRKY8WT) were used for pull-down, Y2H, and BiFC assays (Figure 2D). In the in vitro pull-down assay, mutations within the D domain disrupted the interaction between WRKY8 and SIPK (Figure 2E). In the Y2H assay, the interaction between WRKY81-170 and SIPK was also reduced by mutation in the D domain (see Supplemental Figure 6 online). In the BiFC assay, WRKY8mD did not interact with SIPK (Figure 2F). These results indicate that the D domain is required for effective interaction between WRKY8 and SIPK. However, interaction between WRKY8 and SIPK did not disappear completely by mutation in the D domain. Therefore, we cannot rule out the possibility that an additional interaction motif for SIPK may exist in WRKY8.
We tested whether the D domain–dependent interaction is required for phosphorylation of WRKY8 by MAPK. We performed in vitro phosphorylation assays using WRKY8WT and WRKY8mD as substrates. The in vitro phosphorylation intensities of WRKY8WT by active SIPK were reduced markedly in WRKY8mD (Figure 2G). Thus, the D domain–dependent interaction is critical for efficient phosphorylation of WRKY8 by SIPK. We also investigated the contribution of the D domain to in vivo phosphorylation of WRKY8 by MAPK. WRKY8WT-HA-StrepII and WRKY8mD-HA-StrepII were coexpressed with either FLAG-MEK2KR or FLAG-MEK2DD in N. benthamiana leaves by agroinfiltration (Figure 2H). WRKY8 phosphorylation was monitored by immunoblot analyses using anti-pSer79 and anti-pSer86 antibodies. Phosphorylation of Ser-79 and Ser-86 was only detected in leaves expressing FLAG-MEK2DD with WRKY8WT-HA-StrepII, but not with WRKY8mD-HA-StrepII (Figure 2H). These results indicate that D domain–dependent interaction is necessary for MAPK-mediated phosphorylation of WRKY8.
Phosphorylation of WRKY8 by MAPKs Increases DNA Binding and Transactivation Activities
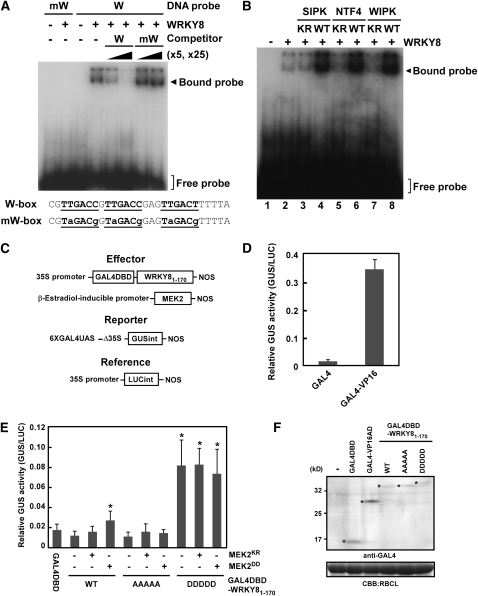
WRKY transcription factors most likely function by binding to cognate W-box cis-elements in the promoter region of target genes (Eulgem et al., 2000; Ciolkowski et al., 2008). To determine if WRKY8 binds specifically to the W-box, binding of WRKY8 recombinant protein to the W-box was assayed using an electrophoresis mobility shift assay (EMSA). A protein-DNA complex with reduced mobility was detected when recombinant WRKY8 was incubated with the W-box probe, but not with a mutated W-box probe (Figure 3A). The specificity of this binding was also confirmed by competition experiments using an unlabeled probe. These results indicate that WRKY8 specifically binds to the W-box sequence. Phosphorylation affects the DNA binding activity of many transcription factors (Yang et al., 2003). Therefore, we tested if MAPK-mediated phosphorylation of WRKY8 altered its ability to bind to the W-box sequence. Recombinant WRKY8 protein was incubated with wild-type, active, or kinase-inactive SIPK, NTF4, or WIPK and then underwent EMSA. The DNA binding activity of WRKY8 was increased by wild-type SIPK, NTF4, and WIPK (Figure 3B, lanes 4, 6, and 8). By contrast, kinase-inactive SIPK, NTF4, and WIPK did not affect the DNA binding activity (Figure 3B, lanes 3, 5, and 7). These results suggest that MAPK-mediated phosphorylation of WRKY8 promotes its DNA binding activity.
Figure 3.
Increased DNA Binding and Transactivation Activities by MAPK-Mediated Phosphorylation of WRKY8.
(A) Binding of recombinant WRKY8 to a W-box (W) sequence, but not to a mutated W-box (mW) sequence, using EMSA. Unlabeled W-box fragments and mutated W-box fragments were used as competitor DNAs.
(B) Increased binding activity of WRKY8 to the W-box sequence by MAPK-mediated phosphorylation. After incubation with WRKY8 and SIPKKR (lane 3), SIPKWT (lane 4), NTF4KR (lane 5), NTF4WT (lane 6), WIPKKR (lane 7), WIPKWT (lane 8), or no recombinant MAPK (lane 2), EMSA was done using a 32P-labeled W-box probe. Lane 1 shows mobility of free probe.
(C) Schematic diagram of effector, reporter, and reference plasmids used in transient assays.
(D) High transactivation activity of GAL4DBD-VP16. Data are means ± sd from three independent experiments
(E) Transactivation of the GUS gene by GAL4DBD-WRKY8 variants in N. benthamiana leaves. Total proteins were extracted from leaves coinfiltrated with Agrobacterium-containing reporter plasmids, effector plasmids, or reference plasmids. GUS activity was normalized to LUC activity. AAAAA and DDDDD indicate that WRKY81-170 mutants mimic the nonphosphorylated form and phosphorylated form, respectively. Data are means ± sd from at least three independent experiments, *P < 0.01 versus the GAL4DBD-WRKY8WT1-170 alone by the two-tailed Student’s t test. WT, wild type.
(F) Protein gel blot analysis of GAL4DBD, GAL4DBD-VP16, and GAL4DBD-WRKY81-170 variants. Anti-GAL4 antibody was used to detect accumulation of GAL4DBD and GAL4DBD-fused proteins. Protein loads were monitored by Coomassie Brilliant Blue (CBB) staining of the bands corresponding to ribulose-1,5-bisphosphate carboxylase large subunit (RBCL). Asterisks refer to the pertinent bands.
To determine the transactivation activity of WRKY8 in plants, we developed an Agrobacterium-mediated transient assay system. The GUS reporter gene containing an intron to avoid expression in Agrobacterium was fused under the control of a synthetic promoter consisting of the −46 minimal CaMV 35S promoter and six copies of the GAL4 upstream activation sequence (GAL4UAS) (Figure 3C). The GAL4 DNA binding domain (GAL4DBD) that was fused to the strong activation domain of herpes simplex virus VP16 (GAL4DBD-VP16) strongly activated the GAL4UAS-GUS reporter gene compared with GAL4DBD only (Figure 3D). Because protein accumulation of full-length WRKY8 fused to GAL4DBD could not be detected by immunoblot analyses using anti-GAL4 antibody, WRKY81-170 containing an SP cluster was used (Figure 3C). To test the function of phosphorylation of the SP cluster, WRKY8AAAAA1-170 (mimicking nonphosphorylated WRKY8) and WRKY8DDDDD1-170 (mimicking constitutively phosphorylated WRKY8), in which Ser-62, Ser-67, Ser-79, Ser-86, and Ser-98 are replaced by Ala or Asp, were used. Variants of GAL4DBD-WRKY81-170 were expressed in the presence or absence of MEK2KR and MEK2DD in N. benthamiana leaves. The expression of MEK2DD induces HR-like cell death in tobacco plants (Yang et al., 2001). To synchronize the expression of effector genes and to exclude cell death–associated effects by MEK2DD on the expression of reporter gene, we used an estradiol-inducible system to make a time lag between the reporter and effector genes after agroinfiltration. At 24 h after agroinfiltration, leaves were injected with estradiol to express MEK2 variants and were incubated for 12 h. Before the onset of visible cell death, the reporter activities were determined. GAL4DBD-WRKY8WT1-170 and GAL4DBD-WRKY8AAAAA1-170 showed no higher GUS activities compared with GAL4DBD (Figure 3E), indicating that WRKY8WT1-170 has no transactivation activity. However, we should interpret these results cautiously because WRKY81-170, but not full-length WRKY8, was used in these experiments, and accumulation of GAL4DBD-WRKY8 variant proteins was much lower than GAL4DBD (Figure 3F). Coexpression of GAL4DBD-WRKY8WT1-170 with MEK2DD, but not with MEK2KR, increased GUS activity (Figure 3E). By contrast, coexpression of GAL4DBD-WRKY8AAAAA1-170 with MEK2DD did not increase GUS activity. GAL4DBD-WRKY8DDDDD1-170 showed massive GUS activities irrespective of MEK2DD. These results indicate that MAPK-dependent phosphorylation of the SP cluster of WRKY8 increases transactivation activity. GAL4DBD-WRKY8DDDDD1-170 showed much higher GUS activity compared with coexpression of GAL4DBD-WRKY8WT1-170 and MEK2DD (Figure 3E). We think that the difference in GUS activities might be due to dephosphorylation of phosphorylated WRKY8WT1-170 by some phosphatases.
MEK2DD expression induces SIPK, NTF4, and WIPK activation (Ren et al., 2006). To clarify which MAPK-mediated phosphorylation increases transactivation activity of GAL4DBD-WRKY8WT1-170, we further analyzed the transactivation activity of GAL4DBD-WRKY8WT1-170 in response to SIPKWT or SIPKKR expression. Coexpression of GAL4DBD-WRKY8WT1-170 with SIPKWT, but not with SIPKKR, increased GUS activity (see Supplemental Figure 7 online), indicating that at least SIPK-mediated phosphorylation increases the transactivation activity of WRKY8.
Knockdown of WRKY8 Enhances Susceptibility to P. infestans and C. orbiculare
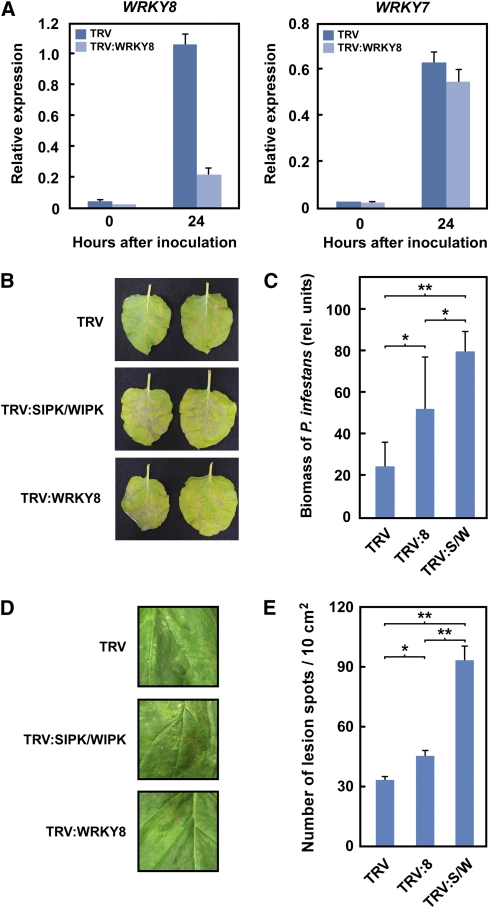
In this study, the roles of WRKY8 in the basal defense against P. infestans and C. orbiculare, potent pathogens of N. benthamiana (Kamoun et al., 1998; Shen et al., 2001; Takano et al., 2006), were investigated using VIGS based on the TRV vector (Ratcliff et al., 2001). The oomycete P. infestans is a near-obligate pathogen (i.e., a potential biotroph) (Fry, 2008), and the ascomycete fungus C. orbiculare, a typical hemibiotroph. The expression level of WRKY8 was markedly reduced in TRV:WRKY8-infected plants compared with the TRV control (Figure 4A). Silencing of WRKY8 appeared to be specific because the mRNA levels of Nb WRKY7, a WRKY closely related to Nb WRKY8 (see Supplemental Figure 1B online), were not affected in WRKY8-silenced plants (Figure 4A). Quantitative RT-PCR (qRT-PCR) analyses indicated that TRV:SIPK/WIPK reduced transcript accumulation of SIPK (81.9%), NTF4 (85.4%), and WIPK (82.4%) compared with TRV control plants at 24 h after inoculation with P. infestans.
Figure 4.
Increased Disease Susceptibility to a Virulent Strain of P. infestans by Silencing of WRKY8.
(A) Specific gene silencing of WRKY8 in TRV:WRKY8-infected plants. Silencing of WRKY8 was monitored by qRT-PCR using specific primers. WRKY7, which shows high homology to WRKY8 (see Supplemental Figure 1B online), was used as a silencing control. Data are means ± sd from three independent experiments.
(B) Susceptibility to P. infestans in silenced plants. Photographs were taken 5 d after the inoculation.
(C) Effects of infection of TRV:WRKY8 (8) or TRV:SIPK/WIPK (S/W) on P. infestans infection. Biomasses were determined by qPCR 5 d after inoculation. Data are means ± sd from three independent experiments.
(D) Susceptibility to C. orbiculare in silenced plants. Photographs were taken 5 d after the inoculation.
(E) Effects of infection of TRV:8 or TRV:S/W on C. orbiculare infection. The number of disease spots in the leaves was counted 5 d after the inoculation. Data are means ± sd from four independent experiments.
*P < 0.05 and **P < 0.01; two-tailed Student’s t test.
We used qPCR technology to examine changes in P. infestans biomass during the interaction with the silenced plants. PCR primers specific to highly repetitive sequences in the P. infestans genome were used to quantify relative levels of P. infestans DNA in infected plant tissues and were found to reflect an accurate and sensitive estimate of the P. infestans biomass (Judelson and Tooley, 2000). Zoospores of a virulent isolate of P. infestans were inoculated on the surface of the silenced leaves of N. benthamiana. Analysis of P. infestans biomass showed that the growth rate of P. infestans increased in WRKY8-silenced plants compared with TRV-control plants (Figures 4B and 4C). SIPK/NTF4/WIPK-silenced plants showed a greater decrease in resistance to P. infestans compared with WRKY8-silenced plants (Figures 4B and 4C).
C. orbiculare conidia were sprayed on the silenced leaves. We counted the number of disease spots, which reflect susceptibility, not HR cell death, on inoculated leaves. The number of disease spots on WRKY8-silenced leaves increased compared with that on TRV-control leaves (Figures 4D and 4E). SIPK/NTF4/WIPK-silenced plants developed more disease spots than WRKY8-silenced plants (Figures 4D and 4E). These results indicate that WRKY8 contributes to the defense response against P. infestans and C. orbiculare, but to a lesser extent than the contribution of SIPK, NTF4, and WIPK, which are the presumed upstream kinases.
Phosphorylation of WRKY8 Increases Expression of Downstream Defense-Related Genes
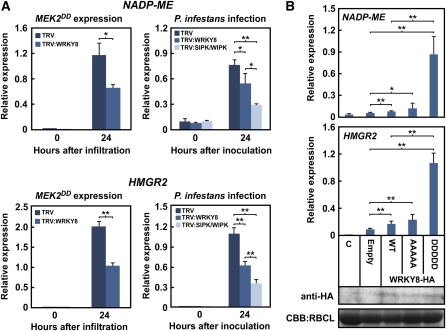
To identify target genes of WRKY8, we used loss-of-function and gain-of-function screening using the suppression subtractive hybridization (SSH) method (Diatchenko et al., 1996). For loss-of-function screening, we constructed an SSH cDNA library from leaves of WRKY8-silenced plants expressing MEK2DD and TRV control plants. Positive clones underwent reverse RNA gel blot analysis for reproducibility and then were analyzed by qRT-PCR. From a total of 1920 randomly selected colonies, we isolated NADP-malic enzyme (NADP-ME) as being downregulated in WRKY8-silenced plants. NADP-MEs catalyze oxidative decarboxylation of malic acid, producing pyruvate, CO2, and NADPH, and are suggested to participate in the plant defense response by providing NADPH for defense-related lignification (see Supplemental Figure 8 online) (Wheeler et al., 2005). For gain-of-function screening, we constructed an SSH cDNA library from leaves transformed with the WRKY8DDDDD vector and the empty vector. We used the same procedures as for loss-of-function screening for the analysis. From a total of 960 randomly selected colonies, we isolated 3-hydroxy-3-methylglutaryl CoA reductase 2 (HMGR2) as an upregulated gene from leaves expressing WRKY8DDDDD. HMGR catalyzes the rate-limiting step in the biosynthesis of all isoprenoid compounds, including capsidiol, the major tobacco phytoalexin accumulated during the defense response (see Supplemental Figure 8 online) (Wu et al., 2006). qRT-PCR was used to validate that these genes (NADP-ME and HMGR2) function downstream of WRKY8. The expression levels of NADP-ME and HMGR2 were significantly upregulated in response to MEK2DD expression and P. infestans infection (Figure 5A). To evaluate the contribution of WRKY8 and SIPK/NTF4/WIPK to the upregulation of these genes, we used the TRV-based VIGS method to silence the target genes. We detected no significant induction of NADP-ME and HMGR2 genes after TRV infection (see Supplemental Figure 9 online). The upregulation of NADP-ME and HMGR2 in response to MEK2DD expression and P. infestans infection was compromised in WRKY8-silenced plants (Figure 5A). The expression levels of both genes decreased more in SIPK/NTF4/WIPK-silenced plants (Figure 5A). These results indicate that accumulation of NADP-ME and HMGR2 mRNAs depends on SIPK, NTF4, and WIPK activation and requires WRKY8. Our data support the previous report by Zhang and Liu (2001) describing that ectopic expression of SIPK induces HMGR in N. tabacum.
Figure 5.
Induction of Target Gene Expression by a Phosphorylation-Mimicking Mutant of WRKY8.
(A) Expression levels of NADP-ME (top) and HMGR2 (bottom) induced by MEK2DD (left) or P. infestans (right) were compromised by TRV:WRKY8 or TRV:SIPK/WIPK. Data are means ± sd from three independent experiments.
(B) Expression of NADP-ME and HMGR2 in response to WRKY8 variants. Total RNAs were extracted from N. benthamiana leaves 48 h after agroinfiltration and were used for qRT-PCR. Data are means ± sd from at least three independent experiments. Anti-HA antibody was used to detect accumulation of WRKY8-HA variants. Protein loads were monitored by CBB staining of the bands corresponding to RBCL. WT, wild type.
*P < 0.05 and **P < 0.01; two-tailed Student’s t test.
[See online article for color version of this figure.]
To examine the effect of WRKY8 phosphorylation on the induction of NADP-ME and HMGR2, we expressed HA-tagged WRKY8 (WRKY8WT) and its mutants, WRKY8AAAAA and WRKY8DDDDD, in N. benthamiana leaves, and we analyzed the expression levels of both genes. WRKY8AAAAA and WRKY8WT induced expression of NADP-ME and HMGR2 to some extent (Figure 5B). In a transactivation assay, GAL4DBD-WRKY8WT1-170 and GAL4DBD-WRKY8AAAAA1-170 showed no higher GUS activities compared with GAL4DBD (Figure 3E). However, accumulation of GAL4DBD-WRKY8 variant proteins was much lower than GAL4DBD (Figure 3F). We think that the nonphosphorylated form of WRKY8 has basal transactivation activity because WRKY8WT bound to the W-box sequence (Figure 3A). WRKY8DDDDD highly induced the expression of both genes compared with WRKY8AAAAA and WRKY8WT (Figure 5B), indicating that the phospho-mimicking mutation of WRKY8 results in the induction of downstream genes. We made expression analyses of NADP-ME, HMGR2, and WRKY8 in response to syringe infiltration of water or to INF1 elicitin, which activates prolonged MBP kinase activities in kinases with a molecular mass of 48 and 44 kD (see Supplemental Figure 10A online). SIPK and NTF4 are 48 kD, and WIPK is 44 kD (Ren et al., 2006). Considering that Agrobacterium-mediated INF1 expression induces SIPK and WIPK activation and that NTF4 might be functionally redundant with SIPK in the defense-related signaling pathway (Ren et al., 2006; Asai et al., 2008), 48-kD MBP kinases are presumed to correspond to SIPK and NTF4 and 44-kD MBP kinase to WIPK. INF1 elicitin treatment may induce activation of SIPK, NTF4, and WIPK. Although no obvious difference was observed in the accumulation profile of WRKY8 mRNA between water treatment and INF1, accumulation of NADP-ME and HMGR2 transcripts increased in response to INF1 treatment (see Supplemental Figure 10B online), suggesting that upregulation of WRKY8 mRNA is not enough to induce expression of NADP-ME and HMGR2. These results together suggest that MAPK-dependent phosphorylation of WRKY8 positively regulates expression of downstream genes NADP-ME and HMGR2.
DISCUSSION
MAPK-Mediated Phosphorylation of WRKY8 Regulates Defense Responses as a Positive Transcription Factor
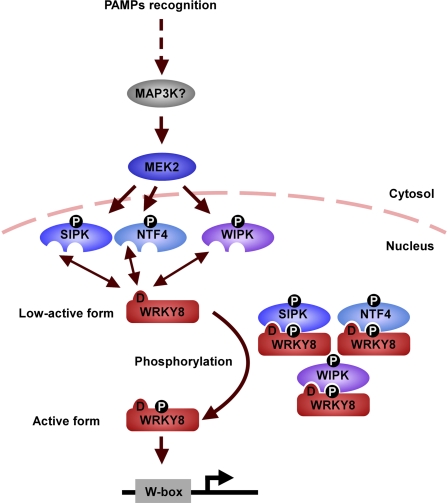
MAPKs have pivotal roles in induced defense responses of plants. However, the regulatory mechanisms by which MAPKs induce defense responses are unclear. In this study, we showed that WRKY8 is phosphorylated by SIPK, NTF4, and WIPK in vivo and that D domain–dependent interaction is necessary for SIPK-mediated phosphorylation of WRKY8. We also showed that MAPK-mediated phosphorylation of WRKY8 activates its DNA binding and transactivation activities and that WRKY8 induces downstream defense genes (Figure 6). D domain–mutated WRKY8 was not phosphorylated in response to MEK2DD expression, which induces SIPK, NTF4, and WIPK activation (Figure 2H). Because NTF4 and WIPK interacted with and phosphorylated WRKY8 in the same way as SIPK (Figures 1 and 2), we hypothesize that phosphorylation of WRKY8 by NTF4 and WIPK also requires D domain–dependent interaction (Figure 6).
Figure 6.
Model of the Regulatory Mechanism of WRKY8 by MAPK-Dependent Phosphorylation.
Pathogen recognition leads to activation of MEK2 by unidentified MAP3K(s). Active MEK2 phosphorylates and activates SIPK, NTF4, and WIPK. Active SIPK interacts with WRKY8 in a D domain–dependent manner and phosphorylates WRKY8. Active NTF4 and WIPK may act in a way similar to SIPK. Phosphorylated and activated WRKY8 binds to W-box sites within its target genes and upregulates the expression of these genes.
One major mechanism to effect changes in gene expression appears to be through MAPKs altering the activity of transcription factors and hence transcription of their cognate target genes (Yang et al., 2003). Phosphorylation of transcription factors by MAPK can alter (1) cellular localization, (2) protein stability, (3) DNA binding activity, (4) transactivation or repression activity, and (5) remodeling of nucleosome structure (Yang et al., 2003; Tootle and Rebay, 2005). We showed that SIPK can phosphorylate Ser residues within the SP cluster and that SIPK-mediated phosphorylation of WRKY8 increases both DNA binding and transcriptional activities (Figures 1 and 3; see Supplemental Figure 7 online). Ectopic expression of the SP cluster phospho-mimicking mutant WRKY8DDDDD induced downstream target genes NADP-ME and HMGR2 (Figure 5B). These results support the idea that phosphorylation of WRKY8 by SIPK functions in (3) and (4) above. Even though we could not indicate individual functions of MAPKs for WRKY8, silencing of WIPK is required to eliminate SP cluster phosphorylation of Ser-79 and Ser-86 by MEK2DD (Figure 1F), and overexpression of SIPK or NTF4 induced Ser-79 and Ser-86 phosphorylation (Figure 1G). NTF4- or WIPK-mediated phosphorylation of WRKY8 also promoted its DNA binding activity (Figure 3B). GAL4DBD-WRKY8DDDDD1-170 showed massive transactivation activity irrespective of MEK2DD (Figure 3E). These results suggest that SIPK, NTF4, and WIPK may phosphorylate similar sites and have roles similar to WRKY8 in (3) and (4) above. In mammals, Elk-1 transcription factor, a substrate of MAPKs, increases both DNA binding and transactivation activities that depend on phosphorylation at multisite residues within the transcriptional activation domain (TAD) (Yang et al., 1999, 2003; Tootle and Rebay, 2005). The TAD of Elk-1 binds directly to the ETS domain, which is a helix-turn-helix DNA binding domain, limiting interaction with DNA until the TAD is phosphorylated (Yang et al., 1999). In this case, phosphorylation by MAPKs triggers release of autoinhibition in Elk-1. Future studies will examine if WRKY8 is regulated in a way similar to Elk-1.
In this study, silencing of WRKY8 decreased expression levels of NADP-ME and of HMGR2 induced by MEK2DD expression and P. infestans infection (Figure 5A). WRKY8-silenced plants showed increased disease susceptibility to the potential biotroph P. infestans and the hemibiotroph C. orbiculare. These results indicate that WRKY8 regulates broad-spectrum disease resistance through activation of target defense genes. However, the decrease in the transcript level of both genes by WRKY8 silencing was much less than by SIPK/NTF4/WIPK silencing (Figure 5A). Similarly, SIPK/NTF4/WIPK-silenced plants showed a greater increase in susceptibility than WRKY8-silenced plants (Figure 4). Redundancy of genes encoding plant transcription factors complicates loss-of-function analyses that aim to identify the biological functions of large families that contain members with strongly conserved DNA binding domains (Mitsuda and Ohme-Takagi, 2009). Structurally closely related Arabidopsis WRKY11 and WRKY17 act as negative regulators of basal resistance to bacterial pathogens (Journot-Catalino et al., 2006). Expression analysis of genes regulated by WRKY11 or WRKY17 or both showed that WRKY11 and WRKY17 act partially redundantly to a subset of target genes (Journot-Catalino et al., 2006). These observations suggest that WRKY8 functions redundantly with other group I WRKYs that are not silenced by TRV:WRKY8 because of mismatching of its nucleotide sequence and that silenced plants show weak phenotypes. Alternatively, because MAPKs are potentially capable of phosphorylating various transcription factors in vitro (Popescu et al., 2009), such factors could also regulate target genes directly or indirectly.
The SP Cluster of the N-Terminal Region of WRKY Is a Target Sequence of MAPK
WRKY8 was phosphorylated by SIPK, NTF4, and WIPK (Figures 1 and 2). Other studies showed that Arabidopsis MPK3 and MPK6 (WIPK and SIPK orthologs, respectively) have a high level of functional redundancy (Pitzschke et al., 2009), suggesting that MPK3 and MPK6 might share some of their substrates. ETHYLENE INSENSITIVE3, PHOS32, and SPEECHLESS are substrates of both MPK3 and MPK6 (Lampard et al., 2008; Merkouropoulos et al., 2008; Yoo et al., 2008). Extensive in vitro analysis showed that MPK6 shares 40% of in vitro substrates with MPK3 (Popescu et al., 2009). In this study, we showed that WRKY8 is a target of SIPK, NTF4, and WIPK and that WRKY8 appears to be phosphorylated within the SP cluster, is activated by these MAPKs, and participates in defense-related signaling pathways. To our surprise, the SP cluster is highly conserved in the N-terminal region of several group I WRKYs, including Arabidopsis WRKY25 and WRKY33 (see Supplemental Figure 1B online), which are phosphorylated by MPK4 in vitro (Andreasson et al., 2005), and Nt WRKY1, which is phosphorylated by SIPK in vitro (Menke et al., 2005). Eulgem and Somssich (2007) described the possibility that the D-motif, a conserved pattern of SP dimers within group I WRKYs, is a consensus phosphoacceptor site for MAPKs. Because Ser-79 and Ser-86 of WRKY8 correspond to SP dimers within the D-motif, our study provides evidence that the D-motif can be a MAPK target sequence. The Ala scanning analysis indicated that adjacent SP sequences could be phosphorylated by MAPKs (Figure 1B). We propose that WRKYs containing an SP cluster might be a substrate of certain MAPKs. Although the phosphorylation site of Nt WRKY1 by SIPK was not identified, SIPK-mediated phosphorylation of Nt WRKY1 increased its DNA binding activity in vitro (Menke et al., 2005). Transient coexpression of Nt WRKY1 with SIPK indicated that Nt WRKY1 positively participates in SIPK-dependent cell death (Menke et al., 2005). We hypothesize that the activation method of the SP cluster of Nt WRKY1 might be similar to that of Nb WRKY8.
Some SP cluster–containing WRKYs have a D domain–like sequence (see Supplemental Figure 1B online). In this study, we provided biochemical evidence that Nb WRKY8 directly interacts with SIPK, NTF4, and WIIPK and that interaction between Nb WRKY8 and SIPK requires a D domain (Figure 2). Because the phosphorylation intensity of the D domain–mutated Nb WRKY8 was reduced markedly (Figures 2G and 2H), D domain–mediated high-affinity interaction might contribute to the specific and highly efficient phosphorylation by certain MAPK proteins. It is interesting that part of the WRKY-type transcription factor used a typical MAPK-docking motif, which has been identified in several eukaryotic MAPK substrates, to ensure effective phosphorylation.
Advances in mass spectrometry–based technologies have permitted high-throughput, large-scale in vivo phosphorylation site mapping, and several plant phosphoproteome studies have been reported (Kersten et al., 2009). However, we believe no SP cluster of group I WRKY has been identified as a phosphorylated peptide. This may be due to the deficiency of Arg and Lys residues in the SP cluster (see Supplemental Figure 1B online). Because the most commonly used protease in proteome analysis is trypsin, which cleaves the C-terminal to Arg and Lys residues, characterizing a protein region with very few or even with no such basic residues is difficult. Although we made a mass analysis of trypsin-digested WRKY8 that was phosphorylated by MAPK in vitro, using liquid chromatography–tandem mass spectrometry, fragments corresponding to the SP cluster were not detected.
MAPKs Have Dual Functions in Phosphorylation-Dependent Modulation and Sequestration of the Transcription Factor
Bacterial PAMP flg22 and pathogens cause dissociation of an MPK4/MKS1/WRKY33 complex in Arabidopsis (Qiu et al., 2008). Phosphorylation of MKS1 by activated MPK4 triggers the release of MKS1 and WRKY33; thus, WRKY33 can activate the expression of target genes containing PHYTOALEXIN DEFICIENT3 (PAD3), which encodes cytochrome P450 monooxygenase and is required for the last step of the synthesis of camalexin. In this situation, MPK4 indirectly interacts with WRKY33. No interaction has been detected between WRKY33 and MPK4 by Y2H assay (Andreasson et al., 2005). By contrast, in this study, Nb WRKY8 directly interacts with SIPK, NTF4, and WIPK, and the interaction between WRKY8 and SIPK requires a D domain (Figure 2). Our study suggests that both the D domain– and the SP cluster–containing WRKY proteins, such as WRKY33, might be phosphorylated by certain MAPKs and positively regulate defense responses. The MPK3/MPK6 cascade regulates camalexin synthesis through transcriptional activation of the PAD3 gene, which is the downstream target of WRKY33 (Ren et al., 2008). We hypothesize that unassociated WRKY33 may be recognized by MPK3/MPK6 directly after WRKY33 is induced by pathogen signals (Lippok et al., 2007), possibly because the stoichiometric imbalance resulting from the synthesis of WRKY33 does not allow the recruitment of WRKY33 into the MKS1 complex. Expression of WRKY8 was also upregulated by pathogen attack and mechanical stress (Figure 4A; see Supplemental Figure 10B online), suggesting that WRKY8 may exist in an unassociated form, even although a counterpart of MKS1 is unknown from N. benthamiana.
MPK4/MKS1/WRKY33 forms a ternary complex at steady state, which dissociates upon MPK4 activation (Qiu et al., 2008). Transcription factor ERF104 interacts with MPK6 at steady state, but this interaction is disrupted in protoplasts by MPK6 activity or ethylene signaling in response to flg22 (Bethke et al., 2009). In these models, MAPK has a role not only in substrate phosphorylation, but also in the sequestration and release of transcription factors, which allows access to target gene promoters. By contrast, activation of WRKY8 by MAPKs may be caused by phosphorylation-dependent structural change. In this study, DNA binding analyses using WRKY8 recombinant protein and a probe containing a cognate W-box cis-element showed that WRKY8 only weakly binds to the probe (Figures 3A and 3B). DNA binding activity of WRKY8 was activated by incubation with wild-type active MAPKs, but incubation with the kinase-inactive mutant of MAPKs had no effect on its activity (Figure 3B). These results show that binding of SIPK, NTF4, and WIPK to WRKY8 does not participate in the regulation of its DNA binding activity. We propose that sequestration of transcription factors by MAPK is not important in the function of WRKY8.
WRKYs Containing the SP Cluster Participate in Diverse Biological Processes in Plants
MAPKs regulate a diverse set of processes, including abscission, stomatal and ovule development, signals for various abiotic stresses, and defense responses against bacterial and fungal pathogens (Pitzschke et al., 2009; Rodriguez et al., 2010). At WRKY2, which contains the SP cluster, has been implicated in seed germination and postgermination arrest of development by abscisic acid, which activates MPK4 and MPK6 (Xing et al., 2008; Jiang and Yu, 2009). Likewise, At WRKY10 is expressed in developing endosperm and participates in seed development (Luo et al., 2005). Overexpression of rice WRKY53, which is the closest homolog of Nb WRKY8 in rice, upregulates several defense-related genes in rice cells (Chujo et al., 2007). Nt WRKY1 appears to participate in the initiation of HR-like cell death (Menke et al., 2005). These studies suggest that WRKYs containing an SP cluster may contribute to the regulation of many biological processes; thus, MAPK-mediated phosphorylation within the SP cluster may participate in responses other than defense responses. Currently, we do not know how certain MAPKs regulate a wide variety of events in response to biotic stimuli or in plant growth. Temporal and spatial expression profiles of group I WRKYs, in particular cell types at particular developmental stages or under particular environmental conditions, and comprehensive biochemical analyses will provide insight into the detailed mechanisms underlying the pleiotropic functions of MAPK.
METHODS
Plant Growth Conditions
Nicotiana benthamiana plants were grown at 25°C and 70% humidity under a 16-h photoperiod and an 8-h dark period in environmentally controlled growth cabinets.
Pathogen Inoculation
Phytophthora infestans zoospore inoculation was done as described by Yoshioka et al. (2003). For expression analysis, leaves were inoculated with P. infestans race 1.2.3.4 zoospore suspension (1 × 106 zoospores/mL). For pathogen growth assays, leaves were inoculated with P. infestans race 1.2.3.4 zoospore suspension (1 × 104 zoospores/mL). Determination of P. infestans biomass was done as described by Asai et al. (2010). Colletotrichum orbiculare strain 104-T conidia suspension was prepared and inoculated as described by Asai et al. (2008).
Plasmid Constructs for Recombinant Proteins
PCR fragments of WRKY8 were inserted into the MscI/HindIII sites of the pET-32a vector (Novagen). The following primer sets were used: WRKY8 (5′-TTCTGGCCATATGGCAGCTTCTTCAACAATCATAG-3′and 5′-CCCAAGCTTGCAGAGCAATGTCTCCATAAAC-3′), WRKY81-170 (5′-TTCTGGCCATATGGCAGCTTCTTCAACAATCATAG-3′ and 5′-CCCAAGCTTATCATTCTGCTTTCTTGATTCTTG-3′), and WRKY8171-538 (5′-TTCTGGCCATCAATCATCTGAAAATGCTAATC-3′ and 5′-CCCAAGCTTGCAGAGCAATGTCTCCATAAAC-3′). The PCR fragments of Nb SIPK, Nb NTF4, Nb WIPK, and Nb NTF4 were inserted into the SpeI/BamHI sites of the pET-41a vector (Novagen). The following primers were used: Nb SIPK (5′-GGACTAGTATGGATGGTTCTGGTCAGC-3′ and 5′-CGGGATCCTCACATGTGCTGGTATTCAGG-3′), Nb NTF4 (5′-GGACTAGTATGGATGGTCCAGCTCATCA-3′ and 5′-CGGGATCCTCACATGTGCTGGTATTCAGG-3′), Nb WIPK (5′-GGACTAGTATGGCTGATGCAAATATGGG-3′ and 5′-CGGGATCCTTAAGCATATTCAGGATTCAGCG-3′), and Nb NTF6 (5′-GGACTAGTATGGAAAACGAAACCAATGA-3′ and 5′-CGGGATCCTCACTTCATTGTATTAGGATCAAAC-3′). Amino acid substitutions were introduced by PCR-based, site-directed mutagenesis using a PrimeSTAR mutagenesis basal kit (Takara Bio).
The PCR fragment of Nb MEK1DD, which was amplified from plasmids containing full-length cDNA as a template, was inserted into the SacI/XhoI site of the pET-32a vector. The following primer was used: Nb MEK1DD (5′-CCGAGCTCATGAAGACGACGAAGCCAT-3′ and 5′-CCGCTCGAGTTATGCGTAGTCTGGTACGTCG-3′). pET-32a vector containing St MEK2DD was described by Katou et al. (2005).
Expression and Purification of Recombinant Proteins
pET-32a vectors containing various mutants of full-length WRKY8, WRKY8mD, or WRKY81-170 were transformed into Escherichia coli BL21-CodonPlus (DE3)-RIPL (Stratagene). The overnight culture at 37°C was transferred to 100-fold Luria-Bertani (LB) medium containing 100 μg/mL ampicillin and incubated to OD600 1.0 at 37°C. Expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C. Cells were collected and resuspended in 50 mM HEPES-NaOH, pH 7.5. The suspension was sonicated and centrifuged at 12,000g for 10 min at 4°C. The pellet was washed twice with 50 mM HEPES-NaOH, pH 7.5, and 0.5% Triton X-100 and twice with distilled deionized water. The pellet was resuspended in 50 mM HEPES-NaOH, pH 7.5, and 8 M urea and was centrifuged at 12,000g for 10 min at 4°C. The supernatant was dialyzed against 25 mM HEPES-KOH, pH 7.6, and 1 mM DTT. pET-32a vector containing WRKY8171-538 was transformed into E. coli BL21-CodonPlus (DE3)-RIPL. E. coli was incubated in LB medium containing 100 μg/mL ampicillin at 25°C using the Overnight Express Autoinduction System 1 (Novagen), according to the manufacturer's instructions. The C-terminal region of WRKY8 proteins was extracted and purified using Ni Sepharose 6 Fast Flow (GE Healthcare), according to the manufacturer’s instructions. pET-41a vectors containing wild-type or kinase-inactive MAPKs were transformed into E. coli BL21-CodonPlus (DE3)-RIPL. The overnight culture at 37°C was transferred to 100-fold LB medium containing 50 μg/mL kanamycin and was incubated to OD600 1.0 at 37°C. Protein synthesis was induced by adding 0.2 mM IPTG for 4 h at 23°C. Each MAPK was extracted and purified using Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions. pET-32a vectors containing N. benthamiana MEK1DD and Solanum tuberosum MEK2DD were transformed into E. coli BL21-CodonPlus (DE3)-RIPL. The bacteria were cultured overnight at 37°C, transferred to 100-fold LB medium containing 50 μg/mL kanamycin, and then incubated to OD600 1.0 at 37°C. Protein synthesis was induced by adding 0.2 mM IPTG for 4 h at 23°C. Each constitutively active MAPKK was extracted and purified using Ni Sepharose 6 Fast Flow according to the manufacturer’s instructions.
Preparation of Active MAPKs
In vitro phosphorylation of wild-type or kinase-inactive Nb SIPK, Nb NTF4, and Nb WIPK by St MEK2DD, and Nb NTF6 by Nb MEK1DD, was performed in a phosphorylation buffer (20 mM HEPES-KOH, pH 7.6, 10 mM MgCl2, and 1 mM DTT) containing 100 mM Na3VO4, 5 mM NaF, 5 mM β-glycerophosphate, 500 μM ATP, 0.3 mg MAPK, and 0.075 mg constitutively active MAPKK at 30°C for 30 min. To remove MAPKK, S-protein-agarose (Novagen) equilibrated with phosphorylation buffer was added. After 30 min incubation at room temperature with gentle shaking, the agarose-protein complex was removed by brief centrifugation. The resulting supernatants containing active forms of MAPKs were concentrated using Microcon YM-10 (Millipore) and were stored at −80°C in phosphorylation buffer containing 50% glycerol.
In Vitro Phosphorylation
Recombinant WRKY8 protein (0.025 μg/μL) or MBP (0.1 μg/μL; Sigma-Aldrich) was incubated with 0.01 μg/μL of active MAPKs in phosphorylation buffer (20 mM HEPES-KOH, pH 7.6, 1 mM DTT, and 10 mM MgCl2) containing 50 μM ATP and 50 μCi/mL [γ-32P]ATP at 30°C for 30 min. The reactions were stopped by adding 2× SDS-PAGE sample buffer. Kinase activities were analyzed by SDS-PAGE followed by autoradiography.
Antibody Production and Immunoblotting
The peptides for pSer-79 (CIPPGLS[PO3H2]PTELL) and pSer-86 (CTELLDS[PO3H2]PLLLS) were synthesized and were conjugated to keyhole limpet hemocyanin carrier, and the polyclonal antisera were raised in rabbits (BioGate). The pSer-79–specific antibody and pSer-86–specific antibody were purified by affinity chromatography using pSer-79 and pSer-86 peptide coupled to Hitrap NHS-activated columns (GE Healthcare). The eluates were then passed through an affinity column coupled to a nonphosphorylated peptide. FLAG- and HA-tagged proteins were detected by monoclonal anti-FLAG antibody (clone M2; Sigma-Aldrich) and monoclonal anti-HA antibody (clone HA-7; Sigma-Aldrich), respectively. Fusion proteins of GAL4 DBD were detected by monoclonal anti-GAL4 antibody (clone RK5C1; Santa Cruz).
For immunoblotting, equal amounts of proteins were separated on an SDS-polyacrylamide gel and were transferred to a nitrocellulose membrane (Whatman). After blocking in TBS-T (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) with 5% nonfat dry milk or 3% BSA for 1 h at room temperature, the membranes were incubated with anti-pS79, anti-pS86, anti-FLAG, anti-GAL4, or anti-HA antibodies diluted with TBS-T at room temperature for 1 h. After washing with TBS-T, the membranes were incubated with horseradish peroxidase–conjugated anti-rabbit Ig or anti-mouse Ig antibody (GE Healthcare) diluted with TBS-T for 30 min at room temperature. The antibody-antigen complex was detected using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and Light-Capture equipped with a CCD camera (ATTO), and chemiluminescence signals were analyzed using a CS Analyzer 3.0 (ATTO).
Preparation of Protein Extracts and Strep-Tactin Affinity Chromatography
Leaves were ground in liquid nitrogen with a mortar and pestle and were thawed in lysis buffer (20 mM HEPES-NaOH, pH 7.5, 10 mM KCl, 10 mM MgCl2, 10% glycerol, 1% Triton X-100, 40 mM β-glycerophosphate, 10 mM NaF, 1 mM Na3VO4, 5 mM DTT, and 1% Protease inhibitor cocktail for plant cell tissue extracts [Sigma-Aldrich]). The homogenate was incubated at 4°C for 10 min, and then 5 M NaCl was added to a final concentration of 420 mM. The mixture was incubated at 4°C for 1 h with occasional mixing and was then sonicated to reduce viscosity. The mixture was centrifuged at 20,000g for 10 min at 4°C, and the supernatant was used for Strep-Tactin affinity chromatography of StrepII-tagged proteins. After adjusting the protein concentrations, the protein extracts (600 μL) were diluted with 1 mL of dilution buffer (20 mM HEPES-NaOH, pH 7.5, 5 mM DTT, 10% glycerol, 25 μg/mL avidin, and 1% Triton X-100). Forty microliters of MagStrep type 2 Beads (IBA), which were preequilibrated with wash buffer (20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 5 mM DTT, 10% glycerol, and 1% Triton X-100), were added, and the mixture was incubated overnight at 4°C with gentle shaking. The resin was washed three times with wash buffer and finally was resuspended in 30 μL of 2× SDS-PAGE sample buffer. The protein concentration was determined using the Protein Assay Dye Reagent (Bio-Rad Laboratories) with BSA as a standard.
GFP Fusions and BiFC Constructs for Expression in N. benthamiana
For subcellular localization analysis, cDNA fragments of Nb WRKY8, Nb SIPK, Nb NTF4, Nb WIPK, Nb NTF6, and NLS-GUS were cloned into pGWB406, which fused GFP to the N terminus of the protein (Nakagawa et al., 2007). Each plasmid was transformed into Agrobacterium tumefaciens GV3101. Infiltration of Agrobacterium was done at OD600 0.5. For BiFC studies, a cDNA fragment of full-length Nb WRKY8 was cloned into a binary vector, which fused the N-terminal half of YFP (nYFP) to the N terminus of the protein. cDNA fragments of Nb SIPK, Nb WIPK, Nb NTF6, and NLS-GUS were cloned into a pGWB-based binary vector, which fused the C-terminal half of YFP (cYFP) to the N terminus of the protein. Binary plasmids were transformed into Agrobacterium GV3101. Coinfiltration of Agrobacterium containing the BiFC constructs was done at OD600 0.25:0.25.
Microscopy
Epidermal cell layers of N. benthamiana leaves were assayed for fluorescence 48 h after agroinfiltration. Cells were visualized using a fluorescence microscope (Axio Imager M1; Carl Zeiss) with fluorescent filters: filter set 38HE (excitation BP 470/40, beam splitter FT 395, emission BP 525/50) for GFP and YFP and filter set 49 (excitation G 365, beam splitter FT 395, emission BP 445/50) for 4',6-diamidino-2-phenylindole. Images were collected using a CCD camera (AxioCam HRc; Carl Zeiss).
In Vitro Pull-Down Assay
For pull-down assays, 3 μg of N-terminal GST-tagged SIPK, NTF4, WIPK, NTF6, or GST alone was bound to 10 μL of 50% Glutathione Sepharose 4B and incubated with 1 μg C-terminal His6-tagged WRKY8WT or WRKY8mD in pull-down (PD) buffer (20 mM HEPES-KOH, pH 7.6, 80 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.1% Triton X-100, and 1 mM DTT) containing 1% protease inhibitor cocktail for plant cell tissue extracts at 4°C under gentle rotation. After 2 h, the samples were washed three times with PD buffer. After the final wash, the samples were analyzed by SDS-PAGE. After electrophoresis, the gels were stained with Coomassie Briliant Blue or underwent immunoblot analysis using anti-His6 antibody (clone H-3; Santa Cruz).
Y2H Assay
cDNA fragments of WRKY81-170 variants were cloned into the pGADT7 vector in-frame with the GAL4 activation domain. cDNA fragments of Nb SIPK and Nb NTF4 were cloned into the pGBKT7 vector in-frame proximal to the binding domain. These vectors were cotransformed into yeast AH109 (Clontech) using the protocol Matchmaker GAL4 Two-Hybrid System 3 (Clontech). Cotransformants were selected on SD-LW (synthetic dropout medium lacking Leu and Trp). The interactions were tested on selective medium (SD-LWAH) lacking Leu, Trp, adenine, and His, according to the manufacturer’s instructions. Serial 1:10 dilutions were prepared in water, and 4 μL of each dilution was used to yield one spot. Plates were incubated at 30°C for 2 d before scoring and taking photographs. SV40 T-antigen with p53 or Lamin C was used as the positive and negative control, respectively.
EMSA
Double-stranded synthetic oligonucleotides were end-labeled with 32P by Megalabel (Takara Bio). The probe was purified using a MicroSpin G-50 column (GE Healthcare). Phosphorylation of WRKY8 recombinant proteins was described in the section entitled Expression and Purification of Recombinant Proteins. The EMSA reaction mixture comprised 25 mM HEPES-KOH, pH 7.6, 40 mM KCl, 1 mM DTT, 10% glycerol, 5 μg poly (dI-dC) (GE Healthcare), 0.4 μg recombinant WRKY8, and 1 μL (600,000 cpm/μL) of the probe in a final volume of 16 μL. DNA-protein complexes were allowed to form for 20 min at 30°C and were separated on a 10% polyacrylamide gel in 0.5× TBE at 4°C. Bands were visualized by autoradiography.
Assays of Transactivation Activity
For reporter plasmids, six tandem repeats of the GAL4 upstream activation sequence (5′-CGGAGTACTGTCCTCCG-3′) were fused upstream of the minimal 35S promoter (−46) and Ω sequence and were cloned into pGreen. The cDNA of the GUS reporter gene containing one intron was subsequently cloned behind the synthetic promoter. For effector plasmids, the N-terminal cDNA fragments of WRKY81-170 variants or the VP16 activation domain (amino acid positions 413 to 490) were fused downstream of the GAL4 DNA binding domain (amino acid positions 1 to 147) and were cloned into pGreen under the control of the 35S promoter. HA-NtMEK2KR and HA-NtMEK2DD were cloned into pER8 behind the 17-β-estradiol–inducible promoter. For reference plasmids, the cDNA fragment of luciferase (LUC) containing one intron was cloned into pGreen under the control of the 35S promoter.
Transient assays were performed using an agroinfiltration method. Agrobacterium strains contained GAL4-WRKY8 variants, Nt MEK2 variants, reporter plasmids, and reference plasmids at a ratio 5:1:10:5 (final OD600 0.25). At 24 h after agroinfiltration, leaves were injected with 10 μM 17-β-estradiol and were incubated for 12 h. To determine the activation of GUS reporter gene expression by the effector proteins, leaves were ground in liquid nitrogen and were thawed in extraction buffer (100 mM potassium phosphate, pH 7.8, 1 mM EDTA, 7 mM 2-mercaptoethanol, 1% Triton X-100, and 10% glycerol). The homogenate was centrifuged at 20,000g for 10 min at 4°C. The supernatant was used to assay GUS and LUC activities. GUS activity was assayed by fluorometric quantitation of 4-methylumbelliferone produced from the glucuronide precursor, according to the protocol described by Jefferson et al. (1987), using a multiplate reader (355-nm excitation filter and 460-nm emission filter; TriStar LB941; Berthold Technologies). LUC activity was assayed using a Pica Gene Luminescence Kit (TOYO INK) according to the manufacturer’s protocol with a multiplate reader.
VIGS
VIGS was performed as previously described by Ratcliff et al. (2001). The following primers were used to amplify cDNA fragments of WRKY8 from an N. benthamiana cDNA library (Yoshioka et al., 2003), and restriction sites were added to the 5′-ends of the forward and reverse primers for cloning into the TRV vector pTV00 (RNA2): WRKY8-F-BamHI (5′-CGGGATCCTTCATTTTTCCTTTTAATTTCCC- 3′) and WRKY8-R-ClaI (5′-CCATCGATGAAGATCAGTGAATGAGAAACTTG-3′). Amplification using these primers produced a 180-bp fragment. pTV00 vectors containing the cDNA fragments of Nb SIPK, Nb WIPK, or Nb SIPK/Nb WIPK were described by Tanaka et al. (2009). N. benthamiana was transfected by viruses by means of Agrobacterium-mediated transient expression of infectious constructs. The vectors pBINTRA6 and pTV00, containing the inserts RNA1 and RNA2, respectively, were transformed separately by electroporation into Agrobacterium GV3101, which includes the transformation helper plasmid pSoup (Hellens et al., 2000). A mixture of equal parts of Agrobacterium suspensions containing RNA1 and RNA2 was inoculated into 2- to 3-week-old N. benthamiana seedlings. The inoculated plants were grown under a 16-h photoperiod and an 8-h dark period at 23°C. The upper leaves of the inoculated plants were used for assays 3 to 4 weeks after inoculation.
Agrobacterium-mediated Transient Expression in N. benthamiana
cDNA fragments of Nt MEK2DD, Nt MEK2KR, Nb SIPKWT, Nb SIPKKR, Nb NTF4WT, and Nb NTF4KR with a FLAG tag at the N terminus were cloned into pEL2Ω-MCS (Ohtsubo et al., 1999). cDNA fragments of WRKY8 and its variants were cloned into pEL2Ω-MCS with an HA-StrepII tag at the C terminus or into pGreen (Hellens et al., 2000) with an HA tag at the C terminus. All inserts were preceded by the 35S promoter of CaMV and the Ω sequence from Tobacco mosaic virus. The nopaline synthase terminator region was at the 3′ end of the gene. pGreen containing either GUS or INF1 was described by Asai et al. (2008). Transformation of Agrobacterium GV3101 by electroporation and infiltration of Agrobacterium suspensions were done as described by Asai et al. (2008). Immunodetection of WRKY8-HA-StrepII and WRKY8-HA variants was done by coinfiltration of Agrobacterium expressing p19, the suppressor of posttranscriptional gene silencing of Tomato bushy stunt virus (Voinnet et al., 2003).
Screening for Genes Downstream of WRKY8
For loss-of-function screening, total RNA was extracted from WRKY8-silenced or TRV control leaves 24 h after inoculation with Agrobacterium containing St MEK2DD. For gain-of-function screening, WRKY8DDDDD-HA-StrepII was cloned into pER8 behind the 17-β-estradiol–inducible promoter and was transformed into Agrobacterium GV3101. At 48 h after agroinfiltration (OD600 0.05), leaves were treated with 10 μM 17-β-estradiol to induce expression of WRKY8DDDDD. Total RNA was extracted from leaves expressing WRKY8DDDDD or the empty vector (pER8) at 3 h after 17-β-estradiol treatment. Poly(A)+ RNA was purified using Oligotex -dT30 <Super> (Takara Bio). A PCR-based cDNA subtraction method was used to selectively amplify differentially expressed cDNA fragments using the PCR-Select cDNA Subtraction Kit (Clontech). Subtractive cDNA libraries were constructed by following the manufacturer’s instructions. Subtracted cDNAs were cloned into pGEM-T easy vector (Promega) and transformed into E. coli DH5α. Individual clones were amplified using PCR with nested PCR primer 1 (5′-TCGAGCGGCCGCCCGGGCAGGT-3′) and nested PCR primer 2R (5′-AGCGTGGTCGCGGCCGAGGT-3′) and were dotted onto Hybond-XL nylon membranes (GE Healthcare). Duplicated membranes were hybridized with 32P-labeled, oligo(dT)-primed cDNA probes. For loss-of-function screening, the cDNA probes derived from WRKY8-silenced or TRV control leaves 24 h after inoculation with Agrobacterium containing St MEK2DD were used. For gain-of-function screening, total RNA was extracted from leaves expressing WRKY8DDDDD or the empty vector (pER8) at 3 h after 17-β-estradiol treatment. Each cDNA probe was synthesized from 1 μg mRNA using ReverTra Ace -α- (TOYOBO). The mRNA was removed by RNaseH treatment. After hybridization, the membranes were exposed to x-ray film. From the results of dot blot hybridization, differentially expressed clones were selected and sequenced. The expression level of each clone was analyzed by qRT-PCR using specific primers, and false-positive clones were excluded.
RNA Isolation and qRT-PCR
Total RNA from N. benthamiana leaves was prepared using TRIzol reagent (Invitrogen) according to the manufacturer’s procedure. Reverse transcription was done using ReverTra Ace -α-. qRT-PCR analysis was done using the StepOnePlus Real-Time PCR system (Applied Biosystems) with Power SYBR Green PCR Master Mix (Applied Biosystems). The expression of Nb WRKY7, Nb WRKY8, NADP-ME, or HMGR2 was normalized to the expression of EF-1α. For RT-PCR, the PCR reaction was done using the TaKaRa Ex Taq Hot Start Version (Takara Bio) with denaturing, annealing, and extension at temperatures of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, respectively. PCR products were separated on a 1.8% agarose gel and were visualized by ethidium bromide staining. EF-1α was used as a control. Gene-specific primers of each sequence were as follows: Nb WRKY8 (5′-AACAATGGTGCCAATAATGC-3′ and 5′-TGCATATCCTGAGAAACCATT-3′), Nb WRKY7 (5′-CACAAGGGTACAAACAACACAG-3′ and 5′-GGTTGCATTTGGTTCATGTAAG-3′), HMGR2 (5′-CCAGGGATAACAATGATGATTC-3′ and 5′-CAGCCAAGCGTAGTTGCG-3′), NADP-ME (5′-GTCTTCCTCTGTCACCTGACC-3′ and 5′-AAACATTTACGAAGGAAGTCGTG-3′), and EF-1α (5′-TGTGGAAGTTTGAGACCACC-3′ and 5′-GCAAGCAATGCGTGCTCAC-3′).
In-Gel Kinase Assay
In-gel kinase assays were performed as described by Katou et al. (1999). Briefly, 20 mg of N. benthamiana soluble proteins were separated on a 10% SDS-polyacrylamide gel in the presence of 0.25 mg/mL MBP. After electrophoresis, SDS was removed by washing the gel in 20 mM Tris-HCl, pH 8.0, and 20% 2-propanol four times for 30 min each. The gel was washed in buffer A (20 mM Tris-HCl, pH 8.0, and 1 mM DTT) twice for 30 min each and then was denatured in buffer A containing 6 M guanidine hydrochloride twice for 30 min each. The proteins were renatured overnight at 4°C by incubating the gel in buffer A containing 0.03% Tween 20 with four changes of the solution. The gel was equilibrated in 20 mM HEPES-KOH, pH 7.6, 10 mM MgCl2, and 1 mM DTT and then incubated in the same buffer containing 25 μM ATP and 0.5 μCi/mL [γ-32P]ATP (4000 Ci/mmol) for 1 h at room temperature. The reaction was stopped by washing the gel in 5% trichloroacetic acid and 1% sodium pyrophosphate. The gel was washed extensively with this solution and was washed in 20% methanol, dried under vacuum, and autoradiographed.
Phylogenetic Analysis
For the phylogenetic analysis, the program MEGA 4.1 (Tamura et al., 2007) was used. The protein sequences aligned using the ClustalW method in MEGA 4.1 (pairwise alignment: gap opening penalty 10, gap extension penalty 0.1; multiple alignment: gap opening penalty 10, gap extension penalty 0.2, protein weight matrix using Gonnet). The residue-specific and hydrophilic penalties were ON, whereas the End Gap separation and the Use negative separation were OFF. Gap separation distance used was 4, and the delay divergence cutoff was at 30. This alignment (available as Supplemental Data Set 1 online) was then used for a bootstrap test (3000 replicates, seed = 35,342) for phylogeny using the neighbor-joining method (p-distance model, pairwise deletion of gaps).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers AB552919 (Nb NADP-ME), AB552920 (Nb HMGR2), AB445391 (Nb WRKY7), AB445392 (Nb WRKY8), AB373026 (Nb NTF4), AB360634 (Nb NTF6), AB373025 (Nb SIPK), AB098729 (Nb WIPK), AB360635 (Nb MEK1), and AB091780 (St MEK2). Accession numbers of the amino acid sequences included in the phylogenic tree are presented in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SP Cluster and D Domain Widely Conserved in the N-Terminal Region of Group I WRKY Proteins.
Supplemental Figure 2. In Vitro Phosphorylation of MBP by Recombinant MAPKs.
Supplemental Figure 3. The N-Terminal Region of WRKY8 Is Phosphorylated by SIPK.
Supplemental Figure 4. In Vitro Phosphorylation Analysis of WRKY8 by Recombinant MEK2DD.
Supplemental Figure 5. Y2H Analysis of the Interaction between WRKY81-170 and SIPK.
Supplemental Figure 6. Y2H Analysis of the Interaction between WRKY81-170mD and SIPK.
Supplemental Figure 7. Increased Transactivation Activity of GAL4DBD-WRKY8WT1-170 by SIPK-Mediated Phosphorylation.
Supplemental Figure 8. Schematic Representation of Defense-Related Metabolic Pathways Induced by WRKY8.
Supplemental Figure 9. Expression Analysis of NADP-ME and HMGR2 in N. benthamiana in Response to TRV Infection.
Supplemental Figure 10. Activation of MBP Kinase Activity and Expression of Downstream Genes of WRKY8 Induced by INF1 Treatment.
Supplemental Table 1. Accession Numbers of the Amino Acid Sequences Included in the Phylogenic Tree.
Supplemental Data Set 1. Text File of Alignment Used to Generate Figure 1B.
Supplementary Material
Acknowledgments
We thank Jonathan D.G. Jones for pSLJ4K1 vector, Phil Mullineaux and Roger Hellens for pGreen vector, Sophien Kamoun for INF1 construct, David C. Baulcombe for pTV00 vector and p19 construct, Yuko Ohashi and Ichiro Mitsuhara for pEL2Ω vector and Nt MEK2 clones, Tsuyoshi Nakagawa for pGWB vectors, Nam-Hai Chua for pER8 vector, Yasuyuki Kubo and Yoshitaka Takano for C. orbiculare, and the Leaf Tobacco Research Center, Japan, for N. benthamiana seeds. We also thank members of the Radioisotope Research Center, Nagoya University, for technical assistance. This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), by a Grant-in-Aid for Scientific Research (A) from the Japan Society of the Promotion of Science, and by a Grant in-Aid to N.I. for Scientific Research for Plant Graduate Student from Nara Institute Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
References
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S., Mase K., Yoshioka H. (2010). A key enzyme for flavin synthesis is required for nitric oxide and reactive oxygen species production in disease resistance. Plant J. 62: 911–924 [DOI] [PubMed] [Google Scholar]
- Asai S., Ohta K., Yoshioka H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L., Lau Y.-F.C., Campbell A.P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E.D., Siebert P.D. (1996). Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93: 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Fry W. (2008). Phytophthora infestans: The plant (and R gene) destroyer. Mol. Plant Pathol. 9: 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Yu D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Axtell M.J., Dahlbeck D., Ekwenna O., Zhang S., Staskawicz B., Baker B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3: 291–297 [DOI] [PubMed] [Google Scholar]
- Jin H., Liu Y., Yang K.Y., Kim C.Y., Baker B., Zhang S. (2003). Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 33: 719–731 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson H.S., Tooley P.W. (2000). Enhanced polymerase chain reaction methods for detecting and quantifying Phytophthora infestans in plants. Phytopathology 90: 1112–1119 [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V.G., de Groot K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou S., Senda K., Yoshioka H., Doke N., Kawakita K. (1999). A 51 kDa protein kinase of potato activated with hyphal wall components from Phytophthora infestans. Plant Cell Physiol. 40: 825–831 [Google Scholar]
- Katou S., Yoshioka H., Kawakita K., Rowland O., Jones J.D.G., Mori H., Doke N. (2005). Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol. 139: 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B., Agrawal G.K., Durek P., Neigenfind J., Schulze W., Walther D., Rakwal R. (2009). Plant phosphoproteomics: an update. Proteomics 9: 964–988 [DOI] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lippok B., Birkenbihl R.P., Rivory G., Brümmer J., Schmelzer E., Logemann E., Somssich I.E. (2007). Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20: 420–429 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2004). Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38: 800–809 [DOI] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke F.L., Kang H.G., Chen Z., Park J.M., Kumar D., Klessig D.F. (2005). Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 18: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G., Andreasson E., Hess D., Boller T., Peck S.C. (2008). An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Ohtsubo N., Mitsuhara I., Koga M., Seo S., Ohashi Y. (1999). Ethylene promotes the necrotic lesion formation and basic PR gene expression in TMV–infected tobacco. Plant Cell Physiol. 40: 808–817 [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C. (2001). Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Ren D., Yang K.Y., Li G., Liu Y., Zhang S. (2006). Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 141: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Romeis T., Piedras P., Zhang S., Klessig D.F., Hirt H., Jones J.D.G. (1999). Rapid Avr9- and Cf-9 -dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Okamoto M., Seto H., Ishizuka K., Sano H., Ohashi Y. (1995). Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270: 1988–1992 [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D., Yang S.H., Galanis A. (2000). Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25: 448–453 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen S., Goodwin P.H., Hsiang T. (2001). Infection of Nicotiana species by the anthracnose fungus, Colletotrichum orbiculare. Eur. J. Plant Pathol. 107: 767–773 [Google Scholar]
- Takano Y., Takayanagi N., Hori H., Ikeuchi Y., Suzuki T., Kimura A., Okuno T. (2006). A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol. Microbiol. 60: 81–92 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Ishihama N., Yoshioka H., Huser A., O’Connell R., Tsuji G., Tsuge S., Kubo Y. (2009). The Colletotrichum orbiculare SSD1 mutant enhances Nicotiana benthamiana basal resistance by activating a mitogen-activated protein kinase pathway. Plant Cell 21: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T.L., Rebay I. (2005). Post-translational modifications influence transcription factor activity: A view from the ETS superfamily. Bioessays 27: 285–298 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D.C. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wheeler M.C.G., Tronconi M.A., Drincovich M.F., Andreo C.S., Flügge U.I., Maurino V.G. (2005). A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 139: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.Q., Schalk M., Clark A., Miles R.B., Coates R., Chappell J. (2006). Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 24: 1441–1447 [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54: 440–451 [DOI] [PubMed] [Google Scholar]
- Yamamizo C., Kuchimura K., Kobayashi A., Katou S., Kawakita K., Jones J.D.G., Doke N., Yoshioka H. (2006). Rewiring mitogen-activated protein kinase cascade by positive feedback confers potato blight resistance. Plant Physiol. 140: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Sharrocks A.D., Whitmarsh A.J. (2003). Transcriptional regulation by the MAP kinase signaling cascades. Gene 320: 3–21 [DOI] [PubMed] [Google Scholar]
- Yang S.-H., Shore P., Willingham N., Lakey J.H., Sharrocks A.D. (1999). The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18: 5666–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D.G., Doke N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1998). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95: 7225–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y. (2001). Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20: 10–16 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.